Abstract
Many microbes can be cultured as single-species communities. Often, these colonies are controlled and maintained via the secretion of metabolites. Such metabolites have been an invaluable resource for the discovery of therapeutics (e.g. penicillin, taxol, rapamycin, epothilone). In this article, written for a special issue on imaging mass spectrometry, we show that MALDI-imaging mass spectrometry can be adapted to observe, in a spatial manner, the metabolic exchange patterns of a diverse array of microbes, including thermophilic and mesophilic fungi, cyanobacteria, marine and terrestrial actinobacteria, and pathogenic bacteria. Dependent on media conditions, on average and based on manual analysis, we observed 11.3 molecules associated with each microbial IMS experiment, which was split nearly 50:50 between secreted and colony-associated molecules. The spatial distributions of these metabolic exchange factors are related to the biological and ecological functions of the organisms. This work establishes that MALDI-based IMS can be used as a general tool to study a diverse array of microbes. Furthermore the article forwards the notion of the IMS platform as a window to discover previously unreported molecules by monitoring the metabolic exchange patterns of organisms when grown on agar substrates.
INTRODUCTION
Many microbes can be cultured as single-species communities. The microbial communities, or colonies, curate their environment via metabolic exchange factors such as released natural products. To date, there are very few tools available that can monitor, in a systematic and informative fashion, the metabolic release patterns by microbes grown in a pure or mixed culture. There are significant challenges in the ability to monitor the metabolic secretome from growing microbial colonies. For example, the chemistries of such molecules can be extremely diverse, ranging from polyketides (e.g. erythromycin), non-ribosomal peptides (e.g. penicillin), isoprenoids (e.g. artemisinin), fatty acids (e.g octanoic acid), microcins (e.g. Nisin), to peptides (e.g microcin C7), poly-nucleotides and proteins [1-6]. Because of this chemical diversity, most of these molecules are extracted prior to analysis and studied one at a time and apart from the native spatial context of a microbial colony. Thus, limited information is obtained about the metabolic output of colonies in a synergetic or multiplexed fashion.
Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) imaging mass spectrometry (IMS) is a powerful tool for simultaneously investigating the spatial distribution of multiple different biological molecules [7-11]. The technique offers a molecular view of the peptides, proteins, polymers and lipids produced by a microbial colony without the need of exogenous labels or radioactive trace material [12]. Target compounds can be measured and visualized simultaneously and in a high throughput manner within a single experiment. IMS extends beyond techniques such as MALDI profiling or MALDI intact cell analysis. Although invaluable, these techniques give a broad view of the metabolites produced in reference to a growing colony, where discretely secreted low global concentration but high local concentration metabolites could be missed. IMS entails examining the entire bacterial colony, including the surrounding agar medium, by defining a raster composed of greater than one thousand laser spots (points of data collection), which increases the likelihood of detecting unique, discrete ion distributions patterns and hidden molecular phenotypes that cannot be observed by the naked eye. IMS technology has been widely used in the medical sciences, such as disease pathology and pharmaceutical research [13]. The types of samples analyzed include brain and liver tissues [14] or plant tissue [15]. In these methods, it is necessary that the tissue is cryo-sliced and treated before matrix application [16]. As an extension to the technique, we successfully applied MALDI-TOF imaging for visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sectioned sponges. For this application, single filaments removed from an assemblage of marine cyanobacteria were anchored on a MALDI target plate. The filaments were coated with a matrix for MALDI-TOF analysis and biologically relevant molecules were identified [17]. More recently, our lab developed a further extension in the capabilities of the technique, as it was demonstrated that MALDI-TOF IMS could be used to visualize the metabolic exchange between two competing bacterial populations. The experiment entailed monitoring the chemical exchange between colonies of Bacillus subtilis and Streptomyces coelicolor, grown in proximity on top of a MALDI plate. This experiment enabled the spatial and temporal characterization of metabolites produced during the growth of the microorganisms [18].
