Abstract
Background:
Indwelling medical devices (IMDs) in critical patients are vulnerable to colonization by biofilm producing bacteria. Complex characteristics of bacterial biofilms promote antibiotic resistance, leading to the emergence of resistant device-related infections (DRI), which pose new challenges in their management.
Materials and Methods:
The study was done on 135 hospitalized (Intensive care units) pediatric patients with IMDs (intravascular catheter, urinary catheter, and endotracheal tube) to determine the device-specific infection rates. Biofilm formations were demonstrated by the tube method and by scanning electron microscopy (SEM). Bacteria in biofilms were identified by the standard conventional methods and tested for antibiotic resistance. We also detected the presence of extended spectrum β-lactamases (ESβLs), particularly, blaCTX-M, in gram-negative isolates.
Results:
The rates of biofilm-based catheter-related blood stream infections (CRBSI), catheter-associated urinary tract infections (CAUTI), and Ventilator Associated Pneumonia (VAP), in our study, were 10.4, 26.6, and 20%. Biofilm formation by the tube method correlated well with the SEM findings. A majority of infections were caused by Klebsiella pneumoniae followed by Staphylococcal biofilms. A high percentage (85.7%, 95% confidence interval 64.5 to 95.8%) of biofilm producing bacterial isolates, causing infection, were multidrug resistant. Many biofilm producing gram-negative isolates were ESβLs producers, and a majority particularly harbored blaCTX-M, among the ESβLs genotypes.
Conclusion:
The incidence of resistant device-related infections, predominantly caused by biofilm producing bacteria, is rising. The tube method is an effective screening method to test biofilm production, where sophisticated microscopy facilities are not available. The varying resistance pattern of organisms isolated in our setup, emphasizes the importance of studying the pattern of infection in every setting and providing antibiotic guidelines in the management of such infections.
Keywords: Biofilms, Device-related infection, ESβL, MRSA, Scanning electron microscopy
INTRODUCTION
Indwelling Medical Devices (IMDs) are significant in patient care and due to their routine use in hospitals, critical patients easily become vulnerable to microbial colonization. Colonization of IMDs by biofilm producing bacteria is a universal phenomenon[1] and should not be ignored as it leads to resistant infections. Costerton et al. provided a partial listing of IMDs[2] that have been colonized by biofilms and are a source of persistent infection, unless removed from patient. A device-related infection (DRI) is an infection in a patient with a device (intravascular catheter, endotracheal tube or indwelling urinary catheter) that was in use for at least 48 hours before the onset of infection.[3] The extensive use of indwelling devices in hospitalized patient has increased the incidence of DRI, especially blood stream infections, originating from the microbial colonization of the intravascular catheter. Among the DRI, a majority of infections are catheter-related blood stream infections (CRBSI), followed by catheter associated urinary tract infections (CAUTI), and ventilator associated pneumonia (VAP), as seen in developed countries.[4] However, we have scant literature available in developing countries such as ours, on the infection rates and their resistance pattern. Empirical use of antimicrobial agents and the superimposed complex nature of bacteria in biofilms colonizing IMDs have resulted in resistant DRIs. Management of such infections is now a huge challenge, as they lead to persistent and resistant infections. Although biofilms on various medical devices have been studied extensively, much of the published research has used specialized microscopy such as confocal laser scanning microscopy and scanning electron microscopy (SEM), to visualize the biofilms. However, newer methods like the tube method, tissue culture plate method, and so on, have been recently used to demonstrate the biofilm production ability of microbes and are basic screening techniques, which can be incorporated in the diagnostic clinical laboratory.[5,6]
The aim of this study is to determine the overall rates of DRI (device specific rates), bacteriological profile of the biofilms colonizing the devices, and the associated resistance, with special reference to the prevalent genotypes in our hospital.
