Abstract
Metabolic acidosis is a common clinical condition that is caused by a decrease in blood pH and bicarbonate concentration. Increased extraction and mitochondrial catabolism of plasma glutamine within the renal proximal convoluted tubule generates ammonium and bicarbonate ions that facilitate the excretion of acid and partially restore acid-base balance. Previous studies identified only a few mitochondrial proteins, including two key enzymes of glutamine metabolism, which are increased during chronic acidosis. A workflow was developed to characterize the mitochondrial proteome of the proximal convoluted tubule. Based upon the increase in specific activity of cytochrome c oxidase, the isolated mitochondria were enriched eightfold. Two-dimensional liquid chromatography coupled with mass spectrometry was utilized to compare mitochondrial-enriched samples from control and chronic acidotic rats. Proteomic analysis identified 901 proteins in the control and acidotic samples. Further analysis identified 37 peptides that contain an N-ε-acetyl-lysine; of these, 22 are novel sites. Spectral counting analysis revealed 33 proteins that are significantly altered in abundance in response to chronic metabolic acidosis. Western blot analysis was performed to validate the calculated changes in abundance. Thus the current study represents the first comprehensive analysis of the mitochondrial proteome of the rat renal proximal convoluted tubule and its response to metabolic acidosis.
Keywords: chronic acidosis, proximal convoluted tubule, mitochondria, mass spectrometry, spectral counting
metabolic acidosis is a common clinical condition that results from a decrease in blood pH and the concentration of bicarbonate ions (18, 52). This type of acidosis can occur acutely, lasting for a few hours to a day, or as a chronic condition where acid-base balance is not fully restored. Metabolic acidosis also occurs frequently as a secondary complication, which adversely affects the outcome of patients with various life-threatening conditions. Acid-base homeostasis is achieved, in part, by the renal reabsorption and net synthesis of bicarbonate ions and by the excretion of acids and ammonium ions (55).
The various segments of the nephron exhibit rapid and sustained adaptive responses during the onset of metabolic acidosis (5). For example, the proximal convoluted tubule exhibits a pronounced increase in ammoniagenesis and gluconeogenesis (6). To support this adaptation, the renal proximal convoluted tubule extracts and catabolizes large amounts of plasma glutamine (48). The extracted glutamine is transported into the mitochondrial matrix where it is deamidated by the kidney-type glutaminase (KGA) to yield glutamate and an ammonium ion. Subsequently, glutamate dehydrogenase (GDH) oxidizes the glutamate to α-ketoglutarate and yields a second ammonium ion. Further oxidation by α-ketoglutarate dehydrogenase generates an initial bicarbonate ion (31). The cytosolic phosphoenolpyruvate carboxykinase (PEPCK) channels intermediates of the citric acid cycle to the gluconeogenic precursor, phosphoenolpyruvate, and produces a second bicarbonate ion (50). Activation of the apical Na+/H+ exchanger (NHE3) acidifies the glomerular filtrate and facilitates the removal of cellular ammonium ions (38). The increased secretion of hydrogen ions also promotes bicarbonate reabsorption from the lumen (49). The reabsorbed and newly synthesized bicarbonate ions are then transported across the basolateral membrane by the Na+/3HCO3− cotransporter (NBC1) (38).
Increased expression of the key transporters and enzymes of this pathway sustains the increased renal catabolism of glutamine during chronic metabolic acidosis (6). Various proteins, including KGA (8, 57), are upregulated within the proximal convoluted tubule in response to metabolic acidosis. Increased expression of the mitochondrial KGA is due to selective stabilization of the KGA mRNA (19, 23). This process is mediated by two eight-base AU sequences that function as pH-response elements (pH-RE) (28). During chronic acidosis, the protein levels of the basolateral glutamine transporter (SN1), an unidentified mitochondrial glutamine transporter, GDH, PEPCK, and NHE3 are all increased to facilitate the restoration of acid-base balance (5). The combined adaptations sustain the increased synthesis of ammonium ions, the increased reabsorption and production of bicarbonate ions, and their vectorial transport across the apical and basolateral membranes, respectively. However, recent transcriptome (34) and proteomic analyses (9, 54) indicate that the renal expression of a large number of additional mRNA transcripts and proteins is also altered in response to acidosis.
The sensitivity of proteomic profiling can be significantly enhanced by reducing the focus to highly enriched subcellular fractions. Mitochondria are essential organelles that transduce metabolic energy by electron transport and ATP synthesis. They are also involved in other functions such as apoptosis, ionic homeostasis, carbohydrate and amino acid metabolism, and the β-oxidation of fatty acids. In the current study, mitochondria were purified from proximal convoluted tubules that were isolated from normal and chronically acidotic rats. The mitochondrial proteins were digested with trypsin, fractionated by two-dimensional liquid chromatography, and analyzed by mass spectrometry. Many of the observed changes in protein abundance were subsequently validated by Western blot analysis.
MATERIALS AND METHODS
Animals.
All experiments were performed using male Sprague-Dawley rats (∼200 g) obtained from Charles River Laboratories (Kingston, NY). The rats were fed rodent chow (Harlan-Teklad, Madison, WI). Arterial blood pH and HCO3− concentrations were measured with an i-STAT 1 VetScan (Abaxis). The control rats were provided with tap water for drinking and had an arterial blood pH of 7.37 ± 0.03 and HCO3− concentration of 23.9 ± 2.4 mM. Acute metabolic acidosis was induced by stomach loading rats with 20 mmol NH4Cl/kg body wt and providing 0.28 M NH4Cl as their drinking water for 1 day. This protocol produced pronounced decreases in blood pH (7.11 ± 0.04) and HCO3− concentration (8.8 ± 1.1 mM). Rats that were made acidotic for 3 or 7 days were provided with 0.28 M NH4Cl as their sole source of drinking water. After 7 days of chronic acidosis, blood pH (7.34 ± 0.03) and HCO3− concentration (15.6 ± 0.9 mM) were partially compensated. On average, the control rats consumed 34.3 ± 1.9 ml/day of water and the acidotic rats consumed a 36.4 ± 2.5 ml/day of the NH4Cl solution. All of the procedures were approved by the Institutional Animal Care and Use Committee at Colorado State University.
Isolation of proximal convoluted tubules and mitochondrial fractions.
Rat renal proximal convoluted tubules were isolated as described previously (53). Briefly, freshly isolated renal cortex was digested with collagenase and the proximal convoluted tubules were purified by Percoll gradient centrifugation. The isolated proximal convoluted tubules were pelleted by centrifuging for 10 min at 1,000 g at 4°C and resuspended in homogenization buffer containing 300 mM sucrose, 1 mM EDTA, 12 mM HEPES, and 1 mM phenylmethylsulfonylfluoride (PMSF). The resuspended tubules were homogenized with 20 passes in a 5-ml teflon homogenizer. The homogenate was centrifuged twice at 700 g for 10 min at 4°C to pellet nuclei and cellular debris. The supernatant was centrifuged twice at 7,000 g for 10 min at 4°C to pellet mitochondria and remove soluble cytosolic proteins and smaller organelles. The pellet was resuspended in 400 μl of homogenization buffer and washed twice by centrifugation at 7,000 g for 10 min at 4°C. The final pellet containing mitochondria was resuspended in 1.0 ml of homogenization buffer and layered onto two sucrose step gradients each containing 750 μl of 1.0 M, 1.0 ml of 1.3 M, 1.0 ml of 1.6 M, and 250 μl of 2.0 M sucrose. The sucrose step gradients were centrifuged for 60 min at 134,000 g at 4°C. The mitochondrial fraction was recovered as a dense reddish-brown band, divided into four tubes, and washed three times by centrifugation for 20 min at 74,000 g at 4°C. Three of the pellets were combined and resuspended in 100 μl of 6 M urea with 1 mM PMSF and stored at −80°C. The fourth mitochondrial pellet was resuspended in 40 μl of homogenization buffer and used immediately to assay for cytochrome c oxidase activity (4). Bradford (3) and bicinchoninic acid (46) (Pierce) protein assays were performed to determine protein concentrations.
Cytochrome c oxidase assay.
Mitochondrial enrichment was calculated as the increase in specific activity of cytochrome c oxidase. This activity was determined by measuring the oxidation of reduced cytochrome c at 550 nm. The reaction mixture contained 0.3 mg/ml of cytochrome c dissolved in 30 mM potassium phosphate, pH 7.4. The cytochrome c was reduced by addition of sodium hydrosulfite. The initial rates of oxidation were measured in crude homogenates of proximal convoluted tubules and the purified mitochondrial fractions. The specific activity for each sample was calculated relative to the protein concentration and used to estimate the fold purification.
Proteomic sample preparation.
The study compared mitochondrial fractions of proximal convoluted tubules isolated from control rats (n = 3) and 7-day chronic acidotic rats (n = 3). For proteomic analysis, 50 μg of protein in 6 M urea were bath sonicated for 5 min and vortexed to lyse the organelles. An acetone precipitation was performed before in-solution protein digestion. Briefly, the proteins were resuspended in 8 M urea and 0.2% ProteaseMAX surfactant (Promega), reduced with dithiothreitol, and alkylated with idoacetamide. The samples were digested with trypsin for 3 h at 37°C and then stopped with 0.5% trifluoroacetic acid. The peptides were dried in a speed-vac and purified using a reverse phase C18 TopTip (Glygen). The purified peptides were dried in a speed-vac and reconstituted in 50 μl of 0.1% formic acid/3% acetonitrile.
Mass spectrometry.
