Abstract
Creosote bush-derived nordihydroguaiaretic acid (NDGA), a lipoxygenase inhibitor, possesses antioxidant properties and functions as a potent antihyperlipidemic agent in rodent models. Here, we examined the effect of chronic NDGA treatment of ob/ob mice on plasma dyslipidemia, hepatic steatosis, and changes in hepatic gene expression. Feeding ob/ob mice a chow diet supplemented with either low (0.83 g/kg diet) or high-dose (2.5 g/kg diet) NDGA for 16 wk significantly improved plasma triglyceride (TG), inflammatory chemokine levels, hyperinsulinemia, insulin sensitivity, and glucose intolerance. NDGA treatment caused a marked reduction in liver weight and TG content, while enhancing rates of fatty acid oxidation. Microarray analysis of hepatic gene expression demonstrated that NDGA treatment altered genes for lipid metabolism, with genes involved in fatty acid catabolism most significantly increased. NDGA upregulated the mRNA and nuclear protein levels of peroxisome proliferator-activated receptor α (PPARα), and the activated (phosphorylated) form of AMP-activated kinase. NDGA increased PPARα promoter activity in AML12 hepatocytes and also prevented the fatty acid suppression of PPARα expression. In contrast, PPARα siRNA abrogated the stimulatory effect of NDGA on fatty acid catabolism. Likewise, no stimulatory effect of NDGA on hepatic fatty acid oxidation was observed in the livers of PPARα-deficient mice, but the ability of NDGA to reverse fatty liver conditions was unaffected. In conclusion, the beneficial actions of NDGA on dyslipidemia and hepatic steatosis in ob/ob mice are exerted primarily through enhanced fatty acid oxidation via PPARα-dependent pathways. However, PPARα-independent pathways also contribute to NDGA's action to ameliorate hepatic steatosis.
Keywords: gene array, insulin resistance, hepatic steatosis, dyslipidemia, fatty liver, lipid oxidation, ob/ob mice
nonalcoholic fatty liver disease (NAFLD) is one of the most common causes of chronic liver disease affecting both adults and children in the United States and many other parts of the world (2, 48, 68, 69, 74). NAFLD represents a spectrum of liver disease ranging from simple steatosis, which is relatively benign, to the more severe form, known as nonalcoholic steatohepatitis (NASH), which may progress to advanced fibrosis, cirrhosis, liver failure, and hepatocellular carcinoma (74). NAFLD is also closely associated with obesity, insulin resistance, and Type 2 diabetes (7, 52, 64, 67, 69, 76) and is now considered a hepatic manifestation of the metabolic syndrome (7, 29, 30, 35, 52, 69, 76). In fact, in the majority of cases, development of NAFLD is strongly linked to one or more components of the metabolic syndrome, namely central obesity, insulin resistance, glucose intolerance or diabetes, dyslipidemia, and/or hypertension (29, 30, 35). In the United States and other Western countries, the estimated prevalence of NAFLD is 20 to 30% and that of NASH ∼3%. In patients with obesity or Type 2 diabetes, it is now estimated that up to 85% have NAFLD and over half may have NASH (30, 33, 37).
No validated therapies for NAFLD currently exist (7, 13, 25, 33, 76), except weight reduction by lifestyle modifications (e.g., caloric restriction and increased physical activity) (73), which is well known to have a poor long-term success rate. Given that development of NAFLD is strongly linked to components of the metabolic syndrome, not surprisingly treating components of the metabolic syndrome has become a central therapeutic strategy in the clinical management of NAFLD (7, 13, 25, 33, 76). However, despite several trials of insulin sensitizers (thiazolidinediones, biguanides), antioxidants (vitamin E), and lipid-lowering agents, no highly effective treatment yet exists (7, 13, 25, 33, 76). Moreover, some of these treatments have side effects and have not proven to be effective. Thus there is an urgent need for the development of new, safe, and effective combinations of drugs, more efficacious drugs as well as multifunctional drugs directed at the core components of the metabolic syndrome that can be used as valuable clinical tools in the management of NAFLD.
The desert plant creosote bush, Larrea tridentate (also known as Chaparral), has been used by Native Americans to treat a variety of ailments including infertility, arthritis, diabetes, gallbladder and kidney stones, and inflammation (12, 27, 60). Nordihydroguaiaretic acid (NDGA), a phenolic compound, is the active ingredient of creosote bush; it is found in high concentrations in the leaves and twigs of this shrub. NDGA is a potent lipoxygenase inhibitor and also possesses antioxidant properties. Previous work from our laboratory (12, 27, and references therein) and others (60) has shown that NDGA exerts profound effects on several components of the metabolic syndrome including lowering of blood glucose, free fatty acids (FFA), and triglyceride (TG) levels and attenuation of elevated blood pressure in several rodent models of dyslipidemia, insulin resistance, diabetes, and hypertension.
The present study was initiated to further examine the underlying mechanism by which NDGA exerts its antihyperlipidemic actions. Here we assessed the effects of dietary administration of NDGA on plasma lipids and its impact on hepatic lipid metabolism in leptin-deficient (ob/ob) mice, an animal model of various components of metabolic syndrome including obesity, insulin resistance, dyslipidemia, and mild Type 2 diabetes. We provide evidence that NDGA exerts its hypolipidemic actions predominantly by stimulating the activity of a nuclear hormone receptor, peroxisome proliferator-activated receptor α (PPARα, or NR1C1), the master regulator of all three hepatic fatty acid oxidation systems (58), which in turn improves dyslipidemia by promoting increased channeling of fatty acids toward their oxidation, and thus restricting the crucial supply of fatty acids needed for TG synthesis, TG storage, and very low-density lipoprotein (VLDL)-TG production and secretion. Furthermore, additional PPARα-dependent and PPARα-independent lipid pathways, including NDGA modulation of the IRE1/XBP1 arm of endoplasmic reticulum (ER) stress signaling, may also contribute to NDGA's ability to ameliorate hepatic steatosis.
MATERIALS AND METHODS
Materials.
The following reagents were supplied by Sigma Chemical (St. Louis, MO): ATP, dithiothreitol, oleic acid, palmitic acid, EDTA, 9-cis-RA, and WY-14643. Phospho-AMPKα (Thr172) (40H9) rabbit mAb (no. 2535), AMPKα (23A3) rabbit mAb (no. 2603), phosphor-AMPKβ1 (Ser108) (no. 4181), and AMPKβ1 (no. 4182) antibodies were obtained from Cell Signaling Technology (Danvers, MA). Phospho-AMPKα (Thr172) (40H9) rabbit mAb detects endogenous AMPKα (both α-1 and α-2 isoforms) of the catalytic subunit but does not detect the regulatory β or γ subunits. AMPKα (23A3) rabbit mAb detects endogenous levels of total AMPKα protein. Phospho-AMPKβ1 (Ser108) antibody detects endogenous levels of AMPKβ-1 only when phosphorylated at Ser108. The antibody may cross-react with phosphorylated AMPKβ-2 when phosphorylated at Ser109. AMPKβ-1 antibody detects endogenous levels of total AMPKβ-1. The antibody does cross-react with AMPKβ-2. PPARα (H-98) rabbit antibody (IgG) and PPARα siRNA were supplied by Santa Cruz Biotechnology (Santa Cruz, CA). IRDye800CW-conjugated goat anti-rabbit IgG (H + L) and IRDye680LT-conjugated goat anti-mouse antibodies were the products of LI-COR Biosciences (Lincoln, NE). Glucose, TG, FFA, and cholesterol measurement kits were obtained from Stanbio Laboratory, Boerne, TX. Milliplex Map kits for the quantification of various adipokines and adiponectin were supplied by EMD Millipore (Billerica, MA). Lipofectamine 2000 and [1-14C]-palmitic acid [40–60 mCi (1.48–2.22 GBq)/mmol], were purchased from Invitrogen, Life Technologies (Carlsbad, CA) and PerkinElmer (Waltham, MA), respectively. All other reagents used were of analytical grade.
Animals and design.
All animal experiments were performed according to procedures approved by the VA Palo Alto Health Care System Animal Care and Use Committee (IACUC). Seven-week-old male control C57BL/6J mice, male leptin-deficient (ob/ob) mice, and male Pparα-deficient mutant mice (B6.129S4-Pparatm1Gonz/J) were obtained from the Jackson Laboratory (Bar Harbor, ME). For most studies ob/ob mice were used. Leptin-deficient ob/ob mice exhibit obesity, inactivity, hyperplasia, a diabetes-like syndrome of hyperglycemia, glucose intolerance, elevated plasma insulin, and hyperlipidemia (55, 66). Most importantly, these mice spontaneously develop fatty liver (hepatic steatosis) (20, 48, 66). These mice were fed a standard chow diet (Harlan Laboratories) for 1 wk to allow them to acclimatize to a controlled new environment (25 ± 2°C, 55 ± 5% relative humidity with a 12-h light-dark cycle). Subsequently, one group of ob/ob mice was switched to a chow diet supplemented with either low- (0.83 g/kg chow diet) or high-dose (2.5 g/kg chow diet) NDGA and maintained on this diet for 16 wk. The other groups of ob/ob mice and control mice continued to be fed a normal chow diet for 16 wk. In another set of studies, C57BL/6J mice and Pparα-deficient mutant mice were fed either a high-fat diet (no. TD.06414) (∼60% of total calories come from fat, Harlan Laboratories), or the same high-fat diet supplemented with a high dose of NDGA (NDGA 2.5 g/kg diet). Food intake and body weights were measured once a week throughout the experiment.
GTT and ITT.
Glucose tolerance test (GTT) and insulin tolerance test (ITT) were performed (1 wk apart) on animals that had been fed low-NDGA, high-NDGA, or control chow diet for 14–15 wk. Animals were fasted overnight before GTT experiments, and glucose (2 g/kg body wt) was administered by intraperitoneal injection between 9 and 10 AM. Blood samples were collected from the tail at 30, 60, and 120 min to assess in vivo glucose clearance. Food was removed 6 h before ITT experiments, which were carried out between 2 and 3 PM. Blood samples were collected from the tail at 15, 30, and 60 min after insulin (0.5 U/kg) was administered by intraperitoneal injection, as indicated, and serum glucose levels determined immediately using a glucometer (Precision Xtra, Abbott, IL).
Collection of serum, liver, muscle, and WAT samples.
Before, during, and at the end of the experimental period, blood samples were taken from the tails of the mice for analyses of plasma metabolites in the nonfasted and fasted states. At the end of the 16-wk treatment period, the mice were fasted, blood was collected, and the mice were subsequently euthanized. Blood samples were centrifuged at 4,000 g for 15 min at 4°C and the recovered serum samples were stored at −80°C until analyzed. Liver, muscle, and white adipose tissue (WAT) (epididymal and peripheral WAT) were excised immediately, rinsed with phosphate-buffered saline, weighed, frozen in liquid nitrogen, and stored at −80°C until utilized for various analyses.
Measurement of serum levels of TGs, cholesterol, glucose, adipokines, and adiponectin.
Serum glucose, TG, and total cholesterol levels were determined with commercial assay kits (Stanbio Laboratory). Serum adipokines and adiponectin levels were measured by using Milliplex Map kits (EMD Millipore) according to the manufacturers' protocols.
Fatty acid β-oxidation assay.
