Abstract
The R1185C mutation in WNK4 is associated with pseudohypoaldosteronism type II (PHAII). Unlike other PHAII-causing mutations in the acidic motif, the R1185C mutation is located in the COOH-terminal region of WNK4. The goal of the study is to determine what properties of WNK4 are disrupted by the R1185C mutation. We found that the R1185C mutation is situated in the middle of a calmodulin (CaM) binding site and the mutation reduces the binding of WNK4 to Ca2+/CaM. The R1185C mutation is also close to serum- and glucocorticoid-induced protein kinase (SGK1) phosphorylation sites S1190 and S1217. In addition, we identified a novel SGK1 phosphorylation site (S1201) in WNK4, and phosphorylation at this site is reduced by Ca2+/CaM. In the wild-type WNK4, the level of phosphorylation at S1190 is the lowest and that at S1217 is the highest. In the R1185C mutant, phosphorylation at S1190 is eliminated and that at S1201 becomes the strongest. The R1185C mutation enhances the positive effect of WNK4 on the Na+-K+-2Cl− cotransporter 2 (NKCC2) as tested in Xenopus laevis oocytes. Deletion of the CaM binding site or phospho-mimicking at two or three of the SGK1 sites enhances the WNK4 effects on NKCC2. These results indicate that the R1185C mutation disrupts an inhibitory domain as part of the suppression mechanism of WNK4, leading to an elevated WNK4 activity at baseline. The presence of CaM binding and SGK1 phosphorylation sites in or close to the inhibitory domain suggests that WNK4 activity is subject to the regulation by intracellular Ca2+ and phosphorylation.
Keywords: protein phosphorylation, calmodulin, SGK1, WNK4, NKCC2
pseudohypoaldosteronism ii (PHAII), also known as familial hyperkalemia and hypertension or Gordon's syndrome, is a Mendelian form of disorder featuring hypertension, hyperkalemia, mild metabolic acidosis, and low levels of plasma renin (9, 17). Two genes in the With-No-lysine (K) (WNK) serine/threonine kinase family, WNK1 and WNK4, are associated with PHAII (27). In contrast to the intronic deletion mutations in WNK1, PHAII mutations in WNK4 are missense mutations (27). Most PHAII-causing WNK4 mutations are clustered within an “acidic motif” rich in negatively charged amino acid residues following the kinase domain with the exception of R1185C, which is located in the COOH-terminal region of WNK4 (27). Patients carrying the R1185C mutation manifest hyperkalemia, hyperchloremic acidosis, and normal blood pressure (1). In vitro studies indicate that WNK4 acts as an integrative regulator of renal electrolyte transport pathways including Na+ (28, 31), K+ (13), Cl− (12), and Ca2+ (11). The R1185C mutation partially relieved the suppression of the surface expression of Na+-Cl− cotransporter (NCC) by WNK4, albeit to a lesser extent than the E562K mutation in the acidic motif (3). The distinct location of R1185C mutation indicates that it likely disrupts a unique regulatory element(s) in WNK4. However, it is unclear what regulatory element(s) is involved.
WNK4 has been shown to be regulated by the renin-angiotensin-aldosterone system (5, 22, 24, 25). Infusion of angiotensin II (ANG II) induces phosphorylation of the thiazide-sensitive NCC within 30 min in mice (24). The absence of WNK4 in a WNK4 knockout mouse strain precludes the NCC and SPAK phosphorylation promoted by a low-salt diet or ANG II infusion, suggesting that the ANG II action on the NCC occurs via a WNK4-SPAK-dependent signaling pathway in vivo (5). In the Xenopus laevis oocyte system, ANG II exerts a positive effect on NCC via AT1 through WNK4-SPAK (STE20/SPS1-related proline/alanine-rich kinase) pathway (22). PHAII mutations in the acidic motif abolish the response of WNK4 to ANG II (22). We recently found that WNK4 kinase activity is regulated by Ca2+ ions and the PHAII-causing mutations in the acidic motif alleviate this regulation (14). Because stimulation of ANG II receptor AT1 results in elevation of intracellular Ca2+ concentration ([Ca2+]i), the Ca2+-dependent activation mechanism of WNK4 is a critical link in ANG II signaling. Disruption of this mechanism by PHAII mutations in the acidic motif may permanently “switch on” WNK4, leading to disease. It is unclear, however, whether the R1185C mutation also disrupts a Ca2+-dependent regulatory mechanism in WNK4.
In addition to a direct regulation by ANG II, WNK4 may also be regulated by the serum- and glucocorticoid-induced protein kinase (SGK1), which is induced by aldosterone in the kidney (2). Two SGK1 phosphorylation sites, S1169 and S1196, have been identified in mouse WNK4 (mWNK4) (20, 21). These sites correspond to S1190 and S1217, respectively, in human WNK4 (hWNK4). S1169 is situated within an SGK1 phosphorylation consensus site, RXRXXS/T, and is able to be phosphorylated by SGK1 in vitro (20). Phospho-mimicking S1169D mutation alleviates the inhibition of the epithelial Na+ channel (ENaC) and the renal outer medullary K+ channel (ROMK) by WNK4 (20). The R1185C mutation is situated at the -5 position relative to the SGK1 phosphorylation site S1190 (1185RQRRLS1190 in hWNK4, corresponding to 1164RQRRLS1169 in mWNK4). Thus, this mutation might result in a disruption of phosphorylation of WNK4 by SGK1 and, in turn, a dysregulation of WNK4 targets.
In this study, we aimed at the mechanism disrupted by the R1185C mutation. We identified a calmodulin (CaM) binding site in WNK4 that harbors this mutation. The R1185C mutation reduces CaM binding to WNK4. We also found that the phosphorylation of WNK4 by SGK1 is affected by the R1185C mutation. We chose Na+-K+-2Cl− cotransporter 2 (NKCC2) to assess WNK4 activity because it exhibits a more stable response to WNK4 than NCC, the primary target of WNK4. The R1185C mutation increases the positive effect of WNK4 on NKCC2 in X. laevis oocytes. Functional studies indicate that the R1185C mutation-induced changes in both the CaM binding and the SGK1 phosphorylation sites reflect the perturbation of an inhibitory domain of WNK4 by this mutation.
MATERIALS AND METHODS
cDNA constructs.
The human WNK4 cDNA was a generous gift from Dr. Xavier Jeunemaitre and was used previously (11). The cDNA for mouse NKCC2 (BC016888) was purchased from Open Biosystems (Huntsville, AL). To detect protein expression of NKCC2, a hemagglutinin epitope (HA-tag) was added to its NH2 terminus using a PCR-based approach. The cDNA was subcloned into the X. laevis oocytes expression vector pIN (32). The monomeric red fluorescent protein (mRFP) cDNA was a generous gift from Dr. Roger Tsien. cDNAs for rat CaM and the Ca2+ binding-deficient mutant with D to A mutations in all four EF hands (29) (named as CaM 1–4 in this study) were kindly provided by Dr. John Adelman. They were subcloned into X. laevis oocyte expression vector pIN (11) with HA-tag or GST-tag.
GST fusion proteins.
GST fusion proteins of human WNK4 were generated using a PCR-based approach and mutations were introduced using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) following the manufacturer's instruction. All mutations were confirmed by sequencing. GST fusion WNK4 segments and corresponding mutations were subcloned into pGEX-6p-1 vector (GE Healthcare, Piscataway, NJ). GST and GST-WNK4 fusion proteins were expressed in Escherichia coli BL21. After being induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside, cells were collected and lysed by sonication. GST and GST fusion proteins were purified using Glutathione Sepharose 4B MicroSpin column (GE Healthcare).
