Abstract
Because of the lack of appropriate animal models, the potentially causal contributions of inherited mitochondrial genomic factors to complex traits are less well studied compared with inherited nuclear genomic factors. We previously detected variations between the mitochondrial DNA (mtDNA) of the Dahl salt-sensitive (S) rat and the spontaneously hypertensive rat (SHR). Specifically, multiple variations were detected in mitochondrial genes coding for subunits of proteins essential for electron transport, in mitochondrial reactive oxygen species production, and within the D-loop region. To evaluate the effects of these mtDNA variations in the absence of the corresponding nuclear genomic factors as confounding variables, novel reciprocal strains of S and SHR were constructed and characterized. When compared with that of the S rat, the heart tissue from the S.SHRmt conplastic strain wherein the mtDNA of the S rat was substituted with that of the SHR had a significant increase in mtDNA copy number and decrease in mitochondrial reactive oxygen species production. A corresponding increase in aerobic treadmill running capacity and a significant increase in survival that was not related to changes in blood pressure were observed in the S.SHRmt rats compared with the S rat. The reciprocal SHR.Smt rats did not differ from the SHR in any phenotype tested, suggesting lower penetrance of the S mtDNA on the nuclear genomic background of the SHR. These novel conplastic strains serve as invaluable tools to further dissect the relationship between heart function, aerobic fitness, cardiovascular disease progression, and mortality.
Keywords: Dahl salt-sensitive rat, S rat, spontaneously hypertensive rat, SHR, genetic, polymorphism, gene
in recent years, similar to hypertension, intrinsic aerobic or exercise capacity is also increasingly recognized as a strong prognostic indicator of cardiovascular morbidity and mortality (2–4, 12, 22, 26, 32). Aerobic running capacity (ARC), blood pressure (BP), and longevity are well recognized as complex polygenic traits, but the search for the identities of inherited factors controlling these complex traits and their interrelationships are largely focused on the nuclear genome (5, 9, 10, 16–18, 25, 33, 35, 36, 39, 44–46).
Mitochondrial respiration is not only a source of energy (ATP) but also a major source of reactive oxygen species (ROS) with 0.2% of oxygen consumed, being normally converted into superoxide in a quiescent state (1, 40). ROS cause increased oxidative stress, which is strongly implicated in pathological signaling, leading to elevated BP or hypertension (6, 11, 30, 41, 43). Because several of the subunits of proteins participating in mitochondrial respiration for the generation of both ATP and ROS are encoded by the mitochondrial genome, it is possible that functional variants within the mitochondrial DNA (mtDNA) could serve as genetic determinants of ARC and/or BP, which in turn could affect life span. While there are reports of associations of the mitochondrial genome with exercise capacity and other cardiovascular phenotypes (21, 28, 29, 31, 42), inferences on cause-effect relationships cannot be drawn based on these association studies. To extend such observations beyond mere associations, we generated novel reciprocal conplastic strains using two of the most widely used selectively bred models of cardiovascular disease: the Dahl salt-sensitive (S) rat and the spontaneously hypertensive rat (SHR). The choice of S and SHR was based on three primary factors: 1) the divergence in BP as well as ARC between these two strains; 2) both S and SHR have highly permissive nuclear genomes for the development of high BP, which, in theory, would allow for the experimental detection of any subtle contributions of the reciprocally substituted mitochondrial genomes; and 3) our previous report (24) of complete mitochondrial genome sequencing data wherein 12 nonsynonymous variations were detected within genes coding for subunits of proteins essential for the electron transport chain, for mitochondrial ROS production, and within the D-loop region.
MATERIALS AND METHODS
All animal experiments were conducted as per preapproved protocols by the Institutional Animal Care and Use Committee of the University of Toledo College of Medicine and Life Sciences. The inbred S rat was developed in house at our Institution. The SHR (SHR/Hsd) was originally obtained from Harlan Sprague-Dawley (Indianapolis, IN) and maintained in our colony.
Conplastic strain derivation.
Two reciprocal conplastic strains of S and SHR were generated by taking advantage of the maternal inheritance of mitochondrial genomes. A single SHR female rat was crossed with a male S rat. The resultant F1 female offsprings were backcrossed with male S rats. This backcross procedure was repeated 9–12 additional times to generate S.SHRmt conplastic strains. Similarly, a single S female rat was bred with a male SHR. The F1 female offsprings were backcrossed with male SHR. This backcross procedure was repeated 9–12 additional times to generate SHR.Smt conplastic strains. Unless otherwise mentioned, all studies were conducted using the backcross 13 animals.
Isolation of DNA and genotyping.
DNA was extracted from tail biopsies of S.SHRmt and SHR.Smt and sequenced as detailed in a previous report (24). A total of 162 microsatellite markers were used to assess the nuclear genomes of S.SHRmt and SHR.Smt strains along with their respective control inbred rats.
BP measurements.
The experimental design for BP measurement by the tail-cuff method was as described previously (25). Briefly, at 30 days of age, rats were weaned onto a low-salt (0.3% NaCl) Harlan Teklad 7034 diet. Conplastic strains along with their respective control rats (Dahl S rat and SHR) were housed two to a cage such that two different strains were in each cage. At 40–42 days of age, the rats were switched to a 2% NaCl diet (Harlan Teklad, TD 94217) and maintained on this diet for the duration of the experiment. During days 25–28 on the 2% NaCl diet, each rat had its systolic BP measured by two blinded operators. During BP measurements, rats were restrained and warmed to 28°C. The operators' readings for each rat were averaged and recorded as that animal's systolic BP. BP data were also compared by surgical implantation of radiotelemetric probes, followed by real-time monitoring using the telemetry system from Data Sciences International (25).
Assessment of ARC.
ARC was determined in 10-wk-old rats using a standard ramped treadmill test described by Henderson et al. (14). The equipment used for treadmill running was from Columbus Instruments (Model Exer-4; Columbus, OH). The rats were euthanized after completion of the ARC experiment for collection of heart samples. These samples were used for all biochemical and protein expression studies.
Survival study.
S, S.SHRmt, SHR, and SHR.Smt rats (backcross-10) were raised and administered 2% NaCl containing diet as described under the BP measurements section. These rats were continued on the 2% NaCl diet until their natural death.
Isolation of heart mitochondria.
Heart mitochondria were isolated by using differential centrifugation method (7). Briefly, heart homogenate was obtained using isolation buffer containing 225 mM mannitol, 75 mM sucrose, 1 mM EGTA, 0.1% fatty acid-free BSA, and 10 mM Tris·HCl (pH 7.4). The homogenate was centrifuged at 1,000 g for 10 min, and the supernatant was centrifuged again at 12,000 g for 10 min. Mitochondrial pellet was washed twice, centrifuged at 12,000 g, and resuspended in isolation buffer without EGTA. All centrifugation steps were carried out at 4°C. Protein content was quantified by using the bicinchoninic acid method from Pierce with bovine serum albumin as standard.
Determination of oxidant production.