Liver fatty acid (palmitate) oxidation rate was determined in fresh liver homogenates by a modified method from that described by Mannarets et al. (47). Briefly, ∼50 to 100 mg of liver samples were homogenized on ice in 20 volumes of SET buffer (250 mM sucrose, 1 mM EDTA, 10 mM Tris·HCl, and 2 mM ATP; pH 7.4) in a Potter-Elvehjem homogenizer with a tight-fitting Teflon pestle. The homogenates were centrifuged at 400 g for 10 min at 4°C to sediment cell debris and nuclei and supernatant fractions in each case employed for the measurement of total and peroxisomal β-oxidation activities. All β-oxidation assays were performed by using a trapping device to capture 14CO2 produced during the oxidation reaction, as described previously (47). For the determination of total β-oxidation activity, the reactions were carried out in a final volume of 0.4 ml, containing 0.2 mM palmitate plus 0.2 μCi/ml [1-14C]palmitate (complexed to BSA), 100 mM sucrose, 10 mM Tris·HCl, pH 7.4, 10 mM potassium phosphate, 100 mM KCl, 1 mM 4 MgCl2, 1 mM l-carnitine, 0.1 mM malic acid, 2 mM ATP, 50 μM CoA, and 1 mM DTT. Following addition of 80 μl liver homogenate to the trapping device, the device was sealed immediately and incubated for 60 min at 30°C. Following incubation, the reaction was stopped by the addition of 200 μl of 6 M H2SO4. 14CO2 produced during the incubation was collected in 400 μl of 1 N NaOH and counted for the determination of radioactivity. Peroxisomal β-oxidation assays were carried out under identical conditions as described above (total fatty acid β-oxidation) except in each case the incubation mixture also contained 2 mM KCN, but without 0.5% BSA. The rate of mitochondrial fatty acid β-oxidation was calculated as the difference of total fatty acid β-oxidation minus peroxisomal fatty acid β-oxidation.
Oil red O staining of liver sections for detection of neutral lipids.
Liver samples were collected from mice, embedded in tissue-freezing medium (Leica, Wetslar, Germany), and stored at −80°C until used for sectioning to 8-μm slices. Liver sections were stained with Oil red O for the visualization of neutral lipids stored in the lipid droplets by using a slight modification of the standard procedure. In brief, frozen liver sections (8 μm) were air dried on the slides, fixed in formalin, and briefly washed with running tap water. Next, slides were rinsed with 60% isopropanol, stained with freshly prepared Oil red O working solution (1.5 volume of 0.5% Oil red O in isopropanol mixed with 1.0 volume of distilled water) for 15 min, and rinsed with 60% isopropanol. Following light staining of nuclei with hematoxylin, and a few rinses with distilled water, the slides were mounted in glycerine jelly mounting medium. Slides were viewed under a bright-field microscope (Leica), and photographic images were taken with a digital camera (Diagnostic Instruments).
Measurement of liver TG content.
Suitable aliquots of liver tissue homogenates were extracted with chloroform-methanol according to the procedure of Folch et al. (11), and extracted lipid samples were analyzed for their TG content with an enzymatic assay kit as described above.
Whole genome microarray.
RNA samples for whole genome microarray (Mouse OneArray Microarray v2) were isolated by use of Qiagen reagent (Qiagen USA, Valencia, CA) and quality was verified by using an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA). Total RNA was Cy5-labeled and hybridized to the GeneChip GPL6845 (Mouse OneArray V1) platform (Phalanx Biotech, Belmont, CA) group. This assay is designed to generate amplified and Cy5 sense-strand DNA targets from the entire expressed genome without bias. Three repeats from each group were performed. The microarray data files have been submitted to the Gene Expression Omnibus; the accession number is GSE35075 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35075).
Differential and cluster gene expression analysis.
The raw data from microarrays were analyzed using Partek Genome Suite software, version 6.3 copyright 2008 (Partek, St. Louis, MO). Briefly, GenePix Results (GPR) files containing the hybridization intensity were imported. This step was followed by quintile normalization and log2 transformation to represent gene expression levels. Samples were grouped into animal groups (WT vs. ob/ob), and diets (standard chow vs. standard chow with NDGA). Three-way ANOVA was performed including diet and animal group interaction to generate the lists of differentially expressed genes comparing WT and ob/ob in response to NDGA treatment and a different dose. There were three microarrays for each group. For the comparison between WT-chow and ob/ob-chow mice, probe sets with a fold change of 2.0 and adjusted P value <0.05 were considered differentially expressed. For the comparison between the ob/ob and ob-NDGA group, the analyses were also set with a fold change of 2.0 and adjusted P value of <0.05. The Benjamini-Hochberg false discovery rate (FDR) method was used for false positives. A corrected P value cutoff of 0.05 was used to select the regulated genes with the lowest FDR. Partek Genome Suite was used as the first step for quality control of the data on all the samples with two methods, Pearson correlation and principal component analysis (PCA). PCA was performed as a global view of sample clustering, which is related to the total variance in gene expression for all genes. Normalized expression values for all genes were analyzed. Statistical analysis of metabolic parameters was performed with GraphPad Prism 5.0. Diet-dependent changes were statistically analyzed by two-way ANOVA (repeated measures for within subject samples).
Pathway analysis.
For each comparison a list of differentially expressed genes was generated. The gene lists, along with associated expression or fold-change values, were further analyzed by using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA) to identify differentially expressed pathways that are affected by NDGA in ob/ob. The list of significantly regulated genes, selected by the microarray analysis described above, was loaded in IPA with the following criteria: direct and indirect relationships filtered by species (mouse) and by tissue (liver). Then IPA computed the data to generate significant networks of genes that are associated with particular biological functions, diseases, and signaling pathways.
Total RNA isolation, qRT-PCR.
Total hepatic RNA was extracted by using the Qiagen reagent (Qiagen USA) reverse-transcribed (1 μg total RNA) with a high-capacity RNA-to-cDNA kit (Life Technologies/Invitrogen). Real-time PCR was performed in a final volume of 10 μl containing 50 ng of cDNA template and primers by using a FastStart Universal SYBR Green Master PCR Kit (Roche, Germany) and an ABI Prism 7700 system (Applied Biosystems) according to the manufacturer's protocols. All reactions were carried out employing the following protocol: 95°C for 2 min; 40 cycles of 95°C for 20 s, 60°C for 20 s, 72°C for 40 s and 72°C for 30 s; and a 5-min extension at 72°C with a melting curve. The sequences of the primers used for the quantitative real-time PCR (qRT-PCR) are shown in Table 1. All the samples were normalized by the corresponding expression of 36B4 (acidic ribosomal phosphoprotein P0). The expression level of the gene of interest in the NDGA-treated group relative to its expression level in the vehicle-treated group was calculated by using the formula 2−ΔΔCt, where Ct is defined as the cycle number at which the fluorescence became significantly higher than the background. Specifically, ΔCt = ΔCt interest − ΔCt 36B4 and ΔΔCt = ΔCt of the NDGA-treated group − ΔCt of the vehicle-treated group, which was normalized to 1.0.
Table 1.
Primers used for quantitative real-time PCR gene expression studies
| Gene | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) |
|---|---|---|
| 36b4 | TTTGGGCATCACCACGAAAA | GGACACCCTCCAGAAAGCGA |
| Cpt1 | TCCACCCTGAGGCATCTATT | ATGACCTCCTGGCATTCTCC |
| Cpt2 | CCCAGACATCTCGGTTCTCACT | GCCCTGTGCCCGAGTTT |
| Acox1 | TCCCGATCTGGCGCAAGGAGC | CTGGTGAAGCAAGGTGGGCA |
| Acadl | GGCAAAATACTGGGCATCTGA | CTCCGTGGAGTTGCACACAT |
| Acadvl | ACCTTGCCAGGGCCTGAT | TGGCCTGGTCACCGGTAA |
| Acadm | TGGATCTGTGCAGCGGATT | GGGTCACCATAGAGCTGAAGACA |
| Ucp2 | GCCCCTTCACCTCTTTAGCA | CCAAGCACTGGGAAGGTCTAAC |
| Ppargc1a | TGCGGGATGATGGAGACA | GCGAAAGCGTCACAGGTGTA |
| Ppargc1b | TGGTCACCAGCCACCCGGAA | GCCTTTTCCCGCCGCTTCCT |
| Ppara | TGCCAAGGAGTCGAGGATG | TCGGCACCAGGAACCAA |
Western blot analysis.
Aliquots of tissue homogenates were mixed with an equal volume of 2× lysis buffer (50 mM Tris·HCl, pH7.6, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.25% deoxycholate, 1 mM EDTA, and 1 mM PMSF), and samples were incubated on ice for 10 min. Following centrifugation, the supernatant fractions were analyzed for protein content by use of a BCA protein assay kit (Pierce), and 20 μg of total protein was subjected to 10% SDS-PAGE under denaturing conditions and transferred to nitrocellulose membranes. Blotted membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE) for 1 h and then incubated with either rabbit anti-PPARα (H-98) antibody, phospho-AMPKα (Thr172) rabbit mAb, AMPKα rabbit mAb, phospho-AMPKβ1 (Ser108) antibody, or β-actin mouse mAb for 16 h at 4°C. After three washes with Tris-buffered saline containing 0.1% Tween 20, the membranes were incubated with IRDye 800CW goat anti-rabbit and IRDye 680LT goat anti-mouse secondary antibodies (LI-COR Biosciences) for 1 h. Proteins were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences).
Cell culture.
The model mouse hepatocyte cell line AML12 (catalog number: CRL-2254) was obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured in DMEM and F12 supplemented with 10% heat-inactivated fetal bovine serum and 100 U/ml penicillin A/ streptomycin (all supplied by Life Technologies through its GIBCO Cell Culture Media, Grand Island, NY). Cells were maintained at 37°C in a humidified incubator containing 5% CO2.
Fatty acid loading of mouse hepatocyte cell line.
To investigate the effect of NDGA under fatty acid loading conditions, AML12 hepatocytes were loaded with fatty acids (80 μM oleic and palmitic acid in 1.1% of fatty acid-free BSA) with or without 10 μM NDGA for 48 h (culture medium was changed every 24 h). Subsequently, the cell preparations were quantified for PPARα mRNA and protein levels.
Transfections and luciferase assays.
AML12 hepatocytes were seeded at a density of 1 × 104 cells/well in a 24-well plate containing the specific culture medium as described above. After overnight incubation, cells were transiently transfected with a PPRE-Luc (firefly luciferase) construct (1 μg) ± Pparα plasmid (1 μg) by using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). A control reporter construct containing Renilla luciferase (Rluc, pRL-TK) (500 ng) was cotransfected for normalization of transfection efficiency. After 24 h, cells were treated with or without various concentrations of NDGA for 24 h. 9-cis-RA (10 μM) + WY14643 (10 μM) were used as a positive control. At the end of incubation, cells were harvested by use of cell lysis buffer (Promega, Madison, WI) and subsequently assayed for both firefly and Renilla luciferase activities by use of a Promega Dual-Luciferase reporter kit (E1980) and TD-20/20 Luminometer (Turner Designs). Firefly luciferase activities were normalized to Renilla luciferase reporter activities and shown as fold induction compared with control (empty vector). The results shown are the average of triplicate determinations with the error bars representing ± SE. All the experiments were independently repeated at least three times.
Transfection of hepatocytes with PPARα siRNA.
For siRNA-related experiments, AML12 hepatocytes were plated for 24 h before transfection at ∼60–70% confluence. We used 5 nM of silencer PPARα siRNA (Applied Biosystems) or control (scramble siRNA) was used to transfect cells with Lipofectamine 2000 (Invitrogen). After 24 h, cells were treated with FFA and with or without 10 mM NDGA for another 48 h. At the end of incubation, cells were processed for fatty acid oxidation or harvested using cell lysis buffer for Western blot.
Measurement of cellular fatty acid oxidation.
Cellular fatty acid oxidation was measured by a modification of the procedure of Mannaerts et al. (47). In brief, AML12 hepatocytes were washed with KRP buffer (130 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 1.3 mM MgSO4, 10 mM Na2HPO4, pH 7.4) and subsequently were incubated for 1 h in an incubation medium containing 1% fatty acid-free BSA, 1 mM l-carnitine, 200 μM of unlabeled palmitic acid, and 0.2 μCi [1-14C] palmitate. At the end of the incubation, the medium samples were acidified with 4 M H2SO4 to release CO2 and trapped by filter paper wetted with 2 N NaOH. The dishes were sealed with Parafilm and incubated for an additional 1 h. The filter paper then was removed and transferred into scintillation vials for the determination of radioactivity.