Western blot analysis.
Western blot analyses were performed as described previously (11). Monoclonal anti-HA antibody (product no. H9658, 1:5,000 dilution) was purchased from Sigma (St. Louis, MO). Rabbit anti-WNK4 antibody (WNK41-S, 1:1,500 dilution) was purchased from Alpha Diagnostic International (San Antonio, TX). Rabbit anti-CaM antibody (sc-5537, 1:1,000 dilution) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Chemiluminescence was detected using a SuperSignal West Femto Maximum Sensitivity Substrate kit (Pierce Biotechnology, Rockford, IL) in accordance with the manufacturer's protocol.
In vitro kinase assay.
To assess SGK1 phosphorylation activity, 2 μg of each GST fusion protein were incubated with 20 ng of active SGK1 (δ1–59, S422D, product no. 14–331, Millipore, Billerica, MA) or inactive SGK1 (S422D, product no. 14–332, Millipore) in 25-μl reaction buffer containing 0.5 μCi [γ-32P]ATP (PerkinElmer, Waltham, MA), 50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, at 30°C for 30 min. The reactions were then terminated with 5 μl of 6× electrophoresis sample buffer and the samples were subjected to SDS-PAGE (12% polyacrylamide). After being transferred to PVDF, the incorporation of radioactive phosphate was assessed by autoradiography. Parallel to the kinase assay reaction, equal amounts of GST fusion proteins were subjected to SDS-PAGE and Coomassie Blue staining as equal loading control. The intensities of the bands were analyzed using Gel-Pro Analyzer 4.0.
GST pull-down assay.
X. laevis oocytes were injected with cRNAs encoding GST (as control), GST-CaM, or GST-CaM 1–4 together with HA-WNK4 or HA-WNK4ΔCaM (with deletion of amino acids 1175–1194) and were cultured in 0.5 × L-15 medium at 18°C. Two days after injection, 50 oocytes were lysed with lysis buffer at 20 μl/oocyte. After vigorous vortex, the oocytes were centrifuged at 3,500 g for 10 min at 4°C to remove the cellular debris and yolk proteins. The supernatants were incubated with 50 μl of 1:1 slurry glutathione Sepharose (GE Healthcare). After being rocked at 4°C for 2 h, the Sepharose beads were washed 3 times with 500-μl lysis buffer supplemented with protease inhibitor cocktail. Then, GST, GST-CaM, or GST-CaM 1–4 proteins were eluted from Sepharose beads by incubation with 10 mM l-glutathione reduced for 1 h at 4°C. Both the supernatants and the proteins bound to the Sepharose beads were, respectively, subjected to SDS-PAGE. The proteins were transferred from the SDS-PAGE to PVDF membrane and immunoblotting experiments were carried out with anti-GST (27–4577-01, GE Healthcare, 1:2,000 dilution) and anti-HA (H9658, Sigma, 1:5,000 dilution) antibodies to determine HA-WNK4 constructs associated with GST fusion CaM constructs.
Fluorescent microscopy.
HeLa cells (CCL-2, American Type Culture Collection, Manassas, VA) were seeded and cultured in coverslips and put into six-well plates for 18 to 24 h to reach 90% confluency. Cells were then transfected with 1 μg/well total plasmid DNA containing 0.75 μg pLenti4-FLAG-EGFP-WNK4 and 0.25 μg pmRFP-CaM using GenJet In Vitro DNA Transfection Reagent for HeLa Cell (Ver. II, SignaGen Laboratories, Ljamsville, MD). The former plasmid contains enhanced green fluorescent protein (EGFP) fused with WNK4 in pLenti4 vector (Invitrogen, Carlsbad, CA) and the latter is a homemade vector containing mRFP-fused CaM. Two days later, cells were washed with PBS for three times, fixed by 0.5% paraformaldehyde (in PBS) for 15 min, and then washed again with PBS for five times. Images of EGFP-WNK4 and mRFP-CaM in cells were captured using a Leica DC500 12-megapixel camera and Leica DMIRB fluorescence microscope (Leica Microsystems, Heerbrugg, Switzerland) with a FITC filter (blue, 450–490 nm/LP515) for EGFP and subsequently with a rhodamine filter (green, 515–560 nm/LP590) for mRFP.
Pull-down assay with CaM-agarose beads.
CaM-agarose beads were purchased from Sigma and the experiments were performed following the manufacturer's instruction. Briefly, CaM-agarose beads were equilibrated in binding buffer with 2 mM CaCl2 or 1 mM EGTA. GST and GST-fused WNK4 segments were incubated with CaM-agarose beads for 30 min at room temperature (RT) followed by three washes with the corresponding buffers. Proteins bound to CaM-agarose beads were eluted with elution buffer and then subjected to SDS-PAGE Coomassie Blue staining.
Fluorometric measurements with dansyl-CaM.
Dansyl-CaM was synthesized following the reported protocol (26). Briefly, CaM (1 mg) was dissolved in 1 ml of 20 mM NH4HCO3, pH 7.5, and Ca2+ was added to a final concentration of 1 mM. Dansyl chloride in acetone was added to a final concentration of 50 μM. The mixture was kept at room temperature for 2 h and was vortexed every 20 min. The resulted dansyl-CaM was dialyzed overnight in 20 mM NH4HCO3, pH 7.5. The GST and GST-WNK4 1163–1243 fusion protein were produced using E. coli BL21 and were purified as described earlier. The proteins were also dialyzed overnight in 20 mM NH4HCO3, pH 7.5. Fluorescence emission spectra was monitored at 20°C on an ISS PC1 photon-counting spectrofluorometer (ISS, Champaign, IL) in the presence of 20 mM NH4HCO3, pH 7.5, 400 nM dansyl-CaM, and 400 nM GST-WNK4 1163–1243 or GST (as control) and 1 mM Ca2+ or 2 mM EGTA. The excitation wavelength was 340 nm, and the emission spectra were recorded from 400 to 650 nm using a band pass of 3 nm on both excitation and emission monochromators.
Far-Western overlay and Ca2+ concentration dependency.
[35S]-labeled CaM probe was synthesized using TNT SP6 High-Yield Master Mix following the manufacturer's instruction (Promega, Madison, WI). GST fusion proteins were subjected to SDS-PAGE (12%) and were blotted on PVDF membranes. The membranes were blocked in Hyb 75 (20 mM HEPES, 75 mM KCl, 2.5 mM MgCl2, 1 mM CaCl2, 1 mM dithiothreitol, 0.05% Nonidet P-40, pH 7.5) with 1% nonfat dry milk for 15 min at RT. After being washed with Hyb75 without blocking agent for 5 min 3 times, membranes were cut into 4-mm strips containing 1 μg GST fusion protein each. Strips were incubated with [35S]-CaM probe in the absence and presence of different Ca2+ concentration ([Ca2+]) for 3 h at RT. [Ca2+] was adjusted by using 10 mM EGTA and CaCl2 according to the MAXCHELATOR program (http://maxchelator.stanford.edu/). The membranes stripes were washed for 15 min 3 times in the corresponding binding buffers to remove the unbound [35S]-CaM. The stripes were air dried and exposed to X-ray films. The relative intensities of the bands were analyzed using Gel-Pro Analyzer 4.0 (Media Cybernetics, Bethesda, MD).
Na+ uptake assay.
