Abstract
Many hibernating mammals suspend food intake during winter, relying solely on stored lipids to fuel metabolism. Winter fasting in these species eliminates a major source of degradable substrates to support growth of gut microbes, which may affect microbial community structure and host-microbial interactions. We explored the effect of the annual hibernation cycle on gut microbiotas using deep sequencing of 16S rRNA genes from ground squirrel cecal contents. Squirrel microbiotas were dominated by members of the phyla Bacteroidetes, Firmicutes, and Verrucomicrobia. UniFrac analysis showed that microbiotas clustered strongly by season, and maternal influences, diet history, host age, and host body temperature had minimal effects. Phylogenetic diversity and numbers of operational taxonomic units were lowest in late winter and highest in the spring after a 2-wk period of refeeding. Hibernation increased relative abundance of Bacteroidetes and Verrucomicrobia, phyla that contain species capable of surviving on host-derived substrates such as mucins, and reduced relative abundance of Firmicutes, many of which prefer dietary polysaccharides. Hibernation reduced cecal short-chain fatty acid and ammonia concentrations, and increased and decreased concentrations of acetate and butyrate, respectively. These results indicate that the ground squirrel microbiota is restructured each year in a manner that reflects differences in microbial preferences for dietary vs. host-derived substrates, and thus the competitive abilities of different taxa to survive in the altered environment in the hibernator gut.
Keywords: hibernation, microbes, torpor, fasting, diet
the coevolution of mammals with their gut microbes has produced complex relationships that provide benefits for both partners (38, 39). Microbes shape the biology of their hosts in multiple ways: they enhance resistance to pathogen colonization, influence gastrointestinal structure and function, drive the development of the immune system, and increase energy harvest from the diet (3, 18, 32, 65). In turn, the host provides a nutrient-rich environment that supports the development of diverse microbial communities that inhabit multiple niches (63, 66). The structure of the microbiota is shaped by several factors, including host genetics, maternal effects, interactions with the immune system, interactions among members of the microbial consortium, and the availability of metabolizable substrates. Although host diet provides the major source of substrates to support microbial growth, microbes can also use host-derived substrates, including mucus glycans, nutrients in sloughed epithelial cells, and pancreatic and biliary secretions (4, 31, 37, 45, 59). Gut microbes vary in their abilities to degrade different substrate types, and some species can rapidly adapt their metabolic machinery to use alternative substrates, such as dietary vs. host-derived glycans (34, 59). It is now well established that changes in diet type and amount alter microbiota composition in mammals (38, 49), although most studies have used species that do not experience severe dietary extremes as part of their normal lifestyles.
Circannual cycles of feeding and fasting are a key feature of the biology of many hibernating mammals (13, 27, 43). In these species, hyperphagia drives the accumulation of large fat stores in summer and early fall, followed by voluntary fasting, which can last up to 8 mo (13, 27, 43). This natural cycle of food intake provides the opportunity to examine the response of the gut microbiota to changes in substrate availability free from experimental manipulations, such as imposed fasts, which can increase host stress. In addition to dietary change, other aspects of the hibernation phenotype could potentially alter microbial communities. Most of the hibernation season is spent in a metabolically depressed state known as torpor, when body temperature (Tb) falls to <10°C and metabolism is <4% of active values (13). Although Tb during torpor is below the temperature optimum for most gut microbes, bouts of torpor are interrupted periodically by interbout arousals to normothermia that last for 12–24 h (13), providing sufficient time at high temperature for microbial degradation of any host-derived substrates that may be present in the gut lumen. Hibernation is also accompanied by a remodeling of the intestinal immune system (35), which is an aspect of host biology that is both influenced by, and can itself influence, the microbiota. Here, we demonstrate an annual reorganization of the cecal microbiota in 13-lined ground squirrels that reflects the dominant role of host diet in shaping microbial community structure.
METHODS
Animals.
All procedures were approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee. Six pregnant female 13-lined ground squirrels (Ictidomys tridecemlineatus) were collected in early May, 2010, around Madison, Wisconsin, and housed individually in a 22°C room with a 12:12-h light-dark cycle. In the wild, these squirrels eat a plant-based diet (leaves, flowers, seeds) supplemented with animal material when available (e.g., insects, bird eggs). After capture, water and rat chow (Harlan Teklad no. 7001) were provided ad libitum, and diets were supplemented once per week with fruit (apples, strawberries) and sunflower seeds. After parturition, pups remained with mothers for 5 wk, after which they were moved to individual cages with ad libitum chow + fruit for an additional 2 wk. The pups' food was then restricted to 12 g of chow/day to prevent excessive weight gain, supplemented with 1 g sunflower seeds once per week. Mothers were euthanized in July for sample collection. Pups were randomly assigned to one of six seasonal groups (Fig. 1B), such that each season contained at least one pup from each mother. One group of pups was euthanized in August (summer). The remaining pups were transferred to a 4°C room with constant darkness, except for brief (∼5 min) periods of dim light once per day to check activity states using the sawdust method (52). Food and water were removed after squirrels began using torpor. Hibernating squirrels were euthanized either in early winter (∼30 days of hibernation) or late winter (∼130 days of hibernation) in one of two activity states: torpor (Tb, ∼6°C) or interbout arousal (Tb, ∼36°C). Spring squirrels were returned to the warm room after ∼130 days of hibernation and were euthanized 14 days later. Details on squirrel body masses, Tbs, and hibernation patterns are presented in Table 1.
Fig. 1.
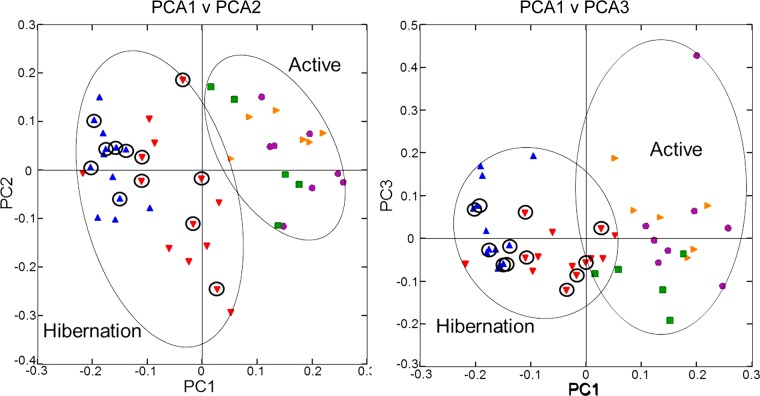
Principal coordinate analysis plots of unweighted UniFrac metrics for squirrel miocrobiotas. Each dot represents individual microbiota samples obtained from Mothers (yellow triangles) and their pups in Summer (purple circles), Early Winter (red triangles), Late Winter (blue triangles), and Spring (green squares). The first three axes are affected by seasonal variation. Left: PCA1 vs. PCA2; Right: PCA1 vs. PCA3. For hibernators, samples from torpid squirrels are circled. Numbers of squirrels in each seasonal group are shown in Table 1.