Statistics.
Statistical analysis was performed using the Prism software (Prism 5.0, GraphPad, San Diego, CA). All data except microarray results are presented as means ± SE. A P < 0.05 was considered statistically significant.
RESULTS
Effects of NDGA treatment on physical and metabolic characteristics of ob/ob mice.
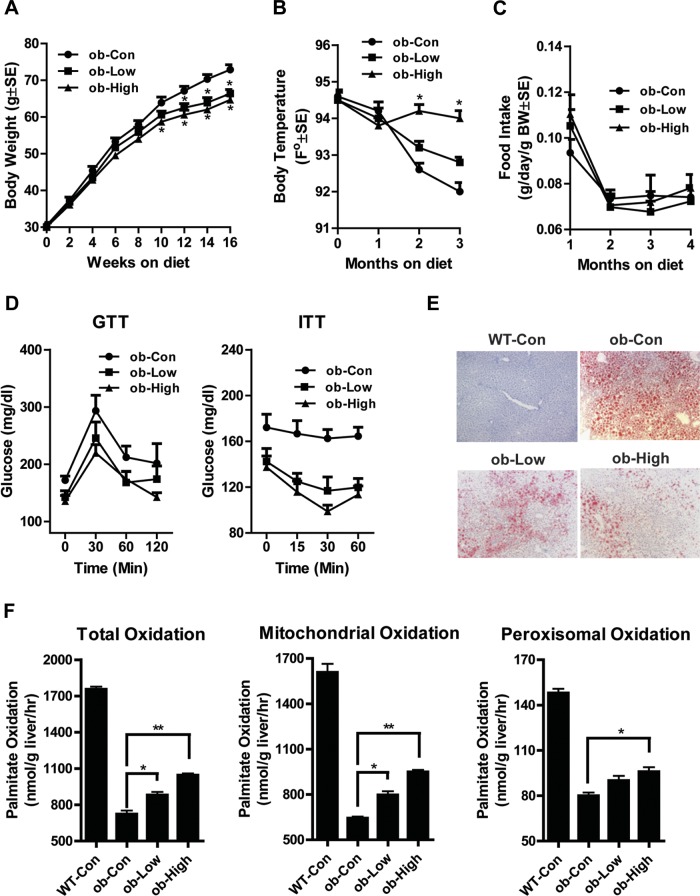
There was no obvious stress or diarrhea observed during the dietary treatment. Compared with the control ob/ob chow diet group, there was a significant reduction of body weight starting from 9 wk treatment in the high-dose NDGA diet group, and from 12 wk in the low-dose group (Fig. 1A). As shown in Fig. 1B, NDGA treatment resulted in higher body (rectal) temperatures of ob/ob mice, especially with the high dose of NDGA. There was no significant difference in food intake between groups (Fig. 1C). These results suggest that suppression of body weight gain in ob/ob mice by NDGA supplementation in the diet is due not to changes in food intake, but to increased energy expenditure.
Fig. 1.
Effect of low- or high-dose dietary nordihydroguaiaretic acid (NDGA) treatment on food intake, body weight, temperature changes, insulin resistance, and hepatic fatty acid oxidation in ob/ob (ob) mice. Groups of 8-wk-old ob/ob mice were maintained on a chow diet, or a chow diet supplemented with either low (0.83 g/kg food; Low) or high-dose (2.5 g/kg food; High) NDGA for 16 wk. WT, wild type; Con control diet. A: body weight. B: body temperature. C: food consumption. D: glucose tolerance test (GTT) and insulin tolerance test (ITT). E: Oil red O staining of liver sections for lipids. F: [14C]palmitate oxidation: total, mitochondrial, and peroxisomal β-oxidation. NDGA treatment decreases body weight gain and increases body temperature without impacting food intake (*P < 0.05 vs. ob/ob-chow control; **P < 0.01 vs. ob/ob-chow control). Data are presented as means ± SE (n = 8).
Serum TG and glucose levels were significantly decreased in mice treated with NDGA (Table 2). However, total cholesterol and nonesterified fatty acids were not different between control and NDGA-treated groups (Table 2). Insulin levels were greatly reduced in response to NDGA (Table 2). As shown in Fig. 1D, glucose intolerance improved in the high-dose NDGA group during GTT. Insulin sensitivity also significantly improved in the high-dose group (Fig. 1D).
Table 2.
Liver weights, liver triglyceride content, and serum parameters
| Parameters | WT-CD | ob/ob-CD | ob/ob-CD-low-NDGA | ob/ob-CD-high-NDGA |
|---|---|---|---|---|
| Liver weight, g | 1.90 ± 0.09 | 5.45 ± 0.15 | 4.63 ± 0.09 | 4.21 ± 0.07* |
| Liver triglyceride, mg/g | 9.64 ± 1.25 | 121.40 ± 7.90 | 83.44 ± 4.72* | 68.80 ± 9.15† |
| Glucose, mg/dl | 176 ± 13 | 242 ± 18 | 171 ± 8* | 176 ± 20 |
| Cholesterol, mg/dl | 101 ± 9 | 123 ± 10 | 128 ± 13 | 130 ± 9 |
| Triglyceride, mg/dl | 75 ± 3 | 89 ± 4 | 78 ± 3 | 67 ± 4* |
| NEFA, mEq/l | 0.75 ± 0.04 | 0.70 ± 0.04 | 0.79 ± 0.09 | 0.54 ± 0.07 |
| Insulin, ng/ml | 11.94 ± 3.37 | 67.14 ± 14.12 | 18.45 ± 3.33 | 18.92 ± 1.46† |
| MCP-1, pg/ml | 18 ± 6 | 99 ± 22 | 29 ± 10† | 34 ± 14† |
| IL-6, pg/ml | 18 ± 5 | 17 ± 2 | 9 ± 2* | 17 ± 5 |
| PAI-1, ng/ml | 4.56 ± 1.22 | 25.65 ± 8.56 | 8.16 ± 3.29 | 15.96 ± 9.52 |
| Resistin, ng/ml | 21.16 ± 2.5 | 24.04 ± 1.75 | 17.93 ± 2.01 | 16.84 ± 1.91* |
WT, wild type; CD, chow diet; ob/ob-CD-low-NDGA, ob/ob mice maintained on a chow diet supplemented with a low dose (0.83 g/kg of diet) of nordihydroguaiaretic acid (NDGA); ob/ob-CD-low-NDGA, ob/ob mice maintained on a chow diet supplemented with a high dose (2.5 g/kg of diet) of NDGA; NEFA, nonesterified fatty acid; MCP-1, monocyte chemoattractant protein-1; PAI-1, plasminogen activator inhibitor-1.
P < 0.05 vs. ob/ob-CD
P < 0.01 vs. ob/ob-CD.
As shown in Table 2, circulating levels of several inflammatory chemokines, monocyte chemoattractant protein-1, IL-6, plasminogen activator inhibitor-1, and resistin, were decreased with the dietary administration of NDGA. These data suggest that NDGA treatment attenuates leptin deficiency-induced hyperlipidemia and insulin resistance and indicates an anti-inflammatory role for NDGA.
Effects of NDGA treatment on hepatic lipid accumulation and fatty acid oxidation in ob/ob mice.
Liver weights in the low-NDGA group were 18% lower than of controls and 27% lower in the high-dose group (Table 2). Liver TG content was 26% lower in low-dose compared with control mice, and 38% lower in high-dose mice (Table 2). Hepatic steatosis (i.e., enhanced accumulation of lipid droplets), as measured by Oil red O staining of liver sections, was significantly improved in ob/ob mice chronically treated with either low or high dose of the NDGA (Fig. 1E).
To begin to investigate the mechanism of NDGA's effects on hepatic lipid metabolism, we first performed hepatic fatty acid oxidation assays. In the liver, fatty acid oxidation occurs in three subcellular organelles: β-oxidation in mitochondria and peroxisomes and ω-oxidation in microsomes (15, 56). Under normal physiological conditions, mitochondrial β-oxidation accounts for most hepatic fatty acid oxidation and is primarily involved in the oxidation of short-chain (<C8), medium-chain (C8–C12), and long-chain (C12–C20) fatty acids. As shown in Fig. 1F, total and mitochondrial palmitate oxidation in the liver homogenates of ob/ob mice was only 41% of wild-type control. Treatment with a low dose of NDGA resulted in an 18% increase in fatty acid oxidation, whereas use of a high dose of NDGA caused a 31% increase in palmitate oxidation. Low levels of peroxisomal palmitate oxidation also showed a similar trend. These data suggest that NDGA may exert its hypolipidemic effect primarily by promoting increased fatty acid oxidation via the mitochondrial β-oxidation system.
Altered global gene expression with NDGA treatment.
To further examine the molecular mechanism by which NDGA promotes enhanced fatty acid oxidation, we employed microarray technology to compare the global gene expression profiles of liver tissues isolated from ob/ob control, or ob/ob mice treated with a high dose of NDGA. C57BL/6J mice were used for baseline gene expression. A total of 31,248 genes were presented on the array chip, and 24,105 genes (identified as “present” by IPA software) were expressed in the liver. A comparison between ob/ob and control (wild-type) mice using the IPA Ingenuity system identified 776 genes that were related to lipid metabolism, of which 198 showed significant changes with a P value of <0.05; among these, 33 genes changed more than twofold. Similarly, a comparison between chow-fed ob/ob mice and ob/ob mice fed a NDGA-supplemented chow diet identified 807 genes that were related to lipid metabolism; 215 of them showed significant changes with a P value of <0.05. Among these 104 registered a change greater than twofold. Changes in some of the lipid metabolism genes are listed in Table 3.
Table 3.