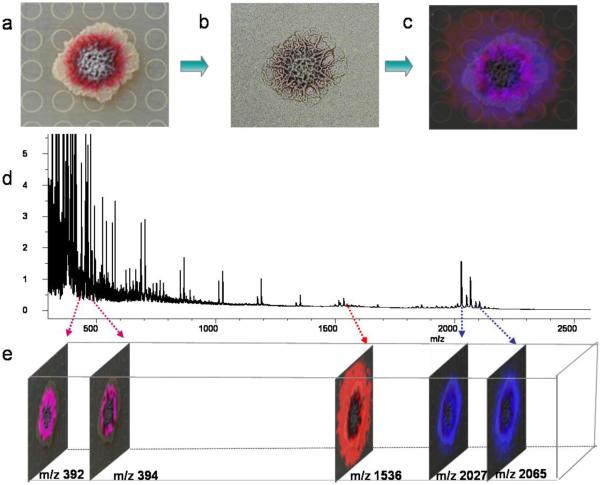
Figure 1 shows an overview of microbial IMS. Microbial colonies are grown in a Petri dish, followed by excision and transfer after reaching a desired stage of growth. The agar is then coated with a solid small molecule crystalline matrix that assists in the ionization process. The MALDI plate is then placed at 37°C to dehydrate the agar and dissolve the matrix, which then acts as an adhesive to stabilize the solid agar. Samples are then introduced into the MALDI-TOF mass spectrometer where a raster grid is designated within or around the microbial colony. The raster is composed of a series of laser spots with micron spacing, directing the MALDI-TOF mass spectrometer to collect data at each point. The result is a series of mass spectra that are processed to display as an average mass spectrum. The Bruker software FlexImaging can then be used to false-color the ion of interest to indicate the spatial resolution within the designated raster.
Figure 1. Overview of IMS of microorganisms.
A. Photograph of a colony grown on the top of the MALDI plate. B. The colony with the agar coated with universal matrix. C. The merged signals with an optical image of the colony. D. The average signal from m/z 300-2300 of all the spectra obtained in the imaging runs. E. Separate signals indicating the m/z ion. Images generated by Bruker Daltonics FlexImaging 2.0.
The work herein describes three aspects of the application of IMS in the investigation of microbial metabolic exchange factors. First, media optimization was used to extend the use of IMS to investigate the metabolic secretome of a large number of different microbial species, establishing MALDI-based IMS as a general platform for the characterization of microbes. Second, using the optimized growth media, it was possible to visualize by IMS that 75% of the studied microbes release metabolites into the surrounding growth media, enabling the enumeration of secreted versus colony-associated metabolites. This observation reinforces the hypothesis that microbes interact with their surroundings to curate or otherwise impact their environmental niches, and that these interactions may have important ecological implications. Finally, we show that a large number of molecular entities can be visualized by IMS from heterogeneous mixtures of organisms isolated from different ecological environments, underscoring the importance of metabolic release for the co-existence of multiple microbes. Together with extraction, tandem mass spectrometry by Fourier Transform Ion Cyclotron mass spectrometry (FT-ICR-MS) and MALDI-TOF/TOF analysis, it was demonstrated that the observed metabolic factors were partially or completely identified. This method has opened a new window into the metabolic output of microbial colonies, and can potentially be used for chemical ecological studies, investigations of microbial biofilms, and the acceleration of the early phases of natural product-based drug discovery.
MATERIALS AND METHODS
Strains, media, and culture conditions
The bacteria and fungi used in this study are listed in Supplementary Table 1. Media used for the different organisms are as follows: ISP-2 medium (yeast extract 4.0 g, malt extract 10.0 g, dextrose 4.0 g, agar 20.0 g, distilled water 1000 mL), A1 medium (starch 10.0 g, peptone 4.0 g, yeast extract 2.0 g, calcium carbonate 1.0 g, agar 18.0 g, natural distilled seawater 1000 mL), PDA medium (Difico™ potato dextrose agar, Becton agar, Becton Dickinson and Company), LB medium (Bacto-tryptone 10 g, Bacto-yeast extract 5 g, NaCl 10 g, distilled water 1000 mL), and BG-11 agar (BGNaNO3 1.5 g, K2HPO4 0.04 g, MgSO4·7H2O, 0.075 g, CaCl2·2H2O 0.036 g, citric acid 0.006 g, ferric ammonium citrate 0.006 g, EDTA (disodium salt) 0.001 g, NaCO3 0.02 g, trace metal mix A5 1.0 mL, agar 10.0 g, distilled water 1000 ml; where trace metal mix A5 contains: H3BO3 2.86 g, MnCl2·4H2O 1.81 g, ZnSO4·7H2O 0.222 g, NaMoO4·2H2O 0.39 g, CuSO4·5H2O 0.079 g, Co(NO3)2·6H2O 49.4 mg, distilled water 1000 mL). All the strains were grown at 28°C except Staphylococcus aureus, which was grown at 37°C.