Table 1.
Ground squirrel body mass, cecal tissue mass, body temperature, and torpor characteristics
| Group | n | Age, days | Body mass, g | Cecal Mass, g | Tb, °C | Total Days Hibernating | Total Days in Torpor | Days in Last Torpor Bout |
|---|---|---|---|---|---|---|---|---|
| Mothers | 6 | >420* | 202 ± 10a | 1.15 ± 0.07a | 35.4 ± 0.5a,b | |||
| Summer | 8 | 98 ± 1a | 156 ± 10b | 1.03 ± 0.04a | 35.0 ± 0.8b | |||
| Early Winter-Aroused | 8 | 157 ± 1b | 158 ± 6b | 0.78 ± 0.11b | 35.8 ± 0.4a, b | 34.3 ± 1.3a | 29.1 ± 1.1a | 6.1 ± 0.4 |
| Early Winter-Torpid | 6 | 155 ± 1b | 157 ± 8b | 0.62 ± 0.05b | 6.5 ± 0.2c | 33.5 ± 0.6a | 28.8 ± 0.6a | 6.2 ± 0.4 |
| Late Winter-Aroused | 7 | 253 ± 3c | 132 ± 7c | 0.68 ± 0.08b | 36.2 ± 0.5a,b | 132.0 ± 2.9b | 115.0 ± 1.5b | 6.1 ± 0.7 |
| Late Winter-Torpid | 6 | 252 ± 1c | 138 ± 4b,c | 0.71 ± 0.04b | 6.5 ± 0.3c | 130.0 ± 1.7b | 116.7 ± 1.5b | 7.2 ± 0.9 |
| Spring | 5 | 307 ± 1d | 160 ± 10b | 0.80 ± 0.02b | 36.5 ± 0.5a |
Values are expressed as means ± SE. For aroused hibernators, days in last torpor bout indicates length of last bout prior to arousal; for torpid squirrels, number of days in continuous torpor.
Age of wild-caught mothers was unknown and assumed to be ≥420 days; therefore, Mother ages were not used in statistical analysis.
Letters that differ within columns show significant differences (P < 0.05).
Sample collection.
Squirrels were euthanized by isoflurane followed by decapitation, except for torpid hibernators, which did not receive isoflurane. Tb was measured immediately by insertion of a thermal probe into the body cavity, and intact ceca were rapidly removed. Cecal contents were harvested and weighed, and one portion frozen immediately in liquid nitrogen followed by storage at −80°C until DNA extraction. The remainder was sonicated and centrifuged (10,000 rpm, 10 min), and one aliquot was stored at −80°C for later analysis of ammonia concentration. The remaining supernatant was acidified with 36N sulfuric acid (2%/volume) and stored −80°C for later analysis of short-chain fatty acids (SCFA). The empty cecal tissue was weighed to determine cecal wet mass.
SCFA and ammonia analyses.
After thawing, an aliquot of the acidified cecal contents was analyzed by gas chromatography for total SCFA concentrations and molar proportions of propionate, acetate and butyrate. A separate aliquot was used to measure ammonia concentrations using a kit (no. AA0100 Sigma-Aldrich).
DNA extraction.
DNA was extracted from thawed cecal contents using the MoBio Powersoil DNA isolation kit with modifications. For each sample, the 16S rRNA gene was amplified (in triplicate) using barcoded V4 primers 515f (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806r (5′-GGACTACHVGGGTWTCTAAT-3′), found to be well suited to the phylogenetic analysis of pyrosequencing reads. Composite samples for pyrosequencing were prepared by pooling the triplicates of each PCR reaction back into one PCR product, which was cleaned, quantified, and then pooled into a composite sample by adding equal amounts of amplicon/DNA for each sample. The replicate PCRs were combined and cleaned with the MoBio UltraClean-htp kit. Samples were quantified using PicoGreen dsDNA reagent. Once quantified, the appropriate volumes of the cleaned PCR amplicons were combined. The composite pool was measured for final concentration, 260/280 ratio, and sent for sequencing. Controls were included in all steps to check for primer or sample DNA contamination. Samples were analyzed by the Environmental Genomics Core Facility at the University of South Carolina-Columbia for pyrosequencing on a 454 Life Sciences Genome Sequencer FLX-Titanium (Roche) machine.
Data processing.
Processing of pyrosequencing reads was performed using the QIIME 1.4.0 software package (10). A total of 82,732 sequences were demultiplexed and passed the default quality filters in QIIME (minimum 497 sequences, maximum 3,772 sequences, and mean 1,798.52 sequences per sample). Demultiplexed sequences were clustered de novo into operational taxonomic units (OTUs) using 97% identity, with chimeric and singleton OTUs filtered using the USEARCH software package (23). Representative sequences were assigned taxonomy via the RDP classifier (67) retrained on the Greengenes reference sequence set. A de novo alignment of these representative sequences was built using PyNast (9), filtered with the Greengenes lanemask, and from this filtered alignment, a phylogenetic tree was constructed with FastTree (56). Alpha diversity was calculated by first filtering the OTU table to remove any samples with less than 1,000 sequences/sample, followed by subsampling the OTU table for 50 repetitions at steps of 50 sequences/sample up to 1,000 sequences/sample. Following this, phylogenetic diversity [PD, defined as the minimum total length of all the phylogenetic branches required to span a given set of taxa on a phylogenetic tree (24)], Chao1, and observed species were calculated to determine alpha diversity for squirrel seasonal groups. Statistical differences among groups in PD and numbers of observed species at 1,000 sequences per sample were determined by Monte Carlo permutations to handle nonparametric data distributions (n = 999, implemented in QIIME GitHub commit 28c0020e05e4d29d9446eb3837e500e6328386bb). Beta diversity was calculated using unweighted and weighted UniFrac (41) on the evenly sampled OTU table at 436 sequences per sample. Taxonomies were grouped at the phylum, class, order, family, and genus levels. We compared sequence counts for each taxonomic level and season from samples rarefied to 436 sequences, using the Kruskal-Wallis rank sum test in R (version 2.12.0). False discovery rate (FDR) corrections were then applied.