The animal protocol used in this study was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Alabama at Birmingham. X. laevis oocytes were collected as previously described (11). Oocytes were microinjected with in vitro transcribed complementary RNAs (cRNAs) of NKCC2 alone or with wild-type (WT) WNK4, R1185C mutant, WNK4 ΔCaM mutant, or mutant with mutations in SGK1 sites at 6.25–12.5 ng/oocyte. Two days after injection, NKCC2 activity was assessed by 22Na+ tracer uptake in isotonic uptake solution in the presence or absence of bumetanide as reported (18), but without Cl−-free treatment. Bumetanide-sensitive Na+ uptake is defined as the difference between uptake values in the presence and in the absence of bumetanide. After being lysed by 10% SDS, radioactivity of each oocyte was determined using a scintillation counter. Statistical significance is defined as P < 0.05 by Student's t-test or Mann-Whitney Rank Sum Test using SigmaPlot 10 (Systat Software, Chicago, IL).
RESULTS
PHAII-causing R1185C mutation is situated within a CaM binding site in WNK4.
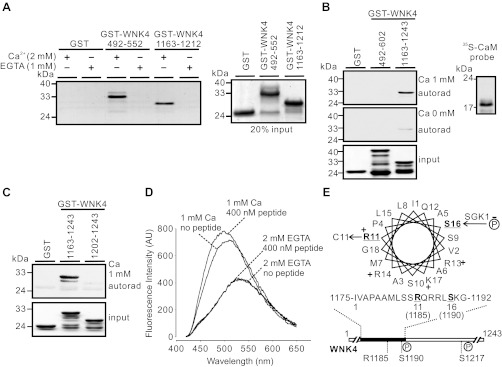
To identify potential regulatory mechanisms relevant to the intracellular Ca2+ signaling pathway, we used an online CaM target database to analyze the sequence of WNK4 (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html). Two potential CaM binding sites in WNK4 were identified. The first (amino acids 505–523) is close to a cluster of PHAII-causing mutations (E562K, D564A, and Q565E). The second (amino acids 1175–1194) contains R1185, the site for a PHAII mutation, and S1190, an SGK1 phosphorylation site (20). GST fusion protein of either WNK4 492–552 or WNK4 1163–1212 was capable of binding to CaM-agarose beads in the presence of Ca2+ (Fig. 1A). The CaM binding abilities of the two sites were further assessed by overlaying 35S-labeled CaM on GST fusion WNK4 segments (Fig. 1B). The WNK4 492–602 segment contained sites for PHAII mutations within the acidic motif, whereas the 1163–1243 segment contained S1190 and S1217, two SGK1 phosphorylation sites (20, 21). The 35S-labeled CaM probe bound to 1163–1243 segment but not to the 492–602 segment in the presence of 1 mM Ca2+, and the binding was greatly reduced in the absence of Ca2+ (Fig. 1B). Ca2+/CaM binding was abolished when an NH2-terminal portion of 40 amino acids containing the CaM binding site was deleted in the construct (Fig. 1C). This supports that 1175–1194 is a CaM binding site in GST-WNK4 1163–1243. The GST-WNK4 1163–1212 construct also bound to dansyl-CaM (Fig. 1D), resulting in an increase in fluorescence intensity and a shift to shorter wavelengths, features of interaction between a protein and dansyl-CaM (26). A helical wheel projection of WNK4 1175–1192 shows a typical feature of a CaM binding site with one side of the helix positively charged and the other side composed of hydrophobic amino acid residues (Fig. 1E) (15). Thus, WNK4 1175–1192 consistently exhibited the features of a bona fide CaM binding site and was further analyzed in this study.
Fig. 1.
Identification of a calmodulin (CaM) binding site in the COOH-terminal region of WNK4. A: GST-WNK4 segments 492–552 and 1163–1212 exhibited ability of binding to CaM-agarose beads in the presence of 2 mM Ca2+ (left). Proteins bound to CaM-agarose beads were stained with Coomassie Blue. No binding was detected in the presence of 1 mM EGTA to chelate trace amount of Ca2+. Twenty percent of the proteins loaded in different groups were also stained by Coomassie Blue to ensure equal loading (right). B: in the presence of 1 mM Ca2+, [35S]-labeled CaM probe (autoradiograpy shown at right) bound to GST-WNK4 1163–1243, but not to GST-WNK4 492–602 (top panel). In the absence of Ca2+, both 492–602 and 1163–1243 of WNK4 lost the CaM binding ability (middle panel). Coomassie Blue staining indicated equal loading of proteins (bottom panel). C: [35S]-labeled CaM probe failed to bind to GST-WNK4 1202–1243, in which the CaM binding site was removed. Results shown in A–C are representatives from at least 3 similar experiments. D: dansyl-CaM fluorescence spectra in the absence or presence of 400 nM GST-WNK4 1163–1212 peptide at 1 mM Ca2+ or 2 mM EGTA. When dansyl-CaM interacted with the peptide in the presence of 1 mM Ca2+, the dansyl-CaM emission spectra displayed a characteristic shift in the absorption of blue light and an increase in fluorescence intensity. AU, arbitrary units. E: helical wheel projection of the CaM binding site (WNK4 1175–1192). The positively charged amino acids are indicated with “+”; disease-causing mutation site R1185 and SGK1 phosphorylation site S1190 are underlined.
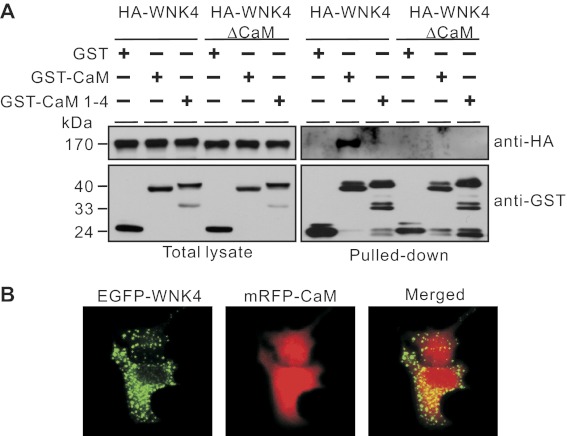
We then used a GST pull-down approach to examine the interaction between GST-tagged CaM and HA-tagged WNK4 constructs. A Ca2+ binding-deficient CaM mutant with mutations in its four EF hands (CaM 1–4) (29) and a WNK4 construct with deletion in amino acids 1175–1194 (WNK4ΔCaM) were also utilized to examine the association between CaM and WNK4. WT GST-CaM pulled down WT HA-WNK4 but not HA-WNK4ΔCaM; GST or GST-CaM 1–4 pulled down neither (Fig. 2A). This indicated that the CaM binding site in WNK4 is necessary for its association with CaM. To confirm that the interaction between WNK4 and CaM occurs in cells, we cotransfected EGFP-tagged WNK4 and mRFP-tagged CaM into HeLa cells. EGFP-WNK4 showed a punctate distribution pattern, whereas mRFP-CaM was ubiquitously distributed in the cell. They exhibited colocalization in the perinuclear region of the cell (Fig. 2B).
Fig. 2.
Association of WNK4 and CaM in Xenopus laevis oocytes and HeLa cells. A: HA-WNK4 was pulled down by GST-CaM but not by GST-CaM 1–4 (Ca2+ binding-deficient mutant) when they were coexpressed in X. laevis oocytes (right, lanes 1–3). After the CaM binding site (amino acids 1175–1194) was removed (HA-WNK4ΔCaM), the association between HA-WNK4 and GST-CaM was abolished (right, lanes 4–6). The presence of GST, GST-CaM, GST-CaM 1–4, HA-WNK4, and HA-WNK4ΔCaM in oocyte total lysates was detected by Western blot using antibodies against GST and HA, respectively (left). Data shown are representative of 2 to 3 experiments. B: enhanced green fluorescent protein (EGFP)-tagged WNK4 (EGFP-WNK4) and monomeric red fluorescent protein-tagged CaM (mRFP-CaM) were transiently cotransfected into HeLa cells. After 24 h, cells were fixed and images were captured by fluorescence microscopy (original magnification ×200).