Sample preparation
In each case, 13 mL of the selected agar medium was poured into a 100 × 15 mm Petri dish and allowed to solidify. Thereafter, each microbial strain to be grown was inoculated on the appropriate agar medium or the medium was poured directly on top of the stainless steel agar plate. Bacteria Lysobacter enzymogenes C3, Bacillus subtilis 3610, Bacillus pumilus CNJ762, Pseudomonas aeruginosa PAO, and Pseudomonas fluorescens PFO-1 were inoculated on LB agar. For Nostoc sp., PCC 7120, a 10 μl inoculum was pipetted onto BG-11 agar that had been poured on top of a square piece (4 cm X 4 cm) of aluminum foil contained in a Petri dish. After 7 days of growth, the foil was removed from the Petri dish and the agar with the culture-medium was placed on the Bruker MSP 96 MALDI target plate. The fungi and spore-producing bacteria were inoculated on ISP-2 or A1 agar until visible spores were produced. The spores were then collected and diluted in water. Either 0.2 μL of the bacteria suspensions or 1 μL of the diluted spores were inoculated onto the surface of Bruker MSP 96 MALDI target plates containing solidified agar media on the surface. All cultures were grown at an appropriate temperature and time. The inoculation times are listed in the Supplementary Table 1. For the organisms that were not grown directly on Bruker MSP 96 target plate but instead on media in Petri dishes, each culture was incubated until it displayed robust colony growth. Then the selected agar was excised with a razor blade and transferred to a Bruker MSP 96 target plate. An optical image using a digital camera was then taken of the colony. All samples were then covered with Fluka Universal MALDI matrix by the sieve method and then dried in a 37°C oven to generate a uniform distribution of the crystalline matrix.
IMS parameters
MALDI time-of-flight (TOF) (Bruker-microflex) was used for IMS analysis. Samples were subjected to IMS with a typical m/z range of 200-3600 Da. Different mass ranges used for each organism are indicated in Supplementary Table 1. Raster grids indicating the laser intervals in both the X and Y direction were typically set, depending on the image, to 200 to 800 μm increments resulting in the collection of ~1,300-20,000 spectra.
IMS data analysis and image generation
The IMS data sets were analyzed using the Bruker FlexImaging software. The mass spectra collected were processed by the software and then displayed as one average mass spectrum. Ions were assessed manually and those of interest were assigned a false color allowing for the spatial distribution of the ion to be displayed within the defined raster (Supplementary Figure 1).
MALDI-TOF/TOF and FT-ICR-MS analysis
Different methods were pursued for the identification of the ions of interest. One method was intact cell MALDI-TOF/TOF, which consists of spotting 1-2 μL of a liquid culture on the MALDI target plate, followed by 1-2 μL of liquid matrix. The sample is allowed to dry and then analyzed. As an alternative method, each microorganism was grown until saturation in the appropriate liquid medium, to a high cell density, extracted with n-butanol or methanol, and dried by SpeedVac. The extracts were then subjected to FT-ICR-MS analysis (Thermo) or MALDI-TOF/TOF (ABI 4800 TOF/TOF) to obtain high-resolution mass spectra for the generation of high mass accuracy molecular formulas and tandem mass spectrometry for structure annotation (Supplementary Table 2 and Supplementary Figure 2).
RESULTS AND DISCUSSION
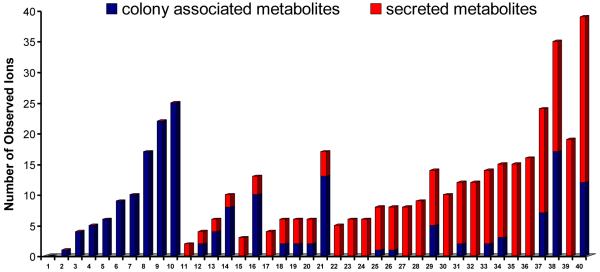
The growth conditions and imaging parameters for several microbial colonies were optimized and are displayed in Supplementary Table 1. The resulting table of mass-to-charge ratios (m/z) and corresponding images obtained with IMS are provided in Supplementary Figure 1. The organisms subjected to IMS ranged from fungi, bacteria to cyanobacteria. These microorganisms were isolated from a wide assortment of habitats such as marine sediments, terrestrial soils, insects, humans and plants. Most of the organisms readily grew on ISP-2 agar except five marine Salinispora strains that grew only on A1 medium, and one cyanobacterium (Nostoc sp.) that grew only on BG-11 agar. In addition, the fungi tested preferably grew on PDA media. In the end, colonies from 40 different organisms were successfully cultured and subjected to IMS, giving information on their respective secretomes, albeit not always in optimized media. On average, we found 11.3 microbial-derived ions were observed within a single IMS run for each organism within the experimental conditions tested. This will be a low estimate of the total metabolic exchange capacity of these microbes as we only capture a small portion of the molecules. Molecules with that are poorly ionized or have no charge, opposite charge of the ionization used or molecules that are present in low quantities are not observed. Nonetheless this number provides an indication of the minimal number of metabolic exchange factors a colony produces. As a means to categorize the observed spatial distributions, ions were denoted as either being colony associated or secreted metabolites (Figure 2).
