Abstract
Phosphoinositides are membrane-bound signaling phospholipids that function in a myriad of cellular processes, including membrane trafficking, cytoskeletal dynamics, ion channel and transporter function, and signal transduction. In order to better understand the role of phosphoinositides in cellular processes, different approaches to study the effects of the presence or absence of these lipids must be devised. Conventional approaches of manipulating phosphoinositide levels such as over-expression or genetic ablation of lipid enzymes cause prolonged exposure of the cells to changes in lipid levels that could result in compensatory actions by the cell or downstream alterations in cell physiology. In this chapter we present an approach used recently by various laboratories, including our own, to acutely manipulate phosphoinositide levels at target locations using chemically induced dimerization (CID) that can be spatially and temporally controlled. We discuss considerations when designing expression constructs for targeting specific cellular compartment membranes and present examples from the literature on different ways of perturbing phosphoinositide levels at particular organelle membranes using CID. In addition, we provide details on image acquisition, data collection and data interpretation. CID technology can be applied to many lipid enzymes to broaden the understanding of the role lipid signaling plays in cell physiology.
I. INTRODUCTION
Phosphoinositides, membrane dynamics and intracellular signaling
Phosphoinositides play prominent roles in the regulation of numerous cellular functions from membrane trafficking, cytoskeletal dynamics, ion channel and transporter function, to signal transduction (Balla et al., 2009; Di Paolo and De Camilli, 2006; Falkenburger et al., 2010; Hilgemann et al., 2001; Suh and Hille, 2005; Yin and Janmey, 2003). Phosphatidylinositol (PI) serves as the precursor of seven phosphoinositide species, which are differentially phosphorylated at the 3, 4 and/or 5 position of the inositol ring. While PI makes up less than 15% of phospholipids found in eukaryotic cells, its phosphorylated derivatives are found at even lower levels, with PI(4,5)P2 and PI4P as the most abundant species of the phosphoinositides (Di Paolo and De Camilli, 2006). Despite their low abundance, phosphoinositides play critical functions in many cellular processes, in part due to their high turnover and the eclectic nature of their phosphorylated head groups.
Phosphoinositides are typically found concentrated at the cytoplasmic face of cellular membranes with their inositol ring, or headgroup, exposed to the cytosolic milieu, available to interact with cytosolic proteins or membrane protein cytodomains. These molecules therefore play an important role in controlling the membrane-cytosol interface. Phosphoinositides can regulate the function of integral membrane proteins at the plasma membrane (PM), such as ion channels and ion transporters (Suh and Hille, 2008). They can also serve as scaffolds that bring together the cytoskeleton or coat proteins with the cytoplasmic membrane surface (Di Paolo and De Camilli, 2006; Haucke, 2005). For example, PI(4,5)P2 participates in the regulation of actin polymerization by binding N-WASP. This causes a conformational change that allows the recruitment and activation of the ARP2/3 complex resulting in the nucleation of actin filaments (Logan and Mandato, 2006; Mao and Yin, 2007; Pollard and Borisy, 2003; Rohatgi et al., 2000).
Conventional methods of manipulating PI levels
Conventional methods of manipulating phosphoinositide levels employed to investigate the role of these lipids in cells include exogenous application of lipids, genetic manipulation and use of pharmacological inhibitors. The most basic of these methods are the exogenous application of phosphoinositides using polyamine carriers which “shuttle” the lipids into the cell in order to reach intracellular membranes (Ozaki et al., 2000) and direct microinjection of lipid micelles into the cell (Golebiewska et al., 2008). A more sophisticated approach is the recently developed membrane-permeant “caged” phosphoinositides that allow release or exposure of the “caged” lipid upon photoactivation (Subramanian et al., 2010). Additionally, metabolically stabilized variants of phosphoinositides have been developed in order to dissect the effects of these lipids from those of their metabolites (Huang et al., 2007; Xu et al., 2006; Zhang et al., 2006a; Zhang et al., 2006b). Such methods result in the rapid entry of phosphoinositides into the cell but do not allow for the facile control of lipid concentrations or destination. A more commonly used approach is to use genetic manipulation to over-express inositide kinases or phosphatases (Kahlfeldt et al., 2010; Kim et al., 2006; Krauss et al., 2003), to silence specific genes through RNAi (Choudhury et al., 2005; Prasad and Decker, 2005; Wang et al., 2004), or to engineer organisms deficient in a gene of interest (for example, see references (Cremona et al., 1999; Di Cristofano et al., 1998; Di Paolo et al., 2004; Gary et al., 1998; Harris et al., 2000; Schu et al., 1993; Stambolic et al., 1998; Verstreken et al., 2003; Zhou et al., 2010)). Modulation of the target phosphoinositide concentration at the desired membrane compartment is achieved using these genetic techniques; however, the effects of prolonged exposure to these changes in lipid levels may result in downstream alterations in cellular processes that are difficult to experimentally resolve from the function directly controlled by the lipid. A more selective system of perturbing lipid levels would be the use of pharmacological inhibitors of lipid enzymes that allow for well-defined suppression of a specific enzyme(s). Inhibitors for the PI 3-kinase family have been well studied with the broadly acting small molecules wortmannin and LY249002 and the more recently developed isoform-specific agents (Knight et al., 2006). Wortmannin and LY294002 (used at higher concentrations) as well as phenylarsine oxide have been used to inhibit PI 4-kinases (Sorensen et al., 1998). However, well-defined, specific pharmacological drugs for other lipid kinases and phosphatases have not been discovered.
Chemical inducers of dimerization (CIDs)
The approach of chemically induced dimerization (CID) has been used in several recent phosphoinositide studies investigating various cellular processes, such as endocytosis, endosomal morphology and cargo sorting, ion channel function and cytoskeletal dynamics (see Table 1). The use of CID allows for the acute and localized regulation of phosphoinositide synthesis or degradation and circumvents issues that arise with standard genetic and molecular approaches, which typically cause prolonged perturbations in phosphoinositide levels. Schreiber, Crabtree and their colleagues first introduced the use of CID in a landmark study on T-cell receptor-mediated signaling pathways in 1993 (Spencer et al., 1993). In their approach, the immunosuppressant molecule FK506, which binds both FK-binding protein 12 (FKBP12) and calcineurin (Liu et al., 1991), was synthetically dimerized (and named FK1012) not only to inactivate its immunosuppressive properties (i.e. block its calcineurin-binding site) but in order to create the ability to bind two molecules of FKBP12 resulting in homodimerization of this protein. The ability of FK1012 to homodimerize the cytoplasmic domains of the T-cell receptor zeta chain fused to FKBP12 was shown to successfully induce downstream signaling cascades elicited normally by intact T-cell receptor dimerization.
Table 1.