Oxidant generation was monitored in mitochondria isolated from the hearts of S, SHR, and conplastic strains by using the redox-sensitive fluorescent probe 2′,7′-dichlorofluorescein-diacetate (DCFH-DA) according to previously described methods (19, 27). In brief, isolated mitochondria (∼0.2 mg protein) were incubated in the assay media consisting of (in mM) 137 KCl, 2.5 MgCl2, 2 K2HPO4, 10 Tris·HCl (pH 7.4), 5 glutamate, and 5 malate and 5 μM DCFH-DA at 37°C for 10 min to allow DCFH-DA to cross the mitochondrial membrane. The solution was then centrifuged at 12,000 g for 10 min, and the supernatant was discarded. The pellets were resuspended in fresh assay media without DCFH-DA. Oxidant generation was measured at the excitation wavelength of 488 nm and emission wavelength of 525 nm using Molecular device fluorescence microplate reader for 30 min. The amount of oxidant generation was expressed as DCF formed per minute per milligram of protein.
Mitochondrial permeability transition pore assay.
The opening of the permeability transition pore causes mitochondrial swelling and is continuously assayed as a decrease in light scattering of a mitochondrial suspension. Mitochondrial permeability transition pore activity was measured using previously described methods (8, 23). In brief, isolated mitochondrial pellet was suspended at the concentration of 1 mg protein/ml in 200 μl of assay buffer containing (in mM) 225 mannitol, 75 sucrose, 5 glutamate, 5 malate, and 10 HEPES (pH 7.4). The samples were continuously monitored at optical density of 540 nm for 30 min with or without the addition of 15 μmol/mg protein of Ca2+. The rate of fall in absorbance was recorded using a microplate reader (Molecular Devices).
Analysis of mitochondrial respiration and mtDNA copy number.
Mitochondrial respiratory chain activities of complex I, II, IV, and V were assessed in mitochondria isolated from heart samples of all groups of rats. Activities were measured using kits from Mitoscience-Abcam according to the manufacturers' instructions. mtDNA copy number was measured by real-time PCR (Bio-Rad) method using SYBR green. Total DNA was isolated from heart tissue (n = 4). mtDNA copy number was assessed by amplification of the mitochondrial D-Loop versus the nuclear Gapdh gene. The difference in CT between D-loop and Gapdh values was used as the measure of relative abundance of the mitochondrial genome. Fold change was calculated and expressed by using the 2−ΔΔCT method (37).
Protein content of mitochondrial respiratory subunits.
Immunoblotting was performed to measure the mitochondrial respiratory protein levels in isolated mitochondrial pellet from hearts of S, S.SHRmt, SHR, and SHR.Smt rats. Samples were homogenized in ice-cold RIPA lysis buffer with protease inhibitor cocktail (Pierce). Thirty microgram of proteins were boiled with Laemmli loading buffer for 5 min at 95°C. Protein samples were resolved using 10% NuPAGE Bis-Tris gel (Invitrogen) at room temperature and transferred on to polyvinylidene difluoride membrane (Millipore). Mitochondrial protein levels were quantitated by Western blot analysis using mouse monoclonal antibodies purchased from Mitoscience-Abcam for the following proteins: β-subunit of the F1F0 ATP synthase (Atp5a), mitochondrial cytochrome-c oxidase subunit 1 (Cox1), mitochondrial cytochrome c oxidase subunit 3 (Cox3), cytochrome c oxidase subunit 4 (Cox4), core 2 subunit of ubiquinol:ferricytochrome c oxidoreductase (Uqcrc2), 39-kDa subunit of NADH:ubiquinone oxidoreductase (Ndufa9), and flavor protein subunit of succinate dehydrogenase (Sdha). Heart homogenates were used for protein Western blot analysis for nuclear encoded transcription factor proliferator-activated receptor coactivator-1α (Pgc-1α; Abcam) and peroxisome proliferator-activated receptor-γ (Ppar-γ; Santa Cruz). Membranes were blocked with 5% fat-free milk and incubated with primary and secondary antibodies before visualization by the chemiluminescence method (Pierce). ImageJ was used to quantify the protein expression level, and values were expressed as arbitrary densitometry units.
Statistical analyses.
Statistical analyses were conducted by ANOVA using the SPSS software (SPSS, Chicago, IL). Data are presented as means ± SE. A P value of ≤0.05 was used as a threshold for statistical significance. Survival data were analyzed and plotted using the Kaplan-Meier plot.
RESULTS
Genomes of conplastic strains.
The mitochondrial genotypes of the conplastic strains were verified by direct sequencing of the complete mitochondrial sequences as S in the SHR.Smt strain and SHR in the S.SHRmt strain, respectively. The mtDNA sequence of the SHR.Smt strain was identical to the mtDNA sequence of the S rat (Genbank accession number GU997608). Similarly, the mtDNA sequence of the S.SHRmt strain was identical to the reported mtDNA sequence of the SHR (Genbank accession number GU997610). The nuclear genomic DNA of both the conplastic strains were verified by whole genome genotyping conducted using 162 microsatellite markers (Table 1). The average distance between the markers was 14.54 Mb. At all the 162 locations examined, the nuclear genome of the S.SHRmt strain was represented by S alleles and the nuclear genome of the SHR.Smt strain was represented by SHR alleles (Table 1).
Table 1.
Genotypes of conplastic strains
| Animal ID S0688 | Animal ID S0685 | Animal ID S0686 | Animal ID 3501 | Animal ID 3499 | Animal ID 3495 | Animal ID 3518 | Animal ID 3530 | Animal ID 3529 | Animal ID 3548 | Animal ID 3544 | Animal ID 3543 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dahl S | Dahl S | Dahl S | S.SHRmt conplastic | S.SHRmt conplastic | S.SHRmt conplastic | SHR | SHR | SHR | SHR.Smt conplastic | SHR.Smt conplastic | SHR.Smt conplastic | |
| Microsatellite marker | Genotypes | Genotypes | ||||||||||
| D1Rat176 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Rat167 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Rat8 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Rat196 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Mgh6 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Rat45 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Rat357 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Rat159 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Mit34 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D1Rat86 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat3 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat116 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat201 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Mit5 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat273 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat31 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Mit8 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat152 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat61 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat294 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D2Rat250 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D3Rat117 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D3Rat45 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D3Rat108 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D3Rat28 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D3Rat160 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D3Rat213 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D3Rat145 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Rat136 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Rat151 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Rat153 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Mgh4 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Mit12 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Rat148 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Rat190 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Rat198 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Rat67 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Rat207 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D4Mgh13 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D5Mgh17 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D5Rat128 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D5Rat126 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D5Rat82 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D5Rat84 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D5Rat108 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D5Rat97 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D5Rat46 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D6Rat80 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D6Rat86 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D6Rat84 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D6Rat92 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D6Rat64 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D6Rat97 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D6Rat12 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D6Mgh1 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat36 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat113 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat35 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat31 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat92 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat73 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat141 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Mit11 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat7 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D7Rat94 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat77 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat56 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Mit5 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Mgh7 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat62 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat75 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat133 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat19 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat13 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat72 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat119 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D8Rat81 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D9Rat138 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D9Rat36 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D9Rat158 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D9Rat18 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D9Rat9 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D9Rat99 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat51 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat121 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat43 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat38 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat211 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat27 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat93 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat142 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat13 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Mgh3 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat11 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Mgh1 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D10Rat135 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D11Rat19 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D11 Rat17 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D11Mgh5 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D11Rat67 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D11Mgh4 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D11Rat4 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D11Rat37 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D11rat46 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D12Rat59 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D12Rat43 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D12Rat3 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D12Mgh1 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D12Rat38 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D12Rat93 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D12Rat107 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D13Rat93 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D13Rat16 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D13Rat88 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D13Arb7 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D13Rat27 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D13Rat133 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D14Rat73 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D14Rat39 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D14Rat94 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15Rat38 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15Rat69 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15Rat116 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15rat36 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15Rat48 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15Rat126 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15Rat103 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15Rat106 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D15Rat50 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D16Mit2 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D16Rat44 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D16Rat65 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D16Rat57 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D16Rat15 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D17Rat2 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D17Rat112 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D17Mgh5 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D17Rat89 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D17Rat52 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D18Rat113 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D18Mit14 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D18Rat99 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D18Rat57 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D18Rat61 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D18Mgh3 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D18Mit1 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D18Rat69 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D19Rat34 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D19Rat28 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D19Rat97 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D19Rat47 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D19Rat15 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D19Rat29 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D19Rat58 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D19Rat4 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D20Rat21 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D20Rat1 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D20Rat46 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D20Rat31 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D20Rat23 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D20Mit4 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
| D20Rat55 | S | S | S | S | S | S | SHR | SHR | SHR | SHR | SHR | SHR |
Dahl S, Dahl salt-sensitive rat (S); SHR, spontaneously hypertensive rat; mt, mitochondrial.