R1185C mutation reduces CaM binding to WNK4.
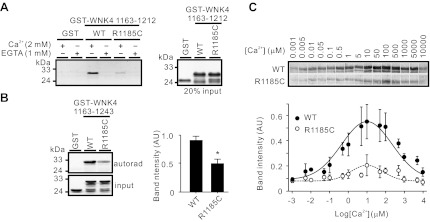
The R1185C mutation eliminates a positive charge carried by arginine (R) and is expected to affect CaM binding ability (Fig. 1E). Indeed, R1185C mutation reduced the binding of GST-WNK4 1163–1212 fragment to CaM-agarose beads (Fig. 3A). Similarly, binding of 35S-labeled CaM to GST-WNK 1163–1243 was decreased by 45.0 ± 8.9% in the presence of R1185C mutation (Fig. 3B). The R1185C mutation resulted in a lower level of CaM binding at any [Ca2+] tested (Fig. 3C).
Fig. 3.
R1185C mutation suppressed CaM binding ability of WNK4 COOH-terminal region. A: R1185C mutation greatly inhibited the binding of GST-WNK4 1163–1212 to CaM-agarose beads (left). Coomassie Blue staining indicated equal loading of proteins between wild-type (WT) and R1185C mutant of GST-WNK4 1163–1212 (right). B: CaM binding ability of GST-WNK4 1163–1243 was significantly suppressed by the R1185C mutation as demonstrated by far-Western overlay with [35S]-labeled CaM probe. Densitometric analyses of far-Western results (n = 3) are shown at right. *P < 0.05 vs. WT group. C: [Ca2+] dependence of [35S]-CaM binding to GST-WNK4 1163–1243 WT and R1185C mutant demonstrated by far-Western blot analyses. Bottom: means ± SE of band intensities from 5 experiments.
R1185C mutation reduces the phosphorylation of WNK4 by SGK1.
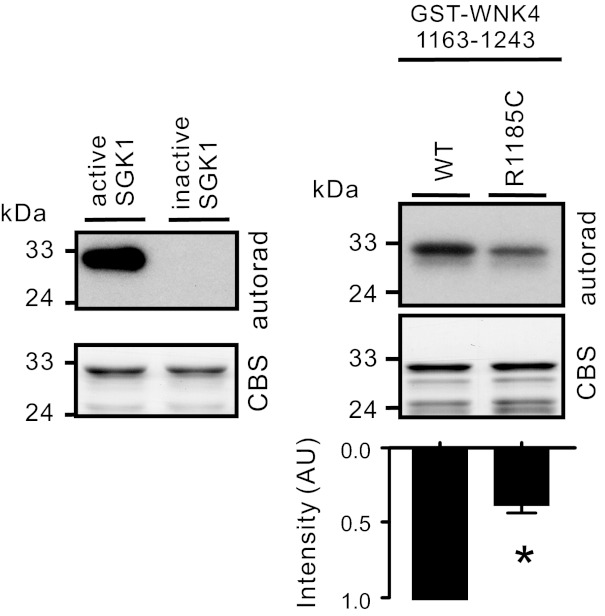
The R1185C mutation is close to SGK1 phosphorylation sites, S1190 and S1217 (hWNK4 numbering, corresponding to S1169 and S1196, respectively, in mWNK4), in the COOH-terminal region of WNK4 (20, 21). We next determined whether the phosphorylation of WNK4 by SGK1 in the COOH-terminal region is altered by the R1185C mutation using in vitro kinase assay. The GST-WNK4 1163–1243 fragment containing both S1190 and S1217 was phosphorylated by active SGK1 but not by inactive SGK1 (Fig. 4, left). A 61.8 ± 4.6% reduction in the phosphorylation intensity was observed in the presence of the R1185C mutation (Fig. 4, right).
Fig. 4.
Phosphorylation of GST-WNK4 1163–1243 by serum- and glucocorticoid-induced protein kinase (SGK1) was decreased by R1185C mutation as demonstrated by in vitro kinase assay. A summary of the band intensities of R1185C normalized to WT from 3 experiments is shown in the bottom panel. *P < 0.01 vs. WT group. Left: GST-WNK4 1163–1243 fragment was phosphorylated by active SGK1 but not by inactive SGK1. Shown are autoradiography of the reaction mixture transferred to PVDF membrane and inputs of the reaction mixtures in SDS-PAGE with Coomassie Blue staining (CBS).
Identification of a novel SGK1 phosphorylation site affected by Ca2+/CaM.
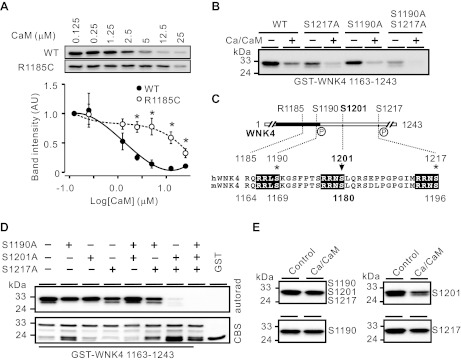
Because the two previously identified SGK1 phosphorylation sites are either in or close to the CaM binding site, we compared the Ca2+/CaM dependence of SGK1 phosphorylation between WT WNK4 and the R1185C mutant (Fig. 5A). In the presence of 50 μM Ca2+, phosphorylation of WT WNK4 1163–1243 by SGK1 was dose dependently suppressed by CaM; in contrast, that of the R1185C mutant was less sensitive to CaM (Fig. 5A). We next examined which of the two SGK1 phosphorylation sites was decreased by Ca2+/CaM by introducing S1190A or S1217A mutation (Fig. 5B). Even when both sites were mutated, GST-WNK4 1163–1243 was still phosphorylated by SGK1 (Fig. 5B). Furthermore, the phosphorylation was still inhibited by Ca2+/CaM in the presence of double mutations (Fig. 5B). This indicated that a novel SGK1 site is present in the 1163–1243 region of WNK4 and its phosphorylation is suppressed by Ca2+/CaM. By examining all the serine/threonine residues within 1163–1243 of WNK4, we found that S1201 is in an RRXS motif (1198RRNS1201) similar to the previously identified SGK1 sites (1187RRLS1190 and 1214RRNS1217; Fig. 5C). Phosphorylation by SGK1 was completely abolished when all the three SGK1 sites in the GST-WNK4 1163–1243 were mutated to alanine (Fig. 5D). This confirmed that S1201 is a novel SGK1 phosphorylation site in the WNK4 COOH-terminal region.
Fig. 5.