MATERIALS AND METHODS
In all, 135 hospitalized (Intensive Care Unit) pediatric patients with IMDs (105 with intravascular catheters, 15 with urinary catheters, and 15 with endotracheal tubes) and suspicion of DRI were studied. Complete blood count, Chest X-rays, and urine culture were done to rule out any infective etiology at the time of admission. The IMDs were immediately removed on the clinical suspicion of DRI and their tip was cut with a sterile blade and sent to the microbiology laboratory in a sterile container. An intravascular catheter tip culture by the roll plate method was done in patients with suspicion of CRBSI.[7] The clinical isolates obtained from various specimens (Blood, urine, and bronchoalveolar lavage (BAL)) of the patient with suspected CRBSI, CAUTI, and VAP, respectively, were processed and identified according to the standard protocol.[8]
Tips of removed IMDs were also fixed in 1% glutaraldehyde to be used later on for examining biofilms on them.
Establishment of device-specific infection
The device-specific infection was established according to a surveillance definition by the Centers for Disease Control (CDC) and Prevention for healthcare-associated infections:[4]
Catheter-related blood stream infection
Is defined as, bacteremia in a patient with an intravascular device, developing after 48 hours of admission, with two positive blood cultures obtained by peripheral vein and clinical manifestations of sepsis in the absence of any source of sepsis apart from the device. The semi-quantitative culture from a catheter segment should have ≥ 15 bacterial colony forming units (CFU), and the same organism should be obtained by peripheral culture.
Catheter-associated urinary tract infection
Is found in patients who have had an indwelling urinary catheter for at least 48 hours before the culture is sent and the clean catch catheterized urine culture shows >105 bacterial CFU, with no more than two species of bacteria by the semi-quantitative method.
Ventilator associated pneumonia
Is said to occur when a patient on mechanical ventilation, for at least 48 hours, exhibits increasing Total leucocyte count (TLC), and new shadows (infiltrates) on a chest x-ray, and the BAL culture shows no more than two species of bacteria.
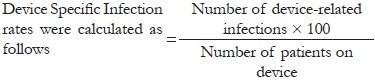
Antibacterial susceptibility disk diffusion method
Antibacterial susceptibility testing for various drugs (Himedia) was performed by the Kirby-Bauer disk diffusion method on Muller Hinton Agar as per the Clinical and Laboratory Standards Institute (CLSI) guidelines.[9]
Extended spectrum beta lactamase detection
Phenotypic detection: The gram-negative isolates were screened and confirmed (by the double disk synergy and double disk combination methods) for Extended Spectrum Beta Lactamase (ESβL) production, as per the CLSI guidelines.[10]
blaCTX-M detection
Polymerase chain reaction (PCR) was performed to detect the presence of blaCTX-M, as done in our earlier study.[11] Bacterial plasmid deoxyribonucleic acid (DNA) was extracted by the phenol chloroform method.[12] PCR was carried out with a first cycle of denaturation at 94°C for four minutes, and then 35 cycles of annealing at 52°C for two minutes, elongation at 72°C for two minutes, and denaturation at 94°C for 30 seconds. The PCR was completed by a final elongation step at 72°C for 10 minutes. The primers designed were from Operon Biotechnologies Nattermannallee 1, 50829 Cologne Germany, which spanned the universal blaCTX-M, F-ATG TGC AGY ACC AGT AAR GT (CTX-MU1) and R-TGG GTR AAR TAR GTS ACC AGA (CTX-MU-2). The PCR products were analyzed by gel electrophoresis.
Detection of methicillin resistance
Methicillin resistance in the strains of Staphylococci was tested by the cefoxitin disk (30 μg) for prediction of mec A gene-mediated methicillin resistance in the Staphylococcus species, as documented in CLSI.[13]
Detection of biofilms
Tube method
Biofilm production of isolates causing DRI was demonstrated with a slight modification of the method, as described by others.[5,6] Briefly 0.5 ml of suspension (0.5 McFarland turbidity standard of saline-washed 24-hour culture isolate) was inoculated into a polystyrene tube containing 4.5 ml of Luria Bertani Broth. The tube was incubated at 37°C for 48 hours without agitation. The culture broth from the tube was then aspirated, washed twice with distilled water, and stained with Crystal violet. Biofilm formation was positive when a blue visibly adherent layer lined the wall and bottom of the tube and was scored as weak (1+), moderate (2+ or 3+) or strong (4+), based on amount of biofilm production. Each isolate was tested at least three times and read independently by two different observers. Biofilm producer Staphylococcus epidermidis ATCC 35984 and non-biofilm producer S. epidermidis ATCC 12228 were used as the positive and negative controls, respectively.
