Abstract
The Zn, Co, and Mn K-edge extended X-ray absorption fine structure (EXAFS) spectra of the N-acetyl-L-ornithine deacetylase (ArgE) from Escherichia coli, loaded with one or two equivalents of divalent metal ions (i.e., [Zn(II)_(ArgE)], [Zn(II)Zn(II)(ArgE)], [Co(II)_(ArgE)], [Co(II)Co(II)(ArgE)], [Mn(II)_(ArgE)], and [Mn(II)Mn(II)(ArgE)]), were recorded. The Fourier transformed data (FT) for [Zn(II)_(ArgE)], [Zn(II)Zn(II)(ArgE)], [Co(II)_(ArgE)] and [Co(II)Co(II)(ArgE)] are dominated by a peak at 2.05 Å, that can be fit assuming five or six light atom (N,O) scatterers. Inclusion of multiple-scattering contributions from the outer-shell atoms of a histidine-imidazole ring resulted in reasonable Debye-Waller factors for these contributions and a slight reduction in the goodness-of-fit value (f′). Furthermore, the data best fit a model that included a M-M vector at 3.3 and 3.4 Å for Zn(II) and Co(II), respectively, suggesting the formation of a dinuclear site. Multiple scattering contributions from the outer-shell atoms of a histidine-imidazole rings are observed at ~3 and 4 Å for Zn(II)- and Co(II)-loaded ArgE suggesting at least one histidine ligand at each metal binding site. Likewise, EXAFS data for Mn(II)-loaded ArgE are dominated by a peak at 2.19 Å that was best fit assuming six light atom (N,O) scatterers. Due to poor signal to noise ratios for the Mn EXAFS spectra, no Mn-Mn vector could be modeled. Peak intensities for [M(II)_(ArgE)] vs. [M(II)M(II)(ArgE)] suggest the Zn(II), Co(II), and Mn(II) bind to ArgE in a cooperative manner. Since no structural data has been reported for any ArgE enzyme, the EXAFS data reported herein represent the first glimpse for ArgE enzymes. These data also provide a structural foundation for the future design of small molecules that function as inhibitors of ArgE and may potentially function as a new class of antibiotics.
The emergence of antibiotic resistant bacterial infections has created a significant and growing medical problem in the United States and throughout the world [1–5]. Antibiotic resistance has been recognized since the introduction of penicillin more than 50 years ago when penicillin-resistant infections caused by Staphylococcus aureus rapidly appeared [3, 6]. Because bacteria have been exposed to many of the currently available antibiotics for years, they have evolved resistance to these drugs [3, 7–10]. In fact, several bacterial infections, some of which were thought to have been eradicated, have made a significant resurgence due to bacterial resistance to antibiotics. While overall per capita mortality rates for infectious diseases declined in the United States from 1980 to 1992, the per capita mortality rate associated with antibiotic resistant bacterial infections increased 58% over this period, making these infectious illnesses the third leading cause of death in the United States [11, 12]. These trends continue and underscore the fact that a significant and growing percentage of infectious disease is caused by bacteria. In fact, according to the CDC several bacterial strains currently exhibit multidrug resistance, including: staphylococci, enterococci, gonococci, streptococci, salmonella, methicillin-resistant Staphylococcus aureus (MRSA), and Mycobacterium tuberculosis. With the resurgence of antibiotic resistant bacterial strains, the search for new antibiotics that target hereto unexplored enzymes involved in metabolic processes exclusive to bacteria is critically important in order to renew our arsenal of antibiotics [13].
Currently available antibiotics work on relatively few targets through mechanisms such as inhibiting protein or cell wall synthesis [14–17]. Therefore, there is an urgent need to identify novel antibacterial targets and discover small molecule inhibitors that can function as antimicrobial agents [18]. Based on bacterial genetic information, the arginine biosynthetic pathway offers several potential antibacterial enzyme targets, some of which have yet to be explored [19–21]. Prokaryotes synthesize arginine through a series of eight enzymatically-catalyzed reactions that differ from those of eukaryotes by two key steps (Figure 1): i) acetylation of glutamate (Step 1) and ii) the subsequent deacetylation of the arginine precursor Nα-acetyl-L-ornithine (L-NAO) by the argE-encoded Nα-acetyl-L-ornithine deacetylase (ArgE) (Step 5) [22–24]. The arginine biosynthetic pathway is found in all Gram-negative and most Gram-positive bacteria including but not limited to Enterobacteriaceae, Haemophilus, Vibrionaceae, Corynebacterium, Campylobacter, Yersinia pestis, Vibrio Cholerae, Bacillus anthracis, E. coli 0157:H7, Shigella flexneri, S. typhimurium, and Neisseria gonorrhoeae [21, 22, 25–30]. Because ornithine is required, not only for the synthesis of arginine in bacteria, but also for polyamines involved in DNA replication and cell division, the L-NAO deacetylation step is critical for bacterial proliferation [31]. Indeed, when Meinnel and coworkers transformed an arginine auxotrophic bacterial strain void of L-NAO deacetylase activity with a plasmid containing argE, an Arg+ phenotype resulted [32]. However, when the start codon (ATG) of argE in the same plasmid was changed to the Amber codon (TAG), the resultant plasmid was unable to relieve arginine auxotrophy in the same cell strain. Therefore, ArgE is required for cell viability. Given the fact that ArgE is only found in prokaryotes and is required for bacterial cell growth and proliferation, it represents an enzymatic target for the development of a new class of antimicrobial agents [31].
Figure 1.
Arginine biosynthetic pathway for prokaryotes.
All ArgE enzymes characterized to date require Zn(II) ions for enzymatic activity [20, 21, 33] and the amino acid residues that function as metal ligands in the carboxypeptidase from Pseudomonas sp strain-RS-16 (CPG2), the aminopeptidase from Aeromonas proteolytica (AAP), the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase from Haemophilus influenzae (DapE)[34], and the N-acetyl-L-citrulline deacetylase (ACD) from Xanthomonas campestris [35–37] are strictly conserved in ArgE enzymes [20]. CPG2, DapE, and AAP possess a (μ-aquo)(μ-carboxylato)dizinc(II) core with one terminal carboxylate and one histidine residue at each metal site suggesting a similar active site exists in ArgE enzymes. The Zn-Zn distance is 3.4, 3.5, and 3.3 Å for DapE, AAP, and CPG2, respectively, with each metal ion residing in a distorted tetrahedral coordination geometry. Recently, kinetic and spectroscopic evidence suggested that the ArgE from E. coli also contains a dinuclear metalloactive site [33]; however, no structural information regarding an ArgE enzyme has yet to be reported. In order to provide evidence for a (μ-aquo)(μ-carboxylato)dizinc(II) core in ArgE enzyme, we have obtained K-edge extended X-ray absorption fine structure (EXAFS) spectroscopic data on Zn(II), Co(II), and Mn(II)-loaded ArgE from E. coli. Since EXAFS is sensitive to heavy atom scatterers in the second coordination sphere, it is an effective tool for identifying dinuclear metal sites.