Differences in SCFA and ammonia levels were analyzed by Kruskal-Wallis rank sum test or with t-test when only two groups were compared. Correlations between SCFAs and microbial taxa were analyzed by Pearson's correlation analysis and corrected for FDR.
RESULTS
Cluster analysis of microbiotas.
Comparison of individual squirrel microbiotas using principal coordinate analysis (PCoA) of the unweighted UniFrac metric showed a distinct clustering by season, with most of the variation explained by the first three coordinates (Fig. 1). Microbiotas from active season groups (Mothers, Summer, and Spring groups) clustered together and were distinct from those in Late Winter, which were tightly clustered. Clustering of the Early Winter group was the most diffuse, and as a group, these microbiotas were intermediate between those from the active season groups and the Late Winter group. The thermal and metabolic state of hibernating squirrels at the time of sampling had little effect on microbiota clustering in either Winter group, as microbiotas from torpid squirrels (Fig. 1, circled points) were evenly distributed among those from aroused hibernators. Because of this, in subsequent microbiota analyses, torpid and aroused hibernators were combined within the respective Early or Late Winter groups. Age of the animals had no effect on microbiota clustering; for example, Spring microbiotas clustered more closely with those from Mother and Summer squirrels than with the Late Winter group, despite Spring and Late Winter squirrels being closest in age (Fig. 1, Table 1). Squirrel body mass was also not a discriminating factor in microbiota clustering, as masses of Early Winter hibernators were not different from those of Spring or Summer pups (Table 1) yet their microbiotas were quite distinct. Clustering of weighted Unifrac results showed a similar pattern, although the separations based on PCoA1 vs. PCoA3 were not as robust (data not shown).
Phylogenetic diversity and observed species.
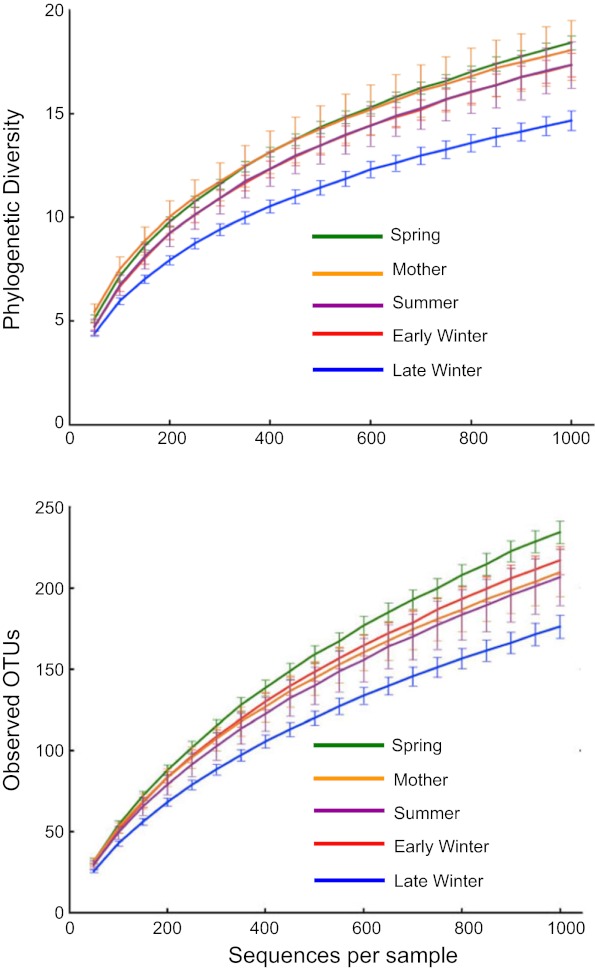
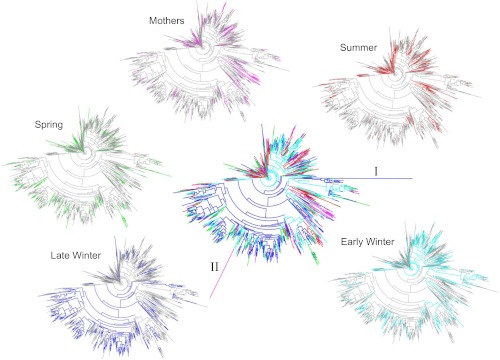
Phylogenetic diversity and numbers of observed species (OTUs) were lowest in microbiotas from Late Winter hibernators and highest in Spring squirrels (Fig. 2). For PD, all groups differed significantly from each other except Early Winter and Spring, and for observed species counts, only Mother and Summer groups were not different (Fig. 2). To better visualize microbiota diversity, we constructed phylogenetic trees using the Topiary Explorer software program (54). Inspection of trees constructed from each seasonal group (Fig. 3, peripheral trees) illustrates the shift in diversity within specific taxa (see below) in Late Winter microbiotas relative to the other seasonal groups and to the entire ground squirrel microbiota data set (Fig. 3, center tree). Although the phylogenetic trees only show presence or absence of particular OTUs and all seasonal groups have some representation across the tree, the trees suggest there is increased (or decreased) diversity within certain clades as environmental selection changes through the hibernation states.
Fig. 2.
Alpha diversity rarefaction plots of squirrel cecal microbiotas. Top: phylogenetic diversity (PD). Bottom: number of observed OTUs based on 97% sequence identity. Points are means ± SE, with numbers of squirrels in each group shown in Table 1. Differences among groups were analyzed at 1,000 sequences/sample. For PD, all groups differed significantly from one another (P < 0.05) except for early winter and summer. For observed OTUs, all groups differed except for the mother and summer groups.
Fig. 3.
Phylogenetic trees of squirrel cecal microbiotas colored by seasonal taxa. Center plot includes all taxa with coloration as follows: Mothers, pink; Summer, red; Early Winter, light blue; Late Winter, dark blue; Spring, green. Branch labeled “I” represents the archeal genus Methanosphera; “II” represents a poorly defined taxa. Perimeter plots show trees for individual seasonal groups. Trees were generated and colored with Topiary Explorer (54).
Taxonomic composition of squirrel microbiotas.