Fractionation by two-dimensional liquid chromatography (2D-LC) was performed to increase the number of peptides identified in the samples. A 10-μg aliquot of each sample was injected onto a strong cation exchange (SCX) column (Agilent, ZORBAX BioSCX Series II, 300 μm × 35 mm, 3.5-μm column). Peptides were eluted from the SCX column using 20-μl salt bumps of 10, 15, 25, 35, 50, 75, 150, and 500 mM NaCl. The eluted peptides were further fractionated on a reverse-phase column (1,100 nanoHPLC, Zorbax C18, 5 μm, 75-μm ID × 150-mm column, Agilent) using a linear gradient from 25 to 55% of 90% acetonitrile and 0.1% formic acid at a flow rate of 300 nl/min. Peptides were eluted from the reverse-phase column directly into the mass spectrometer (LTQ linear ion trap, Thermo Scientific), and spectra were collected over an m/z range of 200–2,000 Da using a dynamic exclusion limit of three tandem mass spectra (MS/MS) of a given peptide mass for 30 s and an exclusion duration of 90 s. Compound lists of the resulting spectra were generated using Xcalibar 2.2 software (Thermo Scientific) with an intensity threshold of 5,000 and 1 scan/group. Duplicate 2D-LC injections were performed for each biological sample. All chromatograms were manually checked for consistency in retention time between injections and replicates. A tryptic digest of bovine serum albumin was injected between samples for quality control.
Bioinformatics.
MS/MS spectra were searched against the Uniprot-KB Rattus norvegicus protein sequence database (74,140 sequence entries) concatenated with a reverse database using both the Mascot (version 2.3.02, Matrix Science) and SEQUEST (version v.27, rev. 11, Sorcerer, Sage-N Research) database search engines. The search parameters were average mass, peptide mass tolerance of 2.5 Da, fragment ion mass tolerance of 1.0 Da, complete tryptic digestion allowing two missed cleavages, variable modifications of methionine oxidation and lysine acetylation, and a fixed modification of cysteine carbamidomethylation. Peptide identifications from both search engine results were combined using protein identification algorithms in Scaffold 3 (Version 3.6.0, Proteome Software). Peptide and protein probability thresholds of 90 and 99%, respectively, were applied with Peptide and Protein Prophet algorithms in Scaffold 3 (25, 33). Proteins containing shared peptides were grouped by Scaffold 3 to satisfy the laws of parsimony. A peptide false discovery rate (FDR) of 0.2% was determined by the target decoy approach of searching the reversed database (12). Only proteins that were identified by a minimum of two unique peptides in at least two biological replicates were considered confidently identified. Gene ontology (GO) terms for cellular locations and processes were extracted from Scaffold 3. Additional functional information on the identified proteins was manually determined from the Uniprot-KB database (www.uniprot.org).
Spectral counting.
Spectral counting analysis was performed to determine the relative protein abundance changes between control and chronic metabolic acidosis. Spectral counts (SpC) correspond to the sum of all MS/MS spectra identified for all of the peptides assigned to an individual protein. The number of spectral counts has been shown to highly correlate to the abundance of a protein in a sample (36). Total spectral counts for all peptides identified for each protein were determined by summing the spectra identified using both the Mascot and SEQUEST database search engines for the analysis of each raw file from each biological replicate. The MS/MS data were normalized between samples in Scaffold 3 by using the sum of spectral counts for each sample to determine a scaling factor that was then applied to each protein in the sample. A pseudovalue of 1 was added to all normalized values to eliminate zero values, and fold-changes were calculated using the mean normalized spectral counts for the three biological replicates for each condition. The following analyses were performed to assess data quality: principal components analysis, box plots, density plots, and scatter plots. Variance in the raw data was assessed by analyzing the number of proteins identified per sample, the total spectra identified, and the total peptides identified. Correlation of sample replicates was tested using Pearson's correlation and Spearman's rank sum correlation tests. All of the statistical analysis was performed using the R v2.13 statistics package (http://r-project.org). To test for significant changes in protein abundance, Student's t-test was performed on the normalized spectral count values using Scaffold 3. Fold-changes ≥1.5 with a P value ≤0.05 were considered significant. Manual validation of MS/MS spectra was performed for protein identifications above the thresholds that were based on two unique peptides for proteins with significant P values and for acetylated peptides. Criteria for manual validation included the following: 1) a minimum of at least three theoretical y or b ions in consecutive order that are peaks >5% of the maximum intensity; 2) an absence of prominent unassigned peaks >5% of the maximum intensity; and 3) indicative residue-specific fragmentation, such as intense ions N-terminal to proline and immediately C-terminal to aspartate and glutamate.
Western blot analyses.
Additional mitochondrial fractions of isolated proximal convoluted tubules were prepared to validate the SpC data. The samples included control, 1-day acute acidotic, 3-day acidotic, and 7-day chronically acidotic rats. For Western blotting, samples containing 10 μg of protein were separated by 8 or 10% SDS-PAGE. Proteins were transferred to polyvinylidenedifluoride membranes (Immobilon-FL, Millipore), and the blots were incubated overnight with the primary antibodies. The following rabbit polyclonal antibodies were used to validate changes identified by SpC: kidney-type glutaminase (KGA) (7); glutamate dehydrogenase (GDH; Rockland); carbonic anhydrase 5B (CA5B; Acris); UDP-glucuronosyltransferase 1A1 (UGT1A1; Millipore); catalase (CAT; Abcam); and acetyl-coenzyme A acyltransferase 1 (ACAA1), enoyl-coenzyme A hydratase/3-hydroxyacyl-coenzyme A dehydrogenase (EHHADH), and 17-β-hydroxysteroid dehydrogenase 4 (HSD17B4; GeneTex). Peroxisomal enrichment was determined using a rabbit anti-peroxisomal biogenesis factor 3 (PEX3) antibody (GeneTex). A rabbit anti-acetylated-lysine (K-Ac2-100) antibody (Cell Signaling) was used to quantify changes in total acetylation. Rabbit antibodies to malate dehydrogenase 2 and aconitase 2 (Aviva Systems Biology) and a mouse monoclonal antibody to the 70-kDa subunit of mitochondrial succinate dehydrogenase (SDH; MitoSciences) were used as loading controls. Either 680 Dylight-conjugated goat anti-mouse IgG or 800 Dylight-conjugated goat anti-rabbit IgG (Pierce) was used as a secondary antibody. The resulting bands were visualized and quantified using an Odyssey Infrared Imager (LI-COR Biosciences).
Renal hypertrophy.
Sham-operated (n = 3) and left side unilateral nephrectomized rats (UNX; n = 3) were purchased from Charles River Laboratories (Kingston, NY). To achieve maximal compensatory hypertrophy, the rats were killed 21 days postoperation (37). The average wet weights of the right kidneys of the sham-operated and UNX rats were 1.35 ± 0.05 and 1.82 ± 0.11 g, respectively. Therefore, unilateral nephrectomy resulted in a 36% increase in the wet weight of the right kidney.
RESULTS
Isolation of mitochondria and peroxisomes from proximal convoluted tubules.
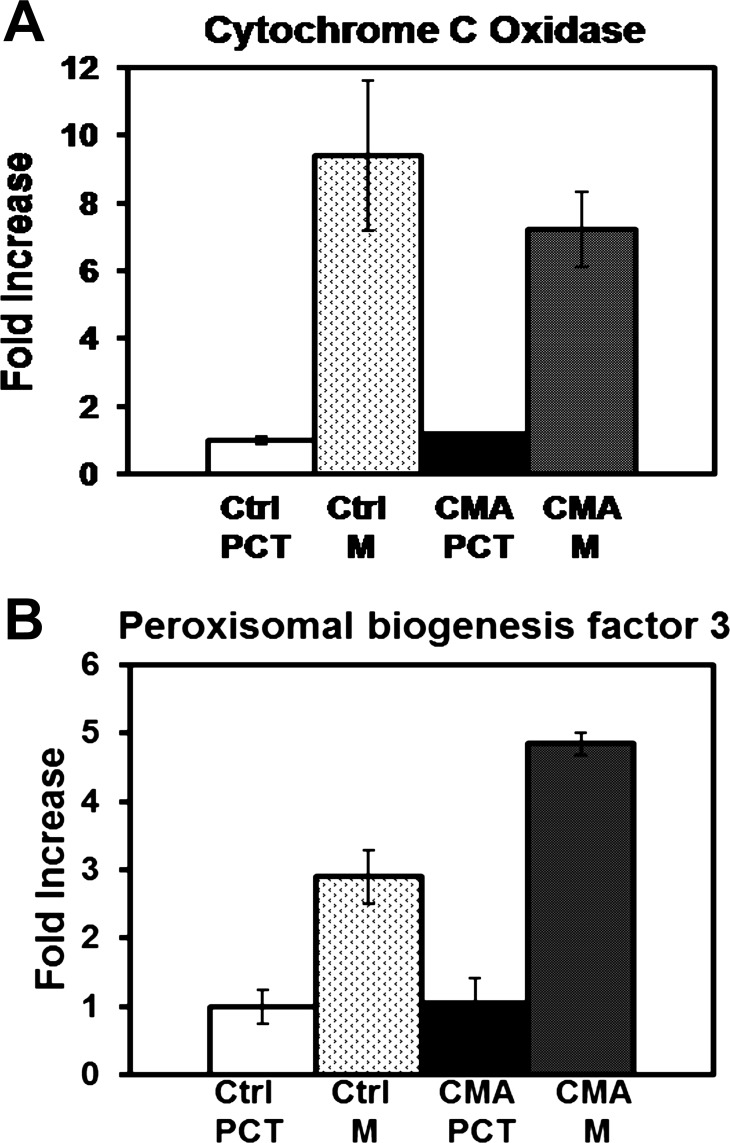
Proximal convoluted tubules were isolated from rat renal cortex by Percoll density gradient centrifugation (10). This preparation was previously shown to be essentially free of other nephron segments and to contain ∼95% proximal convoluted tubules (9, 53). Mitochondria were purified using differential and sucrose density centrifugation procedures similar to the protocol of Reifschneider et al. (39). Cytochrome c oxidase and protein assays were performed to determine the enrichment of mitochondria (Fig. 1A). This analysis indicated that the mitochondria were enriched approximately eightfold compared with homogenates of the corresponding proximal convoluted tubules. Because of their similar densities, it was not possible to obtain mitochondria that were free of peroxisomes. Therefore, PEX3, which is an integral membrane protein that participates in peroxisomal biogenesis (15), was used as a marker to assess the enrichment of peroxisomes in the mitochondrial fractions. Western blot analysis indicated PEX3 was enriched approximately fourfold compared with the homogenates of the proximal convoluted tubules (Fig. 1B). These data confirm that this protocol enriches predominantly for intact mitochondria and to a lesser extent for peroxisomes. Importantly, there was no significant difference in enrichment of control vs. chronic metabolic acidotic samples.