Figure 2. The number of ions observed by IMS in the tested strains.
Red bars: secreted metabolites, and blue bars: colony-associated metabolites. (1) S. pacifica CNS-143 (2) S. pacifica CNT-133 (3) R. leguminosarum S36 (4) Nostoc sp. PCC 7120 (5) F. verticillioides 5777 (6) Thermofungus sp. C03081330 (7) Thermofungus sp. C03081334 (8) Chaetomium chiversii CS-36-62 (9) P. fluorescens (10) S. aureus (11) S.arenicola CNT-088 (12) S. arenicola CNH-643(13) Acremonium sp. CNC890 (14) Fusarium sp. CNL 292 (15) Actinomycete sp. CNS 575 (16)B. pumilus CNJ762 (17) S. clavuligerus ATCC 53653 (18) Fusarium sp. CNC 477 (19) S. coelicolor A3 (20) S. ghanaensis 14672 (21) P. aeruginosa PAO (22) A. teichomyceticus PSK0532 (23) Streptomyces sp. WASP (24) S. viridochromogenes 40736 (25) Streptomyces sp. Mg1 (26) B. subtilis 3610 (27) Streptomyces SPB74 (28) Streptomyces sp. SPB 78 (29) Streptomyces sp. AA#4 (30) S. pristinispiralis ATCC25486 (31) S. roseosporus NRRL15998(32) S. albus J1074 (33) Salinispora arenicola CNS-205 (34) S. griseoverticillatum ATCC 31499(35) S. roseosporus NRRL11379 (36) S. sviceus ATCC 29083 (37) Kutzneria sp. 744 (38) Lysobacter enzymogenes C3 (39) S. hygroscopicus ATCC 53653 (40) B. bassiana ATCC 7159.
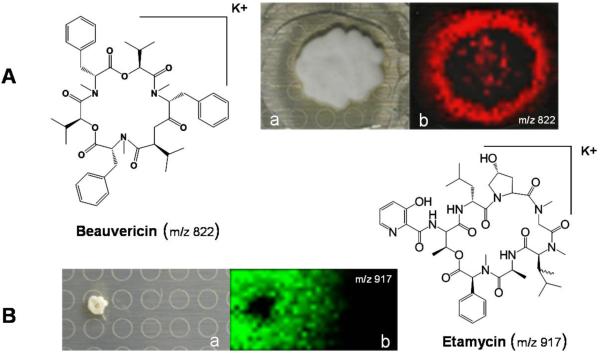
The most challenging colonies to image were the microbes grown on the seawater-based medium A1. Given its high salt contents, this medium appeared to suppress ion formation, resulting in data sets that were inconclusive. The imaging did improve when the A1 media was diluted, reducing the salt concentration to approximately 25% of its original content, which did not affect colony growth or morphology. For example, using this optimized method, the antibiotic etamycin was readily identified from the marine actinomycete strain CNS-575 (Figure 3) which had been isolated from a sediment sample collected from Fiji [19]. On average, the actinomycetes analyzed, secreted 7.5 compounds per strain compared to 1.5 metabolites that were associated with the colony. All other organisms secreted far fewer metabolites and had more colony-associated ions (Figure 2 and Supplemental Figure 1). The observation that most organisms produce colony-bound metabolites may make ecological sense. For example, the cyanobacterium studied, Nostoc sp., only had visible ions associated with the colony. Because this cyanobacterium lives in an aquatic environment, it is metabolically reasonable that it retains important natural products such as the photosynthetic pigment pheophytin A (m/z 871 [M+H]+) [20] or defensive natural products. Examples of the latter include inhibitors of trypsin (used to digest food) or thrombin (responsible for blood clotting) in potential predators. In agreement with this observation, we found no evidence for the release of any metabolites from filamentous cyanobacteria, except those that were dying.
Figure 3. IMS of Beauveria bassiana ATCC 7159 and Actinomycete CNS575.