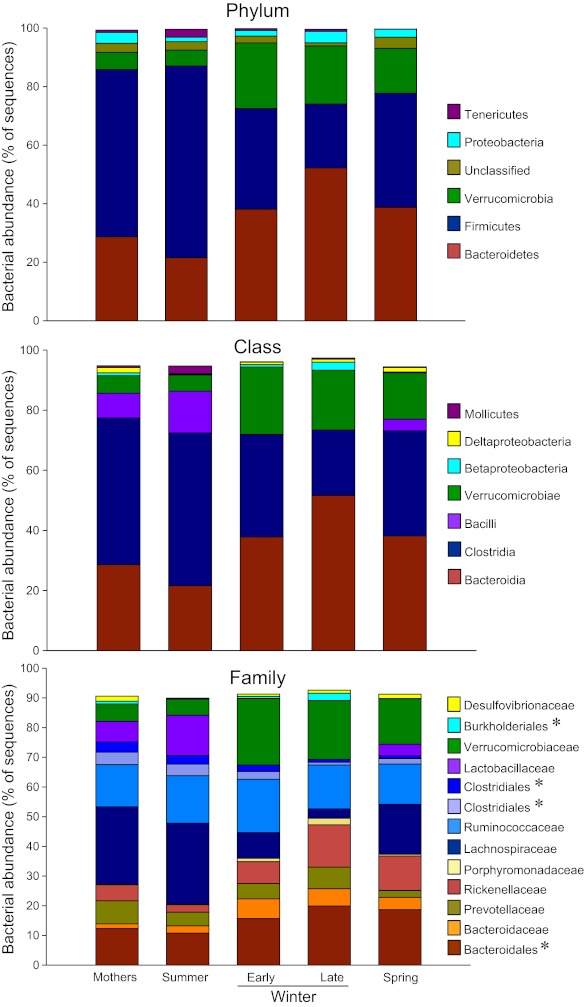
Eight bacterial phyla were identified in the ground squirrel microbiota (Supplemental Table S11), with the majority of sequences classified as Bacteroidetes (22–52%), Firmicutes (22–66%), or Verrucomicrobia (5–22%) (Supplemental Table S2). Other less abundant (1–5%) phyla represented were Proteobacteria, Tenericutes, and Actinobacteria, along with some very low abundance groups (<1%) that included Elusimicrobia and Cyanobacteria (Supplemental Table S1). An unclassified group of sequences (1–4%) accounted for the remainder of the bacterial diversity in the squirrel microbiota. The Archaean phylum Euryarchaeota was represented only in a few of the Mothers' microbiotas at <0.005%.
The majority of Bacteroidetes sequences within the squirrel microbiota matched to the order Bacteroidales and were represented primarily by the families Bacteroidaceae, Porphyromonadaceae, Prevotellaceae, and Rikenellaceae. At the genus level, dominant groups were Bacteroides, Prevotella, and Alistipes (Supplemental Table S1). Firmicutes were dominated by Clostridiales, with most OTUs matching to the families Lachnospiracea and Ruminoccocaceae. Dominant genera included Clostridium, Ruminococcus, Oscillospira, and Coprococcus. The Lactobacillales were the next most dominant Firmicutes group, represented by Lactobacillaceae (genus Lactobacillus) (Supplemental Table S1).
Season strongly affected taxonomic representation of the dominant phyla. Relative to the two summer groups (Summer pups and Mothers), hibernator microbiotas had lower representation of Firmicutes and higher representation of Bacteroidetes (Fig. 4, Supplemental Table S2). These differences were most evident between Late Winter hibernators and Summer squirrels. Spring microbiotas tended to be intermediate between Summer and Late Winter, likely reflecting the early repopulation of a “normal” microbiota once feeding resumed. The same general pattern held for seasonal changes in relative abundance of Verrucomicrobia (Supplemental Table S2). Sequences within this phylum matched to a single species, Akkermansia muciniphila (Supplemental Table S1).
Fig. 4.
Relative abundance of major taxa in squirrel cecal microbiotas. Numbers of squirrels in each seasonal group are shown in Table 1. Asterisks indicate unclassified families within a taxonomic group. Taxa with relative abundances less than 1.5% were not included.
Representation of the order Bacteroidales roughly doubled from Summer to Late Winter, including a six-fold rise in relative abundance of Alistipes (Rickenellaceae) (Fig. 4, Supplemental Table S2). In contrast, the abundance of several Firmicute taxa fell during hibernation, including a 9-fold reduction in Lachnospiraceae from Summer to Late Winter. The family Lactobacillaceae was severely affected by hibernation, with only 4 of the 14 Early Winter samples containing any sequences that matched to Lactobacillus, and no matches within the 13 Late Winter samples (Fig. 4, Supplemental Table S2). A preliminary report using quantitative PCR analysis also showed that hibernation depletes Lactobacilli in the squirrel microbiota (15). Relative abundance of Firmicutes taxa that were reduced during winter showed strong trends for partial or full reversal in Spring microbiotas. Interestingly, the only Firmicutes group that increased significantly during hibernation was one or more unclassified genera in the family Peptococcaceae, although relative abundance of these OTUs represented only 1–1.2% of all sequences.
Season also influenced relative abundance of some of the less common taxa (Supplemental Table S2). Notable among these was an unclassified genus in the Burkholderiales (class Betaproteobacteria) that represented <1% of OTUs in each of the seasonal groups except Late Winter, when it increased to 2.5%.
Short-chain fatty acid and ammonia levels in cecal contents.
We measured SCFA and ammonia concentrations in cecal contents to gain insight into how season affects the metabolic output of squirrel microbial communities. Total SCFA concentrations were highest in Summer squirrels and decreased by about 75% in aroused hibernators (whose Tbs were similar to Summer) (Table 2). Torpor further reduced total SCFA concentrations in Late Winter hibernators. Among the three SCFA, acetate was highest regardless of season, and all SCFA concentrations were lower in Late Winter (aroused) hibernators compared with Summer. Butyrate concentration was most affected by hibernation, falling to nearly undetectable levels in many hibernator samples and particularly those from torpid squirrels. Values from two Spring squirrels shown in Table 2 were not included in the statistical analysis due to low sample size; however, those data suggest that Spring SCFA concentrations remain closer to winter levels than to Summer, despite the 2-wk period of refeeding and constant homeothermy, which would stimulate rapid production of SCFA from dietary substrates. Hibernation also influenced the molar proportions of the individual SCFAs generated by the microbiota. Compared with Summer, the percent composition of acetate increased and butyrate decreased in aroused hibernators (Table 2), and both returned toward summer levels in spring. The proportion of propionate in cecal contents was similar in Summer squirrels and aroused hibernators. Interestingly, compared with aroused hibernators, proportions of acetate and propionate in torpid squirrels were higher and lower, respectively.
Table 2.