Leptin deficiency and NDGA treatment-induced changes in hepatic gene expression: microarray analysis
| Pathways/Genes | Symbol | Accession | ob/ob-WT Ratio | p | NDGA-ob/ob Ratio | p |
|---|---|---|---|---|---|---|
| Fatty acid transport | ||||||
| Long-chain fatty acid transport protein 1 (FATP-1) | Slc27a1 | NM_011977 | 0.558427362 | 0.030106 | 1.245964460 | 0.400773 |
| Very long-chain acyl-CoA synthetase (VLACS) | Slc27a2 | NM_011978 | 0.641598397 | 0.000807 | 1.598747219 | 0.006022 |
| Fatty acid oxidation | ||||||
| Peroxisomal acyl-coenzyme A oxidase 1 (AOX) | Acox1 | NM_015729 | 0.674270919 | 0.071991 | 1.950396981 | 0.008805 |
| Carnitine O-palmitoyltransferase 2 (CPT II) | Cpt2 | NM_009949 | 0.926932013 | 0.60305 | 2.303352084 | 4.04E-11 |
| Long-chain specific acyl-CoA dehydrogenase (LCAD) | Acadl | NM_007381 | 0.987923224 | 0.931455 | 2.097472816 | 0.000002 |
| Medium-chain specific acyl-CoA dehydrogenase, mitochondrial (MCAD) | Acadm | NM_007382 | 0.999478741 | 0.997573 | 2.262311421 | 0.00006 |
| Short-chain specific acyl-CoA dehydrogenase, mitochondrial (SCAD) | Acads | NM_007383 | 0.810900294 | 0.366283 | 3.596045954 | 8.77E-10 |
| Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial (SBCAD) | Acadsb | NM_025826 | 0.856101523 | 0.560594 | 1.842802488 | 0.010863 |
| Very long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (MVLCAD)(VLCAD) | Acadvl | NM_017366 | 1.160732214 | 0.269673 | 1.762036109 | 0.0000041 |
| Enoyl-CoA δ isomerase 1 | Dci | NM_010023 | 0.714660355 | 0.01128 | 2.543055515 | 1.71E-18 |
| Enoyl CoA hydratase, short chain, 1, mitochondrial | Echs1 | NM_053119 | 1.126657079 | 0.143268 | 1.248285258 | 0.003673 |
| Enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase | Ehhadh | NM_023737 | 0.855649975 | 0.264005 | 3.440488733 | 7.03E-14 |
| Enoyl-CoA δ isomerase 2 | Peci | NM_0011103 | 0.568660802 | 0.020355 | 2.21591912 | 0.001466 |
| Fatty acid synthesis/lipogenesis | ||||||
| ATP citrate lyase | Acly | NM_134037′ | 1.145387756 | 0.756348 | 2.181336699 | 0.010493 |
| Long-chain acyl-CoA synthetase 1 (LACS 1) | Acsl1 | NM_007981 | 0.633270359 | 0.00002 | 1.932050309 | 0.00000 |
| Long-chain acyl-CoA synthetase 4 (LACS 4) | Acsl4 | NM_019477 | 1.088355070 | 0.798051 | 1.912374605 | 0.000961 |
| Acyl-CoA synthetase medium-chain family member 3 | Acsm3 | NM_212442 | 1.228703185 | 0.157751 | 1.757779544 | 0.000001 |
| Acyl-CoA synthetase short-chain family member 2 | Acss2 | NM_019811 | 0.483239314 | 0.015197 | 3.398995213 | 5.83E-10 |
| Fatty acid synthase | Fasn | NM_007988 | 1.839170164 | 0.005237 | 1.570196193 | 0.016948 |
| Elongation of very long chain fatty acids protein 2 | Elovl2 | NM_019423 | 0.845673939 | 0.349175 | 1.658868162 | 0.015732 |
| Elongation of very long chain fatty acids protein 5 | Elovl5 | NM_134255 | 1.114574562 | 0.453637 | 1.601036249 | 0.000776 |
| Elongation of very long chain fatty acids protein 6 | Elovl6 | NM_130450 | 1.718544266 | 0.056042 | 1.553898745 | 0.016998 |
| NADP-dependent malic enzyme (NADP-ME) (Malic enzyme 1) | Me1 | NM_008615 | 1.243587130 | 0.269897 | 1.679627212 | 0.001156 |
| NAD-dependent malic enzyme, mitochondrial precursor (NAD-ME) | Me2 | NM_145494 | 0.986553428 | 0.855384 | 0.697631428 | 0.009561 |
| Malonyl-CoA decarboxylase | Mlycd | NM_019966 | 0.923644934 | 0.536297 | 1.729146843 | 0.000001 |
| Glucokinase (Hexokinase type IV, −4, or -D)(HK- IV, −4 or -D) | Gck | NM_010292 | 1.270363260 | 0.216082 | 1.462148348 | 0.022001 |
| Pyruvate kinase isozymes R/l (L-PK) | Pklr | NM_010997 | 0.935555571 | 0.441302 | 0.655938990 | 0.000013 |
| Acyl-CoA desaturase 1 (Stearoyl-CoA desaturase 1) | Scd1 | NM_009127 | 1.144128631 | 0.029428 | 0.589256823 | 2.73E-15 |
| Acyl-CoA desaturase 2 (tearoyl-CoA desaturase 2) | Scd2 | NM_009128 | 3.219702746 | 0.000012 | 2.248591020 | 0.002919 |
| Tryglyceride synthesis and assembly | ||||||
| 1-Acyl-sn-glycerol-3-phosphate acyltransferase α (1-AGPAT 1) | Agpat 1 | NM_018862 | 0.997074155 | 0.981684 | 0.701355523 | 0.013221 |
| 1-Acyl-sn-glycerol-3-phosphate acyltransferase β (1-AGPAT 2) | Agpat2 | NM_026212 | 0.801640709 | 0.185652 | 2.480056483 | 0.000000 |
| Glycerol-3-phosphate acyltransferase 4 precursor (GPAT4/1-AGPAT 6) | Agpat6 | NM_018743 | 0.767911552 | 0.018326 | 1.628685395 | 0.000001 |
| Glycerol-3-phosphate acyltransferase 3 (GPAT3/1-AGPAT 9) | Agpat9 | NM_172715 | 1.244111365 | 0.4096525 | 1.929334823 | 0.000110 |
| Diacylglycerol O-acyltransferase 2 (Diglyceride acyltransferase 2) | Dgat2 | NM_0226384 | 0.821680911 | 0.083083 | 1.51081412 | 0.000002 |
| ADP-ribosylation factor 3 | Arf3 | NM_007478 | 1.122824851 | 0.456788 | 1.340471593 | 0.0075350 |
| Fatty acid desaturase 1 | Fads1 | NM_146094 | 0.794361905 | 0.109064 | 2.438959827 | 2.56E-08 |
| Fatty acid desaturase 2 | Fads2 | NM_019699 | 0.961159614 | 0.875472 | 3.394171825 | 2.79E-12 |
| Monoacylglycerol O-acyltransferase 1 | Mogat1 | Nm_026713 | 1.571736761 | 0.109971 | 3.518969057 | 9.08e-12 |
| Cholesterol metabolism | ||||||
| Acetyl-CoA acetyltransferase 2 | Acat2 | NM_009338 | 1.008976672 | 0.959174 | 2.090073606 | 0.000064 |
| 3-Hydroxy-3-methylglutaryl-CoA reductase | Hmgcr | NM_008255 | 1.02203705 | 0.950538 | 2.523089156 | 0.000058 |
| Insulin induced gene 1 | Insig1 | NM_153526 | 0.899987361 | 0.825931 | 2.230378952 | 0.006641 |
| Insulin induced gene 2 | Insig2 | NM_133748 | 1.377049424 | 0.072652 | 1.545180835 | 0.000519 |
| Low-density lipoprotein receptor | Ldlr | NM_010700 | 1.089668515 | 0.801805 | 2.091661502 | 0.000935 |
| Mevalonate kinase | Mvk | NM_023556 | 0.793588921 | 0.134295 | 1.535068684 | 0.000187 |
| Niemann-Pick disease, type C1 | Npc1 | NM_008720 | 0.549604432 | 0.044905 | 3.247726986 | 2.98E-12 |
| SREBF chaperone | Scap | NM_001001 | 0.659276502 | 0.002658 | 1,504013318 | 0.000656 |
| Lipid clearance | ||||||
| ATP-binding cassette, subfamily A (ABC1), member 4 | Abca4 | NM_007378 | 0.747968138 | 0.047085 | 0.702763205 | 0.023802 |
| ATP-binding cassette, subfamily B (MDR/TAP), member 4 | Abcb4 | NM_008830 | 1.150314676 | 0.055704 | 3.10103514 | 2.48E-17 |
| ATP-binding cassette, subfamily B (MDR/TAP), member 11 | Abcb11 | NM_021022 | 0.907157725 | 0.162527 | 1.381896672 | 0.000739 |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 3 | Abcc3 | NM_029600 | 1.056967218 | 0.692523 | 2.798380559 | 5.69E-10 |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 6 | Abcc6 | NM_018795 | 0.834376659 | 0.545808 | 2.217708393 | 0.029299 |
| ATP-binding cassette, subfamily D (ALD), member 1 | Abcd1 | NM_007435 | 0.909948198 | 0.646187 | 1.430288188 | 0.028496 |
| ATP-binding cassette, subfamily D (ALD), member 3 | Abcd3 | NM_008991 | 0.928259091 | 0.691785 | 2.552608494 | 4.83E-19 |
| ATP-binding cassette, subfamily G (WHITE), member 2 | Abcg2 | NM_011920 | 1.030221607 | 0.842335 | 1.870280574 | 0.000442 |
| ATP-binding cassette, subfamily G (WHITE), member 5 | Abcg5 | NM_031884 | 1.268861114 | 0.097249 | 1.522545861 | 0.007656 |
| Apolipoprotein A-IV | Apoa4 | NM_007468 | 4.088093565 | 5.002E-16 | 0.632168263 | 0.000000 |
| Apolipoprotein B (including Ag(x) antigen) | Apob | NM_009693 | 1.158046684 | 0.056479 | 0.62962321 | 0.000000 |
| Apolipoprotein C-II | Apoc2 | NM_009695 | 1.226624183 | 0.040873 | 1.317998849 | 0.011489 |
| Apolipoprotein C-III | Apoc3 | NM_023114 | 0.762967441 | 0.043733 | 2.354673461 | 1.61E-14 |
| Apolipoprotein E | Apoe | NM_009696 | 1.132474182 | 0.016551 | 0.599126064 | 2.25E-14 |
| Apolipoprotein F | Apof | NM_133997 | 1.045293385 | 0.50485 | 0.758581198 | 6.56E-08 |
| Low-density lipoprotein receptor | Ldlr | NM_010700 | 1.089668515 | 0.801805 | 2.091661502 | 0.000935 |
| Lipoprotein lipase | Lpl | NM_008509 | 1,452922974 | 0.050144 | 1.507945602 | 0.031554 |
| Palmitoyl-protein thioesterase 1 | Ppt1 | NM_008917 | 1.453532886 | 0.000004 | 1.3963282 | 0.000008 |
| Very low-density lipoprotein receptor | Vldlr | NM_013703 | 2.462217746 | 1.17E-10 | 1.634952757 | 2.22E-09 |
| Central metabolic regulators | ||||||
| Hepatocyte nuclear factor 3-α (HNF-3-α)(HNF-3A) (Forkhead box protein A1) | Foxa 1 | NM_008259 | 1.128437 | 0.337461 | 0.667551 | 0.000069 |
| Hepatocyte nuclear factor 4-α (HNF-4-α) (Transcription factor HNF-4) (Nuclear receptor subfamily 2 group A member 1) (Transcription factor 14) | Hnf4a | NM_008261 | 0.973646727 | 0.817565 | 1.279215738 | 0.032602 |
| Williams-Beuren syndrome chromosomal region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like) (ChREBP) | Mlxip1 | NM_021455 | 0.648074 | 0.013632 | 1.845287913 | 0.000112 |
| Oxysterols receptor LXR-α (Liver X receptor α) (Nuclear orphan receptor LXR-α) (Nuclear receptor subfamily 1 group H member 3) | Nr1 h3 | NM_013839 | 0.754751132 | 0.817565 | 2.31252332 | 0.000002 |
| Peroxisome proliferator-activated receptor δ (PPAR-δ) (PPAR-β) (Nuclear receptor subfamily 1 group C member 2) (Nuclear hormone receptor 1) (NUC1) | Ppard | NM_011145 | 1.112874065 | 0.306048 | 0.612536005 | 0.000017 |
| Peroxisome proliferator-activated receptor γ (PPAR-γ) (Nuclear receptor subfamily 1 group C member 3) | Pparg | NM_011145 | 2.231806638 | 0.000033 | 0.795033817 | 0.0166158 |
| Peroxisome proliferator-activated receptor γ coactivator 1-α (PPAR-γ coactivator 1-α) (PPARGC-1-α) (PGC-1-α) | Ppargc1a | NM_008904 | 0.846198481 | 0.307713 | 1.726617121 | 0.000026 |
| Peroxisome proliferator-activated receptor γ coactivator 1-β (PPAR-γ coactivator 1-β) (PPARGC-1-β) (PGC-1-β) (PGC-1β) (ERR ligand 1) | Ppargc1b | NM_133249 | 0.695169765 | 0.004644 | 1.317525256 | 0.009671 |
| Sterol regulatory element binding transcription factor 1 | Srebf1 | NM_011480 | 0.7870365 | 0.0698 | 0.85074 | 0.1345 |
| Sterol regulatory element binding transcription factor 2 | Srebf2 | NM_033218 | 0.830949847 | 0.220886 | 0.622169732 | 0.02801 |
| X-Box binding protein 1 | Xbp1 | NM_013842 | 1.027031945 | 0.772099 | 0.528636163 | 2.61E-10 |
P values <0.05 are underlined
NDGA regulation of hepatic lipid homeostasis in ob/ob mice.