Materials and Methods
Enzyme Expression and Purification
All chemicals used in this study were purchased from commercial sources and were of the highest quality available. ArgE from E. coli was purified as previously described from a stock culture of E. coli BL21 Star™ cells (Invitrogen) hosting the pET-27a(+)argE over-expression construct provided by Professor John S. Blanchard [20]. The purified enzyme manifested a single band, on a 12% SDS gel, which corresponds to its calculated Mr of 42,350, by comparison with molecular weight standards purchased from Sigma. It was subsequently concentrated to >1 mM and stored at 4 °C. Protein concentration was determined using the theoretical value ε280 = 41,250 M−1 cm−1 [38]. Apo-ArgE samples were exchanged into 50 mM Chelex-100 treated HEPES buffer at pH 7.5 and these samples were incubated anaerobically with MCl2 (99.999% Strem Chemicals) where M = Zn(II), Co(II) or Mn(II) for ~20 minutes as previously reported [33].
Enzymatic assay of ArgE
Enzyme kinetic constants were determined in 50 mM Chelex-100 treated sodium phosphate buffer pH 7.5 with L-NAO as the substrate at 25 °C. The rate of NAO deacetylation was monitored as a decrease in absorbance at 214 nm corresponding to the peptide bond (Δε214 = 103 M−1cm−1). Catalytic activities were determined within ±10%. The specific activity of purified ArgE with L-NAO was typically found to be 2,000 units per mg of enzyme, where one unit was defined as the amount of enzyme that releases 1 μmole of ornithine at 25 °C in 60 sec. Initial rates were fit directly to the Michaelis-Menten equation to obtain the catalytic constants Km and kcat.
X-ray Absorption Spectroscopy
XAS data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) with beam current between 80 and 100 mA at 3.0 GeV (Table 1). ArgE EXAFS samples (1 mM), loaded in polycarbonate cuvettes, 24 × 2 mm2 with one 24 × 2 mm2 face consisting of X-ray transparent Mylar tape, were flash-frozen in N2(l) and maintained at 10 K during data collection, using a liquid helium-flow cryostat. XAS fluorescence signals were collected with a 30-element intrinsic GE solid-state detector and Z-1 fluorescence filters with Soller slits. The edge regions for multiple scans obtained on the same sample were compared to ensure that the sample was not damaged by exposure to X-ray radiation. Harmonic rejection was achieved by setting the cutoff energy of the focusing mirror to an energy between the fundamental and second harmonic (10 keV for Mn, 12 KeV for Co and 13 KeV for Zn). The energy was calibrated to the inflection points of the edge of the standard Zn and Co foil, which were assumed to be at 9660.7, 7709.5, and 6539.0 eV for Zn, Co and Mn, respectively.
Table 1.
X-ray absorption data collection.
| Co EXAFS | Zn EXAFS | Mn EXAFS | |
|---|---|---|---|
| SR facility | SSRL | SSRL | SSRL |
| beamline | 9–3 | 9–3 | 9–3 |
| current in storage ring | 80–100 mA | 80–100 mA | 80–100 mA |
| monochromator crystal | Si[220] | Si[220] | Si[220] |
| detection method | fluorescence | fluorescence | fluorescence |
| detector type | solid state arraya | solid state arraya | solid state arraya |
| scan length, min | 15 | 15 | 15 |
| scans in average | 4 | 4 | 4 to 7 |
| temperature, K | 10 | 10 | 10 |
| energy standard | Co foil, 1st inflection | Zn foil, 1st inflection | Mn foil, 1st inflection |
| energy calibration, eV | 7709.5 | 9660.7 | 6539.0 |
| E0, eV | 7715 | 9670 | 6545 |
| pre-edge background | |||
| energy range, eV | 0–7670 | 0–9625 | 0–6500 |
| Gaussian center, eV | 6930 | 8638 | 5898 |
| Gaussian width, eV | 750 | 750 | 750 |
| spline background | |||
| energy range, eV | 7715.0–7947.47 (4) | 9670.0–9898.63 (4) | 6545.00–6778.03 (4) |
| (polynomial order) | 7947.47–8179.95 (4) | 9898.63–10127.3 (4) | 6778.03 –7011.06 (4) |
| 8179.95 –8412.42 (4) | 10127.3–10355.9 (4) | 7011.06 –7244.09 (4) | |
The 30-element Ge solid-state X-ray fluorescence detector at SSRL was provided by the NIH Biotechnology Research Resource.
XAS data reduction was carried out to extract the EXAFS using EXAFSPAK software [39] and the theoretical phase and amplitude functions were calculated with FEFF8.0 [40]. The structural model for FEFF calculation was generated based on the (μ-aquo)(μ-carboxylato)dizinc(II) site in the high-resolution crystal crystallographic data for AAP [36]. The Co model for calculation was based on this Zn-Zn model with Co-O distances around 2.05 Å. Because optimization of distances and Debye-Waller factors for all shells is impossible with such a complicated model, simulations relied on calculated Debye-Waller factors, which required determination of force constants for all bond-atom pairs. The recursion method incorporated into FEFF was subsequently employed to calculate Debye-Waller factors from force constants. All force constants were calculated via the UFF force field [41], except those for Zn-Nimid, Co-Nimid, and Mn-Nimid as well as C-C and C-N bonds within imidazole rings. These required DFT calculations based on vibrational frequencies [42, 43]. For metal-ligand force constant calculations, the values calculated by UFF were halved, as the original values are known to be too large [44]. Multiple scattering analysis was performed as previously described [45]. Both single- and multiple-scattering paths ≤ 4.5 Å from either the Mn(II), Zn(II) or Co(II) atoms were used to identify and quantify imidazole coordination due to histidine. With calculated Debye-Waller factors and the amplitude and phase function of the absorbing and scattering atom, the EXAFS were simulated by allowing only Eo optimization by EXAFSPAK. The structural model was then manually tuned to optimize the comparison of simulated and observed EXAFS.