A. IMS of Beauveria bassiana ATCC 7159, m/z 822 is Beauvericin ([M+K]+). B. IMS of Actinomyces sp. CNS 575, m/z 917 is Etamycin ([M+K] +). (a) Respective photographs of microbial colonies (b) Overlays of the observed specific ions indicated.
Members of the genus Streptomyces have extensive capabilities to produce secondary metabolites and yield the majority of the clinically useful natural antibiotics discovered to date[21]. Fifteen Streptomyces strains were investigated by IMS; to our surprise 11 of these strains displayed an abundance of ions above 1800 m/z. Ions in this mass range are typically predicted to be classes of post-translationally modified ribosomal peptides with thioether, thiazole or oxazole moieties. One of the ions was selected and dereplicated to the known thioether-containing peptide lantibiotic SapB (m/z 2027 [M+H]+), which is required for hyphae formation and sporulation inS. ceolicolor A3. Ten other significant m/z signals greater than 1800 Da were observed in S. albus J1074 (m/z 2282), Streptomyces sp. SPB78 (m/z 2268), S. ghanaensis 14672 (m/z 2014), S. clavuligerus ATCC 53653 (m/z 2197), S. viridochromogenes 40736 (m/z 2169), S. pristinispiralis ATCC25486 (m/z 2014), Streptomyces sp. Mg1 (m/z 2003), S. roseosporus NRRL11379 (m/z 2252) and S. roseosporus NRRL15998 (m/z 2252). Based on genomic evidence, we hypothesized that these signals may correspond to lantipeptide or lasso peptide natural products [22, 23], a large group that are generally missed in traditional natural products isolation procedures. Indeed, many of these ions were recently characterized via peptidogenomics to belong to ribosomal and non-ribosomal peptide natural products [24].
IMS can also be used to study pathogens. Mammals and arthropods alike are continuously exposed to numerous environmental pathogens that are principal sources of infection causing significant morbidity and mortality. IMS was used to monitor the metabolic potential of various pathogens (Supplemental Table 1). It was observed that the insect pathogen Beauveria bassiana and the human pathogen Staphylococcus aureus produced 39 and 25 detectable ions, respectively. Metabolites identified by IMS such as beauvericin (m/z 807 [M+Na]+), bassianalide (m/z 932 [M+Na]+), and δ-toxin (m/z 3005 [M+H]+) have been described to play roles in host immune evasion [25, 26]. We predict IMS can be used to study host-symbiont-pathogen interactions in unprecedented ways leading to a greater understanding of the molecular factors associated with pathogenesis. Figure 3 shows the spatial distribution of the molecule beauvericin in a growing B. bassiana colony.
The IMS data collected as part of this study reinforces these reports by observing ions that correspond to metabolic factors associated with niche establishment or biocontrol processes. For example, sansalvamide (m/z 587 [M+H]+, m/z 609 [M+Na]+, and m/z 625 [M+K]+) produced by the Fusarium sp. CNL-292 [28] is a cytotoxic agent. Other identified molecules have been described to show antimicrobial properties: they may be antibiotics like etamycin (m/z 879 [M+H] +, m/z 901 [M+Na]+, and m/z 917 [M+K]+) from the strain Actinomyces CNS-575 [19], cinnamycin (m/z 2041 [M+H]+, m/z 2063 [M+Na]+, m/z 2079 [M+K]+) from Streptoverticillium griseoverticillatum ATCC 31499 [29], CDA (m/z 1536 [M+H]+) of Streptomyces coelicolor A3[18], surfactin (m/z 1075 [M+K]+) and plipastatin (m/z 1545 [M+K]+) from Bacillus subtilis strain 3610 [30], or antifungal agents like maltophilin (m/z 511 [M+H]+) and dihydromaltophilin (m/z 513 [M+H]+) of Lysobacter enzymogenes C3 [31] (Supplemental Table 2 and Supplemental Figure 2). Visualizing the spatial distributions of secondary metabolites produced by heterogeneous mixtures of microorganisms provides an in situ platform to study the chemical interactions in complex microbial communities from the soil or other important niches (e.g. the human microbiome).