To examine which component(s) of hepatic lipid homeostasis was impacted by the NDGA treatment of ob/ob mice, we classified the genes involved in lipid metabolism into eight groups: 1) fatty acid uptake; 2) acyl-CoA synthases; 3) fatty acid oxidation (catabolism); 4) fatty acid synthesis (lipogenesis); 5) TG synthesis and VLDL-TG assembly (fatty acid export); 6) cholesterol metabolism; 7) lipid clearance; and 8) transcription regulators of lipid metabolism. As can be seen from the data presented in Table 3, the fatty acid oxidation pathway was most robustly affected by the NDGA treatment. Expression levels of key fatty acid β-oxidation genes such as Acox1, Cpt2, Acadl, Acadm, Acads, Acadsb, Dci, Echs1, Ehhadh, and Peci were significantly upregulated with NDGA treatment. Similarly, NDGA-mediated increases in mRNA levels of Ppargc1a (PGC-1α) and Ppargc1b (PGC-1β) [the members of the peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family of transcriptional coactivators that serve as inducible coregulators of nuclear receptors in the control of cellular energy metabolic pathways] (10, 43) were observed in liver samples from NDGA-treated ob/ob mice. In addition, enzymes/transporters that facilitate fatty acid transport (e.g., Acsvl4, Acsl1, and Acsl4) or utilization (e.g., Acl, Acsm3, Acss2, Elovl2, Elovl5, Mlycd) showed a pattern of significant upregulation of mRNAs similar to the enzymes involved in fatty acid β-oxidation in response to NDGA treatment (Table 3).
Expression levels of genes involved in fatty acid synthesis (e.g., Fasn, Scd1, Scd2, Me2, Elovl6, Pklr, and L-Pk) were upregulated in ob/ob mice, but significantly downregulated by NDGA treatment. In contrast, the expression levels of other genes such as Acly, Acsl1, Acsl4, Acsm3, Acss2, Elovl2, Elovl5, Elovl6, Me1, Mlycd, and Gck, which participate in lipogenesis, were upregulated in response to NDGA feeding. Likewise, the expression of most of the TG synthesizing genes (e.g., Agpat2, Agpat6, Agpat9, Dgat2, Fads1, Fads2, and Mogat1) and Arf3, a key gene whose protein product is involved in VLDL assembly, were upregulated in NDGA-treated ob/ob mice. Among the genes involved in the regulation of VLDL metabolism, the expression of the gene for a major apolipoprotein, Apoa4, responsible for hepatic VLDL-TG export was markedly upregulated in ob/ob mice, but greatly reduced following treatment of mice with NDGA. The expression of the other VLDL-related genes such as Apob, Apoe, ApoF, and VLDR was suppressed by NDGA. In contrast, the mRNA levels of Apoc3 and Lpl were increased significantly following NDGA treatment of ob/ob mice. Likewise, NDGA treatment increased (two- to threefold) the expression of other genes involved in lipid clearance such as Abcb4, Abcb11, Abcc3, Abcc6, Abcd1, Abcd3, Abcg2, and Abcg5. Also, the expression of key genes involved in cholesterol metabolism, Acat2, Hmgcr, Ldlr, Mvk, Insig1, Insig2, Npc1, and Scap were significantly altered by NDGA treatment of ob/ob mice (Table 3).
Several key transcription factors regulate hepatic lipid metabolism. The expression of Foxa1 (HNF-3A), Xbp1 (XBP1), Ppard (PPARδ), and Srebf2 (SREBP-2) was reduced ∼50–70% in ob/ob mice that were chronically fed an NDGA supplemented chow diet. In contrast, NDGA treatment led to variable (25–300%) upregulation of genes of several transcription factors including HNF4a, Mlxipl (ChREBP, MONDOB, or WBSCR14), Nr1h3 (LXRα), Ppargc1a (PGC-1α), and Ppargc1b (PGC-1β). The high-glucose-responsive carbohydrate response element binding protein (ChREBP) along with its interacting partner (Mlx; heterodimer partner max-like factor-X) induces transcription of genes involved in glycolysis [liver pyruvate kinase (L-Pk)], lipogenesis (Acc, Fasn, Scd1, Elovl6), and gluconeogenesis [G6pc (glucose-6-phosphatase)] (46, 56, 75). Liver X receptor (LXR), as a LXR/retinoic acid X receptor (RXR) heterodimer, directly regulates expression of a number of genes important for de novo fatty acid synthesis (e.g., Acc1 and Acc2, Fasn, Scd1) (3, 70) but also regulates the expression of transcription factors such as SREBP-1c (3, 62) and ChREBP (5). Transcription factor XBP1, a key regulator of the unfolded protein response, is also involved in the regulation of hepatic lipogenesis (38).
Cluster analysis and confirmation of microarray by qRT-PCR.
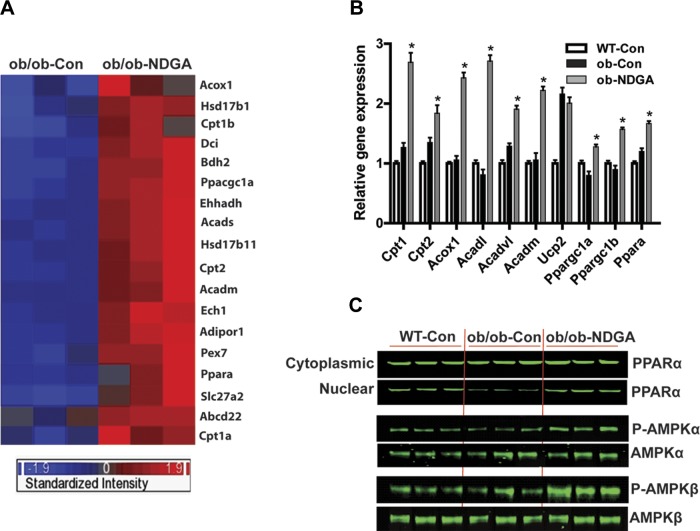
Next, we performed cluster analyses on the 18 fatty acid oxidation related genes that significantly correlated with NDGA treatment (see Fig. 2A, the heat map and clustering diagram of the differentially expressed genes). These genes are overexpressed in the livers of NDGA-treated mice compared with ob/ob control mice. We selected a set of 10 genes involved in hepatic fatty acid β-oxidation for their validation by qRT-PCR employing the same hepatic RNA samples previously used in cDNA microarray studies. Figure 2B shows that qRT-PCR analyses of hepatic mRNA expression of Cpt1, Cpt2, Acox1, Acadl, Acadvl, Acadm, Ucp2, Ppargc1a (PGC-1α), and Ppargc1b (PGC-1β) were upregulated significantly, which was consistent with the microarray data. Although gene array analysis showed no significant changes in response to NDGA treatment in Ppara (PPARα), the master regulator of various metabolic pathways involved in hepatic fatty acid metabolism including cellular fatty acid uptake, intracellular fatty acid transport, ketogenesis, mitochondrial and peroxisomal fatty acid β-oxidation, microsomal fatty acid ω-oxidation, and peroxisomal fatty acid uptake (58, 59), qRT-PCR measurements indicated that NDGA treatment led to significantly increased expression of PPARα mRNA levels.
Fig. 2.
Changes in hepatic gene expression in ob/ob-control and ob/ob-NDGA. Effect of NDGA treatment of ob/ob mice on hepatic gene expression protein levels of peroxisome proliferator-activated receptor α (PPARα) and total- and phospho- (active) forms of AMP-activated kinase (AMPK)-α (AMPKα-1 and AMPKα-2) catalytic and AMPKβ-1 regulatory subunits. A: heat map. Each row represents 1 gene and each column represents 1 sample. Red and blue colors represent upregulated and downregulated genes, respectively. The global gene expression profiles were compared after the high-dose NDGA treatment in ob/ob mice. B: confirmation of the gene expression by real-time quantitative PCR (qRT-PCR) analysis. Ten selected fatty acid oxidation-related genes were verified; 36b4 was used as control housekeeping gene. Values were presented as relative fold of changes over 36b4 (*ob-NDGA vs. ob-Con, P < 0.05). C: NDGA treatment of ob/ob mice restores the nuclear content of PPARα protein to the level seen in chow-fed control (C57BL/6J) mice. Likewise, levels of phosphorylated forms of AMPKα (AMPKα-1 and AMPKα-2) and AMPKβ-1 are significantly increased in response to NDGA treatment. No changes in the total protein levels of either AMPKα or AMPKβ are detected. Ob/ob-control, ob/ob mice fed a chow diet alone for 16 wk; ob/ob-NDGA, ob/ob mice fed a chow diet supplemented with a high dose (2.5 g of NDGA/kg of diet) for 16 wk.
Western blot analysis of PPARα and AMPK protein levels.
Western blot analysis further demonstrated that nuclear protein content of PPARα was reduced in ob/ob mice compared with wild-type (control mice) and that NDGA treatment almost completely restored its expression to the levels of control mice (Fig. 2C). We also evaluated the phosphorylation status of 5-AMP-activated protein kinase (AMPK). AMPK is a heterotrimeric protein comprising α (63 kDa), β (38 kDa), and γ (38 kDa) subunits. The AMPKα is the catalytic subunit, whereas AMPKβ and AMPKγ are regulatory (noncatalytic) subunits. AMPK, a serine-threonine kinase member of the SNF1 (sucrose nonfermentor) kinase family, is critically involved in the regulation of energy homeostasis in various tissues including the liver. When activated by phosphorylation, AMPK in the liver stimulates energy-producing processes such as fatty acid oxidation and glycolysis and inhibits energy by utilizing processes such as lipogenesis, amino acid synthesis, and gluconeogenesis (77, 79). As can be seen from Fig. 2C, the dietary administration of NDGA caused significant activation of AMPK through enhanced phosphorylation of both catalytic (AMPKα) (both AMPKα-1 and α-2) and regulatory (AMPKβ-1) subunits (Fig. 2C).
Effects of NDGA on hepatocyte cell line in vitro.
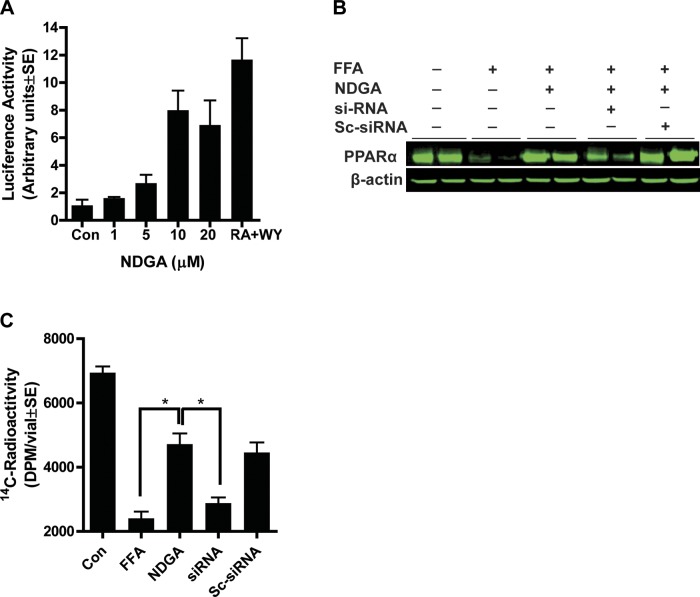
To test whether NDGA has an effect on Pparα promoter activity, we transfected AML12 hepatocytes with PPRE reporter and Pparα plasmid constructs in pCMX vector. Culture dishes were treated with different concentrations of NDGA ranging from 1 μM to 20 μM. NDGA exposure of cells stimulated PPAR promoter activity up to sevenfold with a maximum effect observed at 10 μM (Fig. 3A). To mimic the obese environment in vitro, a mixture of oleic and palmitic acid was used to load cells with lipid. Fatty acid loading significantly inhibited PPARα expression in hepatocytes, and 10 μM NDGA almost completely abrogated the negative effect of fatty acid loading on cellular PPARα levels. At the protein level, FFA loading of cells decreased PPARα protein levels by about 90%; treatment with NDGA abrogated the negative effect of FFA on PPARα protein (Fig. 3B).
Fig. 3.