Results
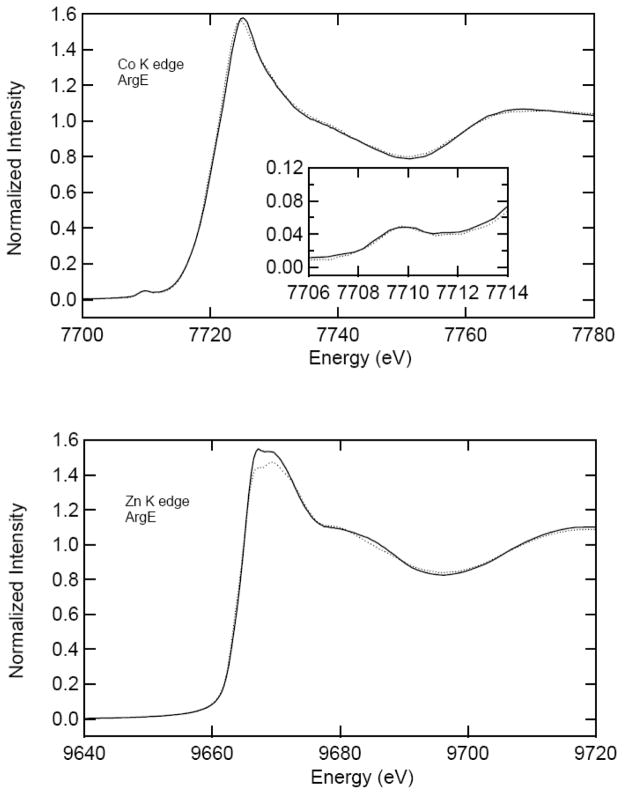
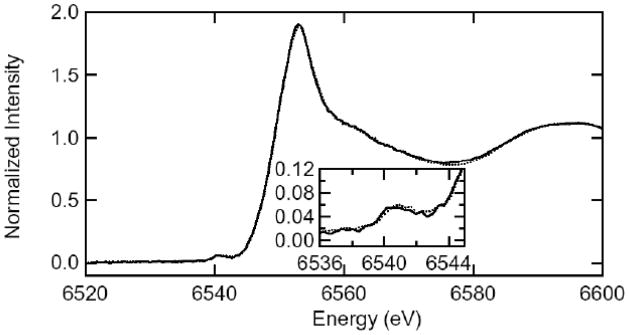
Zinc, cobalt and manganese K-edge X-ray absorption spectra (XAS) of 1 mM samples of ArgE with one or two equivalents of added Co(II) (i.e. [Co(II)_(ArgE)] and [Co(II)Co(II)(ArgE)]), Zn(II) (i.e. [Zn(II)_(ArgE)] and [Zn(II)Zn(II)(ArgE)]) or Mn(II) (i.e. [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)]) are shown in Figures 2 and 3. For fully loaded samples (e.g. [Co(II)Co(II)(ArgE)], [Zn(II)Zn(II)(ArgE)], or [Mn(II)Mn(II)(ArgE)]), the EXAFS data reveal an average of both metal ion environments. For [Co(II)_(ArgE)] and [Co(II)Co(II)(ArgE)], the spectra reveal 1s→3d pre-edge transitions which occur at 7,709 eV with a normalized peak intensity of 0.048 for both (Figure 2, inset). For both [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)], the 1s→3d pre-edge transitions reside in nearly the identical position and exhibit identical shapes but differ in amplitude (Figure 3, inset). The pre-edge feature (6,541 eV) is weak, indicating a high coordination number and symmetric geometry like the Co edge. Since 1s→3d pre-edge transitions are Laporte-forbidden in centrosymmetric environments (e.g., octahedral, but not tetrahedral), the intensity of the 1s→3d pre-edge transitions is inversely proportional to coordination number (assuming tetrahedral four-coordination). The intensities of the observed transitions for both [Co(II)_(ArgE)] and [Co(II)Co(II)(ArgE)] and [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)] are consistent with, on average, five- or six-coordinate divalent metal sites [46, 47].
Figure 2.
X-ray absorption K-edge spectra for [Co(II)_(ArgE)] (solid) and [Co(II)Co(II)(ArgE)] (dotted) (top) and [Zn(II)_(ArgE)] (solid) and [Zn(II)Zn(II)(ArgE)] (dotted) (bottom). The inset in the top panel highlights the preedge 1s → 3d transitions for [Co(II)_(ArgE)] (solid) and [Co(II)Co(II)(ArgE)] (dotted).
Figure 3.
X-ray absorption K-edge spectra for [Mn(II)_(ArgE)] (solid) and [Mn(II)Mn(II)(ArgE)] (dotted). The inset highlights the preedge 1s→3d transitions for [Mn(II)_(ArgE)] (solid) and [Mn(II)Mn(II)(ArgE)] (dotted).
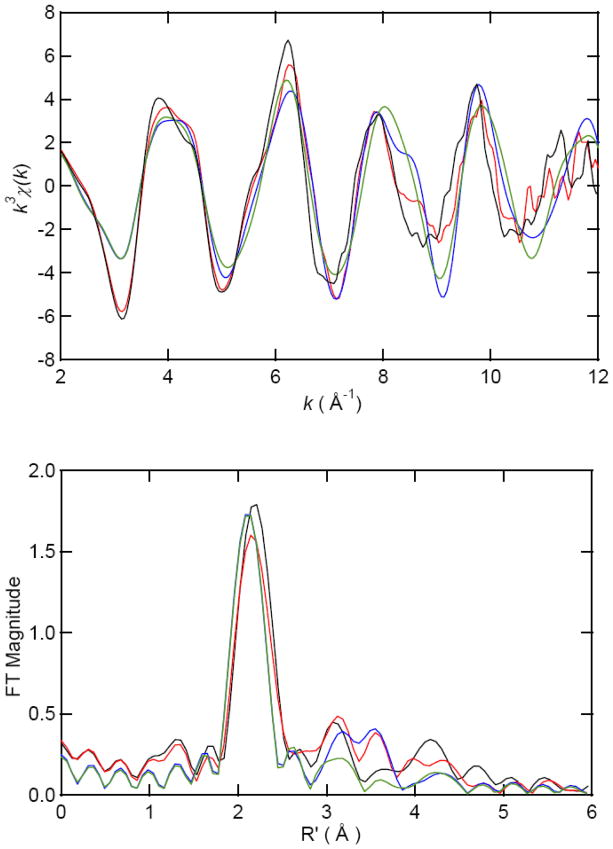
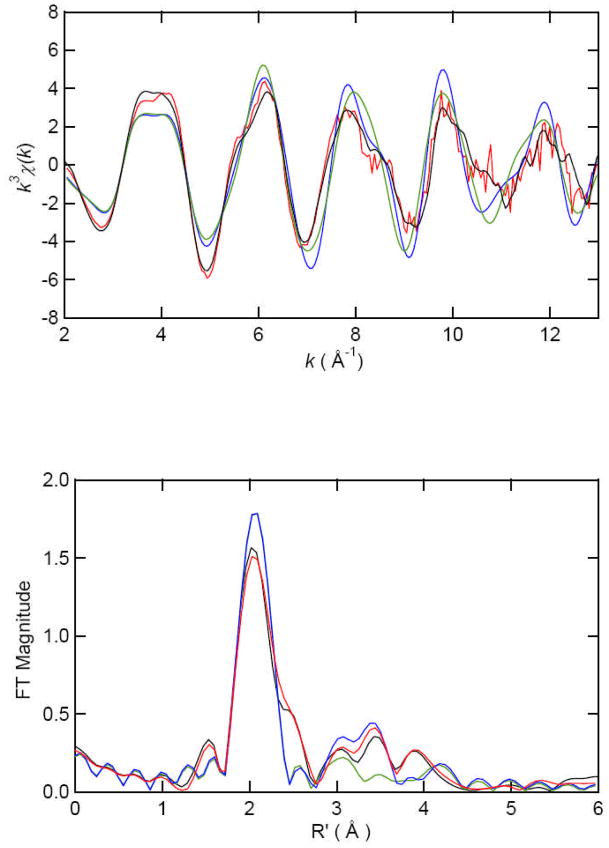
The dominant peak in the Fourier transformed (FT) EXAFS data for [Co(II)_(ArgE)] is centered at 2.11 Å and at 2.10 Å for [Co(II)Co(II)(ArgE)] with amplitude values of 1.78 and 1.61 respectively (Figure 4; Table 2). Smaller peaks were also noted at 3.1 Å, 3.4 Å and 4.1 Å with amplitude values of 0.47, 0.14 and 0.32, respectively, for [Co(II)_(ArgE)]. Additional peaks were also observed in the FT EXAFS data for [Co(II)Co(II)(ArgE)] at 3.2, 3.4 and 4.1 Å with amplitude values of 0.49, 0.38 and 0.22, respectively. Excellent single-shell fits of EXAFS spectra for both [Co(II)_(ArgE)] and [Co(II)Co(II)(ArgE)] were obtained with 5 ± 1 N/O scatterers at 2.05 Å. A second shell feature at ca. 3.4 Å appears in the [Co(II)Co(II)(ArgE)] EXAFS spectrum. Fits incorporating a Co-Co interaction at 3.4 Å for [Co(II)Co(II)(ArgE)] exhibit reasonable Debye-Waller factors (Table 2) and resulted in a significant improvement in goodness-of-fit values (f′). Failure to incorporate a Co-Co vector in the curve-fitting parameters resulted in a large disagreement between the data and simulation. Therefore, the second-shell FT feature observed at 3.4 Å is consistent with the formation of a dinuclear Co(II) active site in ArgE.