Scanning electron microscopy
Device tips fixed in 1% glutraldehyde were used to examine the biofilm formation by SEM, only if device-related infection was established. The device segments were rinsed in 0.1 M phosphate buffer (twice, three minutes each) and then placed in 1% Zetterquist's osmium for 30 minutes. The samples were subsequently dehydrated in a series of ethanol washes (70% for 10 minutes, 95% for 10 minutes, and 100% for 20 minutes), treated (twice, five minutes each) with hexamethyldisilizane (Polysciences Inc., Warrington, Pa.), and finally air-dried in a desiccator. The specimens were coated with gold-palladium (40% / 60%). After processing, the samples were observed with a scanning electron microscope (Leo 435 VP) in high vacuum mode at 15 kV. The images were processed for display using Photoshop software (Adobe Systems Inc., Mountain View, Calif.).
RESULTS
The rates of biofilm-based CRBSI, CAUTI, and VAP in our study were 10.4, 26.6, and 20%, respectively [Table 1]. The patients who developed DRI also had biofilms on their IMDs, as visualized on scanning electron microscopy, which were seen as bacterial cells embedded in an extracellular matrix [Figure 1]. The bacterial isolates causing DRI showed grade 3 (moderate) or 4 (strong) adherences, confirming their moderate-to-strong biofilm producing ability. All except three isolates (Staphylococcus epidermidis, Klebsiella pneumonia, and Acinetobacter baumanii) causing CRBSI, showed moderate biofilm producing ability by the tube method.
Table 1.
Infection producing biofilm isolates obtained from patients with IMDs
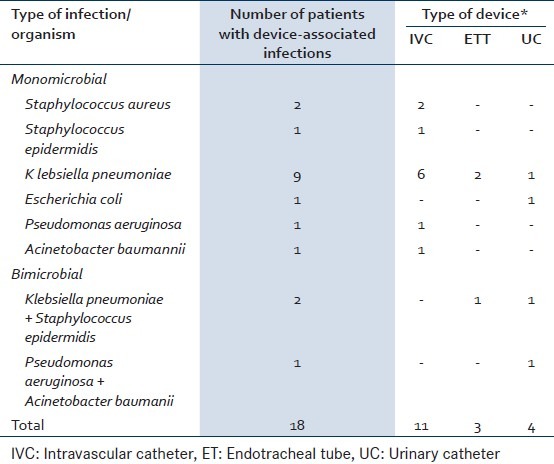
Figure 1.
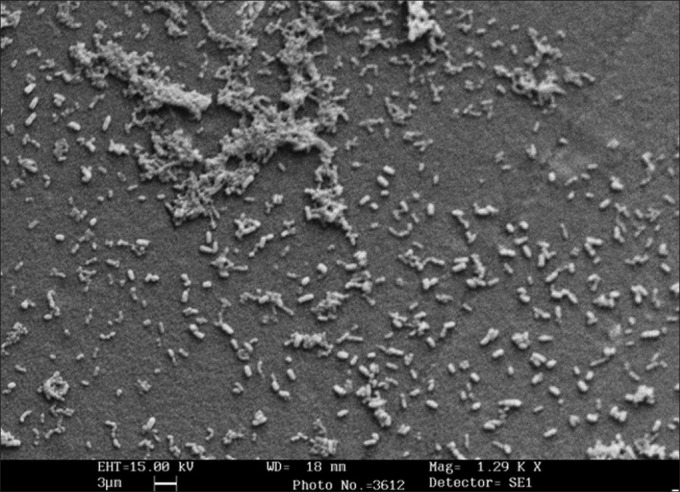
SEM micrograph of urinary catheter showing mixed bacterial biofilm
Antimicrobial susceptibility testing was done against 13 drugs for 16 gram-negative bacilli and eight drugs for 5 gram-positive cocci [Table 2]. Eighteen of the twenty-one (85.7%, 95% confidence interval 64.5 to 95.8%) biofilm producing bacterial isolates, causing infection, were multidrug-resistant.