Examples in the literature of CID use to manipulate phosphoinostide levels.
| Lipid Manipulated |
Enzymatic Tool | Targeted Membrane/Compartment; Targeting Sequence/Protein |
Dimerizer Used | Phenotypes | Reference |
|---|---|---|---|---|---|
| PI(4,5)P2 | type IV 5-phosphatase domain | PM; palmitoylation sequence of GAP43 | rapamycin, AP21967 | termination of ATP-induced Ca2+ signal, inactivation of TRPM8 channels, blockade of TfnR and EGFR endocytosis;, | (Varnai et al., 2006) |
| PI(4,5)P2 | Inp54p phosphatase | PM; Lyn11 | iRAP | current of KCNQ ion channels fall to 0 | (Suh et al., 2006) |
| PI(4,5)P2 | Inp54p phosphatase | PM; Lyn11 | iRAP | with simultaneous blockade of PIP3 production, proteins with polybasic clusters translocated from PM | (Heo et al., 2006) |
| PI3P | myotubularin 1 | endosomes; Rab5a | AP21967 | TfnR accumulation in Rab5a-positive endosomes, endosome positive tubularization | (Fili et al., 2006) |
| PI(4,5)P2 | type IV 5-phosphatase domain | PM; palmitoylation sequence of GAP43 | iRAP | loss of endocytic CCPs, dissociation of endocytic adaptors, dynamin and Arp2/3 complex from PM | (Zoncu et al., 2007) |
| PI(4,5)P2 | Inp54p phosphatase domain | PM; Lyn11 | iRAP | decreased TfnR endocytosis, increased TfnR concentration on PM, dissociation of AP-2 from PM | (Abe et al., 2008) |
| PI(4,5)P2 | p110 catalytic subunit of type I PI 3-kinase | PM; palmitoylation sequence of GAP43 | iRAP | PIP3 production, membrane ruffles, accelerated CCP maturation and turnover | (Nakatsu et al., 2010) |
| PI(4,5)P2 | 5-phosphatase domain of Synj1 | PM; endophilin N-BAR domain | rapamycin, AP21967 | membrane tubule fission | (Chang-Ileto et al., 2011) |
CCP: clathrin-coated pit; EGFR: epidermal growth factor receptor; PM: plasma membrane; Synj1: synaptojanin 1; Tfn: transferrin; TfnR: transferrin receptor; TRPM8: methanol-activated transient receptor potential melastatin 8.
A common variant to this original approach is the use of another immunosuppressive agent, rapamycin. Like FK506, this molecule also binds FKBP12 but instead of calcineurin, its second binding target is the FKBP-rapamycin binding (FRB) domain of mammalian target of rapamycin (mTOR) (Chen et al., 1995). Rapamycin (or its chemical analogs) in CID can be used to induce heterodimerization of two distinct proteins/domains, engineered as a fusion partner with either FKBP12 or FRB.
CID can be used to induce complexes of the same protein (homodimerization) as well as complexes of two distinct proteins (heterodimerization) depending on the dimerizer compound (such as FK1012 and rapamycin) and the protein fusions used (FKBP12 alone or FKBP12 in conjunction with FRB). Variants of dimerizers and of the protein modules FRB and FKBP have been developed to overcome undesirable interactions with endogenous molecules as well as to create the ability to do orthogonal studies in which control of multiple enzymes is desired (Bayle et al., 2006; Belshaw et al., 1996; Choi et al., 1996; Chong et al., 2002; Clackson et al., 1998; Inoue et al., 2005; Liberles et al., 1997). Native interactions between proteins as well as novel interactions and functions can be studied using such reagents. A reverse dimerization strategy uses a self-dimerizing FKBP mutant so that the initial condition of fusion protein partners is an aggregated complex that can be disassembled upon addition of a synthetic monomeric FKBP ligand (Rollins et al., 2000). Localization and mislocalization of protein targets can be achieved using appropriate or inappropriate targeting signals or protein pairs. Protein stability can also be controlled using CID (Banaszynski et al., 2006; Stankunas et al., 2003; Stankunas et al., 2007).
CID and phosphoinositides
Many of the phosphoinositide studies employing CID have focused on the consequences of degradation of the PI(4,5)P2 population at the PM (see Table 1). In these studies, a PI(4,5)P2 5-phosphatase domain fused to FKBP is recruited to the PM via PM-anchored peptides or proteins fused to FRB upon the addition of rapamycin or one of its analogs, iRAP (Inoue et al., 2005) or AP21967 (Chong et al., 2002), resulting in the rapid elimination of PI(4,5)P2. Using the palmitoylation sequence of GAP43 as the PM-anchor and the phosphatase domain of type IV 5-phosphatase, Varnai, et al. showed that upon dephosphorylation of PI(4,5)P2 at the PM multiple processes were affected including Ca2+ signaling, TRPM8 channel function and receptor internalization/endocytosis. Using this same combination of phosphatase and PM-anchor, Zoncu, et al. investigated the effect of PI(4,5)P2 loss on clathrin coat dynamics. PI(4,5)P2 depletion resulted in the disappearance of clathrin-coated pits (CCPs) at the cells surface along with various endocytic adaptors, such as AP-2 and epsin, as well as the dissociation of dynamin and Arp2/3 from the membrane indicating a role for PI(4,5)P2 not only in endocytosis, but also in actin nucleation and actin-mediated motility at the cell edge. The use of Lyn11 as the PM-anchor and Inp54p to degrade PI(4,5)P2 was used to demonstrate that KCNQ ion channel function is dependent on the presence of this phosphoinositide (Suh et al., 2006) and that transferring receptor (TfnR) endocytosis was more sensitive to changes in PI(4,5)P2 levels than TfnR recycling (Abe et al., 2008). In order to study the role of PI(4,5)P2 in targeting proteins with polybasic clusters to the PM, Heo, et al. used the Lyn11/Inp54p combination to selectively decrease PI(4,5)P2 levels and showed that this lipid in concert with PI(3,4,5)P3 was responsible for targeting of proteins with polybasic clusters to the PM. We used the CID approach to demonstrate that recruitment of the 5-phosphatase domain of synaptojanin 1 to endophilin-induced tubules at the cell surface and the resulting PI(4,5)P2 turnover at these structures resulted in membrane fission (Chang-Ileto et al., 2011).
CID has also been used to investigate the role of other phosphoinositides in the cell. In a study on the role of PI(3,4,5)P3 and the inositol 5-phosphatase SHIP2 on CCP dynamics, recruitment of the p110 catalytic subunit of type I PI 3-kinase to the PM was used to induce acute phosphorylation of PI(4,5)P2 and PI(3,4,5)P3 production that resulted in the acceleration of CCP assembly and turnover (Nakatsu et al., 2010). Since SHIP2 has been shown to be a negative regulator of PI(3,4,5)P3-dependent signaling, this result was in agreement with SHIP2’s role in dampening insulin signaling through its regulation of CCP dynamics via PI(3,4,5)P3 (and PI(4,5)P2) turnover. An examination of the role of PI3P on endosomal function and cargo traffic used Rab5 to localize the inositol lipid phosphatase myotubularin 1 to early endosomal membranes in order to decrease PI3P (and PI(3,5)P2) in this compartment (Fili et al., 2006). This study revealed that the phosphoinositide PI3P had an important influence on endosomal morphology and cargo passage through these Rab5-positive endosomes.