Figure 4.
k3-weighted Co EXAFS (top) and Fourier transforms (bottom, over k = 2–12 Å−1) for [Co(II)_(ArgE)] experimental (black) and simulation (green) and for [Co(II)Co(II)(ArgE)] experimental (red) and simulation (blue).
Table 2.
Parameters for simulation of Co and Zn ArgE EXAFSa
| Shell | Ras (Å) | σas2 (Å2) | ΔE0 (eV) | f′b | |
|---|---|---|---|---|---|
| CoCo(II)_ArgE | Co-NHis | 2.04 | 0.0028 | 10.5 | 0.100 |
| CRC0B, 2–12 Å−1 | Co-Owater | 2.05 | 0.0033 | ||
| Δk3χ = 11.37 | Co-Obridged carboxylate | 2.04 | 0.0026 | ||
| Co-Ocarboxylate | 2.04 | 0.0033 | |||
| Co- Ocarboxylate | 2.32 | 0.0035 | |||
| Co-Co | 3.37 | 0.0042 | |||
| ZnZn_ArgE | Zn-NHis | 2.01 | 0.0030 | 8.4 | 0.136 |
| ZRZ0A, 2–12 Å−1 | Zn-Owater | 2.00 | 0.0027 | ||
| Δk3χ = 10.51 | Zn-Obridged carboxylate | 2.05 | 0.0036 | ||
| Zn-Ocarboxylate | 1.99 | 0.0036 | |||
| Zn-Ocarboxylate | 2.32 | 0.0039 | |||
| Zn-Zn | 3.28 | 0.0044 |
Here a small part of paths used in simulation are listed. The simulation of Co ArgE includes 54 single and multiple scattering paths while 57 scattering paths in Zn ArgE. Ras is the metal-scatterer distance from corresponding molecular model. σas2 is a mean square deviation in Ras, calculated using UFF model. ΔE0 is the shift in E0 for the theoretical scattering functions.
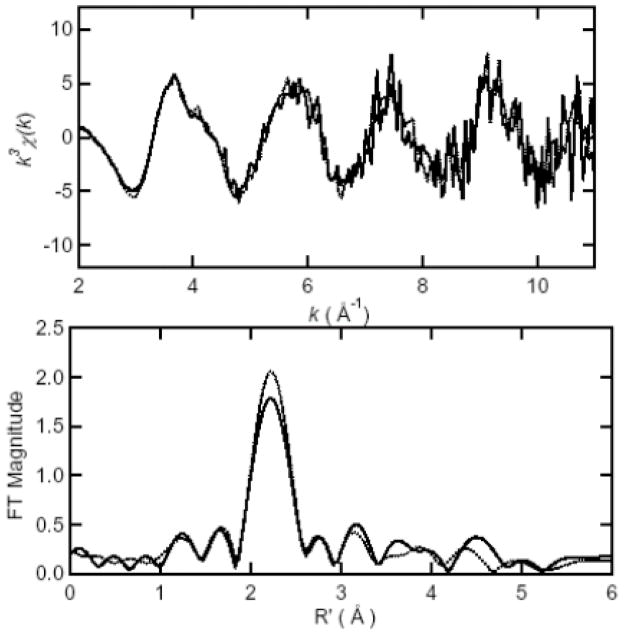
Similar to [Co(II)_(ArgE)] and [Co(II)Co(II)(ArgE)], the FT EXAFS spectra of [Zn(II)_(ArgE)] and [Zn(II)Zn(II)(ArgE)] are dominated by a peak at 2.05 Å (Figure 5; Table 2). However, the amplitude values for this peak are quite similar for [Zn(II)_(ArgE)] and [Zn(II)Zn(II)(ArgE)] (1.56 and 1.50 respectively), suggesting that Zn(II) ions bind to ArgE in a similar manner as Co(II). Additional peaks are also observed at 3.0 Å, 3.5 Å and 3.9 Å for [Zn(II)_(ArgE)] and [Zn(II)Zn(II)(ArgE)], with intensity values of 0.29, 0.33 and 0.26 for [Zn(II)_(ArgE)] and 0.29, 0.40 and 0.26 for [Zn(II)Zn(II)(ArgE)], respectively. Excellent simulations of the observed EXAFS spectra for both [Zn(II)_(ArgE)] and [Zn(II)Zn(II)(ArgE)] were obtained with 5 ± 1 N/O scatterers at 2.00 Å. A second shell feature at ca. 3.3 Å is also present in the EXAFS spectrum of [Zn(II)Zn(II)(ArgE)]. Fits incorporating a Zn-Zn interaction at 3.3 Å for [Zn(II)Zn(II)(ArgE)] exhibited reasonable Debye-Waller factors (Table 2) and resulted in a significant improvement in goodness-of-fit values (f′). Failure to incorporate a Zn-Zn vector in the curve-fitting parameters resulted in a large disagreement between the data and simulation. Therefore, the second-shell FT feature observed at 3.3 Å is consistent with a dinuclear Zn(II) active site in ArgE. Interestingly, the FT EXAFS spectra of [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)] are dominated by a peak at 2.19 Å (Figure 6; Table 3) which resides at a significantly longer distance than that observed for either Zn(II) or Co(II)-bound ArgE. The amplitudes of this peak for both [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)] are quite similar (Figure 6) suggesting that Mn(II) ions also bind to ArgE in a similar manner as Zn(II) and Co(II). Simulations of the observed EXAFS spectra for both [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)] were obtained with 6 ± 1 N/O scatterers at 2.19 Å. However, compared to the EXAFS data obtained for Zn(II) and Co(II)-loaded ArgE, the EXAFS data for Mn(II) is of lower quality making it difficult to identify a possible Mn-Mn interaction or histidine ligation (notice the similar height of FT peaks in the 3–4 Å region and the noise). It should be noted, that there is a visible shoulder at ~ 4 Å−1 in the EXAFS data for both [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)], which suggests the presence of histidine ligands.
Figure 5.
k3-weighted Zn EXAFS (top) and Fourier transforms (bottom, over k = 2–12 Å−1) for [Zn(II)_(ArgE)] experimental (black) and simulation (green) and for [Zn(II)Zn(II)(ArgE)] experimental (red) and simulation (blue).
Figure 6.
k3-weighted Mn EXAFS (top) and Fourier transforms (bottom, over k = 2–12 Å−1) for [Mn(II)_(ArgE)] (solid) and [Mn(II)Mn(II)(ArgE)] (dotted).
Table 3.