Table 2.
Antibiotic resistance pattern of biofilmbased device-related infections
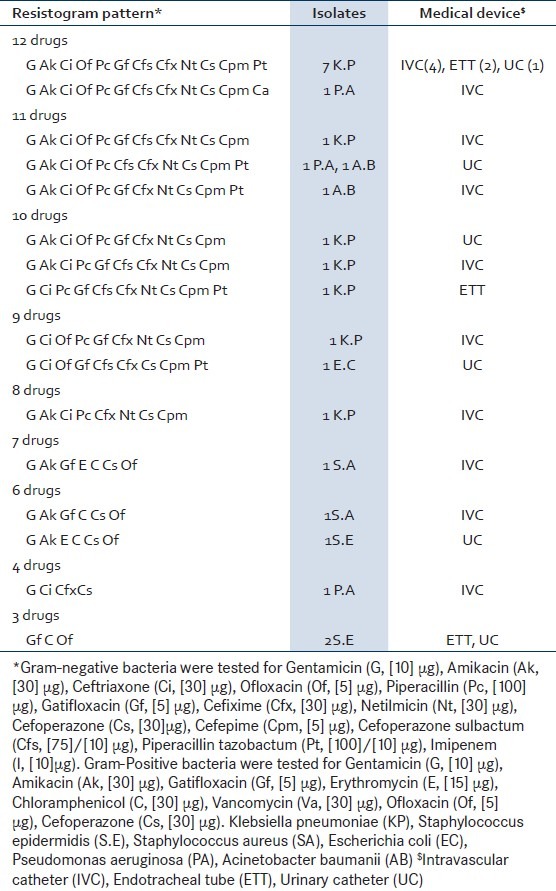
Fifteen out of sixteen (93.8%, 95% confidence interval 69.7 to 99.9%) biofilm producing gram-negative bacilli and three out of five (60%, 95% confidence interval 22.9 to 88.4%) Staphylococcal biofilms, causing DRI, were multidrug- resistant. Thirteen of sixteen (81.3%, 95% confidence interval 56.2 to 94.2%) gram-negative bacterial biofilm isolates were confirmed ESβL positive by phenotypic methods [Table 3]. eleven of sixteen (68.7%, 95% confidence interval 44.2 to 86.1%) gram-negative bacterial biofilm isolates on genotyping showed presence of the blaCTX-M gene (593 base pairs) [Figure 2].
Table 3.
Phenotypic and genotypic ESβL detection in various gram-negative bacilli
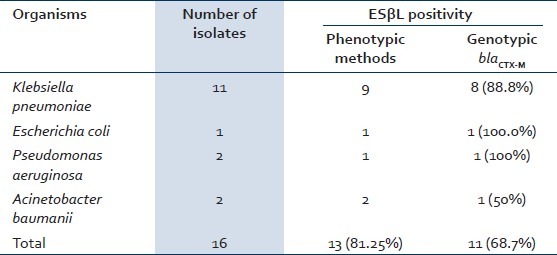
Figure 2.