Fig. 1.
Enrichment of cytochrome c oxidase and peroxisomal biogenesis factor 3 in isolated mitochondrial samples. A: homogenates of proximal convoluted tubules (PCT) and the mitochondrial fractions (M) isolated from three control (Ctrl) and three 7-day chronic acidotic (CMA) rats were assayed for cytochrome c oxidase activity and protein concentration. Values are means ± SE of the fold-increase in specific activities of the triplicate acidotic PCT or M samples, normalized to the control PCT samples. B: corresponding increases in peroxisomal biogenesis factor 3 were quantified by Western blot analysis. Values are means ± the range of fold-increases measured in duplicate biological samples.
Proteomic profiling of mitochondrial fractions from control and 7-day acidotic rats.
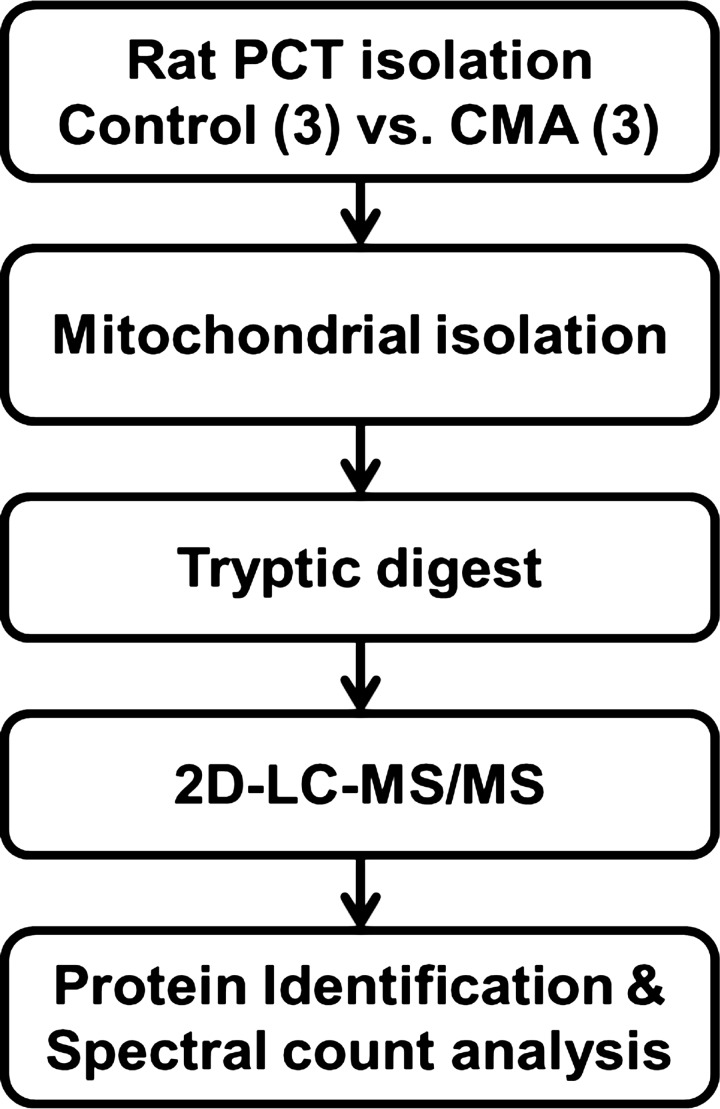
A proteomic approach was utilized to profile the proteins that are expressed in the mitochondria of the proximal convoluted tubule and to identify those that exhibit altered abundance in response to chronic metabolic acidosis (Fig. 2). Initially, triplicate biological samples of mitochondrial fractions were isolated from proximal convoluted tubules of control and 7-day chronic acidotic rats. Duplicate injections of each sample were analyzed by online two-dimensional LC/MS/MS, employing strong cation exchange and C18-reverse phase chromatography for peptide separation. Mass spectra were searched using both Mascot and SEQUEST search engines, and the protein identifications were compiled and validated using Scaffold 3. The two-dimensional LC/MS/MS analyses of the mitochondrial fractions identified a total of 901 proteins in all samples with a peptide FDR of 0.2% calculated based on hits to a decoy database. A complete list of the identified proteins along with their mean spectral counts and transmembrane domain predictions are available at http://helixweb.nih.gov/ESBL/Database/PCT/ (22). There are 183 proteins or, 20% of the total identified proteins, that contain at least one predicted transmembrane helix (47). The presence of predicted transmembrane domains indicates that the mitochondrial isolation and digestion protocols were conducive to retaining and solubilizing membrane proteins.
Fig. 2.
Proteomics workflow. PCT were isolated from 3 control and three 7-day CMA rats. The tubules were homogenized, and the mitochondria were isolated by differential and sucrose gradient centrifugations. Following tryptic digestion, duplicate injections of the resulting peptides were analyzed by 2-dimensional liquid chromatography (2D-LC) coupled directly with tandem mass spectrometry (MS/MS). Proteins were identified by searching the mass spectra vs. the Uniprot-KB Rat protein sequence database. Spectral counting was performed to determine significant changes in protein abundance.
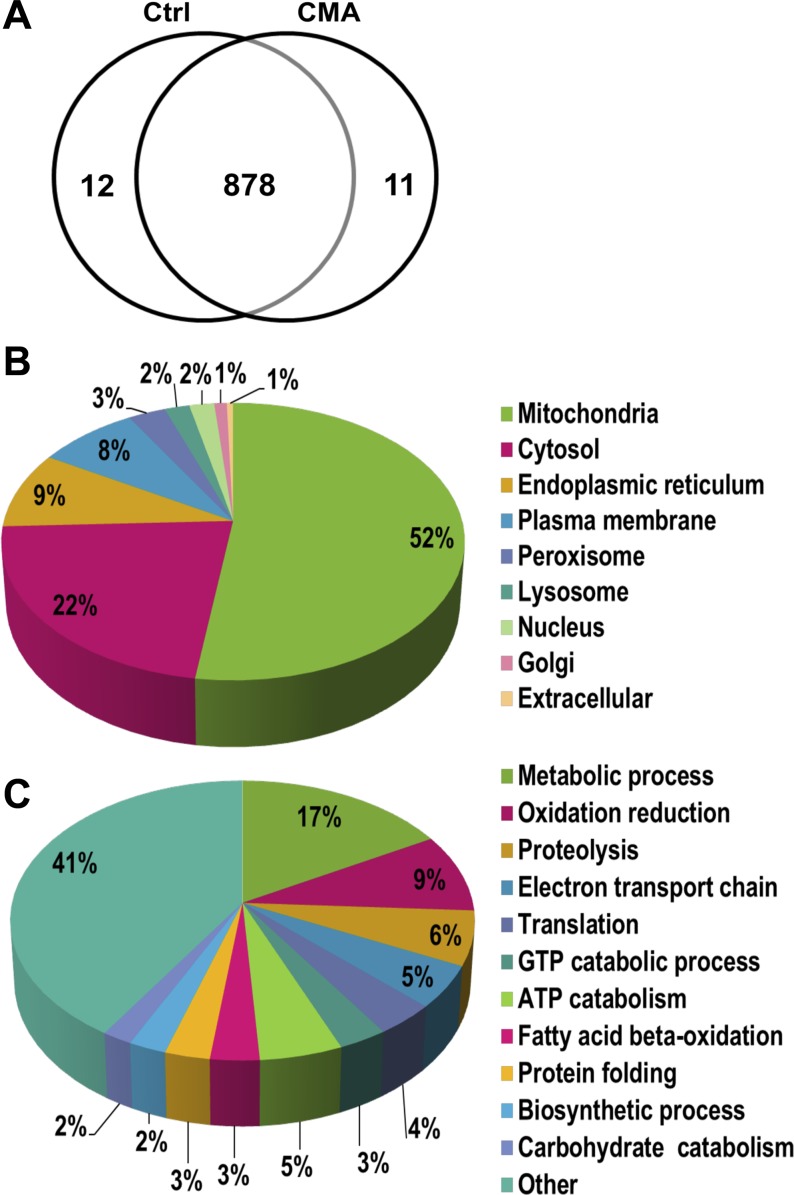
Of the 901 proteins identified, 11 were unique to the control samples, 12 were unique to the chronic acidotic samples, and 878 were common to both based on their presence in at least one biological sample (Fig. 3A). The current version of the MitoProteome Database (http://www.mitoproteome.org/data/proteome/index.html) lists 780 proteins that pass stringent criteria and are well-characterized mitochondrial proteins. Two hundred seventy-two of the proteins identified in this study (35%) are listed in the MitoProteome Database. Information on the cellular location and function of the identified proteins was also derived from the GO terms in Scaffold 3. Cellular locations are represented as a percentage of the 625 proteins that were annotated with a cellular location GO term (Fig. 3B). As expected, the majority of the proteins (325 or 52%) were identified as mitochondrial. The next most prominent localization was cytosolic proteins (22%), followed by the endoplasmic reticulum (9%), which has an interface with the mitochondrial membrane (20). There were 8% plasma membrane proteins, which is not surprising because the mitochondria of the proximal convoluted tubule are positioned adjacent to the apical membrane (27). Thus far only 105 peroxisomal proteins have been identified in the rat (41). As a result, even though peroxisomes were enriched in the mitochondrial fraction, only 16 proteins or 3% of the total were annotated as peroxisomal. Cellular processes were annotated for 455 of the proteins, which were classified into 131 different processes (Fig. 3C). The general annotation of metabolic process (17%) contained the largest cluster of proteins. Major mitochondrial processes such as oxidation/reduction (9%), electron transport chain (5%), and fatty acid β-oxidation (3%) clustered a large portion of the proteins. Many clusters of central cellular processes contained five or fewer proteins. Collectively, they accounted for 41% of the annotated proteins and they were categorized as “Other” in Fig. 3C. Overall, mitochondrial proteins were the most frequently annotated location and mitochondrial processes were the most prominent clusters.