Curve fitting results for ArgE Mn K-edge EXAFSa
| Sample, filename (k range) Δk3 χ |
Fit | Shell | Ras (Å) | σas2 (Å2) | ΔE0 (eV) | f′b |
|---|---|---|---|---|---|---|
| [Mn(II)_(ArgE)] (k = 2 – 11 Å−1) k3 χ = 14.05 |
1 | Mn-O5 | 2.18 | 0.0061 | 1.39 | 0.229 |
| 2 | Mn-O6 | 2.18 | 0.0082 | 1.33 | 0.248 | |
| [Mn(II)Mn(II)(ArgE)] (k = 2 – 11 Å−1) k3 χ = 13.89 |
3 | Mn-O5 | 2.18 | 0.0044 | 1.75 | 0.141 |
| 4 | Mn-O6 | 2.18 | 0.0063 | 1.65 | 0.161 |
Shell is the chemical unit defined for the multiple scattering calculation. Subscripts denote the number of scatterers per metal. Ras is the metal-scatterer distance. σas2 is a mean square deviation in Ras. ΔE0 is the shift in E0 for the theoretical scattering functions.
The fits for Mn ArgE use the smoothed data.
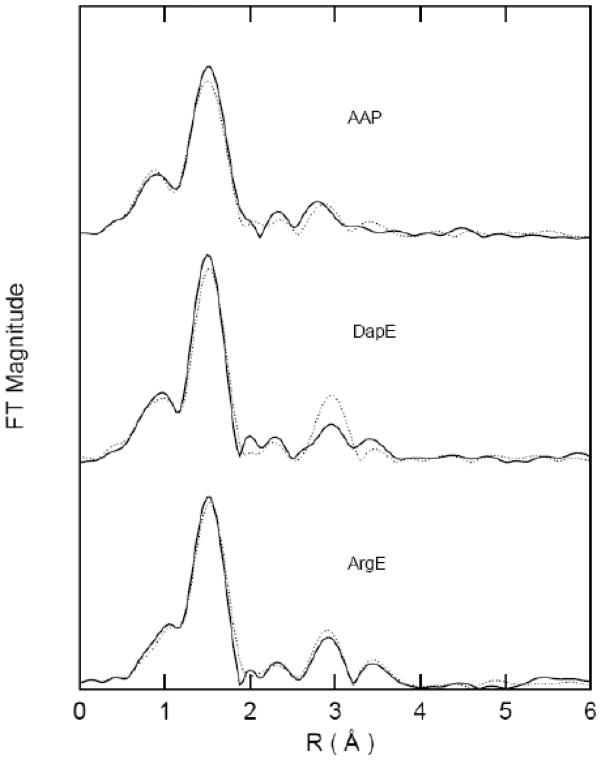
Inclusion of multiple-scattering contributions from the outer-shell atoms of a histidine-imidazole ring for the FT features observed at ~3 and 4 Å for [Co(II)_(ArgE)], [Co(II)Co(II)(ArgE)], [Zn(II)_(ArgE)], and [Zn(II)Zn(II)(ArgE)], resulted in reasonable Debye-Waller factors for these contributions and a slight reduction in the goodness-of-fit values (f′) (Table 2). These fits are similar to those reported for the FT EXAFS spectra of AAP and DapE [48]. The X-ray crystal structures of AAP and DapE reveal a (μ-aquo)(μ-carboxylato)dizinc(II) active site containing one histidine residue bound to each Zn(II) ion [34, 49]. The amplitude values for the features observed at ~3 and 4 Å are distinct for [Zn(II)_(DapE)] vs. [Zn(II)Zn(II) (DapE)] and [Zn(II)_(AAP)] vs. [Zn(II)Zn(II) (AAP)] [48], but are similar for [Zn(II)_(ArgE)] vs. [Zn(II)Zn(II)(ArgE)] (Figure 7) suggesting that divalent metal ions bind to ArgE in a positive cooperative manner.
Figure 7.
Non-phase corrected fourier transforms over k = 2–12 Å−1 for [Zn(II)_(ArgE)] (bottom; solid) and [Zn(II)Zn(II)(ArgE)] (bottom; dotted), for [Zn(II)_(DapE)] (middle; solid) and [Zn(II)Zn(II)(DapE)] (middle; dotted) and for [Zn(II)_(AAP)] (top; solid) and [Zn(II)Zn(II)(AAP)] (top; dotted).
Discussion
The evolution of bacterial resistance to antibiotics poses an imminent health threat to humans. The arginine biosynthetic pathway offers several potential anti-bacterial targets that have yet to be explored (Figure 1) [22, 23, 32, 50]. Since ArgE is critical to bacterial cell survival and proliferation, and no ArgE homolog is produced by humans, this enzyme holds great promise as a new antibiotic target. To date, no X-ray crystallographic data have been reported for any ArgE enzyme and no catalytically important amino acid residues have been identified. Therefore, the catalytically competent mono- and di-Zn(II), Co(II) and Mn(II)-loaded forms of ArgE were examined by extended X-ray absorption fine structure (EXAFS) spectroscopy. EXAFS is particularly well suited to clarify the structures of dinuclear metalloenzymes since EXAFS data are sensitive to heavy atom scatterers in the second coordination sphere providing direct evidence for dinuclear sites, if they exist [51, 52].
The EXAFS 1s→3d pre-edge transitions observed Co(II)-loaded ArgE at 7,709 eV for both the mono- and dinuclear Co(II) ArgE enzymes, are consistent with Co(II) residing in five or six-coordinate geometries [46, 47]. These data are in good agreement with previously reported electronic absorption spectra of Co(II)-loaded ArgE [33]. For [Co(II)_(ArgE)], three distinct peaks at 560, 619 and 705 nm with molar absorptivities (ε) of ε560 = 114 M−1 cm−1, ε619 = 119 M−1 cm−1 and ε705 = 52 M−1 cm−1 were observed. Addition of a second equivalent of Co(II) to [Co(II)_(ArgE)] increased the intensity of each of the observed absorption bands providing ε values of ε560 = 229 M−1 cm−1, ε619 = 290 M−1 cm−1 and ε705 = 121 M−1 cm−1. These data suggest that both Co(II) binding sites in ArgE are five coordinate. Similarly, EPR spectra of [Co(II)_(ArgE)] and [Co(II)Co(II)(ArgE)] revealed the presence of three species termed the Rhombic, Axial, and Dicobalt signals [33]. The predominant signal observed from [Co(II)_(ArgE)] is the Rhombic signal which exhibits high rhombic distortion of the axial zero-field splitting, suggestive of a highly axially asymmetric ligand field. The Rhombic signal also exhibits resolved 59Co hyperfine structure, indicative of low strains in the spin Hamiltonian parameters, i.e. of a site with a low degree of conformational freedom. Taken together EXAFS, UV-Vis, and EPR data suggest a low symmetry site, likely five-coordinate, with a geometry constrained by inflexible protein bound ligands. For [Co(II)Co(II)(ArgE)], a second, broad signal (the Dicobalt signal) is observed and this signal increases in intensity between the addition of one and two equivalents of Co(II); at the same time, both the intensity of the Rhombic signal and the total intensity of the spectrum decrease with added Co(II). The increase in the Dicobalt signal and concomitant decrease in the Rhombic signal are indicative of the formation of a dinuclear center and a consequent change in the electronic structure of the first-Co(II) binding site. These data are also similar to those reported for two dinuclear metallohydrolases from family M28, AAP and DapE [53–56]. For both AAP and DapE, UV-Vis, EPR, and EXAFS data indicate that the first metal binding site resides in a five-coordinate geometry while the second is either five or six-coordinate.