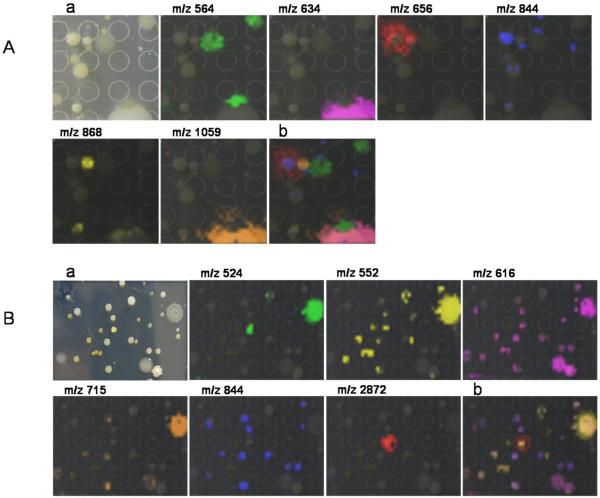
Finally to highlight the microbial complexity of samples that IMS can tackle we looked at cultured environmental samples. It is estimated that one gram of soil, sand or water contains over a million microorganisms [27]. Inevitably complex communities have evolved over millennia by interacting with each other and have evolved to curate their surrounding environment to maximize niche utilization. Undoubtedly, the secondary metabolites produced by microbes in these communities contribute to colony growth, microbial competition, and community dynamics. Figure 4 shows two examples in which IMS was used to visualize the metabolic factors associated with microbial communities. The data demonstrate how IMS can be used to simultaneously visualize the metabolic output from different organisms in a community isolated from a Los Angeles garden soil and a marine sediment from La Jolla Shores beach, San Diego. Although the identity of the ions shown in Figure 4 remain unresolved and some are currently under investigation, microbes originating from soil have been extensively studied, and it has been shown that secreted molecules from these organisms are associated with their ecological roles as environmental curators.
Figure 4. IMS of multiple complex samples.
A. Los Angeles garden soil. B. La Jolla Shores beach marine sediment. (a) Respective photographs of microbial communities (b) Overlay of multiple ions observed within the microbial community.
CONCLUSION
IMS proves to be a powerful discovery tool that allows us to observe and identify secreted or cell-bound metabolic factors, associated with a specific microorganism during colony formation. Many unknown ions were detected by IMS, indicating that traditional extraction-purification methods inevitably overlook many important compounds. The well-known influence of culture conditions and medium components on the metabolic output of microorganisms was also observed. Mass spectrometry parameters such as ionization (here we primarily used positive ion mode), limits of detection, and media types obviously play an important role in the types of metabolites that can be visualized. Within the study herein, different number of metabolites were observed for the same organism, which was dependent on the media type (Supplemental Table 1). Furthermore, given sample preparation conditions for IMS (the use of a large amount of organic matrix to coat the sample and nutrient media composition), in many instances the parent ion [M+H]+ was not the most abundant ion. Instead, salt adducts (primarily sodium or potassium) were observed. Given the dynamic nature of microbial metabolic output and the numerous variables associated with the IMS platform, we predict no one set of conditions is likely to yield all of the compounds that a strain is capable of producing. For this reason, some known compounds of the specific organisms investigated were not detected in this study. Nevertheless, a vast number of ions were observed when microorganisms were grown on ISP-2 media, including many uncharacterized molecules. Organisms that needed a specific type of media, less conducive to IMS, were easily manipulated by media dilution to yield viable signals in the mass spectrometer. This observation indicates that a vast number of media types can be used to visualize metabolites from different microbes by IMS. The well-endowed producers of natural antibiotics, Streptomyces, were highly amenable towards IMS. Several of the observed ions stemming from the Streptomyces strains tested had patterns indicating their release into the surrounding agar in a multiplex or synergistic fashion. Exciting to noteis that the majority of the observed ions have not been described in the literature suggesting that many more compounds await discovery from this well studied genus. Monitoring multiplexed metabolic exchange patterns by IMS can potentially lead to a better understanding of the type of natural products organisms use concretely in vivo. Improvements in technology or innovative methodologies have proven to be the cornerstone of scientific advancement. Considering that the success rate of the discovery of novel antibiotics has declined precipitously in the last few decades [32], IMS shows promise as a front-end tool for identifying novel antibiotic leads. Indeed, we recently demonstrated that IMS can be used as a discovery tool to find antibiotics by monitoring metabolic exchange patterns [33].
Complex microbial communities such as the human microbiome [34] are new biological frontiers. Our work demonstrates that IMS has the ability to simultaneously capture multiple molecular signals from a community of bacteria. To complement the taxonomic efforts being put forth by the Human Microbiome Sequencing Project [35], we predict that IMS will provide an important tool to decipher the molecular basis of interspecies interactions. Understanding “how” microbes interact is a logical progression after identifying the composition of the human microbiome or any complex microbial community. Ultimately, IMS has the capacity to advance our understanding of the hidden microbial world by visualizing the spatial distribution or patterns of multiple compounds derived from colonies grown directly on agar media.