Identification of a new SGK1 phosphorylation site (S1201) whose phosphorylation was affected by Ca2+/CaM. A: CaM dose dependently inhibited phosphorylation of WT GST-WNK4 1163–1243 by SGK1 in the presence of 50 μM Ca2+. In contrast, phosphorylation of R1185C mutant by SGK1 was less sensitive to CaM. Data from 3 independent experiments are normalized to the value of CaM at 0.125 μM in the bottom. *P < 0.05 vs. WT group. B: suppression of SGK1 phosphorylation by Ca2+/CaM (50 μM/5 μM) was unaltered by S1190A, S1217A, or both mutations in GST-WNK4 1163–1243. Double mutation of the 2 SGK1 sites did not completely eliminate the phosphorylation of this segment by SGK1. C: S1201 is within an RRXS motif, in which the 2 known SGK1 phosphorylation sites (S1190 and S1217, marked with *) are located. These motifs are conserved between human (h) and mouse (m) WNK4. D: triple mutation in S1190, S1201, and S1217 abolished the phosphorylation of GST-WNK4 1163–1243 segment by SGK1. E: Ca2+/CaM (50 μM/5 μM) greatly suppressed the phosphorylation of S1201 by SGK1 compared with the other 2 SGK1 phosphorylation sites (S1190 and S1217). In vitro SGK1 phosphorylation experiments were performed with WT (S1190/S1201/S1217) and mutant GST-WNK4 1163–1243 constructs. WT contained no mutations, S1190 contained S1201A and S1217A mutations, S1201 contained S1190A and S1217A mutations, and S1217 contained S1190A and S1201A mutations. Results were confirmed in 2 other experiments.
To determine phosphorylation at which of the three SGK1 phosphorylation sites is affected by Ca2+/CaM, we performed in vitro phosphorylation using GST-WNK4 1163–1243 constructs containing double serine mutations that allow phosphorylation in only one of the three SGK1 sites. Because each site exhibited different SGK1 phosphorylation intensity, we exposed X-ray films in different durations to maintain the intensity at comparable level. As shown in Fig. 5E, SGK1 phosphorylation of S1201 was suppressed greatly by Ca2+/CaM. In contrast, phosphorylation at S1190 or S1217 was only slightly affected by Ca2+/CaM (Fig. 5E). It is unclear why S1201 was more affected by Ca2+/CaM than the other two sites.
R1185C mutation suppresses phosphorylation at S1190 and S1217 and enhances that at S1201.
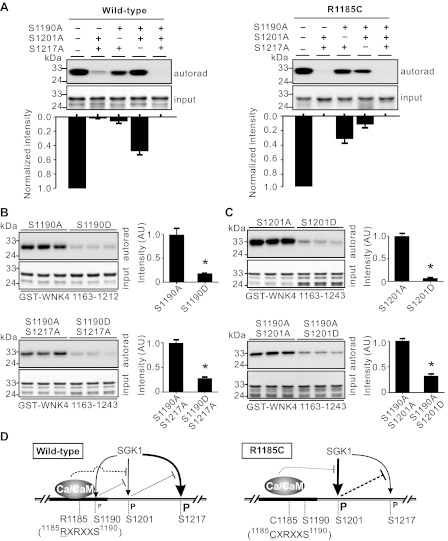
SGK1-mediated phosphorylation at WNK4 COOH-terminal region was decreased by the R1185C mutation (Fig. 4); however, it was unclear how this mutation alters phosphorylation at individual SGK1 sites. To clarify this issue, we performed in vitro phosphorylation assays using GST-WNK4 1163–1243 constructs containing double serine mutations that allow SGK1 phosphorylation at only one of the three SGK1 sites (Fig. 6A). In the WT constructs, the level of phosphorylation increased in the following order: S1190, S1201, and S1217 (Fig. 6A, left). In the presence of R1185C mutation, phosphorylation at S1190 was completely eliminated and S1201 became the most intensely phosphorylated among the three sites (Fig. 6A, right). The R1185C mutation eliminates the arginine (R) at -5 position of S1190 in the classic SGK1 phosphorylation consensus sequence (1185RQRRLS1190); this is likely the reason for the absence of phosphorylation at S1190 in the R1185C mutant. It is unclear how the level of phosphorylation intensity at S1201 or S1217 was altered by the R1185C mutation.
Fig. 6.
R1185C mutation affected SGK1 phosphorylation sites of WNK4 COOH terminal differently. A: SGK1 phosphorylation intensity of S1217 site was the strongest among the 3 SGK1 sites and S1190 was the weakest in WT GST-WNK4 1163–1243 fragment in the absence of Ca2+/CaM (left). In the presence of the R1185C mutation, phosphorylation of S1190 was undetectable, phosphorylation of S1201 was significantly increased, and phosphorylation of S1217 was greatly inhibited (right). The band intensities normalized to controls (without mutations in the 3 SGK1 sites) from 3 experiments are shown in the bottom panels. B: phospho-mimicking mutation S1190D suppressed S1201 phosphorylation by SGK1 in either GST-WNK4 1163–1212 or GST-WNK4 1163–1243 segment. Summaries of the band intensities from 3 experiments are shown at right. C: phospho-mimicking mutation S1201D significantly inhibited the phosphorylation of S1217 by SGK1 in GST-WNK4 1163–1243 segment. Summaries of the band intensities from 3 experiments are shown at right. *P < 0.05 between the 2 groups. D: summary of Ca2+/CaM binding and phosphorylation by SGK1 in the COOH terminal of WNK4. The degree of phosphorylation is indicated by the size of letter “P.” The thickness of lines indicates the relative strength of action.
We next examined whether the disruption of phosphorylation at S1190 affects the phosphorylation at other sites. We mimicked the nonphosphorylated or phosphorylated states using alanine (A) or aspartate (D) substitution at S1190, respectively, and evaluated the phosphorylation of S1201 by SGK1 (Fig. 6B). In either GST-WNK4 1163–1212 (without S1217 site; Fig. 6B, top) or GST-WNK4 1163–1243 (with the S1217A mutation) segment (Fig. 6B, bottom), the level of phosphorylation at S1201 by SGK1 was significantly higher with the S1190A mutation than with the S1190D mutation. Similarly, the phosphorylation of S1217 by SGK1 was much higher with the S1201A mutation than with S1201D mutation (Fig. 6C). Based on these results, it is likely the R1185C mutation blocks the phosphorylation of S1190, which will in turn alleviate the inhibitory effect on the phosphorylation of S1201. The increased phosphorylation at S1201 further leads to the suppression of phosphorylation at S1217 (Fig. 6D). This coincides with the phosphorylation intensities observed at the three sites without or with the R1185C mutation (Fig. 6A).
CaM binding site suppresses WNK4-mediated regulation on NKCC2.
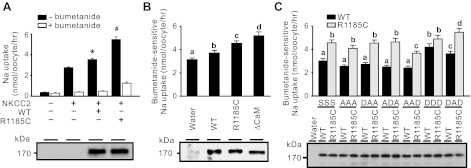
We next evaluated the alterations caused by the R1185C mutation on WNK4-mediated regulation. Because NKCC2 colocalizes with WNK4 in the cortical thick ascending loop of Henle (16), we examined whether NKCC2 is regulated by WNK4. WT WNK4 increased NKCC2-mediated Na+ uptake when expressed in X. laevis oocytes, and this effect was further increased by the R1185C mutation (Fig. 7A). We thus used this system to evaluate the CaM binding site and the SGK1 phosphorylation sites on WNK4 activity.
Fig. 7.