Targeting of CID protein modules
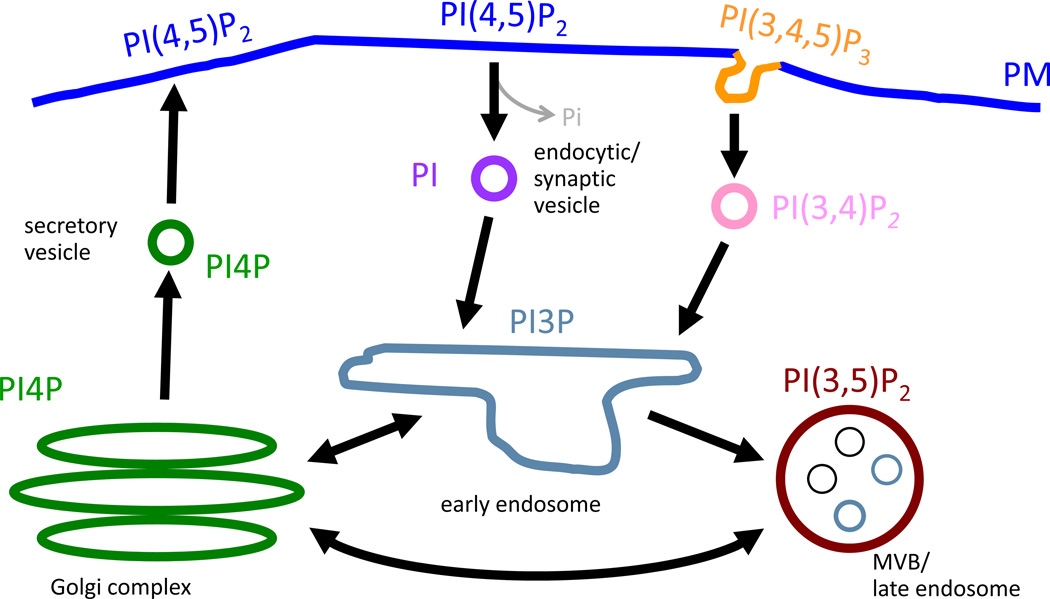
Because of their heterogeneous subcellular distribution, phosphoinositides can serve as markers for different organelles and control the targeting of proteins with phospholipid-binding domains to their destinations (Behnia and Munro, 2005; Di Paolo and De Camilli, 2006). A map of cellular membranes can be drawn according to the distribution of phosphoinositides to illustrate the role of these lipids in organelle identity (Figure 1). Several different approaches can be used to localize your protein(s) of interest to the desired cellular membrane/organelle (Varnai and Balla, 2007). Many proteins that interact with phosphoinositides possess specific phospholipid-binding domains that mediate the recruitment of these proteins to specific membrane domains in the cell (Lemmon, 2008). For example, since the pleckstrin homology domain (PH) of phospholipase Cδ1 (PLCδ1) recognizes PI(4,5)P2, a phosphoinositide concentrated at the PM, the PH domain targets PLCδ1 to the PM (Garcia et al., 1995; Lemmon et al., 1995). Meanwhile, the PH domain of the four-phosphate-adaptor protein 1 (FAPP1) is specific for PI4P thereby targeting this protein to the trans-Golgi network where PI(4)P is predominant (Godi et al., 2004). Another phospholipid-binding domain, FYVE (Fab1, YOTB, Vac1, EEA1) specifically recognizes PI3P and can be found in endosomal proteins such as Hrs (hepatocyte growth factor regulated tyrosine kinase substrate) (Gillooly et al., 2001) and early endosome antigen 1 (EEA1). Organelle targeting motifs for fusion to either FRB or FKBP were evaluated by the Inoue group for the Golgi using a peptide from the giantin protein; for the mitochondria using a peptide from either Tom20 or monoamine oxidase; for the ER using cytochrome b5; and for lysosomes using LAMP1 (Komatsu et al., 2010).
Fig. 1. Phosphoinositides and Organelle Identity.
Phosphoinositides are heterogeneously distributed throughout the cell and can be used as markers for different organelles as illustrated by their discrete subcellular distribution. Arrows indicate the flow of membrane in membrane trafficking routes. PI5P localization in cells is unclear.
II. Rationale
Phosphoinositides reside in the lipid bilayer and not only contribute to the chemical properties of the membrane bilayer but play an essential role in regulating protein recruitment and activity at cellular membranes. Perturbations to the levels of these lipids in the membrane can provide valuable insights into the functions of phosphoinositides in cellular processes. However, prolonged disturbance of phosphoinositide levels via standard genetic or molecular genetic approaches can provide an inaccurate picture of singular processes since it may give rise to compensatory responses either through adjustments of lipid metabolism or through the modulation of phosphoinositide-binding effector complexes in the cells’ attempt to correct the initial defect. Consequently, phenotypes obtained through these prolonged manipulations of phosphoinositide levels may be indirect and difficult to interpret. Additionally, studies suggest that phosphoinositide conversion (i.e., the phosphorylation/dephosphorylation of one species into another) can be “catastrophic” in nature, in that it can occur within time scales of seconds or minutes. While this concept is well-established in signal transduction areas (where the PI(4,5)P2-to-PI(3,4,5)P3 conversion is known to acutely turn on signaling pathways), it is less established in the area of membrane trafficking. However, recent work from a variety of groups, including our own, suggests that acute phosphoinositide conversions can govern key membrane trafficking processes (Chang-Ileto et al., 2011; Rusk et al., 2003; Terebiznik et al., 2002). It has thus become clear in the past few years that the field was in need of novel approaches allowing for acute manipulation of phosphoinositide levels in cells. Based on the many precedents utilizing CID in various fields, this methodology became the approach of choice for the direct spatial and temporal control of phosphoinositide level manipulation (Table 1).
In this chapter, we describe a variant of the CID approach to study changes in PI(4,5)P2 levels specifically on endocytic membranes and their effect on membrane fission.