Supplementary Material
Highlights.
-
-
Imaging mass spectrometry can be performed on a large number of microbes.
-
-
An avg. of 11.3 metabolic exchange factors were observed from a microbial IMS experiment.
-
-
Complex microbial samples can be characterized at the molecular level using IMS.
-
-
IMS can be used to discover therapeutic leads from microbes.
ACKNOWLEGEMENTS
We greatly appreciate the support from: (i) Bruker therapeutic discovery mass spectrometry center at UCSD (ii) DJG acknowledges the A.P. Giannini Medical Research Foundation (iii) PRJ acknowledges NIH grant R01GM086261 (iv) The Swedish Research Council (AE) (iv) PCD acknowledges R01GM086283 (v) Jack E. Dixon for helpful discussions
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Cane DE. Programming of erythromycin biosynthesis by a modular polyketide synthase. J Biol Chem. 2010;285:27517–27523. doi: 10.1074/jbc.R110.144618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Qiao K, Zhou H, Xu W, Zhang W, Garg N, Tang Y. A Fungal Nonribosomal Peptide Synthetase Module that can Synthesize Thiopyrazines. Org. Lett. 2011;7:1758–1761. doi: 10.1021/ol200288w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mok S, Imwong M, Mackinnon MJ, Sim J, Ramadoss R, Yi P, Mayxay M, Chotivanich K, Liong KY, Russell B, Socheat D, Newton PN, Day NP, White NJ, Preiser PR, Nosten F, Dondorp AM. Artemisinin resistance in Plasmodium falciparum is associated with an altered temporal pattern of transcription. BMC Genomics. 2011;12:391. doi: 10.1186/1471-2164-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Teichmann B, Labbé C, Lefebvre F, Bölker M, Linne U, Bélanger RR. Identification of a biosynthesis gene cluster for flocculosin a cellobiose lipid produced by the biocontrol agent Pseudozyma flocculosa. Mol Microbiol. 2011;6:1483–1495. doi: 10.1111/j.1365-2958.2010.07533.x. [DOI] [PubMed] [Google Scholar]
- [5].Willey JM, van der Donk WA. Lantibiotics: Peptides of Diverse Structure and Function. Ann. Rev. of Micro. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- [6].Regni CA, Roush RF, Miller DJ, Nourse A, Walsh CT, Schulman BA. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 2009;13:1953–1964. doi: 10.1038/emboj.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Seeley EH, Schwamborn K, Caprioli RM. Imaging of intact tissue sections: moving beyond the microscope. J Biol Chem. 2011;286:25459–66. doi: 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schwamborn K, Caprioli RM. MALDI imaging mass spectrometry--painting molecular pictures. Mol Oncol. 2010;6:529–38. doi: 10.1016/j.molonc.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dill AL, Eberlin LS, Ifa DR, Cooks RG. Perspectives in imaging using mass spectrometry. Chem Commun (Camb) 2011;47:2741–6. doi: 10.1039/c0cc03518a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrom. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- [11].Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–94. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sauer S, Lange BM, Gobom J, Nyarsik L, Seitz H, Lehrach H. Miniaturization in functional genomics and proteomics. Nat. Rev. Genet. 2005;6:465–476. doi: 10.1038/nrg1618. [DOI] [PubMed] [Google Scholar]
- [13].Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nature Methods. 2007;2:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- [14].Acquadro E, Cabella C, Ghiani S, Miragoli L, Bucci EM, Corpillo D. Matrix-assisted laser desorption ionization imaging mass spectrometry detection of a magnetic resonance imaging contrast agent in mouse liver. Anal. Chem. 2009;81:2779–2784. doi: 10.1021/ac900038y. [DOI] [PubMed] [Google Scholar]
- [15].Anderson DMG, Carolan VA, Crosland S, Sharples KR, Clench MR. Examination of the distribution of nicosulfuron in sunflower plants by matrix-assisted laser desorption/ionisation mass spectrometry imaging. Rap. Comm. Mass Spec. 2009;23:1321–1327. doi: 10.1002/rcm.3973. [DOI] [PubMed] [Google Scholar]
- [16].Francese S, Dani FR, Traldi P, Mastrobuoni G, Pieraccini G, Moneti G. MALDI mass spectrometry imaging, from its origins up to today: the state of the art. Comb. Chem. High Throughput Screen. 2009;12:156–174. doi: 10.2174/138620709787315454. [DOI] [PubMed] [Google Scholar]