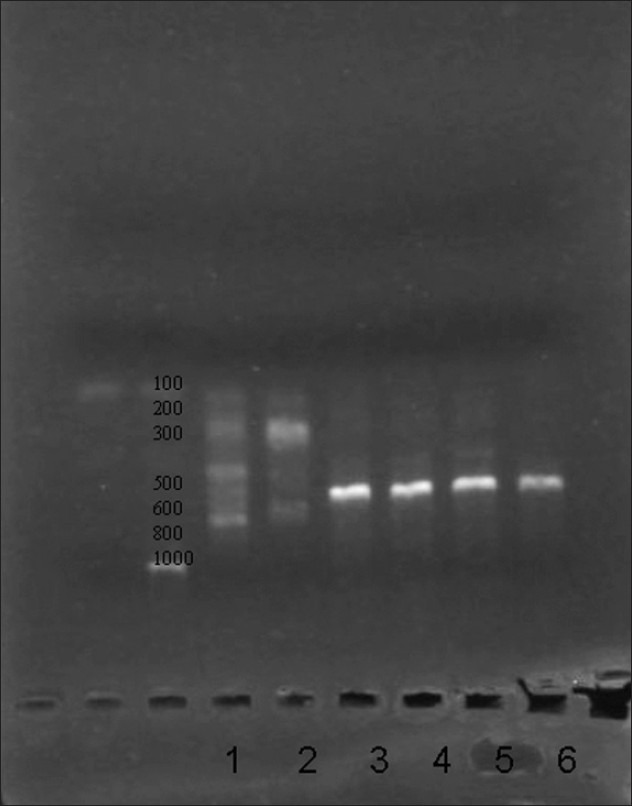
Lane1: MM lane Lane 2: negative, Lane 3 – gram negative bacilli (bla-CTX-M 593 bp)
DISCUSSION
The rates of device-specific infection caused by bacterial biofilms found in hospitalized pediatric patients of our institution were almost similar to those found in other studies;[14–16] however, the overall device-associated infection rates in any hospital can be determined only if an institutional surveillance study is designed, enrolling all hospitalized patients. DRI infections are known to be responsible for higher morbidity and therapeutic failure independent of the initial severity of illness.[17] Moreover, limited data is available on the profile and resistance rates of such infections in our country; therefore, active surveillance of hospital-associated infections from time to time is necessary.
Another noteworthy finding in our study was the biofilm producing ability of bacteria causing DRI. Biofilm formation could be demonstrated as surface adhesion on a bioprosthetic surface by the tube method and could be commonly used in clinical practice to demonstrate biofilms, where sophisticated microscopy techniques are not available. The tube method correlated well with other methods for biofilm demonstration of moderate-to-strong biofilm producing isolates, but the difficulty arose in discriminating between the weak and biofilm-negative isolates.[5,6] Detection of biofilms by the SEM and tube method was well correlated in this study, as all isolates were moderate-to-strong biofilm producers.
The most common bacterial biofilms causing DRI were Klebsiella pneumoniae (61.1%). Pseudomonas aeruginosa, Acinetobacter baumanii, Escherichia coli, Staphylococcus spp. Prominent pathogens most often associated with biofilm-based infections of IMDs wre either normal commensal flora or nosocomial in origin.[2,18] The gram-positive Staphylococcus aureus, Coagulase Negative Staphylococci (CoNS), and the gram-negative Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, and Pseudomonas aeruginosa might form biofilms and could cause DRI whenever the human immune response was compromised, as seen in hospitalized patients.[1] Nelson etal. (2003) also reported that K. pneumoniae was often involved in biofilm-related infections.[19] Prevalent gram-positive cocci, which often included Staphylococcus sp., were also found in our study and their ubiquity as skin flora and their adherence to IMD's surface was also documented by others.[1,5] Bimicrobial biofilms were found on urinary catheters and endotracheal tubes only. Biofilms on urinary catheters and endotracheal tubes might initially be composed of a single species, but longer exposures inevitably lead to multispecies biofilms.[20,21]
The drug resistance was alarming in the clinical isolates obtained from patients with DRI in our setup. Resistance was found to all the drugs checked, except for vancomycin, among gram-positive and imipenem among gram-negative strains. Gram-negative biofilm producers of 93.8%, causing DRI, were multidrug-resistant, 81.3% biofilm producing gram-negative bacilli were ESβL producers, and 68.7% of the isolates particularly showed a presence of blaCTX-M alleles although other genotypes needed to be evaluated. The prevalence of blaCTX-M in gram-negative bacilli was high in our hospital strains and due to financial constraints we could check only their presence among the ESβL genotypes.[22] Of late, few workers also discovered similar findings about biofilm-positive Klebsiella pneumoniae strains, which had the ability to produce ESβLs.[19,23] Staphylococci were important etiological agents of DRI, and a variety of drug-resistant mechanisms were investigated in biofilm isolates.[24] In our study Methicillin-resistant Staphylococci biofilms causing DRI were also found. A significant correlation was found between the exhibition of drug resistance and the biofilm producing ability, especially of gram-negative bacteria, in our hospital. High resistance and biofilm production among isolates of DRI, in our study, showed that bacteria in biofilms displayed altered character, and high resistance could be attributed to the biofilm producing ability of isolates.