III. Preparation of Expression Constructs
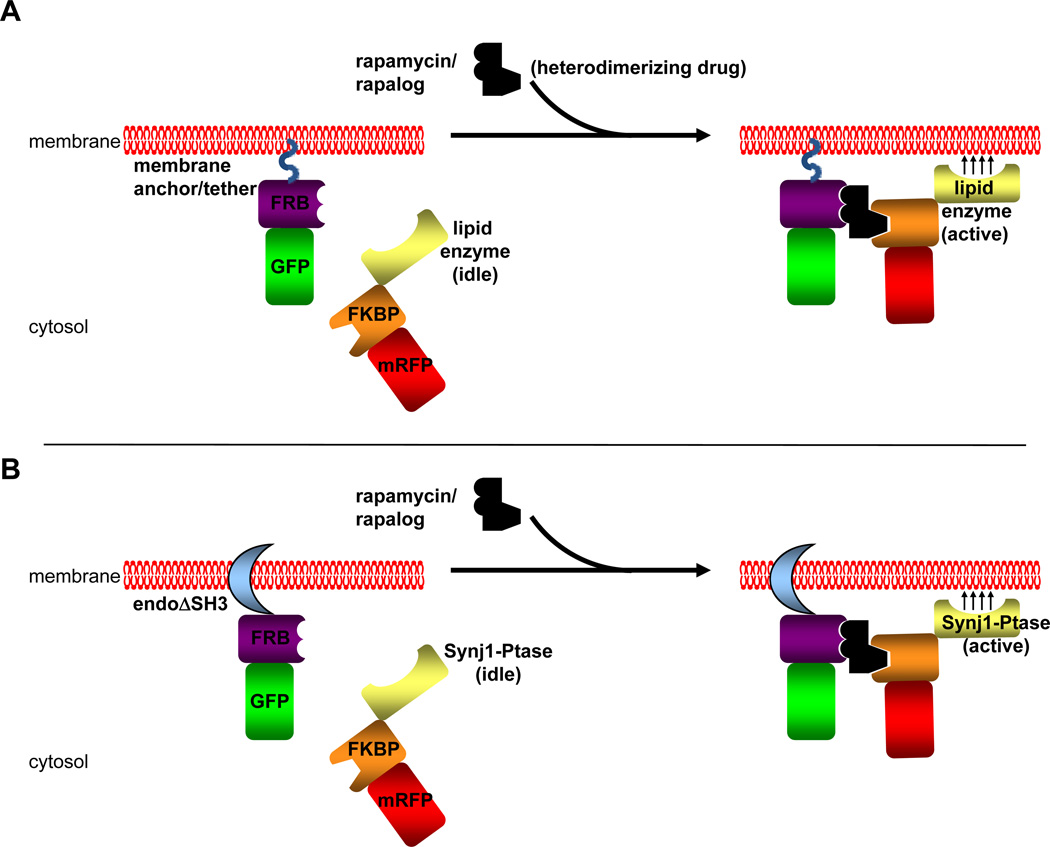
Dimerizers can be classified as either homodimerizers or heterodimerizers. Homodimerizers are symmetric dimerizers that bind and bring together two identical ligand binding domains. There can be a single protein/protein domain of interest designed as a fusion partner to the ligand binding domain or multiple protein/protein domains. Thus, the homodimerization system can be used to oligomerize a single fusion protein or multiple ones. However, in the latter case, when the homodimerizer is added to a system of two different fusion proteins, there will be multiple complexes formed with approximately half of the complexes made up of the two different fusion proteins and a quarter of each dimer of the individual fusion proteins. Use of heterodimerizers would ensure the induction of complexes of two fusion proteins without the side product of homodimers. In this chapter, we describe the use of rapamycin and a rapamycin analog, AP21967 (Clackson, 2006), to induce dimerization of the ligand binding domains FK506 binding protein (FKBP) and the rapamycin binding fragment (FRB) of mTOR (Fig. 2A). The use of AP21967 presents an important control of phenotypes observed using rapamycin since this rapalog can only be recognized by an engineered FRB molecule (with a T2098L mutation) and not by endogenous FRB (Clackson, 2006). This mutant FRB molecule, however, is still able to interact with rapamycin.
Fig. 2. Chemically Induced Dimerization Strategy.
A) A general schematic of CID using the protein modules FRB and FKBP and heterodimerizer rapamycin or one of its chemical analogs (rapalog). The FRB module is fused to a fluorescent tag (GFP in this example) and is targeted to a membrane compartment by a membrane anchor or tether. This membrane anchor/tether can be either a peptide sequence such as a myristoylation signal or a protein/protein domain that either spans or binds to a target membrane. The FKBP module is fused to another fluorescent tag (RFP in this example) and a catalytic domain of a lipid enzyme without any targeting sequences that the full length enzyme may contain. In the pre-rapamycin state (left side), the FKBP-lipid enzyme is soluble. Upon application of rapamycin or a rapalog, this heterodimerizers bring together the FRB and FKBP domains and the FKBP chimera is recruited to the membrane at which the FRB chimera is located. This brings the lipid enzyme into close proximity of the target membrane, allowing for synthesis or turnover of the desired phosphoinositide located at the target membrane. B) The CID strategy used in our study to analyze the effects of PI(4,5)P2 turnover by the endophilin-Synj1 partnership on membrane fission. In this study, the endophilin N-BAR domain was used as the membrane tether. This endophilin domain is also able to induce membrane tubulation similar to full length protein. The SH3 domain is omitted in order to avoid any recruitment of endogenous synaptojanin protein and other PRD domain-containing partners. The inositol 5-phosphatase domain from Synj1 was used to hydrolyze at PI(4,5)P2 at the endophilin-induced membrane tubules upon recruitment to these membranes by the addition of rapamycin/rapalog. The Sac1 domain of Synj1 was omitted in this study in order to study only the enzyme’s ability to dephosphorylate PI(4,5)P2 on the 5' position since the Sac1 domain has been reported to dephosphorylate several different phosphoinositide substrates in vitro (Guo et al., 1999). As with endophilin, the PRD domain of Synj1 was omitted in order to avoid any recruitment of Synj1 to membranes via interaction with SH3 domain-containing proteins.
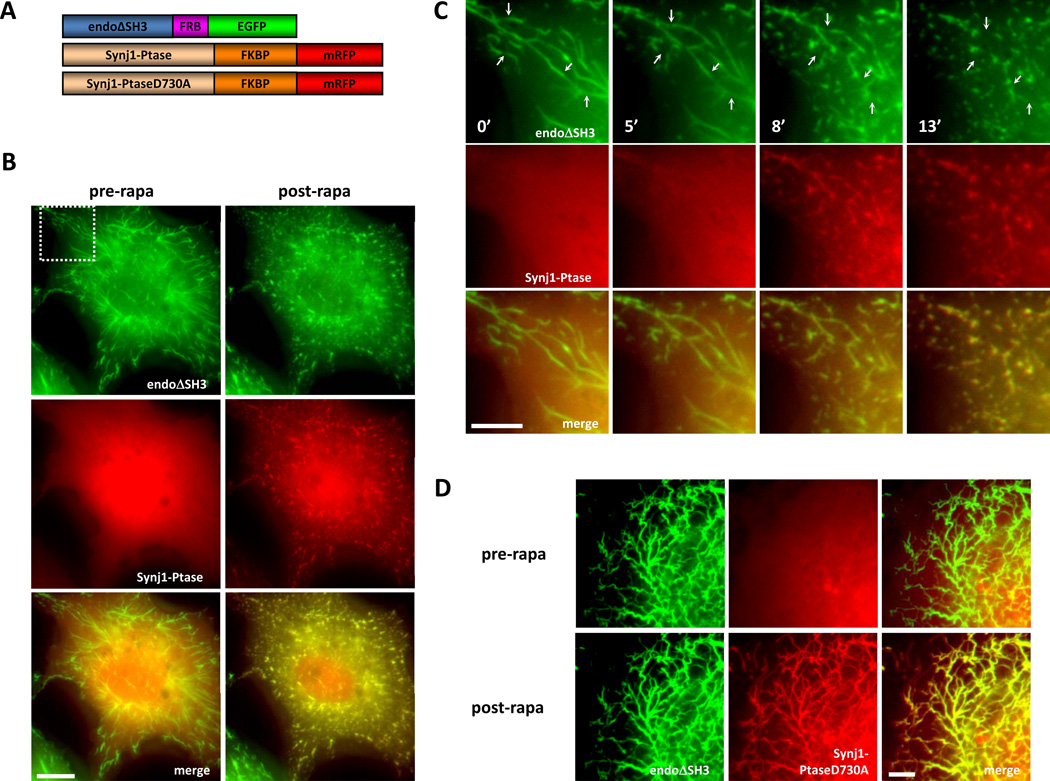
PI(4,5)P2 plays a fundamental role in clathrin-mediated endocytosis. This lipid has been shown to be critical for the recruitment of key endocytic adaptor and accessory proteins to the plasma membrane and is thus important for both the initiation and the progression of endocytosis. In order to dissect out a potential role of PI(4,5)P2 in membrane fission, we used endophilin-induced membrane tubules to model endocytic structures and more specifically bud necks and CID to induce an acute PI(4,5)P2-to-PI(4)P conversion at these membrane sites and analyzed ensuing changes in membrane dynamics using fluorescence microscopy. We were interested in whether membrane fission is modulated by the interaction between the BAR domain-containing protein endophilin and the inositol 5-phosphatase synaptojanin and the PI(4,5)P2 turnover mediated by synaptojanin. Using expression constructs originally available through Ariad Pharmaceuticals (and now Clontech), the 5-phosphatase domain of synaptojanin 1 was fused to the FKBP domain along with the fluorescent tag mRFP while endophilin, without its SH3 domain, was fused to the mutant FRB domain along with the fluorescent tag EGFP (Fig. 2B and 3A). The protein domains responsible for controlling the interaction between these two endocytic proteins (PRD for synaptojanin and SH3 for endophilin) were omitted from these constructs so that the interaction between these proteins could be precisely controlled by the addition of rapamycin or the rapalog AP21967. Overexpression of endophilin, and in this case, endophilin without its SH3 domain, in COS7 cells results in the massive tubulation of the PM (Fig. 3, pre-rapa state). In our system, we were able to take advantage of this phenomenon for two purposes. The first purpose was to use endophilin to localize the FRB domain to the PM where PI(4,5)P2 is enriched and thereby using this construct to localize synaptojanin to these tubular membranes once rapamycin/AP21967 was added. The second purpose for using endophilin overexpression was to create the tubular membranes that would serve as a model for endocytic bud necks, the site at which membrane fission occurs during the endocytic process.