Similar to Co(II)-loaded ArgE, the EXAFS 1s→3d pre-edge transitions observed for Mn(II)-loaded ArgE at 6,541 eV for both the mono- and di-nuclear Mn(II) and Co(II) ArgE enzymes are consistent with both Mn(II) sites in ArgE residing in five or six-coordinate geometries (30). These data are consistent with previously reported EPR data on Mn(II)-loaded ArgE where the EPR spectra of [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)] are similar except that [Mn(II)Mn(II)(ArgE)] exhibits a more intense six-line pattern centered around 3,500 G (A = 8.8 mT) and additional features at 1,200 and 2,100 G [57]. These EPR spectra are characteristic of mononuclear Mn(II) ions [57]. In addition, no appreciable change in any of the spectra over the temperature range 4 – 70 K was observed and no features in the parallel mode spectra were observed that could not be accounted for by transitions due to isolated mononuclear Mn(II). Moreover, the EPR spectrum of [Mn(II)Mn(II)(ArgE)] does not contain a well-resolved 45 G hyperfine splitting, typical of dinuclear Mn(II)-Mn(II) centers when recorded at pH 7.5, but such a pattern is apparent in the spectrum of [Mn(II)Mn(II)(ArgE)] in 50 mM CAPS buffer at pH 10.0 [57]. These data clearly indicate the formation of a dinuclear active site but only at high pH values, suggesting coordination of a hydroxide ion is required to couple the Mn(II) ions. Such behavior is unique compared to that of [Co(II)Co(II)(ArgE)], which was shown to be coupled by a hydroxide ion at physiological pH [33].
The phase-corrected FT EXAFS spectra of [Co(II)_(ArgE)] and [Co(II)Co(II)(ArgE)] are similar to those of Zn(II)-loaded ArgE, which are dominated by a peak at ca. 2.05 Å. On the other hand, the phase-corrected FT EXAFS spectra of [Mn(II)_(ArgE)] and [Mn(II)Mn(II)(ArgE)] are dominated by a peak at ca. 2.19 Å, which is significantly longer than that observed for either Zn(II) or Co(II). Excellent single-shell fits of these first shell features for [Mn(II)_(ArgE)], [Co(II)_(ArgE)] and [Zn(II)_(ArgE)] were obtained and indicate that 5 or 6 N/O scatterers per Mn(II), Co(II) or Zn(II) ion exists at 2.19, 2.11 or 2.05 Å, respectively (Tables 2 and 3). Similarly, excellent single-shell fits of the first shell features observed in the EXAFS spectra of [Co(II)Co(II)(ArgE)], [Mn(II)Mn(II)(ArgE)], and [Zn(II)Zn(II)(ArgE)] were obtained and also indicate 5 or 6 N/O scatterers per Co(II), Mn(II), or Zn(II) ion at 2.19, 2.19, and 2.05 Å, respectively (Tables 2 and 3). These fits provide the average metal-ligand distances at the first metal site for [Co(II)_(ArgE)], [Mn(II)_(ArgE)], and [Zn(II)_(ArgE)], but are the average metal-ligand distances for both sites in [Co(II)Co(II)(ArgE)], [Mn(II)Mn(II)(ArgE)], and [Zn(II)Zn(II)(ArgE)]. The 0.09 Å difference in the dominant peak position in the phase-corrected FT spectra for Mn(II)-loaded ArgE vs. Co(II)- or Zn(II)-loaded ArgE, indicates that the Mn(II) bond distances are slightly longer than those observed for either Co(II) or Zn(II). This increase in bond distance likely represents the increased ionic radius for Mn(II) (0.97 Å) vs. Co(II) (0.89 Å) and Zn(II) (0.87 Å) in octahedral geometries. In addition, a higher coordination number and greater static order for Mn(II)-loaded ArgE vs. Co(II)- and Zn(II)-loaded ArgE is consistent with the thermodynamic data generated from Arrhenius plots for [Mn(II)_(ArgE)], [Zn(II)_(ArgE)] and [Co(II)_(ArgE)]. A more negative entropy values were observed for [Zn(II)_(ArgE)] and Co(II)_(ArgE)] (−106 and −66.0 J/mol· K, respectively) than for [Mn(II)_(ArgE)] (−34.5 J/mol K), suggesting more order is imposed on Zn(II)- and Co(II)-loaded ArgE active sites during catalysis compared with the Mn(II)-loaded form [33, 57]. Furthermore, the rhombic EPR signal observed for [Mn(II)_(ArgE)] exhibits well-resolved 55Mn hyperfine structure, indicating low strains in the spin Hamiltonian parameters [57]. Weak 59Co hyperfine structure is also observed for [Co(II)_(ArgE)] suggesting that Co(II) also binds with a somewhat low degree of conformational freedom [33].
The phase corrected FT EXAFS spectra of [Co(II)Co(II)(ArgE)] and [Zn(II)Zn(II)(ArgE)] include a feature at ~3.3 Å, which is similar to that observed in the phase corrected FT EXAFS spectra of the crystallographically characterized enzymes urease [58], AAP, and DapE [48] that contain dinuclear active sites. This feature was modeled as a M-M interaction suggesting each of these enzymes contain a dinuclear active site. Indeed, when a M-M vector is omitted from the EXAFS fits, significant disagreement between the observed data and the simulation occurs. For both [Co(II)Co(II)(ArgE)] and [Zn(II)Zn(II)(ArgE)], excellent simulations of EXAFS spectra were obtained with a M-M distance of 3.40 Å and 3.28 Å, respectively. Sequence alignment of several ArgE genes with those of the crystallographically characterized AAP, DapE, and CPG2 enzymes indicate that all of the amino acids that function as metal ligands in AAP, DapE, and CPG2 are strictly conserved in ArgEs [20, 34, 59, 60]. The X-ray crystal structures of CPG2, DapE, and AAP reveal a (μ-aquo)(μ-carboxylato)dizinc(II) active site with one terminal carboxylate and histidine residue bound to each metal ion [34–36]. Both Zn(II) ions in AAP, DapE, and CPG2 reside in distorted tetrahedral or pentacoordinate geometries, with Zn-Zn distances of 3.5, 3.4, and 3.3 Å for AAP, DapE, and CPG2, respectively. EXAFS data obtained for [Zn(II)Zn(II)(AAP)] and [Zn(II)Zn(II)(DapE)] also contains features at 3.27 and 3.39 Å, respectively, that can be modeled as a Zn-Zn vector [48]. Combination of these data, indicate that ArgE enzymes contain a dinuclear Zn(II) active site. In addition, based on the X-ray crystal structures and first shell EXAFS fits for [Zn(II)Zn(II)(AAP)] and [Zn(II)Zn(II)(DapE)] vs. those of Zn(II)-, Co(II)-, and Mn(II)-loaded ArgE, we propose that one carboxylate and one histidine residue resides at each Zn(II) site in ArgE and that the two Zn(II) ions are bridged by one carboxylate residue and a water molecule (Figure 8).
Figure 8.