The rates of DRI infection in our study are almost similar to those found in hospitals in developed parts of world, but the astonishing fact is the resistance associated with them is much higher when compared with the others.[17,25–27] The countries with preliminary studies carried out in hospitals in the developing parts of the world, where regulations for the implementation of infection control programs and antibiotic prescribing policies are lacking, have had the same scenario as ours.[28,29] The wide difference in resistance rates associated with DRI may be attributed to lack of infection control programs plus the injudicious and inappropriate use of antibiotics in our country. The intricate nature of bacteria is changed due to such widespread misuse of antibiotics in our country, plus, the biofilm producing ability of the bacteria further complicates the resistance problem.
The tube method can be an effective alternative to sophisticated microscopy techniques for screening the biofilm producing ability of bacteria, in resource-constrained countries. The device-related infections and associated antibiotic resistance by biofilm producing isolates is now an emerging problem. The varying resistance pattern of organisms isolated in our setup emphasizes the importance of studying the pattern of infection in every setting and providing antibiotic guidelines in the management of such infections.
ACKNOWLEDGMENT
Scanning electron microscopy was done by the kind and generous support of Sophisticated Analytical Instrumentation Facility (DST), Department of Anatomy, All India Institute of Medical Sciences, New Delhi, India.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Raad I. Intravascular-catheter-related infections. Lancet. 1998;351:893–8. doi: 10.1016/S0140-6736(97)10006-X. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections. In: Olmsted RN, editor. APIC Infection Control and Applied Epidemiology: Principles and Practice. St. Louis: Mosby; 1996. pp. A1–20. [Google Scholar]
- 4.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–32. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: An evaluation of three different screening methods. Indian J Med Microbiol. 2006;24:25–9. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- 6.Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37:318–26. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maki DG, Weise CE, Sarafin HW. A semiquantitative culture method for identifying intravenous-catheter-related infection. N Engl J Med. 1977;296:1305–9. doi: 10.1056/NEJM197706092962301. [DOI] [PubMed] [Google Scholar]
- 8.Colle JG, Miles RS, Watt B. Tests for the identification of bacteria. In: Collee JG, Marmion BP, Fraser AG, Simmons A, editors. Mackie & McCartney Practical Medical Microbiology. 14th ed. New Delhi: Churchill Livingstone; 1996. pp. 131–45. [Google Scholar]
- 9.Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. CLSI document M100–S16. Wayne (PA): The Institute; 2006. Clinical and Laboratory Standards Institute. [Google Scholar]
- 10.Screening and Confirmatory test for ESBLs.; sixteenth informational supplement. CLSI document M100–S16. Wayne (PA): The Institute; 2006. Clinical and laboratory Standard Institute. [Google Scholar]
- 11.Shahid M, Malik A, Adil M, Jahan N, Malik R. Comparison of beta-lactamase genes in clinical and food bacterial isolates in India. J Infect Dev Ctries. 2009;3:593–8. doi: 10.3855/jidc.550. [DOI] [PubMed] [Google Scholar]
- 12.Heilig J, Elbing KL, Brent R. Large-scale preparation of plasmid DNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, editors. Current Protocols in Molecular Biology. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2001. pp. 1.7.1–1.716. [DOI] [PubMed] [Google Scholar]
- 13.Disc diffusion test for prediction of mec A mediated resistance in Staphylococci; sixteenth informational supplement. CLSI document M100–S16. Wayne (PA): The Institute; 2006. Clinical and laboratory Standard Institute. [Google Scholar]