- [17].Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH, Dorrestein PC. Visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sponges via MALDI-TOF imaging. Molecular Biosystem. 2008;4:562–570. doi: 10.1039/b720018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang YL, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nature Chem. Bio. 2009;5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Haste NM, Perera VR, Maloney KN, Tran DN, Jensen P, Fenical W, Nizet V, Hensler ME. Activity of the streptogramin antibiotic etamycin against methicillin-resistant Staphylococcus aureus. J. Antibiot. 2010;63:219–24. doi: 10.1038/ja.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dembitsky VM, Rezanka T. Metabolites produced by nitrogen-fixing Nostoc species. Folia microbiologica. 2005;50:363–391. doi: 10.1007/BF02931419. [DOI] [PubMed] [Google Scholar]
- [21].Barbe V, Bouzon M, Mangenot S, Badet B, Poulain J, Segurens B, Vallenet D, Marlière P, Weissenbach J. Complete genome sequence of Streptomyces cattleya NRRL 8057, a producer of antibiotics and fluorometabolites. J Bacteriol. 2011;18:5055–5056. doi: 10.1128/JB.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rosengren KJ, Clark RJ, Daly NL, Göoransson U, Jones A, Craik DJ. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 2003;125:12464–12474. doi: 10.1021/ja0367703. [DOI] [PubMed] [Google Scholar]
- [23].Willey JM, van der Donk WA. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- [24].Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;11:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Giese B, Glowinski F, Paprotka K, Dittmann S, Steiner T, Sinha B, Fraunholz MJ. Expression of δ-toxin by Staphylococcus aureus mediates escape from phago-endosomes of human epithelial and endothelial cells in the presence of β-toxin. Cell Microbiol. 2011;13:316–29. doi: 10.1111/j.1462-5822.2010.01538.x. [DOI] [PubMed] [Google Scholar]
- [26].Valencia JW, Gaitán Bustamante AL, Jiménez AV, Grossi-de-Sá MF. Cytotoxic activity of fungal metabolites from the pathogenic fungus Beauveria bassiana: an intraspecific evaluation of beauvericin production. Curr Microbiol. 2011;63:306–12. doi: 10.1007/s00284-011-9977-2. [DOI] [PubMed] [Google Scholar]
- [27].Phelan VV, Liu WT, Puglia K, Dorrestein PC. Microbial metabolic exchange-the chemotype-to-phenotype link. Nat Chem Biol. 2011 Dec;8:26–35. doi: 10.1038/nchembio.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Belofsky GN, Jensen PR, Fenical W. Sansalvamide: A new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Letters. 1999;40:2913–2916. [Google Scholar]
- [29].Kaletta C, Entian KD, Jung G. Prepeptide sequence of cinnamycin: the first structural gene of a duramycin-type lantibiotic. European. J of Biochem. 1991;199:11–415. doi: 10.1111/j.1432-1033.1991.tb16138.x. [DOI] [PubMed] [Google Scholar]
- [30].Gonzalez DJ, Haste N, Hollands A, Fleming T, Hamby M, Pogliano K, Nizet V, Dorrestein PC. Microbial competition between Bacillus Subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiol. 2011;157:485–2492. doi: 10.1099/mic.0.048736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li X, Du L. Structure and biosynthesis of heat-Sstable antifungal Factor (HSAF), a Broad-Spectrum antimycotic with a Novel Mode of Auction. Antimicrob. Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu WT, Kersten RD, Yang YL, Moore BS, Dorrestein PC. Imaging mass spectrometry and genome mining via short sequence tagging identified the anti-infective agent arylomycin in Streptomyces roseosporus. J Am Chem Soc. 2011;133:18010–3. doi: 10.1021/ja2040877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–9. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, Cole JR, Ding Y, Dugan S, Field D, Garrity GM, Gibbs R, Graves T, Han CS, Harrison SH, Highlander S, Hugenholtz P, Khouri HM, Kodira CD, Kolker E, Kyrpides NC, Lang D, Lapidus A, Malfatti SA, Markowitz V, Metha T, Nelson KE, Parkhill J, Pitluck S, Qin X, Read TD, Schmutz J, Sozhamannan S, Sterk P, Strausberg RL, Sutton G, Thomson NR, Tiedje JM, Weinstock G, Wollam A, Genomic Standards Consortium Human Microbiome ProjectJumpstart Consortium. Detter JC. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326:236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.