Fig. 3. Acute Recruitment of the Inositol 5-Phosphatase Domain of Synj1 to Endophilin-Induced Tubules Results in Membrane Fragmentation and Condensation.
A) Diagram of the heterodimerization constructs: rat endophilin1 without the SH3 domain (endoΔSH3) is fused to the NH2-terminal side of the FRB domain followed by EGFP; the inositol 5-phosphatase domain of Synj1, either wild type or catalytically dead mutant D730A (Synj1-Ptase, Synj1-PtaseD730A respectively), was fused to the NH2-terminal side of two FKBP domains followed by mRFP. B) In transfected COS-7 cells, Synj1-Ptase shows a diffuse signal before rapalog treatment (pre-rapa). Rapalog treatment results in Synj1-Ptase recruitment to endophilin-induced membrane tubules and fragmentation and condensation of the membrane tubules take place (post-rapa). C) A time-lapse view of the fragmentation and condensation events upon rapalog treatment from a magnified field of the cell seen in B) as indicated by the dotted square. Example fragmentation sites are indicated by the arrows. D) In transfected COS-7 cells, Synj1-PtaseD730A showed diffuse signal before rapalog treatment (pre-rapa). Rapalog addition results in Synj1-PtaseD730A recruitment to endophilin-induced membrane tubules but no fragmentation/condensation events are observed (post-rapa). Scale bars represent B) 10 µm, C) 5 µm, and D) 5 µm. [Reprinted from Developmental Cell, Vol. 20, Chang-Ileto, B., Frere, S.G., Chan, R.B., Voronov, S.V., Roux, A., and Di Paolo, G., Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission, Pages No. 206–218, copyright 2011, with permission from Elsevier.]
Factors to consider when designing expression constructs:
Membrane localization. The final heterodimerized complex will need to be localized to the target membrane. How will this be achieved? One of the proteins of interest may have the intrinsic ability to localize to the target membrane/organelle. In our case, endophilin could be used as the targeting signal to the PM. Another possibility is the use of a localization peptide fused to either the FKBP or FRB construct, such as the myristoylation signal included in the pC4M-F2E construct encoding the FKBP, that targets its fusion partners to the cytoplasmic face of cellular membranes.
Position of fusion proteins in the expression constructs. The organization of the fusion proteins (protein/domain of interest, FKBP/FRB, fluorescent or reporter tag) must be considered with respect to the accessibility of the different domains and potential effects on structure and/or activity of the proteins/domains. For example, if an enzyme or enzyme domain is being studied, constructs with different arrangement of the fusion domains may have to be engineered to assess whether placing another protein on its NH2-terminus or COOH-terminus affects the catalytic activity. In our case, we replaced the SH3 domain of endophilin and the proline-rich tail of synaptojanin 1 with the FRB and FKBP domains, respectively, so as to preserve the targeting modules at the COOH terminus of these proteins.
Fluorescent tags. Which proteins will be tracked using a fluorescence tag and any fluorescence tags needed for controls must all be carefully considered and coordinated. Generally, monomeric RFP can be combined with eGFP, but CFP and YFP pairs are also possible. In addition, various other color fluorescent proteins, including blue, have been developed (Day and Davidson, 2009) and offer even more possibilities for color pairing of proteins.
Controls and probes. For every lipid enzymes utilized in the CID approach, it is essential to use a catalytically inactive counterpart in order to make sure the observed effects are dependent on the catalytic activity. Generally, this is achieved by generating point mutant versions of the wild-type construct. Such mutants are often reported in the literature for most lipid enzymes, and if this is not the case, appropriate enzymatic assays should be conducted to verify that the mutations abolish the enzymatic activity. Aside from following the protein(s) of interest, the phosphoinositide and/or membrane should also be monitored as controls for the experiment. For example, we tracked the presence or disappearance of PI(4,5)P2 from the PM using the PH domain from the enzyme PLCδ1. Membrane markers used for the plasma include exogenously added styryl dyes (e.g., FM4-64, FM1-43) and fluorophore labeled-wheat germ agglutinin (WGA). Additionally, fluid phase tracers, such as fluorophore labeled dextrans could be used to label the tubules, although they are not membrane probes per se, unlike styryl dyes and WGA.
IV. Expression of Fusion Proteins and Cell Maintenance
Expression of Fusion Proteins
When more than one construct is used to encode for various recombinant proteins, issues of stoichiometry between the various proteins may arise. For example, in initial co-transfections of our endophilin and synaptojanin contructs at equal concentrations, we did not obtain membrane tubulation. By varying the ratio of endophilin and synaptojanin DNA constructs used for transfections, we were able to find an acceptable range of DNA used that resulted in membrane tubulation and detectable levels of synaptojanin. Specifically, we lowered the amount of synaptojanin construct used alongside the endophilin construct until we were able to produce the membrane tubulation. Presumably the cause of the lack of membrane tubulation in the initial experiments was due to the high concentration of synaptojanin that could either catabolize a significant population of the PI(4,5)P2 molecules so that endophilin no longer had a high affinity for the PM or the excess synaptojanin was able to hydrolyze enough PI(4,5)P2, so that the fission process in many cases probably already occurred.