- 14.Mehta A, Rosenthal VD, Mehta Y, Chakravarthy M, Todi SK, Sen N, et al. Device-associated nosocomial infection rates in intensive care units of seven Indian cities. Findings of the International Nosocomial Infection Control Consortium (INICC) J Hosp Infect. 2007;67:168–74. doi: 10.1016/j.jhin.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Singh S, Pandya Y, Patel R, Paliwal M, Wilson A, Trivedi S. Surveillance of device-associated infections at a teaching hospital in rural Gujarat--India. Indian J Med Microbiol. 2010;28:342–7. doi: 10.4103/0255-0857.71830. [DOI] [PubMed] [Google Scholar]
- 16.Deep A, Ghildiyal R, Kandian S, Shinkre N. Clinical and microbiological profile of nosocomial infections in the pediatric intensive care unit (PICU) Indian Pediatr. 2004;41:1238–46. [PubMed] [Google Scholar]
- 17.Gopal Katherason S, Naing L, Jaalam K, Kamarul Imran Musa K, Nik Abdullah NM, Aiyar S, et al. Prospective surveillance of nosocomial device-associated bacteremia in three adult intensive units in Malaysia. Trop Biomed. 2010;27:308–16. [PubMed] [Google Scholar]
- 18.Davey ME, O’toole GA. Microbial biofilms: From ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–67. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson EC, Segal H, Elisha BG. Outer membrane protein alterations and blaTEM-1 variants: Their role in beta-lactam resistance in Klebsiella pneumoniae. J Antimicrob Chemother. 2003;52:899–903. doi: 10.1093/jac/dkg486. [DOI] [PubMed] [Google Scholar]
- 20.Nickel JC, Costerton JW, McLean RJ, Olson M. Bacterial biofilms: Influence on the pathogenesis, diagnosis and treatment of urinary tract infections. J Antimicrob Chemother. 1994;33(Suppl A):31–41. doi: 10.1093/jac/33.suppl_a.31. [DOI] [PubMed] [Google Scholar]
- 21.Inglis TJ, Lim TM, Ng ML, Tang EK, Hui KP. Structural features of tracheal tube biofilm formed during prolonged mechanical ventilation. Chest. 1995;108:1049–52. doi: 10.1378/chest.108.4.1049. [DOI] [PubMed] [Google Scholar]
- 22.Ensor VM, Shahid M, Evans JT, Hawkey PM. Occurrence, prevalence and genetic environment of CTX-M beta-lactamases in Enterobacteriaceae from Indian hospitals. J Antimicrob Chemother. 2006;58:1260–3. doi: 10.1093/jac/dkl422. [DOI] [PubMed] [Google Scholar]
- 23.Yang D, Zhang Z. Biofilm-forming Klebsiella pneumoniae strains have greater likelihood of producing extended-spectrum beta-lactamases. J Hosp Infect. 2008;68:369–71. doi: 10.1016/j.jhin.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Grinholc M, Wegrzyn G, Kurlenda J. Evaluation of biofilm production and prevalence of the icaD gene in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains isolated from patients with nosocomial infections and carriers. FEMS Immunol Med Microbiol. 2007;50:375–9. doi: 10.1111/j.1574-695X.2007.00262.x. [DOI] [PubMed] [Google Scholar]
- 25.Cooke EM, Coello R, Sedgwick J, Ward V, Wilson J, Charlett A, et al. A national surveillance scheme for hospital associated infections in England.Team of the Nosocomial Infection National Surveillance Scheme. J Hosp Infect. 2000;46:1–3. doi: 10.1053/jhin.2000.0801. [DOI] [PubMed] [Google Scholar]
- 26.Gastmeier P, Geffers C, Brandt C, Zuschneid I, Sohr D, Schwab F, et al. Effectiveness of a nationwide nosocomial infection surveillance system for reducing nosocomial infections. J Hosp Infect. 2006;64:16–22. doi: 10.1016/j.jhin.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 27.National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32:470–85. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 28.Starling CE, Couto BR, Pinheiro SM. Applying the centers for disease control and prevention and national nosocomial surveillance system methods in Brazilian hospitals. Am J Infect Control. 1997;25:303–11. doi: 10.1016/s0196-6553(97)90022-5. [DOI] [PubMed] [Google Scholar]
- 29.Rezende EM, Couto BR, Starling CE, Módena CM. Prevalence of nosocomial infections in general hospitals in Belo Horizonte. Infect Control Hosp Epidemiol. 1998;19:872–6. doi: 10.1086/647751. [DOI] [PubMed] [Google Scholar]