Fig. 3.
Proteomic analysis. A: Venn diagram of total proteins identified in the mitochondria prepared from PCT isolated from control and 7-day CMA rats. A total of 901 proteins were identified; 878 were common in both samples, while 12 and 11 were unique to the control and acidotic samples, respectively. Pie charts are shown of the cellular locations (B) and cellular processes (C) as determined by gene ontology (GO) analysis of the identified proteins.
Mitochondrial protein lysine acetylation.
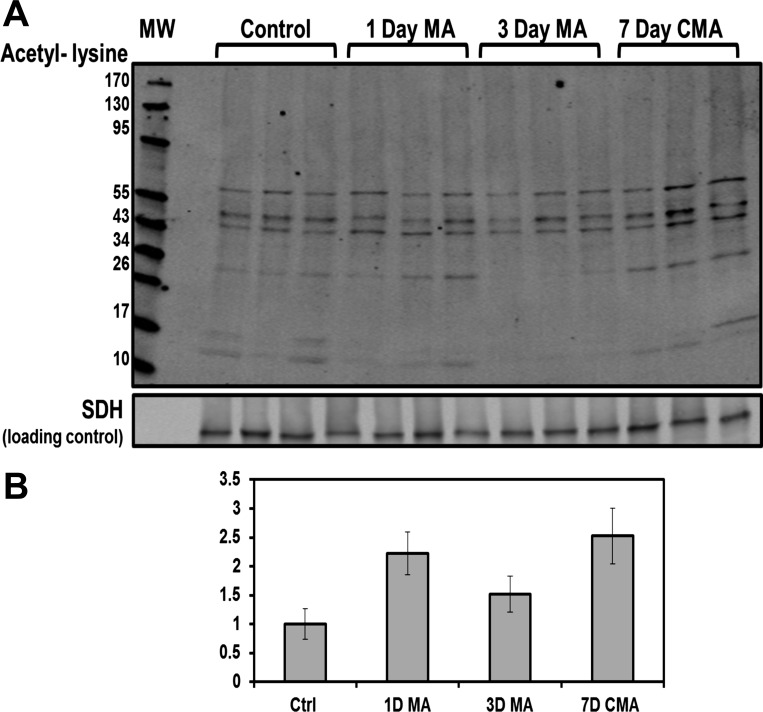
Recently, lysine acetylation has been identified as an important mechanism that regulates cellular metabolism (59). Western blot analysis of the mitochondrial samples obtained from triplicate preparations of control, 1-day acute, 3-day, and 7-day chronic acidotic rat proximal convoluted tubules were performed using an anti-acetyl lysine antibody (Fig. 4A). This analysis indicated that a large number of proteins were acetylated. The signal for all of the acetylated proteins was quantified and normalized to the control samples. The resulting data demonstrate that the level of protein acetylation increased with the progression of acidosis (Fig. 4B). In the 7-day chronic acidotic samples, lysine acetylation was increased 2.5-fold relative to the control mitochondria. Western blot analysis of proximal convoluted tubule homogenates and soluble cytosolic fractions exhibited no change in protein acetylation (data not shown), which indicates that this hyperacetylation is specific to the mitochondria. To further investigate protein acetylation in metabolic acidosis, all MS/MS spectra were searched with the variable modification of lysine acetylation to identify acetylated peptides and the specific lysines that are modified. The mitochondrial data set contained 37 acetylated peptides from 31 different proteins (Table 1). This analysis identified 39 total sites of lysine acetylation, of which 22 were novel sites not previously curated on PhosphoSitePlus (www.phosphosite.org). The identified acetylated proteins are involved in transport, ATP synthesis, and lipid and carbohydrate metabolism. Five acetylated proteins are components of the oxidative phosphorylation pathway; two proteins are contained in complex I, one in complex IV, and two in complex V. Acetylation of lysines has been proposed to prevent protein degradation by blocking sites of ubiquitination (16). Interestingly, eight of the identified acetylation sites were previously identified as sites of ubiquitination (www.phosphosite.org).
Fig. 4.
Western blot analysis of the temporal changes in lysine acetylation of total mitochondrial proteins during development of acidosis. A: mitochondrial samples were isolated from PCT of three control, three 1-day acidotic (MA), three 3-day MA, and three 7-day CMA rats. Western blot analysis was performed using an anti-N-ε-acetyl-lysine antibody. The bands were imaged and quantified with an Odyssey Infrared Imager. B: acetylation of mitochondrial proteins increased during the onset of acidosis. The combined intensities of each lane in A were normalized to the level of succinate dehydrogenase (SDH), which served as a loading control. Values are means ± SE of triplicate samples.
Table 1.
Identification of acetylated peptides
| No. of Modified Spectra |
|||||||
|---|---|---|---|---|---|---|---|
| Protein Name | Accession No. | Gene | Control | 7-Day CMA | Peptide Sequence | Site* | Novel |
| ATP synthase subunit-β | P10719 | Atp5b | 13 | KGSITSVQAIYVPADDLTDPAPATTFAHLDAR | K351 | Yes | |
| ATP synthase subunit-α | P15999 | Atp5a1 | 1 | LKEIVTNFLAGFEP | K541 | No | |
| Glutamate dehydrogenase 1 | P10860 | Glud1 | 1 | KGFIGPGIDVPAPDMSTGER | K212 | Yes | |
| 1 | ISGASEKDIVHSGLAYTMER | K503 | No | ||||
| ADP/ATP translocase 2 | Q09073 | Slc25a5 | 16 | 29 | TDAAVSFAKDFLAGGVAAAISK | K10 | No |
| 22 | 38 | TDAAVSFAKDFLAGGVAAAISKTAVAPIER | K10 or K23 | No | |||
| 6 | KGTDIMYTGTLDCWR | K245 | Yes | ||||
| Malate dehydrogenase | P04636 | Mdh2 | 1 | VNVPVIGGHAGKTIIPLISQCTPK | K203 | No | |
| 10-kDa Heat shock protein | P26772 | Hspe1 | 2 | AGQAFRKFLPLFDR | K8 | No | |
| Cytochrome c oxidase subunit 5A | P11240 | Cox5a | 2 | KGMNTLVGYDLVPEPK | K68 | Yes | |
| Myosin, heavy polypeptide 9 | G3V6P7 | Myh9 | 9 | 10 | AQQAADKYLYVDKNFINNPLAQADWAAK | K8 or K14 | K8-Yes, K14-No |
| 3-Hydroxyacyl-CoA dehydrogenase 2 | F1LNT4 | Hsd17b10 | 4 | 1 | AAACRSVKGLVAVITGGASGLGLSTAK | K9 | No |
| 78-kDa Glucose-regulated protein | P06761 | Hspa5 | 3 | 4 | LGGKLSPEDKETMEK | K585 | No |
| Fructose-1,6-bisphosphatase 1 | P19112 | Fbp1 | 2 | KGNIYSINEGYAK | K206 | Yes | |
| 4F2 cell-surface antigen heavy chain | Q794F9 | Slc3a2 | 8 | SQDTEVDMKDVELNELEPEKQPMNAADGAAAGEK | K10 or K21 | Yes | |
| Protein Myo6 | D4A5I9 | Myo6 | 13 | 5 | MEDGKPVWAPHPTDGFQMGNIVDIGPDSLTIEPLNQK | K5 | Yes |
| Medium-chain-specific acyl-CoA dehydrogenase | P08503 | Acadm | 3 | KGDEYVINGQK | K179 | No | |
| ADP/ATP translocase 1 | Q05962 | Slc25a4 | 18 | 39 | GDQALSFLKDFLAGGIAAAVSK | K10 | No |
| 8 | 8 | GDQALSFLKDFLAGGIAAAVSKTAVAPIER | K10 or K23 | K10-No, K23-Yes | |||
| NADH dehydrogenase 1α-subcomplex 5 | Q63362 | Ndufa5 | 1 | ILDLLK | K36 | No | |
| ATP-binding cassette subfamily D member 3 | P16970 | Abcd3 | 2 | LSGGEKQR | K6 | Yes | |
| Sorbitol dehydrogenase | P27867 | Sord | 13 | 7 | AAPAKGENLSLVVHGPGDIR | K6 | No |
| Uncharacterized protein | D3ZE29 | Unknown | 2 | TDAAVSFAKDFLAGGVAAANSKTAVAPIER | K10 or K23 | Yes | |
| Ndufa7 protein | A9UMV9 | Ndufa7 | 5 | ALVSGKTAESSAVAATK | K80 or K91 | Yes | |
| Glutathione S-transferase-α | P00502 | Gsta1 | 5 | 9 | SGKPVLHYFNAR | K4 | Yes |
| Uncharacterized protein | D4A5M0 | Unknown | 3 | 4 | LGEHNINVLEGDEQFVNAAK | K92 | Yes |
| Protein Map7d2 | D4A1J8 | Map7d2 | 6 | 16 | KSSENLSLDDCNK | K563 or K575 | Yes |
| Uncharacterized protein | F1M7S9 | Unknown | 1 | NLSSTANLKVLEADPYFTVK | K5306 | Yes | |
| 1 | ELPLIFITPLSDVK | K5124 | Yes | ||||
| Uncharacterized protein | D4A5F2 | Cep290 | 1 | 2 | MKAQEVELALEEVEK | K62 | Yes |
| 1 | EVELKVEVSK | K1150 K 1155 | Yes | ||||
| Testis- and ovary-specific PAZ domain-containing 1 | D3ZUC6 | Topaz1 | 1 | AMKKAELPLIPEGNPK | K93, K94, or K106 (2 sites) | Yes | |
| IQ motif containing GTPase-activating protein | G3V7Q7 | Iqgap1 | 1 | KNKEQLSDMMMINK | K940, K942 (2 sites) | Yes | |
| Serine/threonine-protein kinase TNNI3K | Q7TQP6 | Tnni3k | 2 | ILDLQSKLIIAVDVAK | K563 or K572 | Yes | |
| Uncharacterized protein | D4A5V2 | Unknown | 2 | MGYKLQDLTDVQIMAR | K265 | Yes | |
| Tektin-1 | Q99JD2 | Tekt1 | 1 | 3 | IRLERSLESYK | K106 | Yes |
| Enolase (Fragment) | F1LZ68 | Unknown | 1 | MLNNGSHAGNKLAMK | K86 or K90 | No | |
| RCG35999, isoform CRA_a | D4ABA5 | Smtn | 1 | KAMIEKLEK | K771 | No | |
Site determining ion for the acetyl-lysine was not always present, and the acetyl peptide may have more than one possible lysine that could be modified. CMA, chronic metabolic acidosis.
