WWW.RCSB.ORG
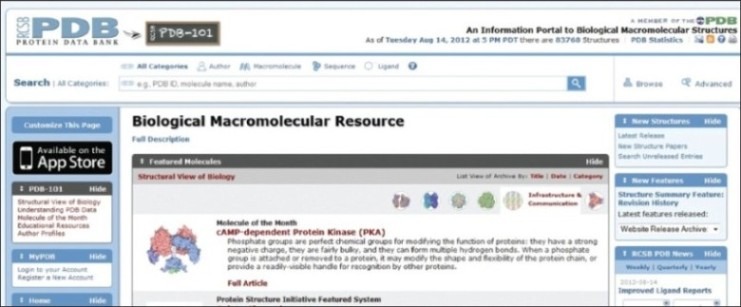
Research Collaborators for structural Bioinformatics Protein Data Bank (RCBS PDB) began in 1970's by group of the young crystallographers, including Edgar Meyer, Gerson Coheon and Helen M Berman.[1] The project was initiated in 1971 to list the structures of all the amino acids using neutron diffraction and has grown as a major international resource for structural biology containing more than 80,000 entries including proteins: 77,529; nucleic acids: 2420; proteins/DNA complexes: 3795 and other: 24 (totally 83,768 entries) as of Tuesday Aug 14, 2012 at 5 PM DST. The website also proclaims that there have been 62987 structure hits and 11915 ligand hits on this date and that the PDB has 24583 citations.
The main function of this database is to organize 3-D structural data of large biological molecules including proteins and nucleic acids of all the organisms including bacteria, yeast, plants, flies, other animals and humans. The three-dimensional structures of the biological macromolecules data available with PDB is determined by experimental methods such as X-ray crystallography, Nuclear magnetic resonance (NMR) spectroscopy, electron microscopy.[2]
The data is freely accessible and the entire site is maintained and managed by group of members from RCBS: Rutgers and UCSD, and is funded by NSF, NIGMS, DOE, NLM, NCI, MINDS and NIDDK. RCBS-Rutgers and RCBS-UCSD groups are located in the Department of Chemistry and Chemical Biology, Biomaps Institute for Quantitative Biology at Rutgers, New Jersey and the San Diego Supercomputer Centre (SDSC), Skaggs School of Pharmacy and Pharmaceutical Sciences (SSPPS) at University of California, San Diego (UCSD), respectively. RCBS PDB website required a modern web browser with JavaScript and cookies enabled. This site can be used using Windows, Apple Mac and Linux operating systems and also on a site with mobile support.
The website can be easily searched for the 3-D protein structure based on various search options which can be limited by authors, by macromolecules, by sequence and by ligand. The search option in all categories provides information under molecule name, structural domains and ontology terms. The selected required file has the PDB identification number and macromolecule name with release date, authors name, protein classification, and instrument used for determine the 3-D structure and citation. The selected/ searched macromolecules are downloadable (download PDB file), viewable (view PDB file) and view as a 3-D file (view in 3-D with Jmol). The file format generated from the database was initially in the PDB format which could be viewed using any one of open source computer programs such as MeshLab, Jmol, QuteMol, ACD/ChemSketch freeware and licensed software such as Chimera, MDL Chime, Pymol, Swiss-PDB viewer, etc. In late 2000, PDB released PDB exchange dictionary and the PDB archival data file in XML format named PDBML.[3]
The website also hosts an educational resource for exploring structural Biology named as PDB-101. This section chooses a molecule of the month and highlights it.Queries can be asked at any time in PDB website by logging in your free registered account. The PDB archive is updated every week on or about Wednesday (00:00 UTC) with new and modified entries and updates.
PDB also has a section called “Deposition”, that has a built in structures deposit option. Here one can upload the macromolecular data (electron microscopy/ x-ray/ NMR data of polypeptides, polynucleotides and polysaccharides). The uploaded file is validated and released based on publisher policies. From the “Tool” option a direct download of the protein macromolecules is possible in any one of PDB, mmCIF, PDBXL/XML formats, Structure factors, NMR restraints and Biological assemblies by entering PDB IDs. The tool option also has the “RCBS PDB protein comparison tool” to calculate pairwise sequence or structure alignments and “RCBS PDB widgets” a resource for web developers.
PDB in a joint program between “Structure Integration with Function, Taxonomy and Sequence” (SIFTS) (http://www.ebi.ac.uk/pdbe/docs/sifts/) and Uniportaims to provide an up-to-date resource for residue level mapping. SIFTS is the authoritative source of residue-level annotation of structures in the PDB data available in Uniport, IntEnz, CATH, SCOP, GO, InterPro, Pfam and PubMed.
The RCBS PDB and other organizations such as Protein Data Bank in Europe (PDBe; web link: www.ebi.ac.uk/pdbe/), Protein Data Bank Japan (PDBj; web link: www.pdbj.org/) and Biological Magnetic Resonance Data Bank (BMRB; web link: http://www.bmrb.wisc.edu/), USA are jointly working on Worldwide Protein Data Bank (wwPDB; web link: www.wwpdb.org/) in a concerted effort since 2003. The RCBS PDB and PDBj presently act as an archive centre for wwPDB .[4] The wwPDB's mission is to maintain a single PDB archive of macromolecular structural data that is freely and publicly available to the global community.[5]
Footnotes
Source of Support: Nil,
Conflict of Interest: None declared
REFERENCES
- 1.Berman HM. The Protein Data Bank: A historical perspective. Acta Crystallogr A. 2008;64:88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- 2.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, et al. Data deposition and annotation at the worldwide protein data bank. Mol Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 3.Westbrook J, Ito N, Nakamura H, Henrick K, Berman HM. PDBML: The representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–92. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 4.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 5.Statement downloaded. [Last accessed on 2012 Aug 20]. Available from: http://www.wwpdb.org/index.html .