Short-chain fatty acid and ammonia concentrations and molar proportions of SCFA in ground squirrel cecal contents
| Acetate | Propionate | Butyrate | Total SCFA | Ammonia | |
|---|---|---|---|---|---|
| Concentration, mM | |||||
| Summer (6,6) | 49.01 ± 9.70a | 10.93 ± 2.71a | 17.92 ± 5.82a | 81.39 ± 18.63a | 7.70 ± 0.54a |
| Early Winter (6,6) | 13.55 ± 2.16b | 2.41 ± 0.40b | 0.94 ± 0.17b | 18.29 ± 2.86b | 3.50 ± 0.81b |
| Late Winter | |||||
| Aroused (6,6) | 15.64 ± 2.45b | 2.53 ± 0.66b | 0.51 ± 0.15b | 19.50 ± 3.31b | 2.54 ± 0.31b |
| Torpid (6,6) | 6.26 ± 0.93* | 0.41 ± 0.07* | 0.12 ± 0.02* | 7.06 ± 1.02* | 2.25 ± 0.18 |
| Spring (2,5) | 17.55 ± 2.12 | 4.64 ± 1.73 | 3.06 ± 0.92 | 27.72 ± 5.60 | 8.65 ± 1.05a |
| Molar Proportions, % | |||||
| Summer (6) | 63.43 ± 2.84a | 13.17 ± 0.50 | 18.71 ± 2.98a | ||
| Early Winter (6) | 74.17 ± 1.01b | 13.09 ± 0.61 | 5.01 ± 0.37b | ||
| Late Winter | |||||
| Aroused (6) | 81.30 ± 2.58b | 12.20 ± 1.72 | 2.52 ± 0.51b | ||
| Torpid (6) | 88.48 ± 1.38* | 5.85 ± 0.57* | 1.75 ± 0.27 | ||
| Spring (2) | 64.39 ± 5.36 | 16.13 ± 2.98 | 10.81 ± 1.13 |
Shown are means ± SE with sample sizes in parentheses [short-chain fatty acids (SCFA), ammonia]. For SCFA, groups analyzed with Kruskal-Wallis test were Summer squirrels and aroused hibernators in Early and Late Winter (no torpid Early Winter samples were available, and the two Spring squirrel samples were not included due to low sample size). For ammonia, groups analyzed were Spring and Summer squirrels and aroused hibernators in Early and Late Winter.
Significant differences in SCFA and ammonia levels between torpid and aroused late winter hibernators, analyzed by t-tests.
Different letters within columns show significant differences. P < 0.05 for all comparisons.
SCFA levels were correlated with relative abundances of some bacterial taxa (Table 3). Concentrations of all three SCFA were positively correlated with Firmicutes (specifically the Class Clostridia and Family Lachnospiraceae). Acetate and propionate concentrations were negatively correlated with Bacteroidetes and Verrucomicrobia, whereas butyrate concentration was negatively correlated only with Bacteroidetes. Concentrations of all SCFA were also positively correlated with relative abundance of the minor taxa Alphaproteobacteria and Mollicutes. The molar proportion of acetate relative to total SCFA concentration was negatively correlated with Firmicutes, and propionate was negatively correlated with Bacteroidia and Verrucomicrobiae (Table 3). Butyrate proportion was positively correlated with Firmicutes and negatively correlated with Bacteroidetes. Several other OTUs that were not classified to specific taxonomic groups were also significantly correlated with SCFA levels (data not shown).
Table 3.
Correlations between SCFA levels and microbial taxa in squirrel cecal contents
| Acetate |
Propionate |
Butyrate |
||||
|---|---|---|---|---|---|---|
| mM | % | mM | % | mM | % | |
| Bacteroidetes | − | − | − | − | ||
| Bacteroidia | − | − | − | − | ||
| Firmicutes | + | − | + | + | + | |
| Clostridia | + | − | + | + | + | |
| Lachnospiraceae | + | − | + | + | + | |
| Verrucomicrobia | − | − | ||||
| Verrucomicrobiae | − | − | ||||
| Verrucomicrobiaceae | − | |||||
| Proteobacteria | ||||||
| Alphaproteobacteria | + | + | + | + | ||
| Tenericutes | + | |||||
| Mollicutes | + | + | + | |||
Shown are significant (P < 0.05) positive (+) and negative (−) correlations between SCFA concentrations (mM) or molar proportions (%) and microbial taxa.
Ammonia concentrations were higher in Spring and Summer squirrels compared with Early and Late Winter aroused hibernators (Table 2). Torpor had no effect on ammonia concentrations in Late Winter squirrels.
DISCUSSION
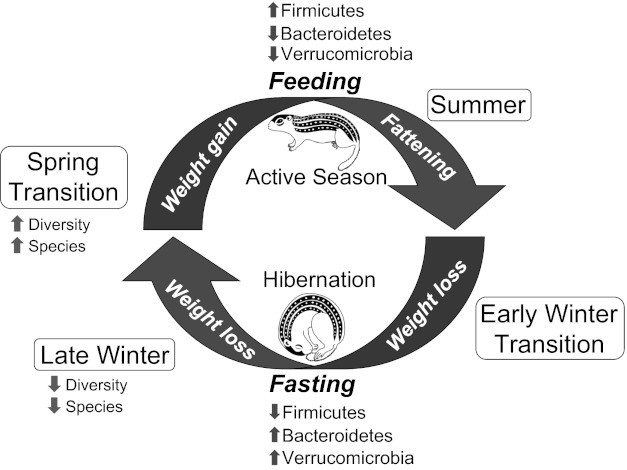
Like other fat-storing hibernators, the 13-lined ground squirrel accumulates large adipose stores during the active season, survives solely on endogenous nutrients (primarily lipids) for the 5–6 mo hibernation season, and then begins refeeding in spring in a relatively lean condition (13). Our goal here was to determine whether this annual cycle of extreme dietary change alters the structure of the gut microbiota. Our UniFrac analysis of 16S rRNA gene sequences confirmed that season plays a dominant role in the structure of ground squirrel microbial communities, as indicated by the close clustering of microbiotas of Spring and Summer pups with those of their Mothers, and the separate clustering of microbiotas in the Winter groups. Moreover, within the hibernation season, the microbiotas of Late Winter squirrels were more tightly clustered than were Early Winter animals, indicating that the Early Winter microbiota is a transitional state that precedes the more severe contraction of the community after 4–5 mo of continuous fasting. This is further illustrated by the rarefaction curves, which show that Late Winter microbiotas had the lowest phylogenetic diversity and numbers of observed species (OTUs) among all seasons. In contrast, microbiotas of Spring squirrels had the highest diversity and numbers of observed species. Thus, emergence from hibernation appears to induce blooms of certain microbial taxa that are adept at rapid expansion into new niches created by the constant supply of dietary substrates in an environment of sustained, high temperature. The result is a transient, diverse community that probably contains members with a range of metabolic capabilities, including those that are able to thrive on endogenous substrates alone, those that prefer (or require) substrates present only in the diet, and those that are able to degrade both substrate types. As time after refeeding progresses, the microbial community becomes more stable and less diverse (as indicated by the lower diversity and number of OTUs of Mother microbiotas compared with Spring), and it is likely dominated by species that are competitively superior in the presence of abundant and continuously available dietary substrates. These cyclical changes in structure of the ground squirrel microbiota are summarized in Fig. 5.