Spectral count analysis.
Spectral counting is not reliable for quantifying differences in proteins with low SpC values (29). Therefore, only proteins with a total of >10 SpC from all 3 biological replicates were included in the analysis to determine changes in relative protein abundance. Additionally, at least two unique spectra had to be identified in at least two biological replicates for a protein to be considered for analysis. Using this cutoff, spectral counting identified 33 proteins that exhibit a significant fold-change (fold-change ≥1.5, P ≤ 0.05) in the control vs. 7-day acidotic samples (Table 2). Two of the identified mitochondrial proteins were KGA and GDH, which are key enzymes of glutamine catabolism. Both KGA (8, 57) and GDH (56) were previously established to be upregulated during metabolic acidosis. UGT1A1 was significantly increased (4.6-fold). CAT has been localized to both the mitochondria and peroxisomes. By spectral counting, CAT was increased 2.2-fold during chronic metabolic acidosis. Additional proteins that are localized to both the mitochondria and peroxisomes that were increased during acidosis include ACAA1, HSD17B4, and EHHADH. Another novel protein of interest, CA5B, was also found to increase 5.7-fold, but with a borderline significant P value of 0.068. This isoform of carbonic anhydrase contains a cleavable mitochondrial targeting signal, and immunofluorescence analysis established that the mature CA5B protein is localized in mitochondria (14, 43).
Table 2.
Significant changes in protein abundance during chronic metabolic acidosis
| Uniprot |
Mean Spectral Counts ± SE |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein Name | Accession No. | Gene | Mass, kDa | P Value | Control | 7-Day CMA | Fold-Change | Potential pH-RE | Location | TMD |
| UDP-glucuronosyltransferase 2B15 | P36511 | Ugt2b15 | 61 | 0.00022 | 1.8 ± 1.0 | 17.0 ± 0.7 | 6.4 | Yes | ER | 1 |
| Carbonic anhydrase 5B, mitochondrial | Q66HG6 | Ca5b | 37 | 0.068* | 0.0 ± 0.0 | 4.7 ± 1.9 | 5.7 | Yes | M | |
| Protein AMBP | Q64240 | Ambp | 39 | 0.024 | 2.0 ± 0.9 | 13.8 ± 3.2 | 5.0 | No | Membrane | |
| UDP-glucuronosyltransferase 1-1 | Q64550 | Ugt1a1 | 60 | 0.00086 | 11.0 ± 1.8 | 54.1 ± 4.5 | 4.6 | Yes | ER | 2 |
| All-trans-13,14-dihydroretinol saturase, CRA_b | G3V7V6 | Retsat | 67 | 0.043 | 0.7 ± 0.4 | 7.5 ± 2.3 | 4.9 | No | Unknown | 1 |
| Ras-related protein Rab-21 | Q6AXT5 | Rab21 | 24 | 0.011 | 1.5 ± 0.8 | 9.5 ± 1.6 | 4.1 | Yes | Membrane | |
| Uncharacterized protein | D4A0Y1 | Cfb | 141 | 0.014 | 1.2 ± 0.7 | 7.7 ± 1.4 | 4.0 | NA | Extracellular | |
| Glutaminase kidney isoform, mitochondrial | P13264 | Gls | 74 | 0.019 | 17.7 ± 2.1 | 59.3 ± 10.7 | 3.2 | Yes | M | |
| Enoyl-CoA hydratase domain-containing protein 3 | Q3MIE0 | Echdc3 | 32 | 0.0061 | 0.4 ± 0.4 | 3.8 ± 0.6 | 3.6 | Yes | M | |
| Epoxide hydrolase 1 | P07687 | Ephx1 | 53 | 0.013 | 11.1 ± 3.3 | 36.3 ± 4.9 | 3.1 | No | ER | |
| Vitamin D-binding protein | P04276 | Gc | 54 | 0.0062 | 15.0 ± 4.4 | 40.2 ± 1.9 | 2.6 | No | Extracellular | |
| Acetyl-coenzyme A acyltransferase 1B | F1LPD6 | Acaa1b | 44 | 0.0049 | 11.3 ± 1.1 | 28.4 ± 2.8 | 2.4 | No | M and P | |
| Catalase | P04762 | Cat | 60 | 0.0095 | 55.1 ± 12.0 | 123.1 ± 8.3 | 2.2 | Yes | M and P | |
| Ribonuclease 4 | O55004 | Rnase4 | 17 | 0.044 | 7.5 ± 1.4 | 17.7 ± 3.2 | 2.2 | Yes | Extracellular | 1 |
| Ectonucleoside triphosphate diphosphohydrolase 5 | Q6P6S9 | Entpd5 | 47 | 0.018 | 8.0 ± 0.8 | 18.2 ± 2.5 | 2.1 | No | ER | 2 |
| Enoyl-coenzyme A hydratase/3-hydroxyacyl-coenzyme A | P07896 | Ehhadh | 79 | 0.014 | 14.3 ± 3.5 | 31.5 ± 2.0 | 2.1 | Yes | M and P | |
| Fumarylacetoacetate hydrolase domain-containing 2 | B2RYW9 | Fahd2 | 35 | 0.011 | 29.6 ± 5.9 | 61.1 ± 3.8 | 2.0 | Yes | M | |
| 17-β-Hydroxysteroid dehydrogenase 4 | P97852 | Hsd17b4 | 79 | 0.0018 | 48.1 ± 3.6 | 95.3 ± 5.3 | 2.0 | No | M and P | |
| Cytochrome P-450 4A2 | P20816 | Cyp4a2 | 58 | 0.023 | 91.2 ± 22.0 | 175.1 ± 7.7 | 1.9 | Yes | ER | 1 |
| Peroxisomal acyl-coenzyme A oxidase 1 | P07872 | Acox1 | 75 | 0.021 | 26.2 ± 6.8 | 51.4 ± 0.5 | 1.9 | No | ER and M | |
| Dimethylglycine dehydrogenase, mitochondrial | Q63342 | Dmgdh | 96 | 0.0024 | 47.4 ± 7.6 | 68.3 ± 2.5 | 1.5 | Yes | M | |
| Apolipoprotein A-IV | P02651 | Apoa4 | 44 | 0.041 | 46.3 ± 10.0 | 83.6 ± 7.6 | 1.8 | No | ER | |
| ATP-binding cassette subfamily D member 3 | P16970 | Abcd3 | 75 | 0.037 | 29.3 ± 6.3 | 52.5 ± 4.0 | 1.8 | Yes | M and P | 3 |
| Cytochrome b5 | P00173 | Cyb5a | 15 | 0.014 | 37.5 ± 5.8 | 64.7 ± 4.8 | 1.7 | Yes | ER and M | 1 |
| Glutamate dehydrogenase 1, mitochondrial | P10860 | Glud1 | 61 | 0.0035 | 431.5 ± 45.2 | 715.7 ± 7.9 | 1.7 | Yes | M | |
| Probable d-lactate dehydrogenase, mitochondrial | F1LVD7 | Ldhd | 52 | 0.037 | 27.0 ± 4.0 | 45.7 ± 4.5 | 1.7 | NA | M | |
| NADPH-cytochrome P-450 reductase | P00388 | Por | 77 | 0.044 | 13.2 ± 1.4 | 21.6 ± 2.6 | 1.6 | Yes | ER and M | 1 |
| GrpE protein homolog 1, mitochondrial | P97576 | Grpel1 | 24 | 0.04 | 10.2 ± 0.9 | 16.3 ± 1.8 | 1.5 | No | M | |
| Enoyl-CoA δ isomerase 1, mitochondrial | P23965 | Eci1 | 32 | 0.035 | 76.7 ± 6.5 | 116.2 ± 10.8 | 1.5 | No | M | |
| Filamin-B | D3ZD13 | Flnb | 278 | 0.023 | 4.2 ± 0.5 | 2.1 ± 0.3 | 0.6 | NA | Cytosol | |
| 4F2 cell-surface antigen heavy chain | Q794F9 | Slc3a2 | 58 | 0.02 | 105.6 ± 13.0 | 50.5 ± 6.8 | 0.5 | NA | Membrane | 1 |
| Calnexin | P35565 | Canx | 67 | 0.0069 | 35.1 ± 3.6 | 14.8 ± 1.5 | 0.4 | NA | ER | 1 |
| Protein FAM151A | Q642A7 | Fam151a | 67 | 0.036 | 29.0 ± 6.1 | 13.0 ± 2.6 | 0.4 | NA | Membrane | 1 |
Proteins in italics were previously shown to be induced during acidosis.
Greater than 0.05 P value. pH-RE, potential pH response element; NA if 3′−UTR not sequenced or fold-decrease; M, location mitochondria; P, peroxisome; ER, endoplasmic reticulum; TMD, no. of transmembrane domains predicted.