Effects of NDGA on a model hepatocyte cell line (AML12). A: dose-dependent stimulation of PPARα promoter activity by treatment of cells with NDGA. B: attenuation of fatty acid loading-induced downregulation of PPARα protein expression by NDGA (10 μM). FFA, free fatty acid. C: PPARα siRNA (5 nM) abrogates NDGA effect on fatty acid oxidation. Sc-siRNA, scramble siRNA (control). Results are means ± SE of 4 experiments. *P < 0.05.
Next, we evaluated the role of PPARα in mediating the effects of NDGA treatment on fatty acid oxidation in mouse hepatocytes (AML12) using siRNA transfection. Transfection of cells with a PPARα-specific siRNA reduced PPARα protein levels ∼60–70%, whereas scrambled siRNA had no effect. Fatty acid loading of cells dramatically reduced mitochondrial and peroxisomal palmitate oxidation, whereas NDGA treatment partially restored the rates of fatty acid oxidation to the levels exhibited by cells without fatty acid loading (Fig. 3C); however, the ability of NDGA to restore the rates of fatty acid oxidation was abrogated by the knockdown of PPARα.
Effects of NDGA treatment in Pparα-deficient mice.
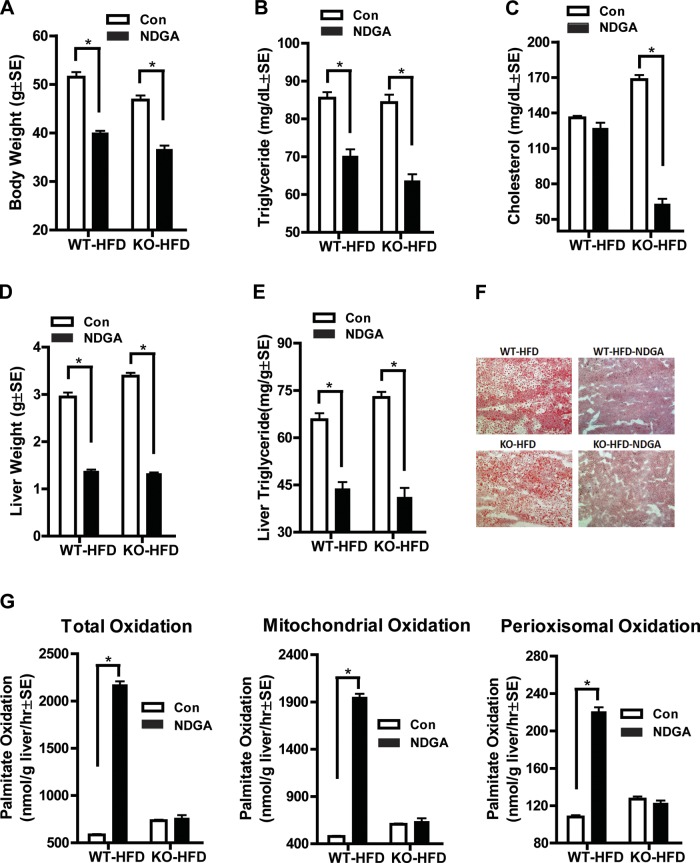
To further investigate whether NDGA stimulates fatty acid oxidation through PPARα, we fed Pparα-deficient mice either a high-fat diet or a high-fat diet supplemented with NDGA. Compared with wild-type mice fed a high-fat diet or NDGA-containing high-fat diet, we found that the increase in palmitate oxidation produced by NDGA was almost totally abrogated in Pparα-deficient mutant livers (Fig. 4F), though both body and liver weights as well as plasma and hepatic TG levels decreased to similar degrees as in wild-type mice (Fig. 4, A–C). These data further suggest that NDGA stimulates fatty acid oxidation primarily through PPARα.
Fig. 4.
Effects of NDGA treatment of Ppar-α-deficient mice on body weight, liver weight, and hepatic fatty acid oxidation. A: body weight. B: serum triglyceride. C: serum cholesterol. D: liver weight. E: liver triglyceride content. F: Oil red O staining of liver sections for lipids. G: [14C]palmitate oxidation: total, mitochondrial, and peroxisomal β-oxidation. Results are means ± SE (n = 8). KO, knockout; HFD, high-fat diet.
DISCUSSION
NAFLD encompasses a spectrum of disease that originates with excessive accumulation of lipid mainly in the form of TGs, or hepatic steatosis. Hepatic steatosis results from an imbalance between synthesis, uptake, export (secretion), and oxidation of fatty acids (58, 64, 69, 74). Previous work from our laboratory (12, 27 and references therein) and others (60) has shown that NDGA exerts profound effects on key components of the metabolic syndrome including lowering of plasma glucose, lipid (FFAs and TG) levels, attenuation of elevated blood pressure, inhibition of lipolysis in relevant rodent models of dyslipidemia, insulin resistance, diabetes, and hypertension. However, little is known about the mechanism of antihyperlipidemic actions of NDGA on dyslipidemia and its potential effect on hepatic steatosis. Thus the present study was conducted to elucidate the molecular mechanism(s) by which NDGA exerts its hypolipidemic actions, including attenuation of hepatic steatosis, using both in vivo models involving ob/ob and Pparα-deficient mice and an in vitro model of a mouse hepatocyte cell line (20, 48, 55, 56).
Increased body weight gain in ob/ob mice was diminished by dietary treatment with NDGA. Moreover, NDGA significantly reduced liver weight and steatosis as evidenced by decreases in Oil red O staining and hepatic TG content. Likewise, plasma levels of TGs and glucose were significantly reduced. The balance between energy intake (calories consumed) and energy expenditure (calories burned) determines body energy (fat) stores. Our data showing no change in food intake, but elevation in body temperature in response to NDGA treatment, suggest that NDGA has an effect on energy expenditure. Interestingly, a major effect of NDGA is to promote hepatic fatty acid utilization through increased oxidation of fatty acids via mitochondrial and peroxisomal β-oxidation pathways, which may account for its antiobesity action and may be responsible for lowering hepatic TG content. Using Pparα-deficient mice, where almost no stimulatory effect of NDGA on hepatic fatty acid oxidation was observed, we established that the ability of NDGA to increase hepatic fatty acid catabolism is mediated via PPARα. Interestingly, similar to control (wild-type) mice, Pparα-deficient mice showed comparable reduction in body and liver weights as well as plasma and hepatic TG (steatosis) content in response to NDGA treatment. Thus the present studies raise the possibility that NDGA most likely attenuates body weight gain (i.e., increased energy expenditure) and hepatic steatosis in a PPARα-independent manner, which is consistent with the recent report that NDGA attenuation of TG accumulation in HepG2 cells is mediated by AMPK (40). Additional studies are needed to delineate the actual mechanism involved. NDGA-mediated decreases in plasma glucose levels appear to result from both improvements in insulin resistance and steatosis and an enhanced rate of whole body glucose clearance. NDGA treatment also decreased the circulating levels of key inflammatory chemokines, indicating that NDGA may also function as an anti-inflammatory agent and suggesting that NDGA may improve hepatic insulin resistance and steatosis by promoting the decreased expression of certain inflammatory chemokines.
Gene expression profiling revealed a complex network of changes unique to hepatic lipid metabolism attributed to NDGA. Among the major hepatic lipid metabolizing pathways, NDGA treatment led to upregulation of PPARα, PGC-1α, PGC-1β, as well as genes involved in various steps of fatty acid oxidation and utilization. PPARα is a ligand-activated nuclear transcription factor (58) and is highly expressed in the liver, where it is most responsible for regulating genes involved in fatty acid uptake, fatty acid activation and transport into mitochondria, mitochondrial and peroxisomal β-oxidation, and ketogenesis (1, 6, 14, 15, 18, 28, 36, 43, 59, 71). Both PGC-1α and PGC-1β potently enhance the expression of genes involved in fatty acid oxidation and ketogenesis by coactivating PPARα (for review see Refs. 10 and 45). PGC-1β, but not PGC-1α, activates the expression of genes involved in lipogenesis, TG synthesis, and VLDL secretion by directly docking and coactivating SREBP-1c and LXR (10, 45), which play an essential role in the control of lipogenesis (17, 62). To what extent NDGA-mediated induction of PGC-1β contributes to hepatic lipogenesis is not known at present, but mounting evidence indicates that induction of both PGC-1α and PGC-1β expression in the liver is a key regulatory event in the activation of energy metabolic pathways, including fatty acid β-oxidation for energy (ATP) production (10, 45). The expression of genes involved in fatty acid catabolism, including those involved in hepatic fatty acid uptake (Slc27a2), activation (Acsl1, Acsl4), and transport into the mitochondria (Cpt1, Cpt2), peroxisomal and mitochondrial β-oxidation (Acox1, Acadl, Acadm, Acads, Acadsb, Acadvl, Dci, Echs1, Ehhadh, Peci), fatty acid elongation (Elovl2, Elovl5, Elovl6), desaturation (Fads1, Fads2), and hepatic clearance of VLDL (Apoc2, Lpl), were increased in response to NDGA treatment, and many of these genes are directly regulated by PPARα (59). We also noted a twofold induction of Mlycd mRNA levels in response to NDGA treatment, the gene encoding malonyl-CoA decarboxylase (MCD). MCD is transcriptionally regulated by PPARα (39), catalyzes the degradation of malonyl-CoA to acetyl-CoA, and is a key regulatory enzyme for fatty acid oxidation (39) through modulation of malonyl-CoA levels, a potent inhibitor of carnitine palmitoyltransferase 1 (CPT1) (16). Thus increased expression of MCD is expected to result in lowering of malonyl-CoA levels and, consequently, enhanced mitochondrial fatty acid β-oxidation. These gene-specific changes in fatty acid oxidation pathways in response to NDGA treatment are in good accord with the observed increases in the rate of total, mitochondrial and peroxisomal hepatic fatty acid β-oxidation. On the other hand, NDGA treatment did not affect hepatic mRNA expression of PPARα-responsive mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase (Hmgcs2), the rate-limiting enzyme in ketone body production (34). When considered together, these various findings lead us to conclude that NDGA-mediated attenuation of liver TG content and suppression of circulating TG levels results primarily through increased removal of fatty acids via their oxidative catabolism. To what extent NDGA mediated inhibition of hepatic steatosis also involves fatty acid elimination through induction of the ketogenesis pathway (58) remains to be explored, but its contribution is likely to be of much lower magnitude than that of hepatic β-oxidation systems. On the basis of these data, it is highly likely that NDGA activated PPARα, PGC-1α, and PGC-1β work in concert to promote increased channeling of fatty acids away from lipogenesis and TG synthesis toward fatty acid β-oxidation (41, 42).
Furthermore, we observed that mRNA levels of key fatty acid synthesizing enzymes such as Fasn, Me2, Elovl6, Scd1, Scd2, and Pklr (a specific target gene of ChREBP) were decreased, whereas the levels of most of the TG-synthesizing (i.e., Agpat2, Agpat6, Agpat9, Dgat2, Arf3, Mogat1) and lipogenic enzymes (Acly, Fads1, Fads2, ME1, Elovl2, and Elovl5) were upregulated in ob/ob mice when fed a high-NDGA supplemented diet. NDGA treatment of ob/ob mice also affected the expression of SREBP-2, ChREBP, and LXR, the three major lipogenic transcription factors (5, 17, 46, 56, 62, 65, 76), and XBP1 (38), all of which are involved in the regulation of fatty acid and TG synthesis (39, 46, 62, 70, 75, 79) and contribute to hepatic steatosis (22, 31, 56, 57, 70, 78). However, the patterns of expression of these genes varied considerably from each other. Since SREBP-1c transcriptional activity is also regulated at the posttranslational level by protease-mediated cleavage of the precursor ER transmembrane form, allowing the cleaved and transcriptionally active amino-terminal peptide to translocate into the nucleus (17), it is possible that NDGA promotes increased proteolytic cleavage and activation of SREBP-1c activity by modulating this process. Interestingly, hepatic mRNA levels of SREBP cleavage activating protein (SCAP), which escorts SREBP-1a/1c and SREBP-2 from ER to Golgi for their proteolytic processing, thereby allowing activated SREBPs to stimulate fatty acid and cholesterol synthesis (17), were significantly increased (∼1.5 fold) in NDGA-treated animals. NDGA treatment also led to increased expression of Insig-1 (insulin-induced gene product-1) and Insig-2, which can inhibit the activity of SREBP-1c (and SREBP-2) and their target genes and, as a result, may limit the functional efficacy of NDGA-induced and SREBP-1c-mediated lipogenesis (8, 17, 72).