Fig. 5.
Schematic illustrating major changes in the ground squirrel gut microbiota over the annual hibernation cycle.
The UniFrac analysis also demonstrated that maternal influences, early dietary history, and host age had minimal effects on squirrel microbiotas compared with the dominant effect of season. The microbiotas of littermates (who ate primarily laboratory chow) and their mothers (who ate laboratory chow only after capture) were similar if they were sampled in the same (active) season, and the two seasonal groups that were closest in age—Late Winter and Spring—had microbiotas that were the most diverse.
In addition to substrate availability, other aspects of the gut environment during hibernation could influence microbial physiology, and thus microbiota structure. A major one is host Tb, which oscillates from ∼36°C during interbout arousals to a few degrees above 0°C, as animals cycle into and out of torpor (13). Gut microbes may differ in their abilities to metabolize substrates and proliferate at the low Tbs of torpor (48), which could provide competitive advantages for some groups over others. The principal coordinates analysis revealed no effect of host Tb on microbiota structure, because samples from torpid and aroused hibernators were interspersed within the Early and Late Winter groups. That said, we may not have been able to detect subtle, but significant, changes in microbiota structure that reflect dominance of certain species that function better than others at low temperatures.
Similar to other mammals, the majority of sequences in the ground squirrel microbiota was assigned to the bacterial phyla Bacteroidetes and Firmicutes, with Verrucomicrobia comprising the next most abundant group. At the phylum level, the dominant changes from active to hibernation seasons were increased representation of Bacteroidetes and Verrucomicrobia and a reduction in Firmicutes (Fig. 5). Thus, on the basis of known substrate preferences of gut microbes (26), the structure of the squirrel microbiota appears to be reorganized in winter in favor of taxa that are either specialists on host-derived substrates (Verrucomicrobia) (21) or generalists that have the ability to switch their complement of carbohydrate-degrading enzymes depending on the availability of dietary vs. host-derived substrates (Bacteroidetes) (45, 57, 59). Conversely, the hibernator microbiota is generally depleted in species that are known to prefer dietary polysaccharides, which include many Firmicutes (26).
The majority of host-derived substrates in the mammalian gut are gel-forming mucins, which are complex glycoproteins produced in large quantities by intestinal goblet cells and, when hydrated, form the protective mucus layer that overlies the epithelium (47). Only a few gut microbes have the ability to completely degrade mucins (20, 33, 45, 55, 57); one is Akkermansia muciniphila, the sole representative of the Verrucomicrobia present in the mammalian gut (21), whose relative abundance increased in hibernator microbiotas. A. muciniphila associates with the mucus layer and is able to grow on mucin as its sole carbon and nitrogen source (20, 21, 64).
Competition for the limited resources in the hibernator gut is likely an important factor that shapes the microbiota, as species less able to utilize endogenous substrates or metabolites produced by primary mucin-degraders will gradually be reduced in number until dietary substrates are available in the spring. For example, relative abundance of Lactobacillus, which prefers simple sugars, falls to nondetectable levels once hibernation begins and does not increase until after refeeding in spring. The Lachnospiraceae, which are typically very abundant in the mammalian gut microbiota is also negatively affected by hibernation. Although many members of this Firmicutes family prefer dietary substrates (26), some can utilize endogenous sugars such as fucose (58), which may explain the continued presence of a small subset of Lachnospiracea in the hibernator gut.
Winter fasting reduced cecal SCFA and ammonia concentrations, which is not surprising given the absence of dietary intake. Interestingly, the relative proportions of individual SCFA changed from active to hibernation seasons, with acetate rising and butyrate falling to very low levels. These shifts were likely due to the reduction in certain types of metabolizable substrates coupled with the altered microbiota composition. The absence of complex plant polysaccharides during hibernation reduces abundance of certain taxa (e.g., many Firmicutes) that specialize on these substrates. This reduces production of butyrate, because most of the major butyrate producers, including Roseburia, Eubacterium, and Faecalibacterium are Firmicutes (22). The negative correlations between butyrate levels and Bacteroidetes abundance, as well as the positive correlations between butyrate and Firmicutes, support this relationship. The lower cecal butyrate concentration during winter may contribute to the changes in intestinal structure and function observed in hibernating squirrels. As the preferred fuel source for epithelial cells, butyrate stimulates epithelial proliferation and restitution, and helps maintain integrity of the intestinal barrier through modulation of apoptosis, permeability, and mucus production (8, 29, 40, 51). Hibernation leads to substantial atrophy of the small intestinal mucosa in ground squirrels (11, 12, 15), and as reported in this study, cecal mass is also reduced. Hibernation also alters expression of several apoptosis-related proteins in the intestine (25), and it increases gut permeability (11, 12, 15).
In contrast to butyrate, the molar proportion of acetate rose during hibernation, which may be due, in part, to the activity of mucolytic bacteria, such as A. muciniphila, which convert mucins to acetate (21). Microbially derived acetate could influence hibernation physiology in several ways. After absorption, it can be metabolized in peripheral tissues to meet energy demands, or it can be converted to ketone bodies in gut epithelial cells or hepatocytes. Ketones play a particularly important role in hibernation, serving as an alternative fuel to glucose, particularly in brain and heart during torpor (1). Although the majority of circulating ketones derive from de novo synthesis in the liver, it is possible that microbially derived acetate contributes to hepatic ketogenesis and thus cellular energetics during the winter fast, as has been shown in mice (18). Microbially derived propionate may also play a role in hibernation, because in addition to serving as a fuel source, propionate is a substrate for gluconeogenesis, another metabolic function that is critical during hibernation to maintain blood glucose levels (28). Although the contribution of microbial SCFA to energy and nutrient balance in ground squirrels is unknown, estimates in other rodents indicate that gut-derived SCFA provide 5–20% of an animal's overall energy budget (7). However, hindgut epithelial cells derive as much as 60–70% of their energy needs from luminal SCFA (2), which raises the possibility that through their production of SCFA from endogenous substrates, the hibernator microbiota may “recycle” some of the energy that is expended during interbout arousal periods in epithelial cell proliferation (14), mucus production, and maintenance of the mucosal immune system (35).