R1185C mutation in WNK4 increased Na+-K+-2Cl− cotransporter 2 (NKCC2)-mediated Na+ uptake by disrupting an inhibitory domain containing the CaM binding site. A: R1185C mutation enhanced the positive regulation of WNK4 on NKCC2-mediated Na+ uptake in X. laevis oocytes. WT WNK4 upregulated NKCC2-mediated 22Na+ uptake (top). R1185C mutation further enhanced the positive regulation of WNK4 on NKCC2 activity. Data are expressed as means ± SE from 4 independent experiments. *P < 0.05 vs. NKCC2 alone group. #P < 0.05 vs. NKCC2 + WT WNK4 group. Open bar, in the presence of 0.1 mM bumetanide; filled bar, in the absence of bumetanide. B: evaluation of the CaM binding site of WNK4 on bumetanide-sensitive 22Na+ uptake mediated by NKCC2. WT, R1185C, and WNK4 mutant with the CaM binding site (amino acids 1175–1194) deletion (ΔCaM) were used in the experiments. Data are derived from 34–49 oocytes in 3 to 4 independent experiments and are expressed as means ± SE. Unless indicated, data are not significantly different (P ≥ 0.05) and are indicated with the same letter. Equal expression of WNK4 constructs was verified by Western blot analyses with WNK4 antibody (bottom). C: evaluation of phospho-mimicking aspartate (D) mutations at S1190, S1201, and S1217 on bumetanide-sensitive 22Na+ uptake mediated by NKCC2. WT and R1185C mutant-containing mutations at the 3 phosphorylation sites are listed in single letter code in the order of their position (S, serine; A, alanine; D, aspartate). NKCC2 was expressed in all groups. Data are derived from 26–57 oocytes in 2 to 4 independent experiments and are expressed as means ± SE. The groups that are not significantly different (P ≥ 0.05) are marked with the same letter. Except for DDD group, WT is significantly different (P < 0.05) from R1185C. Bottom: representative Western blot analysis with antibody against WNK4 showing the equal expression of WNK4 WT and mutants.
CaM is endogenously expressed in X. laevis oocytes (4, 6). Thus, one can evaluate the effect of Ca2+/CaM binding to WNK4 by elevating of [Ca2+]i. However, we chose not to elevate [Ca2+]i because WNK4 possesses a Ca2+ sensor around the acidic motif and WNK4 kinase activity increases as Ca2+ level increases (14). We thus tested the effect of WNK4 on NKCC2 in the absence of the CaM binding site using the WNK4ΔCaM construct (Fig. 7B). The WNK4 exhibited the highest ability to enhance the Na+ uptake activity of NKCC2 in the absence of its CaM binding site (Fig. 7B). Thus, it is likely that the CaM binding site suppresses WNK4 activity and the R1185C mutation disrupts the suppression, leading to an increase in the positive effect of WNK4 on NKCC2.
Phospho-mimicking at multiple SGK1 sites increases WNK4-mediated regulation on NKCC2.
We next examined whether the effect of R1185C mutation on the regulation of NKCC2 by WNK4 is also caused by the differential phosphorylation at the SGK1 phosphorylation sites. When all the three SGK1 sites were mutated to alanine (A), the effect of R1185C mutation to enhance the regulation of WNK4 on NKCC2 was unchanged (Fig. 7C). This indicated that the effect of R1185C on NKCC2 is not due to the difference in the phosphorylation at the three SGK1 phosphorylation sites. None of the individual phospho-mimicking mutations altered the regulation on NKCC2 significantly either in WT or R1185C background (Fig. 7C). However, double (DAD) and triple (DDD) mutations significantly increased bumetanide-sensitive Na+ uptake of NKCC2 over control in WT background (Fig. 7C). The difference between WT and R1185C diminished in the presence of triple mutations (Fig. 7C). Based on the results of Fig. 6A, DAD and ADA may best represent the difference between WT and R1185C in phosphorylation by SGK1, respectively. WT DAD still exhibited significantly higher activity in regulating NKCC2 than R1185C ADA, but the difference became much smaller (Fig. 7C).
DISCUSSION
In this study, we provide evidence that the R1185C mutation disrupts a CaM binding site at the WNK4 COOH-terminal region and alters the phosphorylation of WNK4 by SGK1. The R1185C mutation is located in the middle of the CaM binding site, and the mutation reduces the Ca2+/CaM binding ability of WNK4. The R1185C mutation reduces SGK1 phosphorylation at S1190 and, in turn, increases phosphorylation at S1201 and decreases phosphorylation at S1217 due to the antagonistic effects of phosphorylation at neighboring sites. The increased positive regulation of NKCC2 by the WNK4 R1185C mutant is likely due to the perturbation of an inhibitory domain in WNK4. The CaM binding and SGK1 phosphorylation sites are likely regulatory elements in the inhibitory domain in response to the increase in [Ca2+]i and SGK1, respectively.
Both the CaM binding site (1175–1194) and the SGK1 phosphorylation sites (S1191, S1201, and S1217) are localized in a conserved region in the COOH terminal of WNK4. This region likely plays a key role in regulating WNK4 activity. Deletion of CaM binding site or phospho-mimicking at multiple SGK1 phosphorylation sites all resulted in an augment of the effect of WNK4 on NKCC2 (Fig. 7). This indicates that this region suppresses WNK4 activity at baseline. The suppression might be relieved by Ca2+/CaM binding and/or phosphorylation at the SGK1 sites, although further investigation is necessary in this regard. These regulatory elements are likely instrumental in responding to the activation of the renin-angiotensin-aldosterone system.
The Ca2+/CaM binding site might play a role in mediating the regulation by ANG II. ANG II is one of the important factors that lead to elevation of [Ca2+]i. ANG II binds to ANG II receptor AT1, which couples to Gq/11 to activate phospholipase C and to increase the [Ca2+]i (7). ANG II switches the negative effect of WNK4 on NCC into a positive one in the presence of AT1 in X. laevis oocyte system (22). The underlying mechanism is unclear but it may involve the Ca2+-sensing mechanism around the acidic motif to activate WNK4 kinase in response to the elevation of [Ca2+]i caused by ANG II (14). Interacting with Ca2+ ions by the acidic motif might relieve an inhibitory mechanism on WNK4 kinase activity (14). In addition, the CaM binding site of WNK4 may also sense the elevated [Ca2+]i via interacting with Ca2+/CaM. The binding of Ca2+/CaM may relieve a suppression of WNK4 kinase activity. It is possible that the CaM binding site interacts with the kinase domain to inhibit the kinase activity of WNK4 at baseline. When [Ca2+]i rises, Ca2+/CaM competitively binds to the CaM binding site and relieves the inhibition. Similar mechanisms are present in Ca2+/CaM-dependent kinases, such as CaM KI, CaM KII, and CaM KIV (10). However, the CaM binding site is usually close to the inhibitory domain and the kinase domain in these kinases. In the case of WNK4, there is a potential CaM binding site at amino acids 505–523 (Fig. 1), which is close to the putative autoinhibitory domain (amino acids 466–489) (30). However, further evidence is needed to demonstrate its functionality. The CaM binding site/inhibitory domain at the COOH-terminal region is likely an additional layer of Ca2+-dependent regulatory mechanism in WNK4 in addition to that around the acidic motif. The R1185C mutation may disrupt the interaction between the CaM binding site and the kinase domain, and meanwhile its ability to interact with Ca2+/CaM is also decreased, resulting in a reduced inhibition of WNK4 activity at baseline. This is supported by our results (Fig. 7B) that removal or disruption of the CaM binding site by R1185C mutation increased the regulation of NKCC2 by WNK4.
The SGK1 phosphorylation sites in WNK4 may mediate the effect of aldosterone. In response to ANG II, SGK1 expression is induced by aldosterone (2). Phosphorylation of WNK4 by SGK1 at multiple sites in the COOH-terminal region relieves the inhibition of WNK4 activity, and in turn increases the activity of NKCC2. It is likely phosphorylation at the SGK1 site relieves the inhibition on the WNK4 kinase activity, which is necessary to regulate SLC12 family members via the OSR1/SPAK (8). This is consistent with the observation that aldosterone mediates the long-term effect of ANG II on the phosphorylation of OSR1 and NCC in mice (24). It is possible that the CaM binding site and the SGK1 phosphorylation sites are all parts of the same inhibitory domain. Phosphorylation by SGK1 at multiple sites likely also disrupts the binding of this inhibitory domain to the kinase domain, resulting in an elevation of kinase activity. Thus, the same inhibitory domain of WNK4 might be the target of both acute and long-term actions of ANG II via Ca2+/CaM and SGK1-mediated phosphorylation, respectively. More studies are needed to elucidate the exact mechanisms.