Earlier studies have shown that activation or overexpression of SREBP-1c or LXR in mice leads to the development of extensive hepatic steatosis (31, 57, 65, 66), whereas genetic deletion of SREBP-1c, LXR, or ChREBP attenuates steatosis (20, 31, 51, 57, 62, 70, 78). Thus, despite their involvement in the pathogenesis of hepatic steatosis, we were quite surprised to find that NDGA exerted no inhibitory action on the expression of SREBP-1c, LXR, and ChREBP but at the same time attenuated hepatic steatosis. Even more surprising was the fact that NDGA treatment actually increased the expression of LXR and ChREBP (and possibly SREBP-1c) along with some of their downstream targets (i.e., many lipogenic and most of the TG synthesizing genes). Although currently we are unable to provide an exact explanation about the lack of inhibitory action of NDGA on these three lipogenic transcription factors, several possibilities may explain this phenomenon. First, PPARα can positively impact hepatic lipogenesis (9, 16, 23, 32, 53) by regulating SREBP-1c and LXR (16, 23, 32, 53, 59), as well as certain enzymes involved in polyunsaturated fatty acid or triacylglycerol synthesis (23). Second, fatty acid synthase (FAS) reaction products (i.e., “new” fatty acids) can activate physiologically distinct pools of PPARα in the liver to regulate glucose, lipid, and cholesterol metabolism (6). Finally, genomewide profiling of LXR, PPARα, and their common dimerization partner RXR in mouse liver revealed extensive overlap (71 to 88%) between the binding sites of LXR and PPARα (3) and potential cross-talk between PPARα and LXR or other LXR-regulated lipogenic transcription factors such as SREBP-1c and ChREBP. Our demonstration that NDGA functions as a potent ligand or proligand for and activator of PPARα (either directly or indirectly through its actions as a lipoxygenase inhibitor) suggests that it most likely stimulates LXR, ChREBP (and possibly SREBP-1c), and, consequently, lipogenesis, by a mechanism similar to that employed by other PPARα agonists (16, 23, 32, 53) to stimulate lipogenesis. Overall, the involvement of PPARα in lipogenesis may represent a compensatory mechanism for the continued availability of an adequate amount of fatty acids as a fuel for oxidation and high energy production. In this context, NDGA-mediated increased expression of ChREBP may also be necessary to safeguard the liver against becoming energy deficient (4) and may also upregulate the expression of SREBP-1c both directly (21) and indirectly through other mechanisms (8, 17, 72).
ob/ob mice exhibit high levels of ER stress (24), and NDGA treatment resulted in significant downregulation of the ER stress response protein XBP1 mRNA. XBP1 is downstream of the unfolded protein response (UPR) sensor inositol-requiring kinase-1α (IRE1α) and regulates the expression of a number of genes involved in de novo lipogenesis, including SREBP1, SCAP, FAS, SCD1, ACC2, and DGAT2 (22, 24, 38, 54, 61, 80), and it has been suggested that dysregulated ER stress may be a major contributor to the pathogenesis of hepatic steatosis (61). In our studies, NDGA-mediated repression of XBP1 gene expression suggests that the IRE1α-XBP1 pathway may represent another potential mechanism by which NDGA reduces liver TG content. Overall, these results point to a direct link between NDGA-mediated reduction in XBP1 gene expression to reductions in FAS, SCD1, and SCD2 mRNA levels, hepatic TG content, and lipogenesis, most likely via amelioration of ER stress (24).
Dietary administration of NDGA results in activation of AMPK, i.e., increased phosphorylation of both catalytic (AMPKα-1 and AMPKα-2) and regulatory (AMPKβ-1) subunits. When activated by phosphorylation, AMPK stimulates energy generating processes, such as fatty acid oxidation and glycolysis, and inhibits energy consuming processes, such as lipogenesis and amino acid synthesis (77, 79). This is achieved, in part, through phosphorylation and inactivation of acetyl-CoA carboxylase, leading to a decrease in the concentration of malonyl-CoA (77, 79). Activated AMPK also attenuates hepatic steatosis by restricting the availability of FFAs for TG synthesis through phosphorylation and inactivation of SREBP-1c (44), ChREBP (26), and LXR (19), and inhibition of glycerol-3-phosphate acyltransferase (GPAT), the rate-limiting step in TG synthesis. We found that an increase in activated AMPK with NDGA supplementation was associated with an increase in CPT1 expression and a decrease in the expression of FAS, SCD1, SCD2, and L-PK. Although NDGA treatment increased the expression of ChREBP, LXRα (and a possibly active form of SREBP-1c), the observed decreases in the gene expression of FAS, SCD1, SCD2, and L-PK may be related to the ability of AMPK to inhibit (via phosphorylation) the functional activities of these transcription factors (40). In addition, at present, we cannot exclude the possibility that activated AMPK, along with the reduced expression of XBP1, may be jointly responsible for downregulating these lipogenic genes and restricting lipogenesis. Collectively, these data indicate that NDGA may reduce lipid accumulation in the liver, at least in part, by directly stimulating AMPK activity, which in turn enhances hepatic fatty acid β-oxidation and reduces de novo lipogenesis. These NDGA-mediated reciprocal changes in the rates of fatty acid synthesis and degradation should allow increased channeling of acyl-CoA toward β-oxidation and away from the biosynthesis of TGs and their subsequent accumulation in the liver (steatosis).
In summary, we have provided insights into the molecular mechanisms by which dietary supplementation of NDGA ameliorates hepatic steatosis in ob/ob mice. On the basis of the experimental data obtained, the beneficial actions of NDGA on dyslipidemia and hepatic steatosis in ob/ob mice are exerted primarily through PPARα-dependent and PPARα-independent pathways. Furthermore, NDGA modulation of the IRE1/XBP1 arm of ER stress and AMPK signaling may also contribute to NDGA's ability to attenuate hepatic steatosis.
GRANTS
The work was supported by the Office of Research and Development, Medical Service, Department of Veterans Affairs, and National Heart, Lung, and Blood Institute Grant 1R01HL92473.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by authors.
AUTHOR CONTRIBUTIONS
H.Z., W-J.S., F.B.K., and S.A. conceived and designed the research; H.Z., W-J.S., Y.C., and S.A. performed the experiments; H.Z. and W-J.S. analyzed data; H.Z., W-J.S., F.B.K. and S.A. interpreted results of experiments; H.Z. and W-J.S. prepared figures; H.Z. drafted the manuscript; H.Z., W-J.S., F.B.K., and S.A. edited and revised the manuscript; H.Z., W-J.S., Y.C., F.B.K., and S.A. approved the final version of the manuscript.
REFERENCES
- 1. Akiyama TE, Nicol CJ, Fievet C, Staels B, Ward JM, Auwerx J, Lee SST, Gonzales FJ, Peters JM. Peroxisome proliferator-activated receptor-α regulates lipid homeostasis, but is not associated with obesity: studies with congenic mouse lines. J Biol Chem 276: 39088–39093, 2001 [DOI] [PubMed] [Google Scholar]
- 2. Angulo P. Nonalcoholic fatty liver disease. N Engl J Med 346: 1221–1229, 2002 [DOI] [PubMed] [Google Scholar]
- 3. Boergesen M, Pedersen TÅ, Gross B, van Heeringen J, Hagenbeek D, Bindesbøll C, Caron S, Lalloyer F, Steffensen KR, Neb HI, Gustafsson JA, Stunnenberg HG, Stales B, Mandrup S. Genome-wide profiling of liver X receptor, retinoid X receptor and peroxisome proliferator-activated receptor-α in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol 32: 852–867, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burgess SC, Iizuka K, Jeong NH, Harris RA, Kashiwaya Y, Veech RL, Kitazume T. Carbohydrate-response element-binding protein deletion alters substrate utilization producing an energy-deficient liver. J Biol Chem 283: 1670–1678, 2008 [DOI] [PubMed] [Google Scholar]
- 5. Cha JY, Repa JJ. The liver receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J Biol Chem 282: 743–751, 2007 [DOI] [PubMed] [Google Scholar]
- 6. Chakravarty MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, Turk J, Semenkovich CF. “New” hepatic fat activates PPARα to maintain glucose, lipid and cholesterol homeostasis. Cell Metab 1: 309–322, 2005 [DOI] [PubMed] [Google Scholar]
- 7. Dowman JK, Armstrong MJ, Tomlinson JW, Newsome PN. Current therapeutic strategies in non-alcoholic fatty liver disease. Diabetes Obes Metab 13: 692–702, 2011 [DOI] [PubMed] [Google Scholar]
- 8. Engelking LJ, Kuriyama H, Hammer RE, Horton JD, Brown MS, Goldstein JL, Liang G. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J Clin Invest 113: 1168–1175, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farnández-Alvarez A, Alvarez MS, Gonzalez R, Cucarella C, Muntané J, Casado M. Human SREBP1c expression in liver is directly regulated by peroxisome proliferator-activated receptor α (PPARα). J Biol Chem 286: 21477–21477, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 116: 615–622, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226: 497–509, 1957 [PubMed] [Google Scholar]
- 12. Gowri MS, Azhar RK, Kraemer FB, Reaven GM, Azhar S. Masoprocol decreases rat lipolytic activity by decreasing the phosphorylation of HSL. Am J Physiol Endocrinol Metab 279: E593–E600, 2000 [DOI] [PubMed] [Google Scholar]
- 13. Greenfield V, Cheung O, Sanyal AJ. Recent advances in nonalcoholic fatty liver disease. Curr Opin Gastroenterol 24: 320–327, 2008 [DOI] [PubMed] [Google Scholar]
- 14. Harano Y, Yasui K, Toyama T, Nakajima T, Mitsuyoshi H, Mimani M, Hirasawa T, Itoh Y, Okanoue T. Fentofibrate, a peroxisome proliferator-activated receptor alpha agonists, reduces hepatic steatosis and lipid peroxidation in fatty liver Shionogi mice with hereditary fatty liver. Liver Int 26: 613–620, 2006 [DOI] [PubMed] [Google Scholar]
- 15. Hashimoto T, Cook WS, Qi C, Yeldandi AV, Reddy JK, Rao MS. Defect in peroxisome proliferator activated receptor α-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J Biol Chem 275: 28918–28928, 2000 [DOI] [PubMed] [Google Scholar]
- 16. Hebbachi AM, Knight BL, Wiggins D, Patel DD, Gibbons GF. Peroxisome proliferator-activated receptor α deficiency abolishes the response of lipogenic gene expression to re-feeding: Restoration of the normal response by activation of liver X receptor α. J Biol Chem 283: 4866–4876, 2008 [DOI] [PubMed] [Google Scholar]
- 17. Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109: 1125–1131, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang J, Jia Y, Fu T, Viswakarma N, Bai L, Rao S, Zhu Y, Borensztajn J, Reddy JK. Sustained activation of PPARα by endogenous ligands increases hepatic fatty acid oxidation and prevents obesity in ob/ob mice. FASEB J 26: 628–638, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hwahng SH, Ki SH, Bae EJ, Kim HE, Kim SG. Role of adenosine monophosphate-activated protein kinase-p70 ribosomal S6 kinase-1 pathway in repression of liver X receptor-alpha-dependent lipogenic gene induction and hepatic steatosis by a novel class of dithiolethiones. Hepatology 49: 1913–1925, 2009 [DOI] [PubMed] [Google Scholar]
- 20. Iizuka K, Miller B, Uyeda K. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am J Physiol Endocrinol Metab 291: E358–E364, 2006 [DOI] [PubMed] [Google Scholar]