This is the first report of seasonal changes in the microbiota of a fasting hibernator based on deep sequencing of microbial 16S rRNA genes, and it expands on previous work that used only culture-based techniques (5, 6, 16) or quantitative PCR (15). In a culture-independent study in Syrian hamsters, a hibernating species that eats during periodic arousals, a 4-day fast in nonhibernators had a greater effect on species composition than did hibernation, relative to fed (nonhibernating) animals (60). This underscores the crucial role of diet in the regulation of gut microbial communities, because periodic arousals provide ample time at high Tb for microbes to degrade and utilize newly ingested dietary substrates. In fact, several changes we noted in the microbiotas of hibernating 13-lined ground squirrels were comparable to those in fasted, but not hibernating hamsters; for example, the fasted hamster microbiota had a greater proportion of sequences represented by A. muciniphila, and reduced representation of Firmicutes, including Lachnospiraceae (60). Our results also resemble the effects of natural fasting between meals in the Burmese python microbiota. After a more than 30-day fast, phylogenetic diversity of the python colonic microbiota is relatively low and dominated by Bacteroidetes, with reduced representation by Firmicutes (17). Feeding causes a rapid increase in phylogenetic diversity and numbers of observed OTUs, with proportional representations of Bacteroidetes and Akkermansia falling and Firmicutes rising, similar to our observations in Spring squirrels relative to Late Winter. Even in laboratory mice, fasting for as little as 24 h increases and decreases proportional representation of Bacteroidetes and Firmicutes, respectively (18).
Hibernation is associated with a dramatic remodeling of the intestinal immune system, with increased numbers of intraepithelial and lamina propria lymphocytes, increased IgA expression and elevated mucosal cytokine levels (35). It is likely that seasonal reorganization of the microbiota is a major driver of these immune alterations, because the immune system is the primary sensor of gut microbes and their metabolites (30, 63). Immune changes are often associated with altered host-microbe signaling (30, 36, 53, 61) and may function to maintain a mutually beneficial, tolerant state (19, 46). The elevation in intestinal secretory IgA (sIgA) expression during hibernation (35) provides particularly strong evidence for a seasonal shift in host-gut microbe signaling. Gut microbiota can stimulate sIgA production (61, 62), and there is growing evidence that in addition to preventing bacterial adherence to epithelial cells, sIgA plays a key regulatory role in maintaining the complex, mutualistic interactions between the microbiota, the epithelium, and the immune system (53, 61). Further, hibernation is accompanied by increased levels of mucosal IL-10, a potent anti-inflammatory cytokine that promotes immune tolerance of the microbiota (35, 46). Epithelial defenses are also enhanced in the hibernator gut, including increased expression of antiapoptotic proteins (25), and increased tight junction localization of occludin, which reduces intestinal barrier dysfunction induced by inflammatory cytokines (15, 44). Because gut permeability increases during hibernation in ground squirrels (11, 12, 15), the benefit of heightened immune and epithelial defenses may reside in minimizing translocation of microbial products across the epithelium and in limiting inflammation if microbial translocation occurs, thus preserving intestinal homeostasis (46).
Perspectives and Significance
The extreme dietary change that is part of the annual hibernation cycle can be viewed as a normal, recurring perturbation (42) that is well tolerated by both the ground squirrel host and its microbial symbionts. Our study revealed how the gut microbial community responds to this perturbation in terms of taxonomic structure but did not provide insight into functional changes in the microbiome (i.e., the microbiota and their complement of genes and gene products). Metagenomic analyses, coupled with mRNA, protein, and metabolite profiling will be required to illuminate how metabolic pathways of the community as a whole, and within specific bacterial species, are modified by winter fasting and the return of dietary substrates in spring (42).
Given the wide variety of effects that gut microbes can exert on mammalian hosts (31, 50), it is likely that the coevolution of hibernating mammals with their microbiotas has shaped not only gut physiology and immune function, but multiple aspects of the hibernation phenotype. For example, metabolic activity of gut microbes may contribute to the crucial process of prehibernation fattening, or influence energetic balance and torpor-arousal patterns during the hibernation season. Identification of which hibernation features carry a microbial imprint will ultimately require manipulation of the microbiota and analysis of host responses. Although gnotobiotic approaches are currently not available for hibernating species, carefully designed studies that use antibiotics to deplete the microbiota or prebiotics to stimulate them may be feasible. Incorporating the potential roles of the microbiota provides a new perspective on the physiology of mammalian hibernation and will also expand our understanding of the intimate relationships that have evolved between animals and their gut symbionts.
GRANTS
This work was supported by funds from the University of Wisconsin School of Veterinary Medicine (to H. V. Carey), National Institutes of Health (NIH), and Howard Hughes Medical Institute (to R. Knight) and NIH T32 GM008759 (to W. A. Walters).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: H.V.C. conception and design of research; H.V.C. performed experiments; H.V.C., W.A.W., and R.K. analyzed data; H.V.C., W.A.W., and R.K. interpreted results of experiments; H.V.C. and W.A.W. prepared figures; H.V.C. and W.A.W. drafted manuscript; H.V.C., W.A.W., and R.K. edited and revised manuscript; H.V.C., W.A.W., and R.K. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Glen Broderick and Wendy Radloff for SCFA analyses and Michael Grahn, Jessica Otis, and Amanda Pike for collection and care of animals.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Andrews MT, Russeth KP, Drewes LR, Henry PG. Adaptive mechanisms regulate preferred utilization of ketones in the heart and brain of a hibernating mammal during arousal from torpor. Am J Physiol Regul Integr Comp Physiol 296: R383–R393, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ardawi MS, Newsholme EA. Fuel utilization in colonocytes of the rat. Biochem J 231: 713–719, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718–15723, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science 307: 1915–1920, 2005 [DOI] [PubMed] [Google Scholar]
- 5. Barnes EM. Effect of hibernation on the intestinal flora. Am J Clin Nutr 23: 1519–1524, 1970 [DOI] [PubMed] [Google Scholar]
- 6. Barnes EM, Burton GC. The effect of hibernation on the caecal flora of the thirteen-lined ground squirrel (Citellus tridecemlineatus). J Appl Bacteriol 33: 505–514, 1970 [DOI] [PubMed] [Google Scholar]
- 7. Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev 70: 567–590, 1990 [DOI] [PubMed] [Google Scholar]
- 8. Bugaut M, Bentejac M. Biological effects of short-chain fatty acids in nonruminant mammals. Annu Rev Nutr 13: 217–241, 1993 [DOI] [PubMed] [Google Scholar]