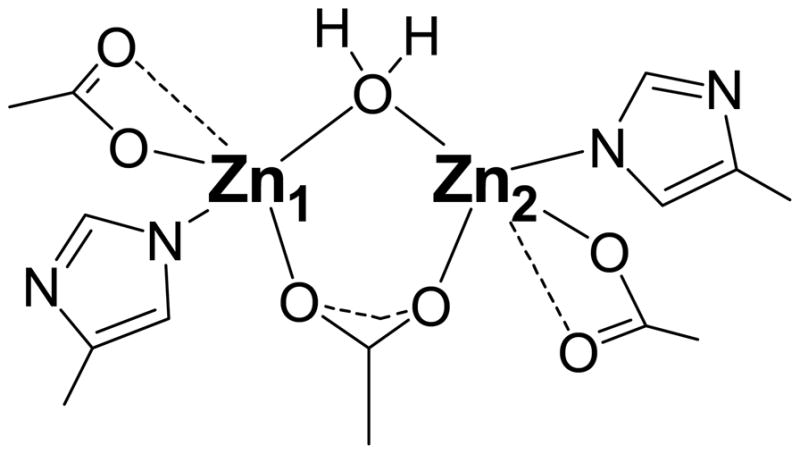
Proposed active site of the ArgE from E. coli based on EXAFS data.
Interestingly, the phase corrected FT EXAFS spectrum of [Zn(II)_(ArgE)] could also be simulated with the inclusion of a 3.28 Å Zn-Zn vector. The amplitude of the feature corresponding to the Zn-Zn vector in the phase corrected FT EXAFS spectra of [Zn(II)_(ArgE)] and [Zn(II)Zn(II)(ArgE)] is striking. The corresponding amplitudes for this feature in the non-phase corrected FT EXAFS spectra of [Zn(II)_(ArgE)] and [Zn(II)Zn(II)(ArgE)] are distinct compared to those of [Zn(II)_(DapE)] vs. [Zn(II)Zn(II)(DapE)] [48] and [Zn(II)_(AAP)] vs. [Zn(II)Zn(II)(AAP)] [48]. For DapE and AAP, the lack of a feature in the 3 to 4 Å for the mono-Zn(II)-loaded form vs. a strong feature in this range for the dinuclear Zn(II) enzymes has been attributed to sequential metal binding for the dinuclear site. Sequential binding has been verified for DapE as X-ray crystal structures for both the mono- and dinuclear Zn(II) DapE enzymes have been reported [34]. On the other hand, similar amplitudes for the feature in the 3 to 4 Å range for [Zn(II)_(ArgE)] and [Zn(II)Zn(II)(ArgE)] suggests positive cooperativity in Zn(II) binding to ArgE. Similarly, the EXAFS features in the 3 to 4 Å range for both Mn(II)- and Co(II)-loaded ArgE also suggest positive cooperativity in divalent metal binding. Taken together, these data suggest that divalent metal ions bind differently to ArgE than AAP or DapE even though the active site ligands appear to be identical [20, 59]. Indeed, electronic absorption and EPR spectra for [Zn(II)Co(II)(ArgE)] vs. [Co(II)Zn(II)(ArgE)] and [Mn(II)Zn(II)(ArgE)] vs. [Zn(II)Mn(II)(ArgE)] are identical, whereas spectral data for [Zn(II)Co(II)(AAP)] and [Co(II)Zn(II)(AAP)] are distinct [54].
In conclusion, ArgE enzymes are potential molecular targets for a novel class of antibiotics. The observation that ArgE is active in the presence of a broad variety of transition metals, including Zn(II), Co(II), or Mn(II), underscores the critical nature of this enzyme to bacterial cell survival (i.e. limiting Zn(II) will not halt ArgE activity because other divalent metal ions can substitute). The EXAFS data reported herein demonstrate that the ArgE from E. coli forms a dinuclear metalloactive site in the presence of Zn(II), Co(II), and Mn(II). In addition, multiple scattering contributions from the outer-shell atoms of a histidine-imidazole rings are observed at ~3 and 4 Å for Zn(II)- and Co(II)-loaded ArgE suggesting at least one histidine ligand at each metal binding site. Since no structural data have been reported for any ArgE enzyme, the EXAFS data reported herein represent the structural first glimpse into the active site of ArgE enzymes. Moreover, EXAFS data obtained for Zn(II)-, Co(II)-, and Mn(II)-loaded ArgE indicate that each of these divalent metal ions binds in a positive cooperative manner, in contrast to the metal binding properties of AAP and DapE [54, 56, 61]. Finally, these data provide a structural foundation for the future design of small molecules that will function as inhibitors of ArgE and may potentially function as a new class of antibiotics.
Footnotes
This work was supported by the National Institutes of Health (R15 AI085559-01A1, RCH) and the XXX.
References
- 1.C.f.D.C.a. Prevention. MMWR Morb Mortal Wkly Rep. 1995;44:1–13. [Google Scholar]
- 2.Howe RA, Bowker KE, Walsh TR, Feest TG, MacGowan AP. Lancet. 1997;351:601–602. doi: 10.1016/S0140-6736(05)78597-4. [DOI] [PubMed] [Google Scholar]
- 3.Levy SB. Sci Am. 1998;278:46–53. doi: 10.1038/scientificamerican0398-46. [DOI] [PubMed] [Google Scholar]
- 4.Chin J. New Scientist. 1996;152:32–35. [Google Scholar]
- 5.Henery CM. C&E News. 2000:41–58. [Google Scholar]
- 6.Nemecek S. Sci Am. 1997;276:38–39. [PubMed] [Google Scholar]
- 7.Miller JB. The Pharma Century. 2000:52–71. [Google Scholar]
- 8.Lesney MS, Frey R. The Pharma Century. 2000:110–129. [Google Scholar]
- 9.Lesney MS, Frey R. The Pharma Century. 2000:92–109. [Google Scholar]
- 10.Lesney MS, Tweedy BD. The Pharma Century. 2000:72–91. [Google Scholar]
- 11.Klugman KP. Clin Microbiol Rev. 1990;3:171–196. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breiman RF, Butler JC, Tenover FC, Elliott JA, Facklam RR. JAMA. 1994;271:1831–1935. [PubMed] [Google Scholar]
- 13.Miller JR, Dunham S, Mochalkin I, Banotai C, Bowman M, Buist S, Dunkle B, Hanna D, Harwood HJ, Huband MD, Karnovsky A, Kuhn M, Limberakis C, Liu JY, Mehrens S, Mueller WT, Narasimhan L, Ogden A, Ohren J, Prasad JVNV, Shelly JA, Skerlos L, Sulavik M, Thomas VH, VanderRoest S, Wang L, Wang Z, Whitton A, Zhu T, Stover CK. Proc Natl Acad Sci U S A. 2009;106:1737–1742. doi: 10.1073/pnas.0811275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh C. Nat Rev Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 15.Leeb M. Nature. 2004;431:892–893. doi: 10.1038/431892a. [DOI] [PubMed] [Google Scholar]
- 16.Schmid M. Nat Biotechnol. 2006;24:419–420. doi: 10.1038/nbt0406-419. [DOI] [PubMed] [Google Scholar]
- 17.Payne DJ. Science. 2008;321:1644–1645. doi: 10.1126/science.1164586. [DOI] [PubMed] [Google Scholar]
- 18.Supuran CT, Scozzafava A, Clare BW. Med Res Revs. 2002;22:329–372. doi: 10.1002/med.10007. [DOI] [PubMed] [Google Scholar]
- 19.Glansdorff N, Xu Y. In: Microbiology Monogr. Wendisch VF, editor. Springer; Berlin Heidelberg: 2007. pp. 219–257. [Google Scholar]