Body weights and relative heart weights.
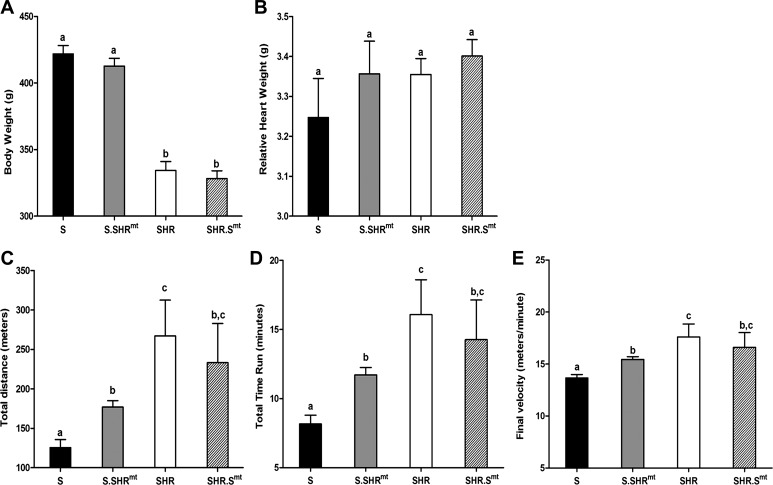
Body weights of S and SHR were significantly different (Fig. 1A). However, the body weights of the conplastic strains were not significantly different from their relative inbred strain comparisons (Fig. 1A). Relative heart weights were not different between any of the experimental groups (Fig. 1B).
Fig. 1.
Body weight (A) and relative heart weight (B) data from inbred and conplastic rats. SHR, spontaneously hypertensive rat; S, Dahl salt-sensitive rat. Data were collected from 101–113-day-old rats. Relative heart weight = heart weight/body weight. Bars with the same letters were not significantly different from each other. Assessment of aerobic running capacity in inbred and their respective conplastic strains was with the following parameters: total distance (C), total time (D), and final velocity (E). All the parameters were calculated as per previously described procedures (14). Bars labeled with the same letters were not significantly different from each other. Bars labeled with different letters were significantly different from each other, P < 0.05.
Intrinsic aerobic capacity.
Inbred SHR had a significantly higher running capacity than inbred S rats (Fig. 1B). The total distance run by SHR (267 ± 45 m) was significantly longer than the distance run by the S rat (126 ± 10 m) (Fig. 1C, P < 0.0006). The total duration for which SHR could run (16 ± 2.5 min) was also higher than that of the S rat (8 ± 0.6 min) (Fig. 1D, P < 0.003). Similarly, the velocity of running of the SHR (17.6 ± 1.3 m/min) was higher than the velocity of the S rats (13.7 ± 0.32 m/min) (Fig. 1E, P < 0.003). Substituting the mitochondrial genome from the SHR into the S rat (S.SHRmt conplastic strain) significantly improved the ARC of S rats (Fig. 1, C–E). The distance, duration, and velocity run by the S rats was significantly improved in the S.SHRmt conplastic rats by 51.4 m (Fig. 1C, P < 0.0004), 3.6 min (Fig. 1D, P < 0.0001), and 1.8 m/min (Fig. 1E, P < 0.0002), respectively. Introducing the mitochondrial genome from the S rat into the SHR in the SHR.Smt conplastic strains did not, however, alter the ARC of the SHR (Fig. 1, C–E).
Blood pressure.
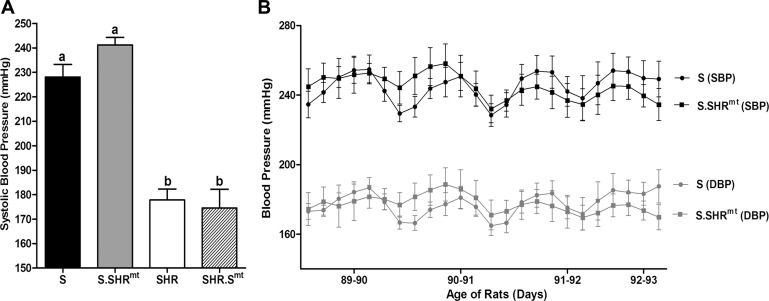
Systolic BP of the S rat (228 ± 5 mmHg) was significantly higher than that of the SHR (178 ± 5 mmHg) (Fig. 2A, P < 0.001). Systolic BP of S.SHRmt (241 ± 3 mmHg) and SHR.Smt (175 ± 8 mmHg) were not statistically different from the systolic BP of S and SHR rats, respectively (Fig. 2A). Although the overall tail-cuff BP data were not significantly different between the inbred and respective conplastic strains, the BP data of S and S.SHRmt were further corroborated by radiotelemetry (Fig. 2B).
Fig. 2.
Blood pressure (BP) measurements. A: BP by the tail-cuff method. Data are means ± SE. BP effect in each strain is defined as the mean systolic BP (SBP) of that strain minus the mean SBP of the S rat. DBP, diastolic blood pressure. Bars labeled with the same letters were not significantly different from each other. Bars labeled with different letters were significantly different from each other, P ≤ 0.05. B: BP by the telemetry method. Rats (n = 4–6 /group) were implanted with radiotelemetry transmitters, and BP was measured as described under materials and methods. Values are 4 h moving averages ± SE.
Survival study.