- 21. Jeong YS, Kim D, Lee YS, Kim HJ, Han JY, Im SS, Chong HK, Kwon JK, Cho YH, Kim WK, Osborne TF, Horton JD. Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS ONE 6: e22544, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Juaczak MJ, Lee AH, Journayvaz FR, Lee HY, Birkenfeld AL, Guigni BA, Kahn M, Samuel VT, Glimcher LH, Shulman GI. Dissociation of inositol-requiring enzyme (IRE1α)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J Biol Chem 287: 2558–2567, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jump DB. Fatty acid regulation of hepatic lipid metabolism. Curr Opin Clin Nutr Metab Care 14: 115–120, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kammoun HL, Chabanon H, Hainault I, Luquest S, Magnan C, Koike T, Ferré P, Foufelle F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest 119: 1201–1215, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kashi MR, Torres DM, Harrison SA. Current and emerging therapies in nonalcoholic fatty liver disease. Semin Liver Dis 28: 396–406, 2008 [DOI] [PubMed] [Google Scholar]
- 26. Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: regulation of carbohydrate responsive element-binding protein by AMP-activated protein kinase. J Biol Chem 277: 3829–3835, 2002 [DOI] [PubMed] [Google Scholar]
- 27. Kelley GL, Allan G, Azhar S. High dietary fructose induces a hepatic response resulting in cholesterol and lipid dysregulation. Endocrinology 145: 548–555, 2004 [DOI] [PubMed] [Google Scholar]
- 28. Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor α mediates the adoptive response to fasting. J Clin Invest 103: 1489–1498, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khashab MA, Liangunsakul S, Chalasani N. Nonalcoholic fatty liver disease as a component of the metabolic syndrome. Curr Gastroenterol Rep 10: 73–80, 2008 [DOI] [PubMed] [Google Scholar]
- 30. Kim CH, Younossi ZM. Nonalcoholic fatty liver disease: a manifestation of the metabolic syndrome. Cleve Clin J Med 75: 721–728, 2008 [DOI] [PubMed] [Google Scholar]
- 31. Knebel B, Haas J, Hartwig S, Jacob S, Köllmer C, Nitzgen U, Muller-Wieland D, Kotzka J. Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass. PLoS ONE 7: e31812, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Knight BL, Hebbachi A, Hauton D, Brown AM, Wiggins D, Patel DD, Gibbons GF. A role for the PPARα in the control of SREBP activity and lipid synthesis in the liver. Biochem J 389: 413–421, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kopec KL, Burns D. Nonalcoholic fatty liver disease: a review of the spectrum of disease, diagnosis, and therapy. Nutr Clin Pract 26: 565–576, 2011 [DOI] [PubMed] [Google Scholar]
- 34. Kostiuk MA, Keller BO, Berthiaume LG. Palmitoylation of ketogenic enzyme HMGCS2 enhances its interaction with PPARα and transcription at the Hmgcs2 PPRE. FASEB J 24: 1914–1924, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kotronen A, Yki-Järvinen H. Fatty liver: a novel component of the metabolic syndrome. Arterioscler Thromb Vasc Biol 28: 27–38, 2008 [DOI] [PubMed] [Google Scholar]
- 36. Later CZ, Yeh MM, Van Rooyen DM, Brooling J, Ghatora K, Farrell GC. Peroxisome proliferator-activated receptor-α, Wy14,643, improves metabolic indices, steatosis and ballooning in diabetic mice with non-alcoholic steatohepatitis. J Gastroenterol Hepatol 27: 341–350, 2012 [DOI] [PubMed] [Google Scholar]
- 37. Lebovics E, Rubin J. Non-alcoholic fatty liver disease (NAFLD): why you should care, when you should worry, what you should do. Diabetes Metab Res Rev 27: 419–424, 2011 [DOI] [PubMed] [Google Scholar]
- 38. Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320: 1492–1496, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee GY, Kim NH, Zhao ZS, Cha BS, Kim YS. Peroxisomal-proliferator-activated receptor α activates transcription of the rat hepatic malonyl-CoA decarboxylase gene: a key regulation of malonyl-CoA level. Biochem J 378: 983–990, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee MS, Kim D, Jo K, Hwang JK. Nordihydroguaiaretic acid protects against high-fat diet-induced fatty liver by activating AMP-activated protein kinase in obese mice. Biochem Biophys Res Commun 401: 92–97, 2010 [DOI] [PubMed] [Google Scholar]
- 41. Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldman HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, Zhang D, Litwin SE, Zaha VG, Fountain KT, Boudina S, Jimenez-Linan M, Blount M, Lopez M, Meirhaeghe A, Bohlooly -YM, Storlien L, Strömstedt M, Snaith M, Oresic M, Abel ED, Cannon B, Vidal-Puig A. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol 4: e369, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol 3: e101, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leone TC, Weinheimer CJ, Kelley DP. A critical role for the peroxisome proliferator-activated receptor α (PPARα) in the cellular fasting response: The PPARα-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA 96: 7473–7478, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, Gao B, Wierzbicki M, Verbeuren TJ, Shaw RJ, Cohen RA, Zang M. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab 13: 376–388, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1: 361–370, 2011 [DOI] [PubMed] [Google Scholar]
- 46. Ma L, Robinson LN, Towle HC. ChREBP·Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem 281: 28721–28730, 2006 [DOI] [PubMed] [Google Scholar]
- 47. Mannaerts GP, Debeer LJ, Thomas J, De Schepper PJ. Mitochondrial and peroxisomal fatty acid oxidation in liver homogenates and isolated hepatocytes from control and clofibrate-treated rats. J Biol Chem 254: 4585–4595, 1979 [PubMed] [Google Scholar]
- 48. Matsusue K, Haluzik M, Lambert G, Yim SH, Gavrilova O, Ward JM, Brewer B, Jr, Reitman M, Gozalez FJ. Liver-specific disruption of PPARγ in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J Clin Invest 111: 737–747, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGarry J, Brown N. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. J Biochem 244: 1–14, 1997 [DOI] [PubMed] [Google Scholar]
- 50. Mencin AA, Lavine JE. Nonalcoholic fatty liver disease in children. Curr Opin Clin Nutr Metab Care 14: 151–157, 2011 [DOI] [PubMed] [Google Scholar]
- 51. Moon YA, Liang G, Xie X, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Brown MS, Goldstein JL, Horton JD. The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab 15: 240–246, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moore JB. Non-alcoholic fatty liver disease: the hepatic consequences of obesity and the metabolic syndrome. Proc Nutr Soc 69: 211–220, 2010 [DOI] [PubMed] [Google Scholar]
- 53. Oosterveer MH, Grefhorst A, van Dijk TH, Havinga R, Staels B, Kuipers F, Groen AK. Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. J Biol Chem 284: 34036–34044, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2α enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab 7: 520–532, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pellemounter MA, Cullen MJ, Baker MB, Hecht R, Winters D, Boone T, Collins F. Effect of obese gene product on body weight regulation in ob/ob mice. Science 269: 540–543, 1995 [DOI] [PubMed] [Google Scholar]
- 56. Postic C, Dentin R, Denechaud PD. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr 27: 179–192, 2007 [DOI] [PubMed] [Google Scholar]
- 57. Postic C, Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest 118: 829–838, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pyper SR, Viswakarma N, Yu S, Reddy JK. PPARα: energy combustion, hypolipidemia, inflammation and cancer. Nucl Recept Signal 8: e002, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rakhshandehroo M, Knoch B, Müller M, Kersten S. Peroxisome proliferator-activated receptor α target genes. PPAR Res 2010: 612089, 2010. Epub 2010 Sep 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reed MJ, Meszaros K, Entes LJ, Claypool MD, Pinkett JG, Brignetti D, Luo J, Khandwala A, Reaven GM. Effect of masoprocol on carbohydrate and lipid metabolism in a rat model of type II diabetes. Diabetologia 42: 102–106, 1999 [DOI] [PubMed] [Google Scholar]
- 61. Ren LP, Chan SMH, Zeng XY, Laybutt DR, Iseli TJ, Sun RQ, Kraegen EW. Differing endoplasmic reticulum stress response to excess lipogenesis versus lipid oversupply in relation to hepatic steatosis and insulin resistance. PLoS ONE 7: e30816, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Repa JJ, Liang G, Bashmakov Y, Lobaccaro JMA, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev 14: 2819–2830, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell 15: 829–840, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Savage DB, Semple RK. Recent insight into fatty liver, metabolic dyslipidemia and their links to insulin resistance. Curr Opin Lipidol 21: 329–336, 2010 [DOI] [PubMed] [Google Scholar]
- 65. Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwender S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes Dev 14: 2831–2838, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem 274: 30028–30032, 1999 [DOI] [PubMed] [Google Scholar]
- 67. Smith BW, Adams LA. Nonalcoholic fatty liver disease and diabetes mellitus: pathogenesis and treatment. Nat Rev Endocrinol 7: 456–465, 2011 [DOI] [PubMed] [Google Scholar]
- 68. Smith BW, Adams LA. Nonlcoholic fatty liver disease. Crit Rev Clin Lab Sci 48: 97–113, 2011 [DOI] [PubMed] [Google Scholar]
- 69. Stefan N, Kantartizis K, Häring HU. Causes and metabolic consequences of fatty liver. Endocr Rev 29: 939–960, 2008 [DOI] [PubMed] [Google Scholar]
- 70. Strable MS, Ntambi JM. Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit Rev Biochem Mol Biol 45: 199–214, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sudgen MC, Bulmer K, Gibbons GF, Knight BL, Holness MJ. Peroxisome-proliferator-activated receptor-α (PPARα) deficiency leads to dysregulation of hepatic lipid and carbohydrate metabolism by fatty acids. Biochem J 364: 361–368, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Takaishi K, Duplomb L, Wang MY, Li J, Unger RH. Hepatic Insig-1 or Insig-2 overexpression reduces lipogenesis in obese Zucker diabetic fatty rats and in fasted/refed normal rats. Proc Natl Acad Sci USA 101: 7106–7111, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Thoma C, Day CP, Trenell MI. Lifestyle interventions for the treatment of non-alcoholic fatty liver disease in adults: a systematic review. J Hepatol 56: 255–266, 2012 [DOI] [PubMed] [Google Scholar]
- 74. Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol 5: 145–171, 2010 [DOI] [PubMed] [Google Scholar]
- 75. Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 4: 107–110, 2006 [DOI] [PubMed] [Google Scholar]
- 76. van Wagner LB, Rinella ME. The role of insulin sensitizing agents in the treatment of nonalcoholic steatohepatitis. Therap Adv Gastroenterol 4: 249–263, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Viollet B, Guigas B, Leclerc J, Hébrad S, Lantier L, Mounier R, Andreelli F, Foretz M. AMP-activated protein kinase in the regulation of hepatic energy metabolism: from physiology to therapeutic perspectives. Acta Physiol (Oxf) 196: 81–98, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yahagi N, Shimano H, Hasty AH, Matsuzaka T, Ide T, Yoshikawa Y, Amemiya-Kudo M, Tomita S, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Osuga J, Hareda K, Gotoda T, Nagai R, Ishibashi S. Absence of sterol regulatory element-binding protein-1 (SREBP-1) ameliorates fatty livers but not obesity or insulin resistance in Lepob/Lepob mice. J Biol Chem 277: 19353–19357, 2002 [DOI] [PubMed] [Google Scholar]
- 79. Zhang BB, Zhou G, Li C. AMPK: an emerging drug target for diabetes and metabolic syndrome. Cell Metab 9: 407–416, 2009 [DOI] [PubMed] [Google Scholar]
- 80. Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ. The unfolded protein response transducer IRE1a prevents ER stress-induced hepatic steatosis. EMBO J 30: 1357–1375, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]