- 9. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carey HV. Effects of fasting and hibernation on ion secretion in ground squirrel intestine. Am J Physiol Regul Integr Comp Physiol 263: R1202–R1208, 1992 [DOI] [PubMed] [Google Scholar]
- 12. Carey HV. Seasonal changes in mucosal structure and function in ground squirrel intestine. Am J Physiol Regul Integr Comp Physiol 259: R385–R392, 1990 [DOI] [PubMed] [Google Scholar]
- 13. Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev 83: 1153–1181, 2003 [DOI] [PubMed] [Google Scholar]
- 14. Carey HV, Martin SL. Preservation of intestinal gene expression during hibernation. Am J Physiol Gastrointest Liver Physiol 271: G804–G813, 1996 [DOI] [PubMed] [Google Scholar]
- 15. Carey HV, Pike AC, Weber CR, Turner JL, Visser A, Beijer-Liefers SC, Bouma HR, Kroese FGM. Impact of hibernation on gut microbiota and intestinal barrier function in ground squirrels. In: Living in a Seasonal World: Thermoregulatory and Metabolic Adaptations, edited by Ruf T, Bieber C, Arnold A, Millesi E. Heidelberg, Germany: Springer, 2012 [Google Scholar]
- 16. Cloud-Hansen KA, Villiard KM, Handelsman J, Carey HV. Thirteen-lined ground squirrels (Spermophilus tridecemlineatus) harbor multi-antibiotic resistant bacteria. J Am Assoc Lab Animal Sci 46: 17–20, 2007 [PubMed] [Google Scholar]
- 17. Costello EK, Gordon JI, Secor SM, Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J 4: 1375–1385, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA 106: 11276–11281, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Derrien M, Van Baarlen P, Hooiveld G, Norin E, Muller M, de Vos WM. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol 2: 166, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1: 254–268, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov, sp nov, a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54: 1469–1476, 2004 [DOI] [PubMed] [Google Scholar]
- 22. Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73: 1073–1078, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461, 2010 [DOI] [PubMed] [Google Scholar]
- 24. Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online 2: 121–128, 2006 [PMC free article] [PubMed] [Google Scholar]
- 25. Fleck CC, Carey HV. Modulation of apoptotic pathways in intestinal mucosa during hibernation. Am J Physiol Regul Integr Comp Physiol 289: R586–R595, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3: 1–18, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Florant GL, Healy JE. The regulation of food intake in mammalian hibernators: a review. J Comp Physiol B 182: 451–467, 2012 [DOI] [PubMed] [Google Scholar]
- 28. Galster W, Morrison PR. Gluconeogenesis in arctic ground squirrels between periods of hibernation. Am J Physiol 228: 325–330, 1975 [DOI] [PubMed] [Google Scholar]
- 29. Gaudier E, Jarry A, Blottiere HM, de Coppet P, Buisine MP, Aubert JP, Laboisse C, Cherbut C, Hoebler C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am J Physiol Gastrointest Liver Physiol 287: G1168–G1174, 2004 [DOI] [PubMed] [Google Scholar]
- 30. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 336: 1268–1273, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Ann Rev Nutr 22: 283–307, 2002 [DOI] [PubMed] [Google Scholar]
- 32. Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291: 881–884, 2001 [DOI] [PubMed] [Google Scholar]
- 33. Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M, Niedermeyer G. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest 75: 944–953, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10: 323–335, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kurtz CC, Carey HV. Seasonal changes in the intestinal immune system of hibernating ground squirrels. Dev Comp Immunol 31: 415–428, 2007 [DOI] [PubMed] [Google Scholar]
- 36. Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330: 1768–1773, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leser TD, Lindecrona RH, Jensen TK, Jensen BB, Moller K. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl Environ Microbiol 66: 3290–3296, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science 320: 1647–1651, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6: 776–788, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294: 1–8, 2009 [DOI] [PubMed] [Google Scholar]
- 41. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 489: 220–230, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lyman CP, Willis JS, Malan A, Wang LCH. Hibernation and Torpor in Mammals and Birds. New York: Academic, 1982 [Google Scholar]
- 44. Marchiando AM, Shen L, Graham WV, Weber CR, Schwarz BT, Austin JR, 2nd, Raleigh DR, Guan Y, Watson AJ, Montrose MH, Turner JR. Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. J Cell Biol 189: 111–126, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4: 447–457, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489: 231–241, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9: 265–278, 2011 [DOI] [PubMed] [Google Scholar]
- 48. Morita RY. Psychrophilic bacteria. Bacteriol Rev 39: 144–167, 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science 336: 1262–1267, 2012 [DOI] [PubMed] [Google Scholar]
- 51. Peng L, He Z, Chen W, Holzman IR, Lin J. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr Res 61: 37–41, 2007 [DOI] [PubMed] [Google Scholar]
- 52. Pengelley ET, Fisher KC. Rhythmical arousal from hibernation in the golden-mantled ground squirrel, Citellus lateralis tescorum. Can J Zool 39: 105–120, 1961 [Google Scholar]
- 53. Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2: 328–339, 2007 [DOI] [PubMed] [Google Scholar]
- 54. Pirrung M, Kennedy R, Caporaso JG, Stombaugh J, Wendel D, Knight R. Topiary Explorer: visualizing large phylogenetic trees with environmental metadata. Bioinformatics 27: 3067–3069, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105: 2420–2428, 2010 [DOI] [PubMed] [Google Scholar]
- 56. Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Salyers AA, Vercellotti JR, West SE, Wilkins TD. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol 33: 319–322, 1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. J Bacteriol 188: 4340–4349, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307: 1955–1959, 2005 [DOI] [PubMed] [Google Scholar]
- 60. Sonoyama K, Fujiwara R, Takemura N, Ogasawara T, Watanabe J, Ito H, Morita T. Response of gut microbiota to fasting and hibernation in Syrian hamsters. Appl Environ Microbiol 75: 6451–6456, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sutherland DB, Fagarasan S. Iga synthesis: a form of functional immune adaptation extending beyond gut. Curr Opin Immunol 24: 1–8, 2012 [DOI] [PubMed] [Google Scholar]
- 62. Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun 67: 1992–2000, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Van den Abbeele P, Van de Wiele T, Verstraete W, Possemiers S. The host selects mucosal and luminal associations of coevolved gut microorganisms: a novel concept. FEMS Microbiol Rev 35: 681–1385, 2011 [DOI] [PubMed] [Google Scholar]
- 64. van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6: e16876, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu PV, Yetukuri L, Islam S, Felin J, Perkins R, Boren J, Oresic M, Backhed F. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res 51: 1101–1112, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol 65: 411–429, 2011 [DOI] [PubMed] [Google Scholar]
- 67. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.