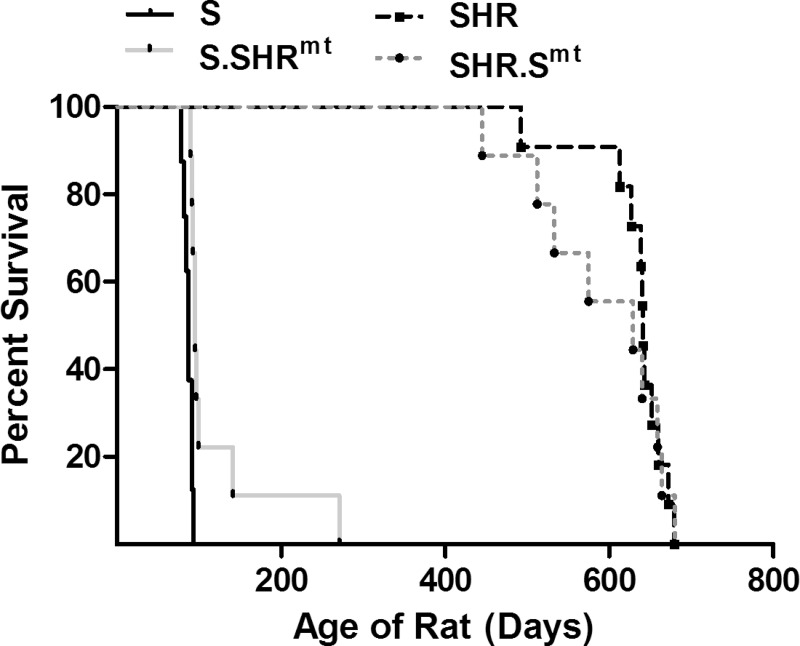
The median survival of S rats was 87 days, which was significantly lower than the median survival of >620 days for the SHR (Fig. 3). The S.SHRmt conplastic strain had a median survival of 95 days, which was significantly higher than that of the S rat (Fig. 3, P < 0.006). The reciprocal conplastic strain, SHR.Smt, had a median survival of >554 days, which was lower than the median survival of the SHR by 66 days, but the difference in median survival was not statistically significant (Fig. 3, P < 0.956).
Fig. 3.
Comparison of survival times between groups. S (n = 8) rat, SHR (n = 11), S.SHRmt (n = 9) and SHR.Smt (n = 9) were studied for survival as described under materials and methods. S vs. SHR (P < 0.0001), S vs. S.SHRmt (P < 0.0012), SHR vs. SHR.Smt (P < 0.96).
Mitochondrial oxidant generation and mitochondrial swelling.
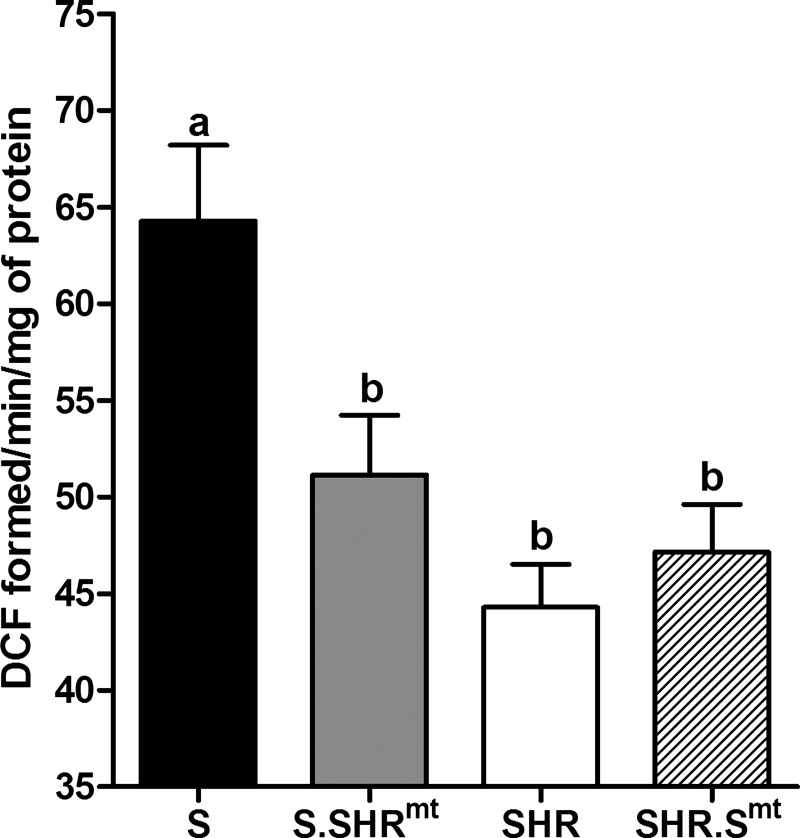
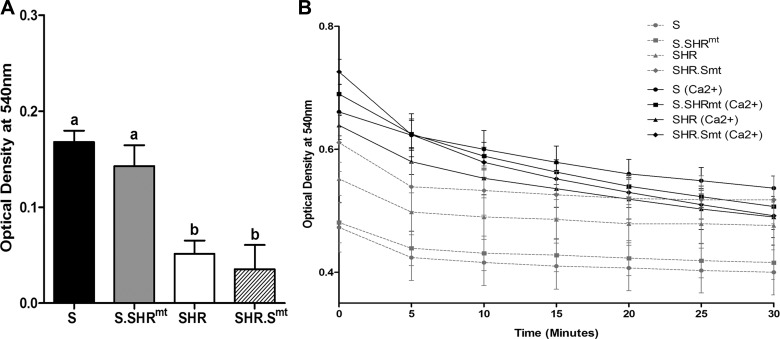
To determine whether the previously reported sequence variation in the mitochondrial genome was responsible for the differential intrinsic aerobic capacity and survival effects described above, we studied the functionality of isolated mitochondria from the SHR conplastic and progenitor inbred strains. Mitochondria from the S rats generated significantly higher ROS compared with the SHR (Fig. 4, P < 0.001). ROS production of the S rats was significantly attenuated by introgression of the SHR mitochondrial genome in S.SHRmt conplastic rats (Fig. 4, P < 0.015). Replacing the SHR mtDNA with that of the S in the SHR.Smt conplastic strain did not change ROS production in the SHR (Fig. 4, P < 0.905). Superoxide dismutase activity was not different among any of the experimental groups (data not shown). Because ROS are known to affect the functionality of the mitochondrial permeability transition pore, we evaluated the extent of mitochondrial swelling in the conplastic and progenitor strains. While there were no differences in basal absorbance measured in the absence of calcium, in the presence of exogenously added calcium, increased mitochondrial swelling was observed in S rats but not in the SHR (Fig. 5A, P < 0.001). Despite the decrease in ROS observed in the S.SHRmt conplastic strain compared with the S rat, the extent of mitochondrial swelling in response to calcium was not different between S.SHRmt and S rats (Fig. 5A, P < 0.609). Basal absorption and mitochondrial swelling in response to calcium was also unaffected in the SHR by the introduction of the S mitochondrial genome into the SHR in SHR.Smt conplastic strain. (Fig. 5A, P < 0.859). Tracings of absorption plotted against time with and without the addition of calcium in all samples is shown in Fig. 5B.
Fig. 4.
Assessment of mitochondrial oxidant generation. Values are expressed as dichlorofluorescein (DCF) fluorescence per minute per milligram of mitochondrial protein. Values are means ± SE; n = 4/group. Bars labeled with the same letters were not significantly different from each other. Bars labeled with different letters were significantly different from each other, P < 0.05.
Fig. 5.
Monitoring basal absorbance and mitochondrial swelling. Mitochondria (200 μg) isolated from hearts of experimental rats were incubated in the presence or absence of Ca2+ (15 mmol/mg protein) in the assay buffer as described under materials and methods. A: bars represent the difference in rate of fall in absorbance/minute observed between the mitochondrial suspension with and without exogenously added Ca2+. Values are means ± SE; n = 4/group. Bars labeled with the same letters were not significantly different from each other. Bars labeled with different letters were significantly different from each other, P < 0.05. B: tracing of mitochondrial swelling monitored by fall in optical absorbance at 54 nm over time (n = 4 rats/group).
Enzyme activities in isolated mitochondria.