Cells to image membrane dynamics
COS7 cells are commonly used for the analysis of membrane tubulation caused by BAR domain-containing proteins (Gallop et al., 2006; Itoh et al., 2005; Peter et al., 2004). These cells have a relatively flat morphology that allows for the visualization of membrane tubulation upon BAR-protein overexpression since a large number of tubular structures derived from the PM are in the same plane, particularly in the peripheral regions of the cell. Visualization of membrane tubules in other cell types, such as CHO and 293, was difficult if not unsuccessful.
Cell culture conditions
COS-7 cells were grown at 37°C at 5% CO2. All media preparations, cell passaging, and cell plating are performed in a sterile laminar flow hood. Media was stored at 4°C in the dark. COS-7 cells were maintained in DMEM Medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% Glutamine or GlutaMAX. Care was taken while maintaining COS-7 cells to prevent overcrowding which often lead to decreased post-transfection cell imaging quality and survival.
Glass coverslips (Warner Instruments) used for growing transfected COS-7 cells were stored in EtOH. Prior to transfection, the coverslips were removed from the EtOH solution and excess alcohol was removed by passing each coverslip quickly through a flame. The coverslips were placed in a tissue culture dish and allowed to cool. COS-7 cells were transfected using a modified reverse transfection technique (Invitrogen) in which the cells were transfected and plated onto the glass coverslips at the same time. COS-7 cells were grown on the glass coverslips for 14–21 hours post-transfection. Time-lapse recordings were performed within this 14–21 hours period since the tubules get very stable and are modestly shortened by the recruitment of Synj1 and the hydrolysis of PI(4,5)P2 after this period.
V. Microscopy
Choice of imaging chamber/conditions (temperature, etc)
For live imaging, transfected coverslips were placed in a Chamlide recording chamber (Life Cell Instruments, South Korea) filled with 300 µl HBS solution (10 mM HEPES [pH 7.4], 136 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1.3 mM MgCl2, 10 mM glucose), and imaging was performed at 37°C using the Olympus IX-81 microscope equipped with a heating incubator (MIU-IBC-IF), connected to a Hamamatsu CCD camera (Hamamatsu Photonics) and mounted with a 60× oil-immersed UIS2 objective (Olympus).
Application of CIDs (rapamycin/rapalog)
Cells were treated with either 5 µM rapalog (200X stock in EtOH, as 200X) or 100 nM rapamycin (200X stock in DMSO) (Spencer et al., 1993). 1.5 µl of the rapamycin/rapalog was added directly to the HBS solution overlaid on the coverslip while in the recording chamber.
Imaging
Images were collected using Slidebook 5.0 software (Olympus). For FM 4–64 (Invitrogen) staining, cells were incubated with the dye in HBS. See Fig. 3 for examples of the membrane tubules in cells before and after rapamycin/AP21967 application with either WT Synj1-Ptase or the catalytically inactive Synj1-PtaseD730A.
Analysis
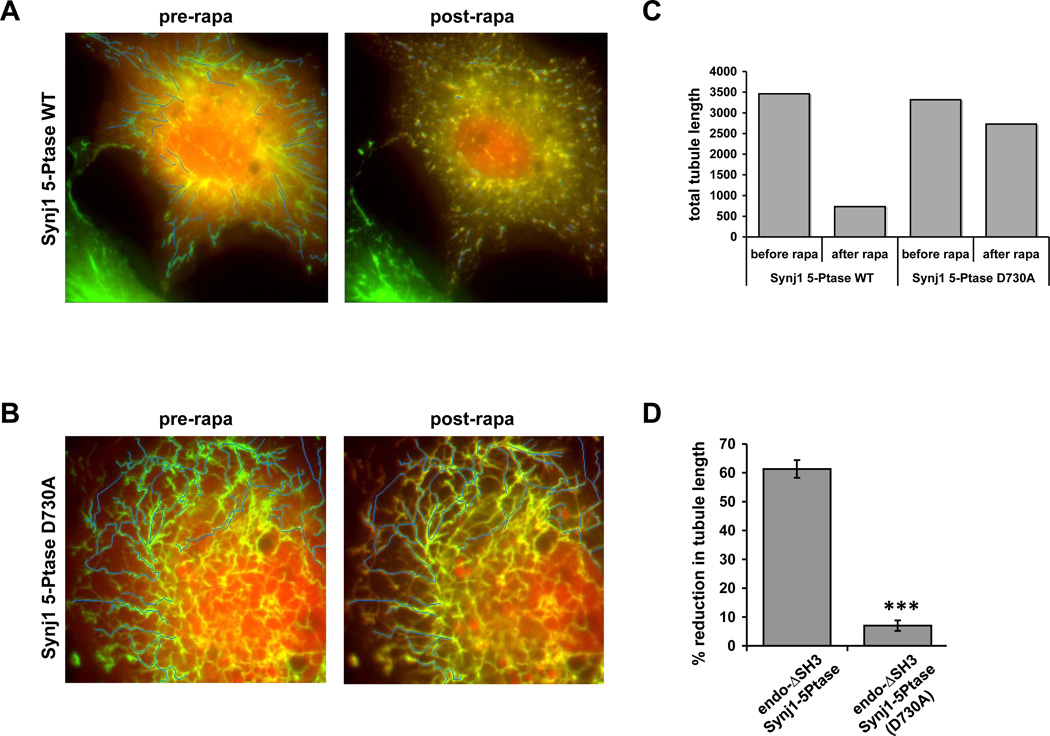
Tubule length before and after rapamycin or rapalog treatment was measured using the ImageJ software (NIH, USA). For each cell recorded, the initial length of tubules before the application of rapamycin/rapalog was measured by drawing a segmented thin line along the tubules (Li). The residual length of the same tubules was measured similarly from the image obtained 2 min after the application of rapamycin (Lr) (Fig. 4A–B). The lengths of the tubules before (SLi) and after (SLr) were summated and the difference (SLi-SLr) was normalized to the total lengths of the tubules before rapamycin treatment (SLi) (Fig. 4C). Tubule length inversely correlated with the extent of fission (Fig. 4D), although a more accurate approach to quantify tubular fission would involve a scoring of the breakpoints per unit of tubule length. This was difficult to achieve because of the intricate network of tubules obtained in these experiments, but would be easier to achieve in a system with low tubule density (for instance, in a cell free assay). We also observed that newly formed and short tubules did not tend to break but had a greater propensity to fully condense.
Fig. 4. Quantification of Membrane Fission and Condensation.