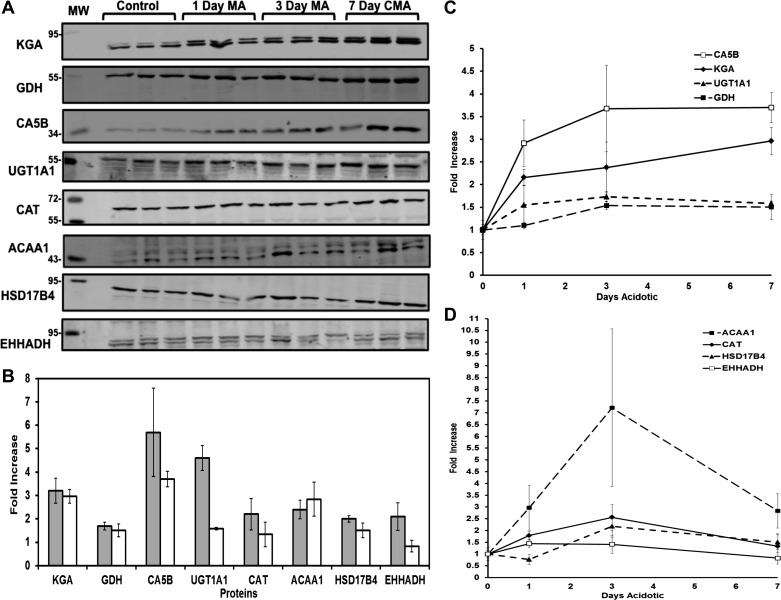
Validation of proteins altered in metabolic acidosis by Western blotting.
The increased expression of eight of the proteins identified by spectral counting was analyzed by Western blotting (Fig. 5A). The fold-increases calculated by spectral counting and Western blot analyses for six proteins, KGA, GDH, CA5B, CAT, ACAA1, and HSD17B4, were statistically equivalent (Fig. 5B). UGT1a1 exhibited a 4.6-fold increase in SpC, but only a 1.6-fold increase by Western blotting. UGT1a1 is a protein of the UGT1 gene family, which has nine alternatively spliced variants. There is significant sequence homology among the variants, which is indistinguishable in the mass spectrometry analysis. The detection of shared peptides can lead to artificial inflation of the SpC due to contributions from the various isoforms. In contrast, the antibody used in Western blotting is specific to the UGT1a1 isoform and should represent a more accurate measurement of abundance of this variant. Based upon SpC, EHHADH was increased 2.1-fold. However, this change was not validated by Western blot analysis. Overall, the results of the spectral counting analysis of relative protein abundance were largely validated by the data obtained from the Western blots.
Fig. 5.
Western blot analysis of the temporal changes in abundance of proteins identified by spectral count analysis. A: separate mitochondrial samples were isolated from PCT of three control, three 1-day MA, three 3-day MA, and three 7-day CMA rats. Western blot analysis was performed using 8 specific antibodies to validate the changes in protein abundance calculated by comparing the spectral counts of control and 7-day CMA samples. MW are molecular weight standards. Each blot was normalized to a mitochondrial protein that served as a loading control. The bands were imaged and quantified with an Odyssey Infrared Imager. B: comparison of the fold-changes calculated from Western blot analyses (open bars) and spectral counts (filled bars). Values are means ± SE of the fold-increase in the triplicate 7-day acidotic samples normalized to the control samples. C and D: time course of the fold-changes in the 8 proteins during development of chronic acidosis derived from the Western blot analyses in A. Values are means ± SE of triplicate samples.
Time course Western blot analysis.
Additional mitochondrial fractions were obtained from triplicate control, 1-day acute, 3-day, and 7-day chronic acidotic rats to evaluate the temporal changes in expression of these proteins. The enrichment of the mitochondrial cytochrome c oxidase activity in all of the samples was similar to that observed in the initial study (data not shown). Western blot analyses were used to quantify the relative abundance of the proteins in the temporal samples. The level of KGA was increased at 1 day and continued to increase for 7 days during the development of chronic acidosis (Fig. 5C). GDH exhibited a more gradual adaptation, was not significantly increased until 3 days, and was expressed at a similar level in the 7-day chronic acidotic samples. The temporal profiles for KGA and GDH are similar to what was previously observed by proteomic analysis of whole cell lysates of isolated proximal convoluted tubules (9). CA5B was increased 2.9-fold after 1 day and then plateaued at a 3.7-fold increase after 3 days. UGT1a1 was increased 1.6-fold after 1 day and then remained at that level through the 7-day period. Catalase was maximally induced by 3 days (2.6-fold increase) (Fig. 5D). ACAA1 was induced rapidly and reached a maximum of 7.2-fold by 3 days but decreased to 2.8-fold by 7 days. EHHADH showed a 1.4-fold increase at 1 and 3 days but no change at 7 days. All of the proteins except for KGA appeared to plateau or even decrease between 3 and 7 days. This may be due to the fact that by 7 days the metabolic acidosis is partially compensated (45).
Hypertrophy.
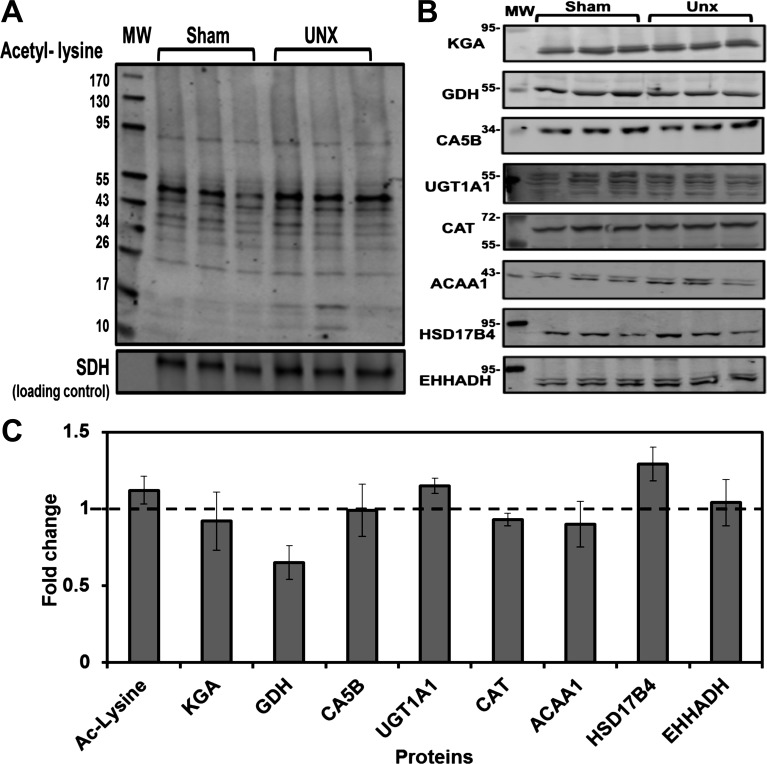
During chronic metabolic acidosis, the proximal convoluted tubule undergoes significant hypertrophy (30). Thus it was important to determine whether the increase in protein expression was due to the hypertrophy and not an adaptation to metabolic acidosis. The loss of one kidney causes a more extensive hypertrophy of the proximal convoluted tubule in the remaining kidney (44). Therefore, mitochondria were prepared from proximal convoluted tubules that were isolated from three sham-operated rats and three UNX rats 21 days postsurgery. The maximal compensatory hypertrophy of the proximal convoluted tubule occurred at 21 days post-UNX (37). UNX rats maintained normal blood pH and HCO3− levels (24). The wet weight of the remaining kidney was increased 36% compared with sham-operated control kidneys. The isolated mitochondrial fractions were analyzed by Western blotting to determine whether the hyperacetylation and increased protein expression observed during acidosis was due, in part, to the hypertrophy (Fig. 6, A and B). There was no significant difference in total protein lysine acetylation between UNX and the sham-operated controls (Fig. 6C). Therefore, increased mitochondrial protein lysine acetylation occurs in response to the changes in acid-base balance and not the hypertrophy that occurs during acidosis. The levels of all the analyzed proteins except for GDH were unchanged in the UNX samples compared with the sham-operated controls (Fig. 6C). GDH showed a significant twofold decrease in the mitochondria isolated from the UNX rats, indicating that the fold-increase observed during metabolic acidosis is an overcompensation for the decrease caused by hypertrophy alone. Overall, Western blot analyses indicate that the increases in lysine acetylation and protein abundance observed during chronic metabolic acidosis are not merely a response to the associated hypertrophy.
Fig. 6.
Effect of unilateral nephrectomy on lysine acetylation and protein abundance of mitochondrial proteins. A: Western blot analysis of homogenates of mitochondria isolated from PCT from 3 sham-operated and 3 uninephrectomized (UNX) rats was performed using an antibody specific for N-ε-acetyl-lysine. B: Western blot analysis was performed using antibodies that are specific for 8 of the proteins that were identified to increase significantly by spectral counting during 7-day CMA. Each blot was normalized to a mitochondrial protein that served as a loading control. MW are molecular weight standards. The bands were imaged and quantified with an Odyssey Infrared Imager. C: comparison of fold-changes in protein expression in UNX rats. Values are means ± SE of the intensities of the triplicate UNX samples normalized to the sham-operated control samples. The dashed line indicates no change in abundance.
DISCUSSION
Previous proteomic analyses have established that mitochondria isolated from rat muscle, heart, and liver exhibit significant differences in protein content (13). The various segments of the renal nephron contain mitochondria that differ in size, subcellular location, and function (27). Therefore, it is very likely that the proteomes of the mitochondria contained in the various nephron segments also differ significantly. This study combined established procedures to isolate rat renal proximal convoluted tubules (9, 53) with a subcellular fractionation protocol to yield a highly enriched preparation of mitochondria specific to the proximal convoluted tubule. Thus the proteomic analysis described in the present study represents the initial characterization of mitochondria from a specific segment of the renal nephron. This organelle constitutes the primary site of the increase in renal ammoniagenesis that occurs during metabolic acidosis (5, 55). Therefore, proteomic profiling of the mitochondria isolated from control and chronic acidotic rats also provides insight into the alterations in mitochondrial metabolism that either accompany or support the increased catabolism of glutamine. In addition, the resulting data expand our knowledge of the proteome of the renal proximal convoluted tubule (9, 53, 54).