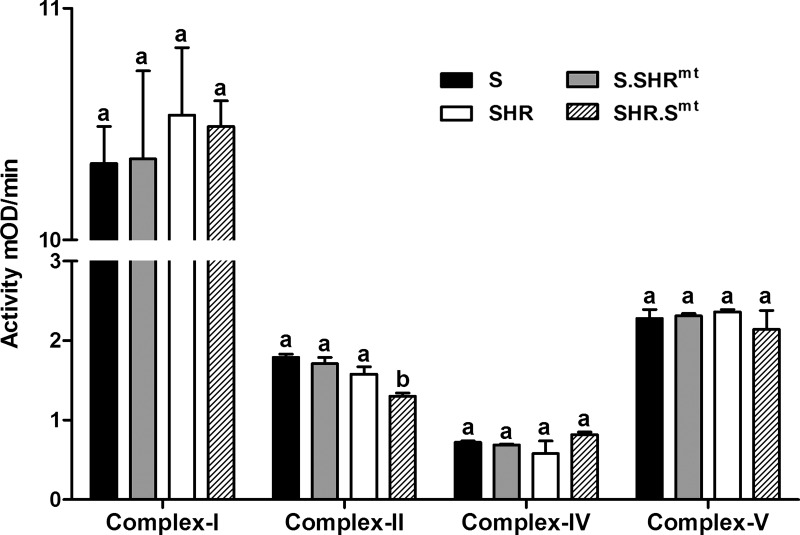
Analysis of enzyme activities of respiratory chain complexes revealed significantly lower activity of complex II in the SHR.Smt conplastic strain compared with the SHR (Fig. 6, P < 0.05). In contrast, we found no strain differences in the activities of complexes I, IV, and V between any of the strain comparisons (Fig. 6).
Fig. 6.
Mitochondrial respiratory chain activity. Mitochondrial complex activities were measured in isolated mitochondrial fractions according to manufacturer's instruction. Each bar represents mean ± SE; n = 4 animal samples/group. mOD, mitochondrial optical density. Bars labeled with the same letters were not significantly different from each other. Bars labeled with different letters were significantly different from each other, P < 0.05.
mtDNA copy number and respiratory enzyme levels.
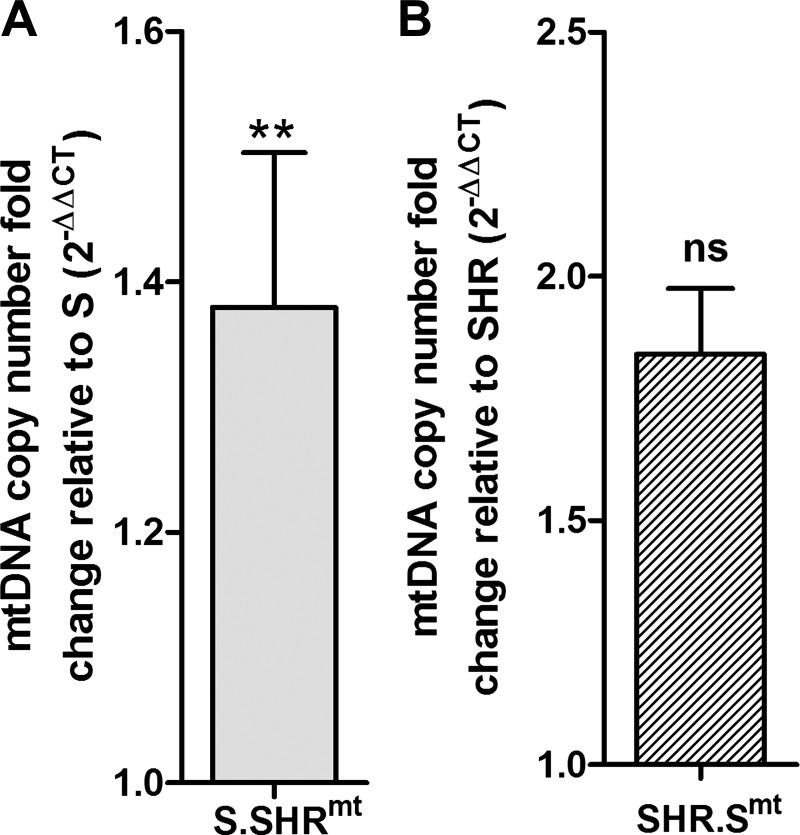
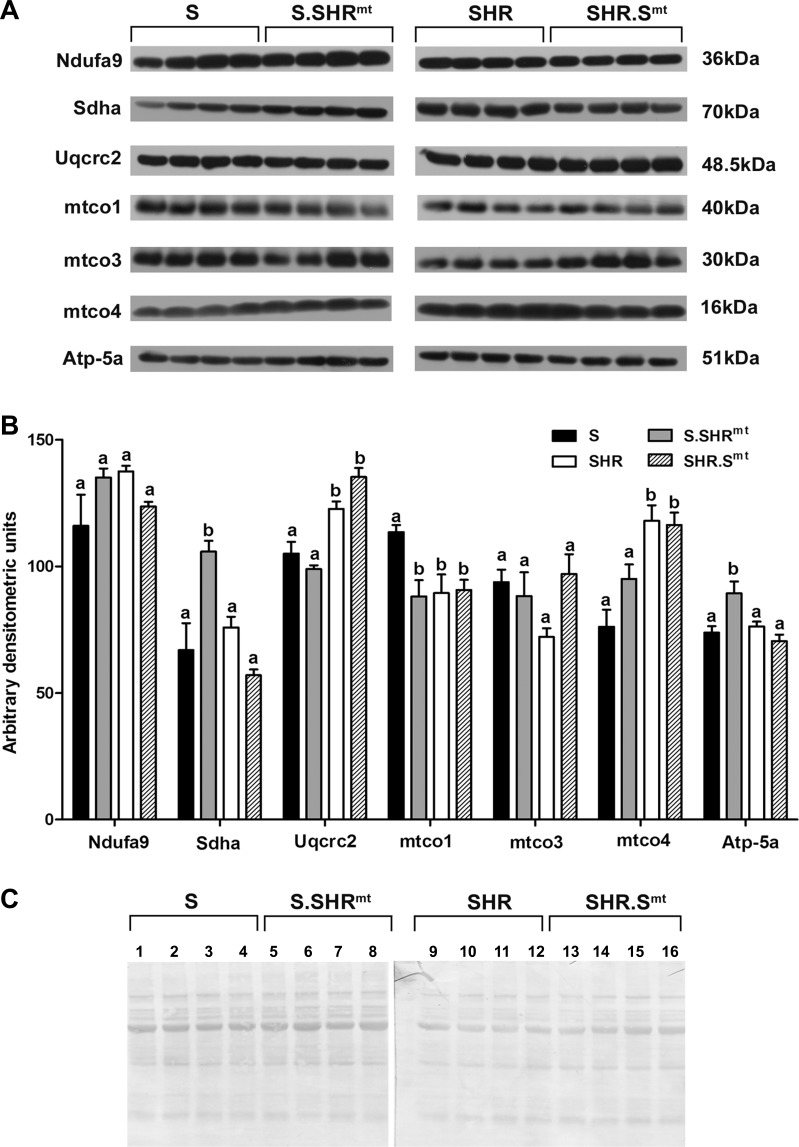
The mitochondrial genome from SHR rat, when introduced into the S rat, resulted in an increase in mtDNA copy number of the S.SHRmt rat (Fig. 7, P < 0.003). The mtDNA copy numbers of the SHR and SHR.Smt conplastic strain were not significantly different from each other (Fig. 7, P < 0.55). In addition, we quantitated several protein subunits involved in mitochondrial respiration. When compared with levels in the S rats, succinate dehydrogenase complex, subunit A (Sdha), and ATP synthase α-subunit gene (Atp-5a) were higher and mitochondrial cytochrome c oxidase 1 was lower in the S.SHRmt strain (Fig. 8). When compared with those of the SHR, none of the proteins were altered in the SHR.Smt rats (Fig. 8). Uncropped Western films were indistinguishable from each other between strains in the regions cropped in Fig. 8.