We observed significant effects on NKCC2 only by mimicking phosphorylation at multiple SGK1 sites. This does not exclude the effect of phosphorylation at single SGK1 site on other electrolyte transport proteins. Phospho-mimicking at S1169 of mWNK4 (corresponding to S1190 in hWNK4) relieves the inhibition of ENaC and ROMK by WNK4 (20). The effects of WNK4 on ENaC and ROMK appear to be independent of the kinase activity of WNK4 (13, 19). Therefore, a different mechanism might be responsible for the effects of phosphorylation at S1169 of mWNK4 on ENaC and ROMK.
The results shown in Fig. 7C also suggest that the WNK4 activity positively correlates with the number of phosphorylated serine residues at the SGK1 phosphorylation sites. This is consistent with the hypothesis that the SGK1 sites are situated within an inhibitory domain of WNK4 and phosphorylation at SGK1 sites relieves the inhibition in a dose-dependent manner. The fact that R1185C mutation is close to the SGK1 sites suggests that this mutation disrupts the same inhibitory domain. The elevated ability of the R1185C mutant to regulate NKCC2 remained in the absence of phosphorylatable serine residues at these SGK1 sites (Fig. 7C), indicating that altered phosphorylation preference at the SGK1 sites caused by the R1185C mutation (Fig. 6A) is not the cause of the elevated activity of R1185C mutant. However, altered phosphorylation status may provide additional changes in response to the induction/activation of SGK1 in the context of R1185C mutation. Further studies using specific antibodies against phosphorylated serine residues at these sites are necessary to confirm the alteration of phosphorylation preference due to R1185C mutation. Additionally, phospho-mimicking serine to aspartate mutation may not fully represent phosphorylation at the SGK1 sites. Again, phosphorylation-specific antibodies for these sites are needed to correlate the phosphorylation status of WNK4 with the activities of NKCC2 and other ion transport proteins.
Loss-of-function mutations in NKCC2 cause Bartter's syndrome featuring hypokalemic alkalosis, normal to low blood pressure (23). As a mirror image of Bartter's syndrome, PHAII caused by the R1185C mutation could also be a consequence of the gain-of-function in NKCC2. The increased activity of NKCC2 by the R1185C mutation is consistent with the hypothesis proposed by Gordon and colleagues (9) that the primary defect in PHAII is the excessive Na+ reabsorption at a site in the nephron proximal to the collecting duct. In addition, the R1185C mutation completely eliminates SGK1 phosphorylation at S1190 (S1169 in mWNK4; Fig. 6A). Phospho-mimicking S1169D mutation relieves the inhibitory effect of mWNK4 on ENaC and ROMK (20). Therefore, the inhibitory effect of WNK4 on ENaC and ROMK is likely maintained in the presence of the R1185C mutation. This will contribute to hyperkalemia.
We utilized the Na+ uptake activity of NKCC2 as a measure of WNK4 activity. This experimental setting may not fully reflect all regulatory actions of WNK4. WNK4 may exert stimulatory and/or inhibitory effects on renal transport proteins. The stimulatory effect involves the activation of SPAK/OSR1, which in turn phosphorylates their substrates such as NCC (8, 24). The inhibitory effect involves a lysosomal degradation pathway (3). In our system, it is likely that the stimulatory effect of WNK4 on NKCC2 plays a dominant role. Our results do not exclude the possibility that the R1185C mutation and its associated changes in CaM binding and SGK1 phosphorylation may have effects on other ion transport proteins, which are not tested in this study. Furthermore, the X. laevis oocyte system used in this study may not fully reflect what occurs in renal tubular cells and one action of WNK4 might be exaggerated over another. Therefore, an integration of results from different systems, including those from biochemical reactions, epithelial cells, and animal models, would be necessary to further illustrate the effects of R1185C mutation on WNK4 activity and how the alterations in WNK4 property result in disordered electrolyte transport.
We did not examine the effects of R1185C and related mutants of WNK4 on NCC because of technical issues. Our NCC construct exhibited low activity without pretreatment with chloride-free solution. Furthermore, we could not obtain stable inhibitory effect of WNK4 on NCC in X. laevis oocytes as reported by others (28, 31). This made it difficult to assess the effects of CaM binding and SGK1 phosphorylation sites of WNK4 using NCC as a measure of WNK4 activity. In contrast, the effect of WNK4 on NKCC2 was more robust and reproducible. This was the reason that we chose NKCC2 over NCC to assess WNK4 activity. This does not mean that the R1185C mutation does not affect NCC, the primary target of WNK4. In fact, the effect of R1185C mutation on NCC has been reported previously (3). The sequence similarity between NCC and NKCC2 indicates that they may share similar mechanisms in response to the regulation of WNK4, although this needs to be verified using an improved experimental system.
In conclusion, the R1185C mutation decreases Ca2+/CaM binding to WNK4 and alters the SGK1 phosphorylation of WNK4. These alterations may simply reflect the perturbation of an inhibitory domain that harbors these regulatory elements in the COOH-terminal region of WNK4.
GRANTS
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases Grant R01DK072154.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: T.N. and J.-B.P. conception and design of research; T.N., G.W., W.Z., W.-J.D., and J.-B.P. performed experiments; T.N., W.-J.D., and J.-B.P. analyzed data; T.N., W.-J.D., and J.-B.P. interpreted results of experiments; T.N., W.-J.D., and J.-B.P. prepared figures; T.N. and J.-B.P. drafted manuscript; T.N. and J.-B.P. edited and revised manuscript; T.N., G.W., W.Z., W.-J.D., and J.-B.P. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Drs. Xavier Jeunemaitre and Juliette Hadchouel for the WNK4 cDNA, Dr. John Adelman for CaM and mutant cDNAs, and Dr. Roger Tsien for mRFP cDNA. Parts of the results in this manuscript were presented at Experimental Biology 2010, Anaheim, CA, April 24–28, 2010, and Experimental Biology 2011, Washington, DC, April 9–13, 2011.