- 20.Javid-Majd F, Blanchard JS. Biochemistry. 2000;39:1285–1293. doi: 10.1021/bi992177f. [DOI] [PubMed] [Google Scholar]
- 21.Velasco AM, Leguina JI, Lazcano A. J Mol Evol. 2002;55:445–459. doi: 10.1007/s00239-002-2340-2. [DOI] [PubMed] [Google Scholar]
- 22.Cunin R, Glansdorff N, Pierard A, Stalon V. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis RH. Microbiol Rev. 1986;50:280–313. doi: 10.1128/mr.50.3.280-313.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ledwidge R, Blanchard JS. Biochemistry. 1999;38:3019–3024. doi: 10.1021/bi982574a. [DOI] [PubMed] [Google Scholar]
- 25.Sakanian VA, Ovespian AS, Mett IL, Kochikian AV, Petrosian PK. Genetika. 1990;26:1915–1925. [PubMed] [Google Scholar]
- 26.Xu Y, Liang Z, Legrain C, Rüger HJ, Glansdorff N. J Bacteriol. 2000;182:1609–1615. doi: 10.1128/jb.182.6.1609-1615.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakanyan V, Petrosyan P, Lecocq M, Boyen A, Legrain C, Demarez M, Hallet JN, Glansdorff N. Microbiol. 1996;142:99–108. doi: 10.1099/13500872-142-1-99. [DOI] [PubMed] [Google Scholar]
- 28.Hani EK, Ng D, Chan VL. Can J Microbiol. 1999;45:959–969. [PubMed] [Google Scholar]
- 29.Valentsev VE, Krupenina VI, Mishan’kin BN. Zh Mikrobiol Epidemiol Immunobiol. 1995;5:48–51. [PubMed] [Google Scholar]
- 30.Picard FJ, Dillon JR. J Bacteriol. 1989;171:1644–1651. doi: 10.1128/jb.171.3.1644-1651.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girodeau J-M, Agouridas C, Masson MPR, LeGoffic F. J Med Chem. 1986;29:1023–1030. doi: 10.1021/jm00156a021. [DOI] [PubMed] [Google Scholar]
- 32.Meinnel T, Schmitt E, Mechulam Y, Blanquet S. J Bacteriol. 1992;174:2323–2331. doi: 10.1128/jb.174.7.2323-2331.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGregor W, Swierczek SI, Bennett B, Holz RC. Journal of the American Chemical Society. 2005;127:14100–14107. doi: 10.1021/ja054081g. [DOI] [PubMed] [Google Scholar]
- 34.Nocek BP, Gillner DM, Fan Y, Holz RC, Joachimiak A. Journal of Molecular Biology. 2010;397:617–626. doi: 10.1016/j.jmb.2010.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowsell S, Pauptit RA, Tucker AD, Melton RG, Blow DM, Brick P. Structure. 1997;5:337–347. doi: 10.1016/s0969-2126(97)00191-3. [DOI] [PubMed] [Google Scholar]
- 36.Chevrier B, Schalk C, D’Orchymont H, Rondeau J-M, Moras D, Tarnus C. Structure. 1994;2:283–291. doi: 10.1016/s0969-2126(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 37.Shi D, Yu X, Roth L, Tuchman M, Allewell NM. Biophys Chem. 2007;126:86–93. doi: 10.1016/j.bpc.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Gill SC, von Hippel PH. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 39.George GN, Garrett RM, Prince RC, Rajagopalan KV. J Am Chem Soc. 1996;118:8588–8592. [Google Scholar]
- 40.Rehr JJ, Mustre de Leon J, Zabinsky SI, Albers RC. J Am Chem Soc. 1991;113:5135–5140. [Google Scholar]
- 41.Rappe AKC, CJ, Colwell KS, Goddard WA, III, Skiff WM. J Am Chem Soc. 1992;114:10024–10035. [Google Scholar]
- 42.Ferreira GC, Franco R, Mangravita A, George GN. Biochemistry. 2002;41:4809–4818. doi: 10.1021/bi015814m. [DOI] [PubMed] [Google Scholar]
- 43.Poiarkova AVaRJJ. Physical Review B. 1999;59:948. [Google Scholar]
- 44.Poiarkova AV. dissertation. University of Washington; 1999. [Google Scholar]
- 45.Cosper NJ, Stalhandske CMV, Saari RE, Hausinger RP, Scott RA. J Biol Inorg Chem. 1999;4:122–129. doi: 10.1007/s007750050295. [DOI] [PubMed] [Google Scholar]
- 46.Wirt MD, Sagi I, Chen E, Frisbis SM, Lee R, Chance MR. J Am Chem Soc. 1991;113:5299–5304. [Google Scholar]
- 47.Zhang JH, Kurtz DM, Maroney MJ, Whitehead JP. vol Inorg Chem. 1991:1359–1366. [Google Scholar]
- 48.Cosper NJ, Bienvenue DL, Shokes J, Gilner DM, Tsukamoto T, Scott* R, Holz* RC. J Am Chem Soc. 2004;125:14654–14655. doi: 10.1021/ja036650v. [DOI] [PubMed] [Google Scholar]
- 49.Desmarais W, Bienvenue LD, Bzymek KP, Holz RC, Petsko AG, Ringe D. Structure. 2002:1063–1072. doi: 10.1016/s0969-2126(02)00810-9. [DOI] [PubMed] [Google Scholar]
- 50.Glansdorff N. American Society for Microbiology. Washington, D.C: 1987. [Google Scholar]
- 51.Scott RA. Methods Enzymol. 1985;117:414–458. [Google Scholar]
- 52.Teo BK. Journal. 1981 Pages. [Google Scholar]
- 53.Bennett B, Holz RC. J Am Chem Soc. 1997;119:1923–1933. [Google Scholar]
- 54.Bennett B, Holz RC. Biochemistry. 1997;36:9837–9846. doi: 10.1021/bi970735p. [DOI] [PubMed] [Google Scholar]
- 55.Prescott JM, Wagner FW, Holmquist B, Vallee BL. Biochemistry. 1985;24:5350–5356. doi: 10.1021/bi00341a012. [DOI] [PubMed] [Google Scholar]
- 56.Bienvenue DL, Gilner DM, Davis RS, Bennett B, Holz RC. Biochemistry. 2003;42:10756–10763. doi: 10.1021/bi034845+. [DOI] [PubMed] [Google Scholar]
- 57.McGregor WC, Swierczek SI, Bennett B, Holz RC. Journal of Biological Inorganic Chemistry. 2007;12:603–613. doi: 10.1007/s00775-007-0211-4. [DOI] [PubMed] [Google Scholar]
- 58.Shengke W, et al. Inorg Chem. 1994;33:1589. [Google Scholar]
- 59.Speno HS, Luthi-Carter R, Macias WL, Valentine SL, Joshi ART, Coyle JT. Mol Pharmacol. 1999;55:179–185. doi: 10.1124/mol.55.1.179. [DOI] [PubMed] [Google Scholar]
- 60.Gillner DM, Bienvenue DL, Nocek BP, Joachimiak A, Zachary V, Bennett B, Holz RC. J Bio Inorg Chem. 2009;14:1–10. doi: 10.1007/s00775-008-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prescott JM, Wagner FW, Holmquist B, Vallee BL. Biochem Biophys Res Commun. 1983;114:646–652. doi: 10.1016/0006-291x(83)90829-x. [DOI] [PubMed] [Google Scholar]