Fig. 7.
Analysis of mitochondrial copy number. Total DNA was extracted and mitochondrial copy number was measured by PCR amplification of the D-loop region vs. GAPDH as internal control. mtDNA, mitochondrial DNA. Values are means ± SE; n = 4/group of rats, 3 replicates/sample. **P < 0.003; ns, not significant.
Fig. 8.
Quantitative measurement of mitochondrial respiratory protein content in isolated mitochondria. Protein levels in mitochondrial fractions isolated from hearts of experimental animals were detected by immunoblotting as described under materials and methods. A: Western blots. Ndufa9, NADH:ubiquinone oxidoreductase; Sdha, succinate dehydrogenase; Uqcrc2, ubiquinol:ferricytochrome c oxidoreductase; mtco1, -3, and -4, mitochondrial cytochrome c oxidase 1, 2, and 3, respectively; Atp-5a, ATP synthase α-subunit gene. B: densitometric quantitation of the image presented in A. Data plotted are means ± SE; n = 4/group. Values for each protein that are labeled with the same letters were not significantly different from each other. Values plotted for each protein that are labeled with different letters were significantly different from each other, P < 0.05. C: ponceau S-stained blots.
Protein content of other nuclear-encoded master regulators of mitochondrial biogenesis.
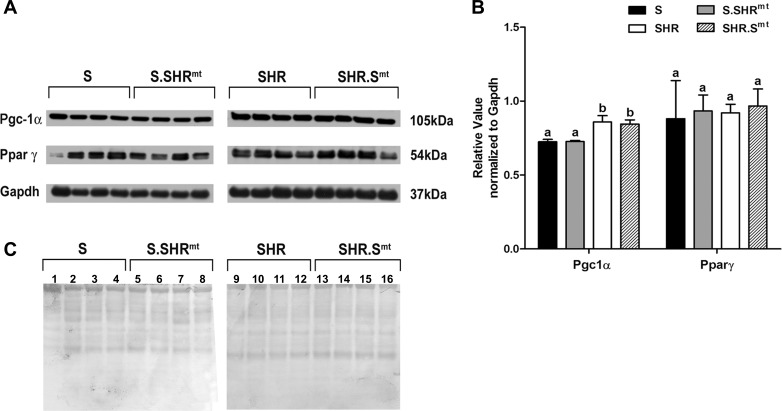
To ascertain that the observed differences between the conplastic and progenitor strains were not due to major changes in the protein content of known regulators of mitochondrial biogenesis, we compared the levels of Ppar-γ and its transcriptional coactivator Pgc-1α. Levels of Pgc-1α were significantly different between S and SHR (P < 0.021), but these differences were not transferred by reciprocal exchange of mitochondrial genomes between S and SHR (Fig. 9). Levels of Ppar-γ were not significantly different between any of the strain comparisons (Fig. 9). Uncropped Western films were indistinguishable from each other between strains in the regions cropped in Fig. 9.
Fig. 9.
Quantitation of major nuclear genome-encoded regulators of mitochondrial respiration. Heart homogenates were subjected to SDS-Tricine PAGE and probed with antibodies to rat proliferator-activated receptor coactivator-1α (Pgc-1α) and peroxisome proliferator-activated receptor-γ (Ppar-γ) and normalized with protein levels of Gapdh. A: Western blots. B: densitometric scans of the Western blot data. Data plotted are means ± SE; n = 4/group. Values plotted for each protein that are labeled with the same letters were not significantly different from each other. Values plotted for each protein that are labeled with different letters were significantly different from each other, P < 0.05. C: ponceau S-stained blots.
DISCUSSION
Although studies on mitochondrial function in health and disease have provided evidence to associate variants of the mitochondrial genome with pathological states, demonstrating cause and effect relationships that go beyond mere associations is made feasible by characterizing conplastic strains (15, 34, 38, 48, 49), which are derived through custom breeding strategies to swap maternally inherited mitochondrial genomes between strains. In the present studies, we found that conplastic rat strains of S and SHR, with virtually identical nuclear genomes but with divergent mitochondrial genomes, demonstrated alterations in cardiac mitochondrial copy numbers and ROS production that were associated with significant differences in ARC and longevity, but not BP.
The S and SHR mitochondrial genomes differ by 106 variations (24). Because the mitochondrial genome is inherited as a single unit, our study points to all of the 106 variants of the mitochondrial genomes of S and SHR, either individually or collectively as haplotypes, as candidate genetic determinants for aerobic capacity and longevity. Out of these 106 total variants, 25 variations are within genes coding for ribosomal or transfer RNA, four of which are within the functionally important D-loop region of mtDNA, with two of them located in the mitochondrial transcription factor-1 binding site (13). Because significant differences in copy numbers were observed between the inbred S and S.SHRmt conplastic strains, it is possible that the variants within the SHR mitochondrial genome transcribing the D-loop region may influence binding of mitochondrial transcription factor 1 and account for increased mitochondrial copy numbers in S.SHRmt. The lack of difference in copy numbers between SHR and SHR.Smt indicates that the nuclear genomic effects of SHR could override the influence of S rat mitochondrial genomic effects on copy numbers.
Besides the variations within the genes coding for various forms of RNA, 12 nonsynonymous variants exist within Nd2, Cox2, Atp6, Nd4, Nd6, and Cytb. Out of these, nine variants are within the genes coding for the complex I protein subunits of the electron transport chain, Nd2, Nd4, and Nd6 genes. Since there was no difference in complex I activity between S and S.SHRmt, it is reasonable to conclude that without affecting the efficiency of the respiratory chain complex, the substitutions occurring as a result of these variants may contribute to the observed reduced ROS production in the S.SHRmt conplastic compared with S by lowering the extent of electron leakage into the mitochondrial matrix. Complex II activity was, however, lower in the reciprocal SHR.Smt conplastic strain compared with S rats, but there were no statistically significant differences in ARC, BP, or longevity between these strains. This suggests that lower complex II activity of the S mitochondrial genome was, per se, insufficient to alter the extent of ARC, BP, and longevity of the SHR. These data point to a substantial nuclear genomic effect of the SHR that may not be permissive to accommodate the phenotypic alterations, if any, imparted by the lower complex II activity observed in the SHR.Smt compared with S rat.