A) A COS-7 cell expressing endoΔSH3-FRB-GFP and Synj1-Ptase-FKBP-RFP is shown before and after rapamycin treatment. Using ImageJ, lines were drawn to follow the endophiilin-induced tubules before application of rapamycin. The evolution of these tubules were then followed after rapamycin treatment to select the broken tubules resulting from the recruitment of Synj1-Ptase-FKBP-RFP to these tubules. The final picture was used to draw the resulting broken tubules. B) The same method was used to draw tubules for a COS-7 cell expressing endoΔSH3-FRB-GFP and Synj1-PtaseD730A-FKBP-RFP before and after rapamycin treatment. C) Quantification of tubule length of the cells shown in A) and B) using ImageJ shows the dramatic decrease in total tubule length in the cell expressing wildtype Synj1-Ptase after rapamycin treatment in comparison to the small decrease experienced by the cell expressing the catalytically inactive Synj1-PtaseD730A. D) Comparison of the % reduction in tubule length in cells expressing either WT or catalytically inactive Synj1-Ptase with a n=24 for WT or n=9 for D730A. Data are represented as mean ± SEM. *** p < 0.0001. [Panel E is reprinted from Developmental Cell, Vol. 20, Chang-Ileto, B., Frere, S.G., Chan, R.B., Voronov, S.V., Roux, A., and Di Paolo, G., Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission, Pages No. 206–218, copyright 2011, with permission from Elsevier.]
VI. Considerations
There are several important considerations when using the CID approach to study the role of a phosphoinositide. Above all, the conversion of a phosphoinositide into another species as a result of CID has two consequences: (i) a reduction in the levels of the substrate; and (ii) an increase in the levels of the product. While these reactions can be monitored using genetically-encoded probes to both the substrate and the product, the biological effects can be potentially caused by either phenomenon or even a combination of both. We thus recommend that whenever possible, alternate manipulations may be conducted to target the same lipid. For instance, PI(4,5)P2 may be dephosphorylated by 5-phosphatases or cleaved by PLC into diacylglycerol and inositol-trisphosphate. If both types of hydrolysis produce the same biological effect, the latter is most likely caused by the loss of PI(4,5)P2 (rather than the generation of PI(4,5)P2 metabolites).
Another important point to consider is how to follow the affected membrane or target phosphoinositide without perturbing the system being studied. Protein domains that recognize phosphoinositides have been widely used as reporters for their target lipids (Lemmon, 2008; Varnai and Balla, 2007). However, the binding of these domains to phosphoinositides may affect the availability of these lipids as substrates for the lipid enzymes or functions being studied. Indeed, over-expression of the PH domain of PLCδ1, commonly used to visualize the localization of PI(4,5)P2 in cells, has been reported to affect cellular functions (Varnai et al., 2005). In our hands, over-expression of PLCδ1-PH fused to a fluorescent tag alongside our endophilin constructs resulted in blockade of endophilin-induced tubule formation. Diluting the concentration of PLCδ1-PH used in transfection remedied this effect and allowed the visualization of PI(4,5)P2-containing membrane and formation of membrane tubules. In addition, fluorescently-labeled membrane and organelle markers, such as FM 4–64 and wheat germ agglutinin for the PM, may also distort experimental results by masking the lipid substrate or protein(s) essential in the cellular process being studied, or may possibly make the membrane too rigid or more stable for fission.
Finally, the FRB protein module has been reported to be subject to a high rate of turnover along with any accompanying fusion partners (Edwards and Wandless, 2007; Stankunas et al., 2003). This protein instability is reversed upon the addition of ligand (i.e., rapamycin or one of its analogs). These features offer the disadvantage of potentially low levels of protein in the target cell particularly if the fusion-FRB chimera is particularly unstable. At the same time, this feature can also be seen as an advantage in that actions of the FRB fusion partner have a lower chance of confounding results until the heterodimerizer is added to the system.
VII. Summary & Conclusion
The use of CID in studying the roles that phosphoinositides play in cellular functions offers many advantages over the existing conventional approaches of exogenous application of lipids, genetic manipulation and use of pharmacological inhibitors. CID allows for the targeted synthesis or elimination of lipids in a spatially- and temporally-controllable manner. Whereas application of lipids to cells and over-expression of lipid enzymes in cells leads to gross or prolonged changes in a cell, CID offers a more sophisticated approach to controlling phosphoinositide levels to reveal their roles in cellular physiology. This method is amenable to a broad variety of lipid enzymes and lipid modifications (i.e., by such enzymes as phospholipases and phosphatidic acid phosphatases) as long as the catalytic domains/regions of these lipid enzymes are not already embedded in the membrane (as is the case for lipid phosphate phosphatases (Sigal et al., 2005)). This techniques is also amenable to all membranous organelles in the cell provided targeting domains are available and lipid substrates are present on these membranes. In addition, this method can be used to deplete the cytosol of soluble lipid enzymes by artificially mistargeting them to cellular compartments where the target substrate are not present in systems operating in a knockdown or null background of the lipid enzyme. As can be seen in phosphoinositide studies using the CID technology, much progress can be gained in the understanding the role of phosphoinositides in the cell. However, there is still a necessity for the identification of pharmacological activators and inhibitors of lipid enzymes and better probes for some phosphoinositides in order to gain even greater strides in our knowledge of the role of phosphoinositides and their lipid enzymes.
Acknowledgements
We thank Robin B. Chan and Aurélien Roux for their work in our original research manuscript (Chang-Ileto et al., 2011). We are also grateful for Dr. Chan for his critical reading of this manuscript. Work on phosphoinositides in the Di Paolo lab is funded by NIH grants R01 NS056049 and R01 HD05547 and by the McKnight Endowment Fund. Belle Chang-Ileto is funded by the NIH (F31 NS058096).