REFERENCES
- 1. Baz M, Berland Y, Dussol B, Jaber K, Boobes Y. Familial hyperkalemia syndrome (Gordon's syndrome). Presse Med 19: 1981–1984, 1990 [PubMed] [Google Scholar]
- 2. Bhargava A, Fullerton MJ, Myles K, Purdy TM, Funder JW, Pearce D, Cole TJ. The serum- and glucocorticoid-induced kinase is a physiological mediator of aldosterone action. Endocrinology 142: 1587–1594, 2001 [DOI] [PubMed] [Google Scholar]
- 3. Cai H, Cebotaru V, Wang YH, Zhang XM, Cebotaru L, Guggino SE, Guggino WB. WNK4 kinase regulates surface expression of the human sodium chloride cotransporter in mammalian cells. Kidney Int 69: 2162–2170, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Cartaud A, Ozon R, Walsh MP, Haiech J, Demaille JG. Xenopus laevis oocyte calmodulin in the process of meiotic maturation. J Biol Chem 255: 9404–9408, 1980 [PubMed] [Google Scholar]
- 5. Castaneda-Bueno M, Cervantes-Perez LG, Vazquez N, Uribe N, Kantesaria S, Morla L, Bobadilla NA, Doucet A, Alessi DR, Gamba G. Activation of the renal Na+:Cl− cotransporter by angiotensin II is a WNK4-dependent process. Proc Natl Acad Sci USA 109: 7929–7934, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chien YH, Dawid IB. Isolation and characterization of calmodulin genes from Xenopus laevis. Mol Cell Biol 4: 507–513, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Gasparo M, Catt KJ, Inagami T, Wright JW, Unger T. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacol Rev 52: 415–472, 2000 [PubMed] [Google Scholar]
- 8. Delpire E, Gagnon KB. SPAK and OSR1, key kinases involved in the regulation of chloride transport. Acta Physiol (Oxf) 187: 103–113, 2006 [DOI] [PubMed] [Google Scholar]
- 9. Gordon RD, Geddes RA, Pawsey CG, O'Halloran MW. Hypertension and severe hyperkalaemia associated with suppression of renin and aldosterone and completely reversed by dietary sodium restriction. Australas Ann Med 19: 287–294, 1970 [DOI] [PubMed] [Google Scholar]
- 10. Hook SS, Means AR. Ca2+/CaM-dependent kinases: from activation to function. Annu Rev Pharmacol Toxicol 41: 471–505, 2001 [DOI] [PubMed] [Google Scholar]
- 11. Jiang Y, Ferguson WB, Peng JB. WNK4 enhances TRPV5-mediated calcium transport: potential role in hypercalciuria of familial hyperkalemic hypertension caused by gene mutation of WNK4. Am J Physiol Renal Physiol 292: F545–F554, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Kahle KT, Gimenez I, Hassan H, Wilson FH, Wong RD, Forbush B, Aronson PS, Lifton RP. WNK4 regulates apical and basolateral Cl− flux in extrarenal epithelia. Proc Natl Acad Sci USA 101: 2064–2069, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kahle KT, Wilson FH, Leng Q, Lalioti MD, O'Connell AD, Dong K, Rapson AK, MacGregor GG, Giebisch G, Hebert SC, Lifton RP. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet 35: 372–376, 2003 [DOI] [PubMed] [Google Scholar]
- 14. Na T, Wu G, Peng JB. Disease-causing mutations in the acidic motif of WNK4 impair the sensitivity of WNK4 kinase to calcium ions. Biochem Biophys Res Commun 419: 293–298, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Neil KT, DeGrado WF. How calmodulin binds its targets: sequence independent recognition of amphiphilic alpha-helices. Trends Biochem Sci 15: 59–64, 1990 [DOI] [PubMed] [Google Scholar]
- 16. Ohno M, Uchida K, Ohashi T, Nitta K, Ohta A, Chiga M, Sasaki S, Uchida S. Immunolocalization of WNK4 in mouse kidney. Histochem Cell Biol 136: 25–35, 2011 [DOI] [PubMed] [Google Scholar]
- 17. Paver WK, Pauline GJ. Hypertension and hyperpotassaemia without renal disease in a young male. Med J Aust 2: 305–306, 1964 [DOI] [PubMed] [Google Scholar]
- 18. Rinehart J, Kahle KT, de los HP, Vazquez N, Meade P, Wilson FH, Hebert SC, Gimenez I, Gamba G, Lifton RP. WNK3 kinase is a positive regulator of NKCC2 and NCC, renal cation-Cl− cotransporters required for normal blood pressure homeostasis. Proc Natl Acad Sci USA 102: 16777–16782, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ring AM, Cheng SX, Leng Q, Kahle KT, Rinehart J, Lalioti MD, Volkman HM, Wilson FH, Hebert SC, Lifton RP. WNK4 regulates activity of the epithelial Na+ channel in vitro and in vivo. Proc Natl Acad Sci USA 104: 4020–4024, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ring AM, Leng Q, Rinehart J, Wilson FH, Kahle KT, Hebert SC, Lifton RP. An SGK1 site in WNK4 regulates Na+ channel and K+ channel activity and has implications for aldosterone signaling and K+ homeostasis. Proc Natl Acad Sci USA 104: 4025–4029, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rozansky DJ, Cornwall T, Subramanya AR, Rogers S, Yang YF, David LL, Zhu X, Yang CL, Ellison DH. Aldosterone mediates activation of the thiazide-sensitive Na-Cl cotransporter through an SGK1 and WNK4 signaling pathway. J Clin Invest 119: 2601–2612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. San-Cristobal P, Pacheco-Alvarez D, Richardson C, Ring AM, Vazquez N, Rafiqi FH, Chari D, Kahle KT, Leng Q, Bobadilla NA, Hebert SC, Alessi DR, Lifton RP, Gamba G. Angiotensin II signaling increases activity of the renal Na-Cl cotransporter through a WNK4-SPAK-dependent pathway. Proc Natl Acad Sci USA 106: 4384–4389, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simon DB, Karet FE, Hamdan JM, DiPietro A, Sanjad SA, Lifton RP. Bartter's syndrome, hypokalaemic alkalosis with hypercalciuria, is caused by mutations in the Na-K-2Cl cotransporter NKCC2. Nat Genet 13: 183–188, 1996 [DOI] [PubMed] [Google Scholar]
- 24. Talati G, Ohta A, Rai T, Sohara E, Naito S, Vandewalle A, Sasaki S, Uchida S. Effect of angiotensin II on the WNK-OSR1/SPAK-NCC phosphorylation cascade in cultured mpkDCT cells and in vivo mouse kidney. Biochem Biophys Res Commun 393: 844–848, 2010 [DOI] [PubMed] [Google Scholar]
- 25. van der Lubbe N, Lim CH, Meima ME, van VR, Rosenbaek LL, Mutig K, Danser AH, Fenton RA, Zietse R, Hoorn EJ. Aldosterone does not require angiotensin II to activate NCC through a WNK4-SPAK-dependent pathway. Pflugers Arch 463: 853–863, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vorherr T, James P, Krebs J, Enyedi A, McCormick DJ, Penniston JT, Carafoli E. Interaction of calmodulin with the calmodulin binding domain of the plasma membrane Ca2+ pump. Biochemistry 29: 355–365, 1990 [DOI] [PubMed] [Google Scholar]
- 27. Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. Human hypertension caused by mutations in WNK kinases. Science 293: 1107–1112, 2001 [DOI] [PubMed] [Google Scholar]
- 28. Wilson FH, Kahle KT, Sabath E, Lalioti MD, Rapson AK, Hoover RS, Hebert SC, Gamba G, Lifton RP. Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc Natl Acad Sci USA 100: 680–684, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xia XM, Fakler B, Rivard A, Wayman G, Johnson-Pais T, Keen JE, Ishii T, Hirschberg B, Bond CT, Lutsenko S, Maylie J, Adelman JP. Mechanism of calcium gating in small-conductance calcium-activated potassium channels. Nature 395: 503–507, 1998 [DOI] [PubMed] [Google Scholar]
- 30. Xu BE, Min X, Stippec S, Lee BH, Goldsmith EJ, Cobb MH. Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J Biol Chem 277: 48456–48462, 2002 [DOI] [PubMed] [Google Scholar]
- 31. Yang CL, Angell J, Mitchell R, Ellison DH. WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J Clin Invest 111: 1039–1045, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang W, Na T, Peng JB. WNK3 positively regulates epithelial calcium channels TRPV5 and TRPV6 via a kinase-dependent pathway. Am J Physiol Renal Physiol 295: F1472–F1484, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]