In rat models that were selectively bred for divergent ARC, a clear dichotomy in emergence of risk for cardiovascular and metabolic diseases was previously observed (47). Recently, with the use of these rat models, intrinsic aerobic capacity was additionally demonstrated to set the divide for aging and longevity (20). Whether the mitochondrial genomes contributed to this divide is an important question that requires proof from conplastic strains. Our study, although with different inbred strains, provides this required evidence to suggest that variants of the mitochondrial genome, independent of the nuclear genome, contribute to both ARC and longevity but may not contribute significantly to cardiovascular risk by way of altering BP. Although our results strongly suggest that ARC is directly related to mitochondrial copy numbers and inversely related to mitochondrial ROS production, the focus of our work was limited to the heart, primarily because of our motivation to assess the function of an organ related to BP control. Given this limitation, our data lend support to the view that the genetic determinants of the emergence of risk for cardiovascular diseases may largely reside within the nuclear genome, whereas genetic determinants of aerobic capacity and longevity are shared between both nuclear and mitochondrial genomes. The conplastic strains developed in this study will serve as important tools to further dissect the potential nuclear and mitochondrial genomic interactions that govern both the genetic and epigenetic control of aerobic capacity and longevity.
GRANTS
This work was supported by grants from National Heart, Lung, and Blood Institute Grants HL-020176, HL-112641, and HL-076709 (to B. Joe).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
S.K., K.G., S.A.-M., R.P.-N., P.F., and B.J. performed experiments; S.K., K.G., and S.A.-M. analyzed data; S.K. and B.J. interpreted results of experiments; S.K. prepared figures; S.K. and B.J. drafted manuscript; S.K., K.G., S.A.-M., R.P.-N., P.F., and B.J. approved final version of manuscript; B.J. conception and design of research; B.J. edited and revised manuscript.
REFERENCES
- 1.Addabbo F, Montagnani M, Goligorsky MS. Mitochondria and reactive oxygen species. Hypertension 53: 885–892, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair SN, Kampert JB, Kohl HW, 3rd, Barlow CE, Macera CA, Paffenbarger RS, Jr, Gibbons LW. Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all-cause mortality in men and women. JAMA 276: 205–210, 1996 [PubMed] [Google Scholar]
- 3.Blair SN, Kohl HW, 3rd, Paffenbarger RS, Jr, Clark DG, Cooper KH, Gibbons LW. Physical fitness and all-cause mortality. A prospective study of healthy men and women. JAMA 262: 2395–2401, 1989 [DOI] [PubMed] [Google Scholar]
- 4.Church TS, Cheng YJ, Earnest CP, Barlow CE, Gibbons LW, Priest EL, Blair SN. Exercise capacity and body composition as predictors of mortality among men with diabetes. Diabetes Care 27: 83–88, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet 7: 829–840, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Feng D, Yang C, Geurts AM, Kurth T, Liang M, Lazar J, Mattson DL, O'Connor PM, Cowley AW., Jr Increased expression of NAD(P)H oxidase subunit p67(phox) in the renal medulla contributes to excess oxidative stress and salt-sensitive hypertension. Cell Metab 15: 201–208, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez-Vizarra E, Lopez-Perez MJ, Enriquez JA. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods 26: 292–297, 2002 [DOI] [PubMed] [Google Scholar]
- 8.Galindo MF, Jordan J, Gonzalez-Garcia C, Cena V. Chromaffin cell death induced by 6-hydroxydopamine is independent of mitochondrial swelling and caspase activation. J Neurochem 84: 1066–1073, 2003 [DOI] [PubMed] [Google Scholar]
- 9.Garrett MR, Saad Y, Dene H, Rapp JP. Blood pressure QTL that differentiate Dahl salt-sensitive and spontaneously hypertensive rats. Physiol Genomics 3: 33–38, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Gopalakrishnan K, Morgan EE, Yerga-Woolwine S, Farms P, Kumarasamy S, Kalinoski A, Liu X, Wu J, Liu L, Joe B. Augmented rififylin is a risk factor linked to aberrant cardiomyocyte function, short-QT interval and hypertension. Hypertension 57: 764–771, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grande MT, Pascual G, Riolobos AS, Clemente-Lorenzo M, Bardaji B, Barreiro L, Tornavaca O, Meseguer A, Lopez-Novoa JM. Increased oxidative stress, the renin-angiotensin system, and sympathetic overactivation induce hypertension in kidney androgen-regulated protein transgenic mice. Free Radic Biol Med 51: 1831–1841, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Gulati M, Pandey DK, Arnsdorf MF, Lauderdale DS, Thisted RA, Wicklund RH, Al-Hani AJ, Black HR. Exercise capacity and the risk of death in women: the St. James Women Take Heart Project. Circulation 108: 1554–1559, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Han CB, Li F, Zhao YJ, Ma JM, Wu DY, Zhang YK, Xin Y. Variations of mitochondrial D-loop region plus downstream gene 1 2S rRNA-tRNA(phe) and gastric carcinomas. World J Gastroenterol 9: 1925–1929, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson KK, Wagner H, Favret F, Britton SL, Koch LG, Wagner PD, Gonzalez NC. Determinants of maximal O2 uptake in rats selectively bred for endurance running capacity. J Appl Physiol 93: 1265–1274, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Houstek J, Hejzlarova K, Vrbacky M, Drahota Z, Landa V, Zidek V, Mlejnek P, Simakova M, Silhavy J, Miksik I, Kazdova L, Oliyarnyk O, Kurtz T, Pravenec M. Nonsynonymous variants in mt-Nd2, mt-Nd4, and mt-Nd5 are linked to effects on oxidative phosphorylation and insulin sensitivity in rat conplastic strains. Physiol Genomics 44: 487–494, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joe B, Garrett MR. Genetic analysis of inherited hypertension in the rat. In: Genetics of Hypertension , edited by Dominiczak A, Amerstam CJ. The Netherlands: Elsevier Science, 2006, p. 177–200 [Google Scholar]
- 17.Joe B, Garrett MR. Substitution mapping: using congenic strains to detect genes controlling blood pressure. In: Cardiovascular Genomics, edited by Raizada MK, Kasparov S, Katovich MJ. Totowa, NJ: Humana, 2005, p. 41–58 [Google Scholar]
- 18.Joe B, Saad Y, Dhindaw S, Lee NH, Frank BC, Achinike OH, Luu TV, Gopalakrishnan K, Toland EJ, Farms P, Yerga-Woolwine S, Manickavasagam E, Rapp JP, Garrett MR, Coe D, Apte SS, Rankinen T, Perusse L, Ehret GB, Ganesh SK, Cooper RS, O'Connor A, Rice T, Weder AB, Chakravarti A, Rao DC, Bouchard C. Positional identification of variants of Adamts16 linked to inherited hypertension. Hum Mol Genet 18: 2825–2838, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JD, McCarter RJ, Yu BP. Influence of age, exercise, and dietary restriction on oxidative stress in rats. Aging (Milano) 8: 123–129, 1996 [DOI] [PubMed] [Google Scholar]