Lysine acetylation and trimethylation are both detected as an addition of 42 Da. We were unable to distinguish between the two modifications due to mass accuracy limitations and the inability to detect low mass fragment ions (⅓ cutoff rule) of ion trap mass spectrometers. However, trimethylation of nonnuclear proteins is uncommon, whereas lysine acetylation of numerous mitochondrial proteins has been characterized in nonrenal tissues (35, 59). Therefore, the identified modifications are likely to be acetylated lysines. Lysine acetylation has been shown to produce conformational changes that affect enzyme activities and protein/protein interactions. In addition, lysine acetylation frequently occurs at sites of or near other posttranslational modifications. Therefore, lysine acetylation may block or induce other regulatory modifications (58). The resulting cross talk between the posttranslational modifications produces a complex set of signaling networks. The identified sites of acetylation in the mitochondrial proteins of the proximal convoluted tubule provide a basis for future mutational and functional analyses to determine their role in the regulation of mitochondrial metabolism and, possibly, in the rapid activation of glutamine catabolism that occurs during an acute onset of metabolic acidosis.
Using stringent criteria, the combined proteomic analyses confidently identified 901 proteins in the mitochondrial preparation isolated from proximal convoluted tubules. Approximately 70% of the identified proteins are previously characterized proteins that have defined GO terms. The analysis of GO terms indicated that the majority of the characterized proteins were previously identified as mitochondrial proteins. Given the incomplete nature of the current assignment of GO terms, it is feasible that many more of the characterized and uncharacterized proteins are also localized in the mitochondria. Spectral counting identified 33 proteins that exhibit a significant adaptive response during chronic metabolic acidosis (Table 2). Only six of these proteins, including KGA and GDH, were previously shown to exhibit an adaptive response to acidosis by either microarray analysis (34) or difference in-gel electrophoresis (9).
The increases in KGA (28) and GDH (42) expression result from selective stabilization of their respective mRNAs. In both cases, the mRNA stabilization is mediated by an eight-base AU-rich sequence within their 3′-untranslated region (UTR) that functions as a pH-RE. Of the 29 proteins that are significantly increased in acidosis, only 26 have mRNAs for which the 3′-UTR has been annotated. Of the characterized mRNAs, 16, or >60%, contain an AU-rich sequence in their 3′-UTR that is >85% identical to either of the pH-REs in the KGA mRNA. The latter group includes two UDP-glucuronosyltransferases (2B15 and 1A1), carbonic anhydrase 5B, Rab-21, catalase, and an enoyl-CoA hydratase. This group constitutes most of the proteins that are increased more than threefold. Therefore, mRNA stabilization may be a common mechanism that mediates the adaptive increases in many proteins in addition to KGA and GDH within the proximal convoluted tubule.
Various isozymes of carbonic anhydrase play an important role in cellular acid-base balance. They catalyze the reversible hydration of carbon dioxide to produce carbonic acid, which spontaneously dissociates to form a bicarbonate ion and a H+ (11). The observed increase in the mitochondrial isoform of carbonic anhydrase (CA5B) during chronic metabolic acidosis may support the associated increase in renal gluconeogenesis by providing bicarbonate ions for the mitochondrial pyruvate carboxylase (11, 21). This enzyme catalyzes an essential reaction in the synthesis of glucose from lactate or pyruvate. Alternatively, the increased expression of this carbonic anhydrase may facilitate the transport of bicarbonate produced by α-ketoglutarate dehydrogenase from the mitochondria to the cytoplasm (43). This translocation is necessary to ensure that bicarbonate ions generated in the mitochondria during acidosis contribute to the net synthesis of bicarbonate ions that is essential to partially restore acid-base balance.
UGTs catalyze the transfer of glucuronic acid to endogenous and exogenous compounds to increase their solubility and facilitate their excretion in the urine (26). The human genome encodes 15 separate UGTs that are categorized into two gene families, 1a and 2B (51). The proteomic analysis identified two UGTs that are significantly increased in the proximal convoluted tubule during acidosis. The primary substrates of UGT1a1 are bilirubin, opioids, and various xenobiotics, whereas UGT2B15 primarily targets bilirubin (26). UGTs are associated with endoplasmic reticulum membrane through a C-terminal transmembrane domain, which anchors the catalytic domain to the cytoplasmic surface. A specialized portion of the endoplasmic reticulum is closely associated with the mitochondria (20). This association of the two organelles may account for the recovery of the two UGTs in the isolated mitochondria.
Catalase is another highly abundant protein in the mitochondrial samples that is significantly increased (2.2-fold) during chronic acidosis. Catalase is typically considered to be a peroxisomal marker. However, a recent study detected catalase in the matrix of rat heart mitochondria and demonstrated that it plays a significant role in reducing oxidative stress (1). Furthermore, the overexpression of mitochondrial catalase reduced the production of reactive oxidative species (17). Western blot analysis of proximal convoluted tubule homogenates using antibodies to 4-hydroxy-2-nonenal and malondialdehyde showed no increase in these markers of oxidative stress during chronic acidosis (data not shown). Therefore, the upregulation of mitochondrial catalase during acidosis may counteract the potential increase in reactive oxygen species that would otherwise accompany an increase in electron transport and ATP synthesis. An additional novel finding is the observation that three enzymes involved in both mitochondrial and peroxisomal fatty acid metabolism are increased in acidosis. Acetyl-CoA acyltransferase, acyl-CoA oxidase, and enoyl-CoA hydratase are all enzymes that contribute to the synthesis of long-chain saturated and unsaturated fatty acids. Their increased expression suggests that acidosis may promote a remodeling of the lipids of the mitochondrial membranes.
The primary functions of the mitochondria-associated endoplasmic reticulum membrane (MAM) are lipid transport, the control of apoptosis, and the efficient shuttling of Ca2+ ions from the endoplasmic reticulum to the mitochondria (20). The mitochondrial influx of Ca2+ ions is known to regulate flux through the TCA cycle by activating the isocitrate and α-ketoglutarate dehydrogenases. Calnexin is a Ca2+-dependent transmembrane chaperone that is compartmentalized to the MAM (32). The association of calnexin with the Ca+2-ATPase may inhibit Ca+2 uptake into the endoplasmic reticulum (40). The level of calnexin associated with the isolated mitochondria was observed to decrease by 0.4-fold during acidosis. This may reflect the dissociation of calnexin from the Ca+2-ATPase and its translocation from the MAM to other regions of the endoplasmic reticulum or to the plasma membrane. Previous studies also established that the level of calmodulin is rapidly increased in proximal convoluted tubule cells during an acute onset of acidosis (9). These findings suggest that increased Ca2+ signaling may contribute to the enhanced mitochondrial catabolism of glutamine during metabolic acidosis.
The proximal convoluted tubule undergoes a significant hypertrophy during chronic metabolic acidosis (30). A similar hypertrophy occurs in the remnant kidney following unilateral nephrectomy (37). Unilateral nephrectomy also causes an increase in metabolism and O2 consumption in the remnant kidney (2). However, Western blot analysis established that the increase in lysine acetylation and the increases in various proteins, including mitochondrial catalase, that occur during chronic acidosis are not due to the associated hypertrophy. Therefore, the reported remodeling of the mitochondrial proteome of the proximal convoluted tubule occurs in response to the changes in acid-base balance during metabolic acidosis.
GRANTS
This research was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK-37124 and DK-75517 awarded to N. P. Curthoys.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: D.M.F. performed experiments; D.M.F. and J.E.P. analyzed data; D.M.F., J.E.P., and N.P.C. interpreted results of experiments; D.M.F. and N.P.C. prepared figures; D.M.F. drafted manuscript; D.M.F., J.E.P., and N.P.C. approved final version of manuscript; J.E.P. and N.P.C. provided conception and design of research; J.E.P. and N.P.C. edited and revised manuscript.
ACKNOWLEDGMENTS
All mass spectrometry was performed in the Proteomics and Metabolomics Facility at Colorado State University (www.pmf.colostate.edu).