References
- Abe N, et al. Dissecting the role of PtdIns(4,5)P2 in endocytosis and recycling of the transferrin receptor. J Cell Sci. 2008;121:1488–1494. doi: 10.1242/jcs.020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, et al. Phosphoinositide signaling: new tools and insights. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, et al. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle JH, et al. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol. 2006;13:99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- Belshaw PJ, et al. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc Natl Acad Sci U S A. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Ileto B, et al. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev Cell. 2011;20:206–218. doi: 10.1016/j.devcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, et al. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, et al. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Chong H, et al. A system for small-molecule control of conditionally replication-competent adenoviral vectors. Mol Ther. 2002;5:195–203. doi: 10.1006/mthe.2002.0531. [DOI] [PubMed] [Google Scholar]
- Choudhury R, et al. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell. 2005;16:3467–3479. doi: 10.1091/mbc.E05-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T. Dissecting the functions of proteins and pathways using chemically induced dimerization. Chem Biol Drug Des. 2006;67:440–442. doi: 10.1111/j.1747-0285.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- Clackson T, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci U S A. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona O, et al. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 1999;99:179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, et al. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, et al. Impaired PtdIns(4,5)P2 synthesis in nerve terminals produces defects in synaptic vesicle trafficking. Nature. 2004;431:415–422. doi: 10.1038/nature02896. [DOI] [PubMed] [Google Scholar]
- Edwards SR, Wandless TJ. The rapamycin-binding domain of the protein kinase mammalian target of rapamycin is a destabilizing domain. J Biol Chem. 2007;282:13395–13401. doi: 10.1074/jbc.M700498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburger BH, et al. Phosphoinositides: lipid regulators of membrane proteins. J Physiol. 2010;588:3179–3185. doi: 10.1113/jphysiol.2010.192153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fili N, et al. Compartmental signal modulation: Endosomal phosphatidylinositol 3-phosphate controls endosome morphology and selective cargo sorting. Proc Natl Acad Sci U S A. 2006;103:15473–15478. doi: 10.1073/pnas.0607040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, et al. The pleckstrin homology domain of phospholipase C-delta1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry. 1995;34:16228–16234. doi: 10.1021/bi00049a039. [DOI] [PubMed] [Google Scholar]
- Gary JD, et al. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, et al. Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem J. 2001;355:249–258. doi: 10.1042/0264-6021:3550249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, et al. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Golebiewska U, et al. Diffusion coefficient of fluorescent phosphatidylinositol 4,5-bisphosphate in the plasma membrane of cells. Mol Biol Cell. 2008;19:1663–1669. doi: 10.1091/mbc.E07-12-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, et al. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Harris TW, et al. Mutations in synaptojanin disrupt synaptic vesicle recycling. J Cell Biol. 2000;150:589–600. doi: 10.1083/jcb.150.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V. Phosphoinositide regulation of clathrin-mediated endocytosis. Biochem Soc Trans. 2005;33:1285–1289. doi: 10.1042/BST0331285. [DOI] [PubMed] [Google Scholar]
- Hilgemann DW, et al. The complex and intriguing lives of PIP2 with ion channels and transporters. Sci STKE. 2001;2001:RE19. doi: 10.1126/stke.2001.111.re19. [DOI] [PubMed] [Google Scholar]
- Huang W, et al. Stabilized phosphatidylinositol-5-phosphate analogues as ligands for the nuclear protein ING2: chemistry, biology, and molecular modeling. J Am Chem Soc. 2007;129:6498–6506. doi: 10.1021/ja070195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, et al. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kahlfeldt N, et al. Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2. J Biol Chem. 2010;285:2734–2749. doi: 10.1074/jbc.M109.074906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, et al. Regulation of transferrin recycling kinetics by PtdIns[4,5]P2 availability. Faseb J. 2006;20:2399–2401. doi: 10.1096/fj.05-4621fje. [DOI] [PubMed] [Google Scholar]
- Knight ZA, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, et al. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M, et al. ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J Cell Biol. 2003;162:113–124. doi: 10.1083/jcb.200301006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, et al. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, et al. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc Natl Acad Sci U S A. 1997;94:7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Logan MR, Mandato CA. Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol Cell. 2006;98:377–388. doi: 10.1042/BC20050081. [DOI] [PubMed] [Google Scholar]
- Mao YS, Yin HL. Regulation of the actin cytoskeleton by phosphatidylinositol 4-phosphate 5 kinases. Pflugers Arch. 2007;455:5–18. doi: 10.1007/s00424-007-0286-3. [DOI] [PubMed] [Google Scholar]
- Nakatsu F, et al. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J. Cell Biol. 2010;190 doi: 10.1083/jcb.201005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki S, et al. Intracellular delivery of phosphoinositides and inositol phosphates using polyamine carriers. Proc Natl Acad Sci U S A. 2000;97:11286–11291. doi: 10.1073/pnas.210197897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Prasad NK, Decker SJ. SH2-containing 5'-inositol phosphatase, SHIP2, regulates cytoskeleton organization and ligand-dependent down-regulation of the epidermal growth factor receptor. J Biol Chem. 2005;280:13129–13136. doi: 10.1074/jbc.M410289200. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, et al. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins CT, et al. A ligand-reversible dimerization system for controlling protein-protein interactions. Proc Natl Acad Sci U S A. 2000;97:7096–7101. doi: 10.1073/pnas.100101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusk N, et al. Synaptojanin 2 functions at an early step of clathrin-mediated endocytosis. Curr Biol. 2003;13:659–663. doi: 10.1016/s0960-9822(03)00241-0. [DOI] [PubMed] [Google Scholar]
- Schu PV, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Sigal YJ, et al. Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem J. 2005;387:281–293. doi: 10.1042/BJ20041771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen SD, et al. A role for a wortmannin-sensitive phosphatidylinositol-4-kinase in the endocytosis of muscarinic cholinergic receptors. Mol Pharmacol. 1998;53:827–836. [PubMed] [Google Scholar]
- Spencer DM, et al. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- Stankunas K, et al. Conditional protein alleles using knockin mice and a chemical inducer of dimerization. Mol Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- Stankunas K, et al. Rescue of degradation-prone mutants of the FK506-rapamycin binding (FRB) protein with chemical ligands. Chembiochem. 2007;8:1162–1169. doi: 10.1002/cbic.200700087. [DOI] [PubMed] [Google Scholar]
- Subramanian D, et al. Activation of membrane-permeant caged PtdIns(3)P induces endosomal fusion in cells. Nat Chem Biol. 2010;6:324–326. doi: 10.1038/nchembio.348. [DOI] [PubMed] [Google Scholar]
- Suh BC, Hille B. Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr Opin Neurobiol. 2005;15:370–378. doi: 10.1016/j.conb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Suh BC, Hille B. PIP2 is a necessary cofactor for ion channel function: how and why? Annu Rev Biophys. 2008;37:175–195. doi: 10.1146/annurev.biophys.37.032807.125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh BC, et al. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terebiznik MR, et al. Elimination of host cell PtdIns(4,5)P(2) by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat Cell Biol. 2002;4:766–773. doi: 10.1038/ncb854. [DOI] [PubMed] [Google Scholar]
- Varnai P, Balla T. Visualization and manipulation of phosphoinositide dynamics in live cells using engineered protein domains. Pflugers Arch. 2007;455:69–82. doi: 10.1007/s00424-007-0270-y. [DOI] [PubMed] [Google Scholar]
- Varnai P, et al. Selective cellular effects of overexpressed pleckstrin-homology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J Cell Sci. 2005;118:4879–4888. doi: 10.1242/jcs.02606. [DOI] [PubMed] [Google Scholar]
- Verstreken P, et al. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- Wang YJ, et al. Critical role of PIP5KI{gamma}87 in InsP3-mediated Ca(2+) signaling. J Cell Biol. 2004;167:1005–1010. doi: 10.1083/jcb.200408008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, et al. Chemical synthesis and molecular recognition of phosphatase-resistant analogues of phosphatidylinositol-3-phosphate. J Am Chem Soc. 2006;128:885–897. doi: 10.1021/ja0554716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- Zhang H, et al. Synthesis and biological activity of PTEN-resistant analogues of phosphatidylinositol 3,4,5-trisphosphate. J Am Chem Soc. 2006a;128:16464–16465. doi: 10.1021/ja065002j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. Synthesis and biological activity of phospholipase C-resistant analogues of phosphatidylinositol 4,5-bisphosphate. J Am Chem Soc. 2006b;128:5642–5643. doi: 10.1021/ja060621d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci U S A. 2010;107:9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]