- 20.Koch LG, Kemi OJ, Qi N, Leng SX, Bijma P, Gilligan LJ, Wilkinson JE, Wisloff H, Hoydal MA, Rolim N, Abadir PM, van Grevenhof EM, Smith GL, Burant CF, Ellingsen O, Britton SL, Wisloff U. Intrinsic aerobic capacity sets a divide for aging and longevity. Circ Res 109: 1162–1172, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokaze A, Ishikawa M, Matsunaga N, Yoshida M, Sekine Y, Sekiguchi K, Harada M, Satoh M, Teruya K, Takeda N, Fukazawa S, Uchida Y, Takashima Y. Longevity-associated mitochondrial DNA 5178 A/C polymorphism and blood pressure in the Japanese population. J Hum Hypertens 18: 41–45, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Kokkinos P, Myers J, Kokkinos JP, Pittaras A, Narayan P, Manolis A, Karasik P, Greenberg M, Papademetriou V, Singh S. Exercise capacity and mortality in black and white men. Circulation 117: 614–622, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Kristal BS, Staats PN, Shestopalov AI. Biochemical characterization of the mitochondrial permeability transition in isolated forebrain mitochondria. Dev Neurosci 22: 376–383, 2000 [DOI] [PubMed] [Google Scholar]
- 24.Kumarasamy S, Gopalakrishnan K, Shafton A, Nixon J, Thangavel J, Farms P, Joe B. Mitochondrial polymorphisms in rat genetic models of hypertension. Mamm Genome 21: 299–306, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumarasamy S, Gopalakrishnan K, Toland EJ, Yerga-Woolwine S, Farms P, Morgan EE, Joe B. Refined mapping of blood pressure quantitative trait loci using congenic strains developed from two genetically hypertensive rat models. Hypertens Res 34: 1263–1270, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakka TA, Venalainen JM, Rauramaa R, Salonen R, Tuomilehto J, Salonen JT. Relation of leisure-time physical activity and cardiorespiratory fitness to the risk of acute myocardial infarction. N Engl J Med 330: 1549–1554, 1994 [DOI] [PubMed] [Google Scholar]
- 27.LeBel CP, Ischiropoulos H, Bondy SC. Evaluation of the probe 2′,7′-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chem Res Toxicol 5: 227–231, 1992 [DOI] [PubMed] [Google Scholar]
- 28.Li R, Liu Y, Li Z, Yang L, Wang S, Guan MX. Failures in mitochondrial tRNAMet and tRNAGln metabolism caused by the novel 4401A>G mutation are involved in essential hypertension in a Han Chinese Family. Hypertension 54: 329–337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Li R, Li Z, Wang XJ, Yang L, Wang S, Guan MX. Mitochondrial transfer RNAMet 4435A>G mutation is associated with maternally inherited hypertension in a Chinese pedigree. Hypertension 53: 1083–1090, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino A, Skelton MM, Zou AP, Roman RJ, Cowley AW., Jr Increased renal medullary oxidative stress produces hypertension. Hypertension 39: 667–672, 2002 [DOI] [PubMed] [Google Scholar]
- 31.Matsunaga H, Tanaka Y, Tanaka M, Gong JS, Zhang J, Nomiyama T, Ogawa O, Ogihara T, Yamada Y, Yagi K, Kawamori R. Antiatherogenic mitochondrial genotype in patients with type 2 diabetes. Diabetes Care 24: 500–503, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Myers J, Prakash M, Froelicher V, Do D, Partington S, Atwood JE. Exercise capacity and mortality among men referred for exercise testing. N Engl J Med 346: 793–801, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Poyan Mehr A, Siegel AK, Kossmehl P, Schulz A, Plehm R, de Bruijn JA, de Heer E, Kreutz R. Early onset albuminuria in Dahl rats is a polygenetic trait that is independent from salt loading. Physiol Genomics 14: 209–216, 2003 [DOI] [PubMed] [Google Scholar]
- 34.Pravenec M, Hyakukoku M, Houstek J, Zidek V, Landa V, Mlejnek P, Miksik I, Dudova-Mothejzikova K, Pecina P, Vrbacky M, Drahota Z, Vojtiskova A, Mracek T, Kazdova L, Oliyarnyk O, Wang J, Ho C, Qi N, Sugimoto K, Kurtz T. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic strains. Genome Res 17: 1319–1326, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapp JP. Genetic analysis of inherited hypertension in the rat. Physiol Rev 80: 135–172, 2000 [DOI] [PubMed] [Google Scholar]
- 36.Regner KR, Harmon AC, Williams JM, Stelloh C, Johnson AC, Kyle PB, Lerch-Gaggl A, White SM, Garrett MR. Increased susceptibility to kidney injury by transfer of genomic segment from SHR onto Dahl S genetic background. Physiol Genomics 44: 629–637, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saad Y, Garrett MR, Manickavasagam E, Yerga-Woolwine S, Farms P, Radecki T, Joe B. Fine-mapping and comprehensive transcript analysis reveals nonsynonymous variants within a novel 1.17 Mb blood pressure QTL region on rat chromosome 10. Genomics 89: 343–353, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sethumadhavan S, Vasquez-Vivar J, Migrino RQ, Harmann L, Jacob HJ, Lazar J. Mitochondrial DNA variant for complex I reveals a role in diabetic cardiac remodeling. J Biol Chem 287: 22174–22182, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel AK, Kossmehl P, Planert M, Schulz A, Wehland M, Stoll M, Bruijn JA, de Heer E, Kreutz R. Genetic linkage of albuminuria and renal injury in Dahl salt-sensitive rats on a high-salt diet: comparison with spontaneously hypertensive rats. Physiol Genomics 18: 218–225, 2004 [DOI] [PubMed] [Google Scholar]
- 40.St.-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem 277: 44784–44790, 2002 [DOI] [PubMed] [Google Scholar]
- 41.Swei A, Lacy F, Delano FA, Parks DA, Schmid-Schonbein GW. A mechanism of oxygen free radical production in the Dahl hypertensive rat. Microcirculation 6: 179–187, 1999 [PubMed] [Google Scholar]
- 42.Takagi K, Yamada Y, Gong JS, Sone T, Yokota M, Tanaka M. Association of a 5178C→A (Leu237Met) polymorphism in the mitochondrial DNA with a low prevalence of myocardial infarction in Japanese individuals. Atherosclerosis 175: 281–286, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Touyz RM. Reactive oxygen species, vascular oxidative stress, and redox signaling in hypertension: what is the clinical significance? Hypertension 44: 248–252, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Ways JA, Cicila GT, Garrett MR, Koch LG. A genome scan for Loci associated with aerobic running capacity in rats. Genomics 80: 13–20, 2002 [DOI] [PubMed] [Google Scholar]
- 45.Ways JA, Smith BM, Barbato JC, Ramdath RS, Pettee KM, DeRaedt SJ, Allison DC, Koch LG, Lee SJ, Cicila GT. Congenic strains confirm aerobic running capacity quantitative trait loci on rat chromosome 16 and identify possible intermediate phenotypes. Physiol Genomics 29: 91–97, 2007 [DOI] [PubMed] [Google Scholar]
- 46.Wendt N, Schulz A, Qadri F, Bolbrinker J, Kossmehl P, Winkler K, Stoll M, Vetter R, Kreutz R. Genetic analysis of salt-sensitive hypertension in Dahl rats reveals a link between cardiac fibrosis and high cholesterol. Cardiovasc Res 81: 618–626, 2009 [DOI] [PubMed] [Google Scholar]
- 47.Wisloff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernstrom M, Rezaei K, Lee SJ, Koch LG, Britton SL. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science 307: 418–420, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Gimsa U, Wester-Rosenlof L, Kanitz E, Otten W, Kunz M, Ibrahim SM. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res 19: 159–165, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X, Wester-Rosenlof L, Gimsa U, Holzhueter SA, Marques A, Jonas L, Hagenow K, Kunz M, Nizze H, Tiedge M, Holmdahl R, Ibrahim SM. The mtDNA nt7778 G/T polymorphism affects autoimmune diseases and reproductive performance in the mouse. Hum Mol Genet 18: 4689–4698, 2009 [DOI] [PubMed] [Google Scholar]