REFERENCES
- 1. Bai J, Cederbaum AI. Mitochondrial catalase and oxidative injury. Biol Signals Recept 10: 189–199, 2001 [DOI] [PubMed] [Google Scholar]
- 2. Benipal B, Lash LH. Influence of renal compensatory hypertrophy on mitochondrial energetics and redox status. Biochem Pharm 81: 295–303, 2011 [DOI] [PubMed] [Google Scholar]
- 3. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254, 1976 [DOI] [PubMed] [Google Scholar]
- 4. Cooperstein SJ, Lazarow A. A microspectrophotometric method for the determination of cytochrome oxidase. J Biol Chem 189: 665–670, 1951 [PubMed] [Google Scholar]
- 5. Curthoys NP. Renal ammonium ion production and excretion. In: The Kidney: Physiology and Pathophysiology (4th ed.), edited by Alpern RJ, Hebert SC. New York: Elsevier, chapt. 56, 2007, p. 1601–1619 [Google Scholar]
- 6. Curthoys NP, Gstraunthaler G. Mechanism of increased renal gene expression during metabolic acidosis. Am J Physiol Renal Physiol 281: F381–F390, 2001 [DOI] [PubMed] [Google Scholar]
- 7. Curthoys NP, Kuhlenschmidt T, Godfrey SS, Weiss RF. Phosphate-dependent glutaminase from rat kidney. Cause of increased activity in response to acidosis and identity with glutaminase from other tissues. Arch Biochem Biophys 172: 162–167, 1976 [DOI] [PubMed] [Google Scholar]
- 8. Curthoys NP, Lowry OH. The distribution of glutaminase isoenzymes in the various structures of the nephron in normal, acidotic, and alkalotic rat kidney. J Biol Chem 248: 162–168, 1973 [PubMed] [Google Scholar]
- 9. Curthoys NP, Taylor L, Hoffert JD, Knepper MA. Proteomic analysis of the adaptive response of rat renal proximal tubules to metabolic acidosis. Am J Physiol Renal Physiol 292: F140–F147, 2007 [DOI] [PubMed] [Google Scholar]
- 10. Doctor RB, Chen J, Peters LL, Lux SE, Mandel LJ. Distribution of epithelial ankyrin (Ank3) spliceoforms in renal proximal and distal tubules. Am J Physiol Renal Physiol 274: F129–F138, 1998 [DOI] [PubMed] [Google Scholar]
- 11. Dodgson SJ, Cherian K. Mitochondrial carbonic anhydrase is involved in rat renal glucose synthesis. Am J Physiol Endocrinol Metab 257: E791–E796, 1989 [DOI] [PubMed] [Google Scholar]
- 12. Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214, 2007 [DOI] [PubMed] [Google Scholar]
- 13. Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics 5: 608–619, 2006 [DOI] [PubMed] [Google Scholar]
- 14. Fujikawa-Adachi K, Nishimori I, Taguchi T, Onishi S. Human mitochondrial carbonic anhydrase VB. cDNA cloning, mRNA expression, subcellular localization, and mapping to chromosome x. J Biol Chem 274: 21228–21233, 1999 [DOI] [PubMed] [Google Scholar]
- 15. Ghaedi KTS, Okumoto K, Matsuzono Y, Fujiki Y. The peroxin pex3p initiates membrane assembly in peroxisome biogenesis. Mol Biol Cell 11: 2085–2102, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene 363: 15–23, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Gurgul E, Lortz S, Tiedge M, Jorns A, Lenzen S. Mitochondrial catalase overexpression protects insulin-producing cells against toxicity of reactive oxygen species and proinflammatory cytokines. Diabetes 53: 2271–2280, 2004 [DOI] [PubMed] [Google Scholar]
- 18. Halperin ML. Metabolic aspects of metabolic acidosis. Clin Invest Med 16: 294–305, 1993 [PubMed] [Google Scholar]
- 19. Hansen WR, Barsic-Tress N, Taylor L, Curthoys NP. The 3′-nontranslated region of rat renal glutaminase mRNA contains a pH-responsive stability element. Am J Physiol Renal Fluid Electrolyte Physiol 271: F126–F131, 1996 [DOI] [PubMed] [Google Scholar]
- 20. Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol 19: 81–88, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henry RP. Multiple roles of carbonic anhydrase in cellular transport and metabolism. Annu Rev Physiol 58: 523–538, 1996 [DOI] [PubMed] [Google Scholar]
- 22. Huling JC, Pisitkun T, Song JH, Yu MJ, Hoffert JD, Knepper MA. Gene expression databases for kidney epithelial cells. Am J Physiol Renal Physiol 302: F401–F407, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hwang JJ, Perera S, Shapiro RA, Curthoys NP. Mechanism of altered renal glutaminase gene expression in response to chronic acidosis. Biochemistry 30: 7522–7526, 1991 [DOI] [PubMed] [Google Scholar]
- 24. Jaramillo-Juarez F, Rodriguez-Vazquez ML, Namorado MC, Martin D, Reyes JL. Acidosis and weight loss are induced by cyclosporin A in uninephrectomized rats. Pediatr Nephrol 14: 122–127, 2000 [DOI] [PubMed] [Google Scholar]
- 25. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392, 2002 [DOI] [PubMed] [Google Scholar]
- 26. King CD, Rios GR, Green MD, Tephly TR. UDP-glucuronosyltransferases. Curr Drug Metab 1: 143–161, 2000 [DOI] [PubMed] [Google Scholar]
- 27. Kriz W, Kaissling B. Structural organization of the mammalian kidney. In: The Kidney: Physiology and Pathophysiology (4th ed.), edited by Alpern RJ, Hebert SC. New York: Elsevier, chapt. 20, 2008, p. 479–564 [Google Scholar]
- 28. Laterza OF, Curthoys NP. Specificity and functional analysis of the pH-responsive element within renal glutaminase mRNA. Am J Physiol Renal Physiol 278: F970–F977, 2000 [DOI] [PubMed] [Google Scholar]
- 29. Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201, 2004 [DOI] [PubMed] [Google Scholar]
- 30. Lotspeich WD. Renal hypertrophy in metabolic acidosis and its relation to ammonia excretion. Am J Physiol 208: 1135–1142, 1965 [DOI] [PubMed] [Google Scholar]
- 31. Lowry M, Ross BD. Activation of oxoglutarate dehydrogenase in the kidney in response to acute acidosis. Biochem J 190: 771–780, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Myhill N, Lynes EM, Nanji JA, Blagoveshchenskaya AD, Fei H, Carmine Simmen K, Cooper TJ, Thomas G, Simmen T. The subcellular distribution of calnexin is mediated by PACS-2. Mol Biol Cell 19: 2777–2788, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658, 2003 [DOI] [PubMed] [Google Scholar]
- 34. Nowik M, Lecca MR, Velic A, Rehrauer H, Brandli AW, Wagner CA. Genome-wide gene expression profiling reveals renal genes regulated during metabolic acidosis. Physiol Genomics 32: 322–334, 2008 [DOI] [PubMed] [Google Scholar]
- 35. Oliver SS, Denu JM. Dynamic interplay between histone H3 modifications and protein interpreters: emerging evidence for a “histone language”. Chembiochem 12: 299–307, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paoletti AC, Washburn MP. Quantitation in proteomic experiments utilizing mass spectrometry. Biotechnol Genet Eng Rev 22: 1–19, 2006 [DOI] [PubMed] [Google Scholar]
- 37. Pfaller W, Seppi T, Ohno A, Giebisch G, Beck FX. Quantitative morphology of renal cortical structures during compensatory hypertrophy. Exp Nephrol 6: 308–319, 1998 [DOI] [PubMed] [Google Scholar]
- 38. Preisig PA, Alpern RJ. Chronic metabolic acidosis causes an adaptation in the apical membrane Na/H antiporter and basolateral membrane Na(HCO3)3 symporter in the rat proximal convoluted tubule. J Clin Invest 82: 1445–1453, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reifschneider NH, Goto S, Nakamoto H, Takahashi R, Sugawa M, Dencher NA, Krause F. Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2D blue-native/SDS-PAGE. J Proteome Res 5: 1117–1132, 2006 [DOI] [PubMed] [Google Scholar]
- 40. Roderick HL, Lechleiter JD, Camacho P. Cytosolic phosphorylation of calnexin controls intracellular Ca2+ oscillations via an interaction with SERCA2b. J Cell Biol 149: 1235–1248, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schlüter A, Real-Chicharro A, Gabaldón T, Sánchez-Jiménez F, Pujol A. PeroxisomeDB 2.0: an integrative view of the global peroxisomal metabolome. Nucleic Acids Res 38: D800–D805, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schroeder JM, Liu W, Curthoys NP. pH-responsive stabilization of glutamate dehydrogenase mRNA in LLC-PK1-F+ cells. Am J Physiol Renal Physiol 285: F258–F265, 2003 [DOI] [PubMed] [Google Scholar]
- 43. Shah GN, Hewett-Emmett D, Grubb JH, Migas MC, Fleming RE, Waheed A, Sly WS. Mitochondrial carbonic anhydrase CA VB: differences in tissue distribution and pattern of evolution from those of CA VA suggest distinct physiological roles. Proc Natl Acad Sci USA 97: 1677–1682, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shechter P, Shi JD, Rabkin R. Renal tubular cell protein breakdown in uninephrectomized and ammonium chloride-loaded rats. J Am Soc Nephrol 5: 1201–1207, 1994 [DOI] [PubMed] [Google Scholar]
- 45. Sleeper RS, Vertuno LL, Strauss F, Preuss HG. Effects of acid challenge on in vivo and in vitro rat renal ammoniagenesis. Life Sci 22: 1561–1571, 1978 [DOI] [PubMed] [Google Scholar]
- 46. Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85, 1985 [DOI] [PubMed] [Google Scholar]
- 47. Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6: 175–182, 1998 [PubMed] [Google Scholar]
- 48. Squires EJ, Hall DE, Brosnan JT. Arteriovenous differences for amino acids and lactate across kidneys of normal and acidotic rats. Biochem J 160: 125–128, 1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tannen RL, Ross BD. Ammoniagenesis by the isolated perfused rat kidney: the critical role of urinary acidification. Clin Sci (Lond) 56: 353–364, 1979 [DOI] [PubMed] [Google Scholar]
- 50. Taylor L, Curthoys NP. Glutamine metabolism: role in acid-base balance. Biochem Mol Biol Educ 32: 291–304, 2004 [DOI] [PubMed] [Google Scholar]
- 51. Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Ann Rev Pharm Toxicol 40: 581–616, 2000 [DOI] [PubMed] [Google Scholar]
- 52. Wagner CA. Metabolic acidosis: new insights from mouse models. Curr Opin Nephrol Hypertens 16: 471–476, 2007 [DOI] [PubMed] [Google Scholar]
- 53. Walmsley SJ, Broeckling C, Hess A, Prenni J, Curthoys NP. Proteomic analysis of brush-border membrane vesicles isolated from purified proximal convoluted tubules. Am J Physiol Renal Physiol 298: F1323–F1331, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Walmsley SJ, Freund DM, Curthoys NP. Proteomic profiling of the effect of metabolic acidosis on the apical membrane of the proximal convoluted tubule. Am J Physiol Renal Physiol 302: F1465–F1477, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weiner ID, Hamm LL. Molecular mechanisms of renal ammonia transport. Annu Rev Physiol 69: 317–340, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wright PA, Knepper MA. Glutamate dehydrogenase activities in microdissected rat nephron segments: effects of acid-base loading. Am J Physiol Renal Fluid Electrolyte Physiol 259: F53–F59, 1990 [DOI] [PubMed] [Google Scholar]
- 57. Wright PA, Knepper MA. Phosphate-dependent glutaminase activity in rat renal cortical and medullary tubule segments. Am J Physiol Renal Fluid Electrolyte Physiol 259: F961–F970, 1990 [DOI] [PubMed] [Google Scholar]
- 58. Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31: 449–461, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]