Abstract
Recent collaborative, large-scale genomic profiling of the most common and aggressive brain tumor glioblastoma multiforme (GBM) has significantly advanced our understanding of this disease. The gene encoding platelet-derived growth factor receptor alpha (PDGFRα) was identified as the third of the top 11 amplified genes in clinical GBM specimens. The important roles of PDGFRα signaling during normal brain development also implicate the possible pathologic consequences of PDGFRα over-activation in glioma. Although the initial clinical trials using PDGFR kinase inhibitors have been predominantly disappointing, diagnostic and treatment modalities involving genomic profiling and personalized medicine are expected to improve the therapy targeting PDGFRα signaling. In this review, we discuss the roles of PDGFRα signaling during development of the normal central nervous system (CNS) and in pathologic conditions such as malignant glioma. We further compare various animal models of PDGF-induced gliomagenesis and their potential as a novel platform of pre-clinical drug testing. We then summarize our recent publication and how these findings will likely impact treatments for gliomas driven by PDGFRα overexpression. A better understanding of PDGFRα signaling in glioma and their microenvironment, through the use of human or mouse models, is necessary to design a more effective therapeutic strategy against gliomas harboring the aberrant PDGFRα signaling.
Keywords: Gliomas, PDGFRα signaling, glioma tumorigenesis
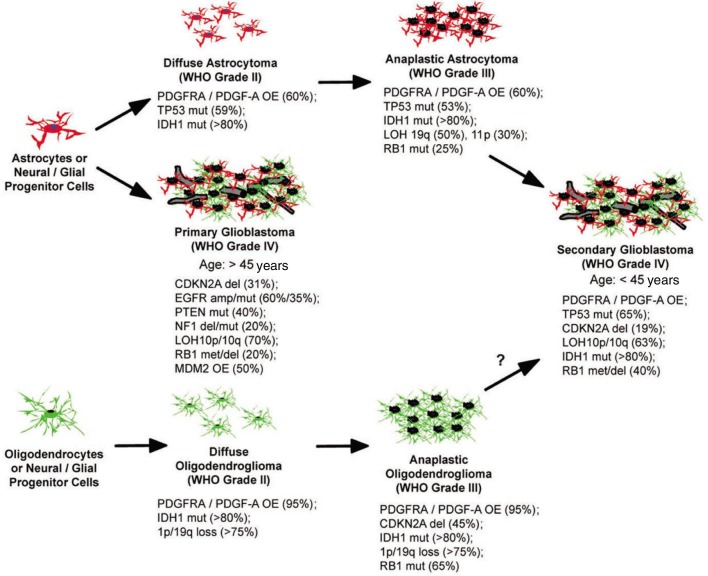
Malignant brain tumors are relatively rare but deadly due to their location and highly invasive capacity, rendering them inaccessible to surgical resection and resistant to current therapies[1],[2]. Clinically, these tumors are classified based on the predominant cell type as determined by histological approaches[3]. Among them malignant gliomas originating from glial cells represent the most common and aggressive type of tumor in the central nervous system (CNS)[2],[4]. Malignant gliomas in adults can be histologically classified into astrocytomas, oligodendrogliomas and mixed oligoastrocytomas. Further, within each type they can be divided into 4 grades corresponding to their malignancy and genetic alterations (WHO grade I–IV) (Figure 1). Despite intensive treatments including maximal surgical resection combined with radiotherapy and concurrent or adjuvant chemotherapy, the median survival of patients with grade IV glioblastoma multiforme (GBM) remains 13 to 16 months after diagnosis[5]. Clinical GBMs are composed of poorly differentiated glial cells with features such as uncontrolled growth, resistance to apoptosis, diffuse infiltration, cellular pleomorphism, nuclear atypia, mitotic abnormalities, microvascular proliferation and focal necrosis[2],[3]. Based on genetic and clinical presentation, GBM has been classified into two subtypes. Primary GBMs, which arise de novo with short to no clinical history, predominantly occur in older patients, whereas secondary GBMs develop from lower-grade gliomas often occur in younger patients[2],[4] (Figure 1).
Figure 1. Progression and genetic alterations in human malignant gliomas.
Astrocytoma may arise from astrocytes or progenitor cells that accumulate genetic alterations and become transformed. IDH1 and TP53 mutations are believed the first alterations occurring in these tumors. Oligodendrogliomas can develop from the same progenitor cells or mature oligodendrocytes that acquire a distinct set of genetic changes. The earliest changes that occur include IDH1 mutation and a 1p/19p loss. Representative genetic changes are shown with the estimated frequency of occurrence within each type of tumor. OE, overexpression; mut, mutation; del, deletion; met, promoter methylation; amp, amplification.
Over the past decade, data have accumulated characterizing genetic alterations in human gliomas, including the activation of oncogenes and the inactivation of tumor suppressor genes[2],[4],[6]–[8]. It appears that in human gliomas, these genetic alterations occur in a pattern corresponding to distinct histologic subtypes and different grades of tumors. For example, it is believed that low-grade astrocytomas and oligodendrogliomas may develop from common glial progenitor cells that acquire features of astrocytic tumors in the presence of TP53 mutations and oligodendrocytic tumors in the presence of 1p/19q chromosomal loss[7] (Figure 1). Additionally, platelet-derived growth factor receptor alpha (PDGFRA)/PDGF-A overexpression and isocitrate dehydrogenase 1 (IDH1) mutations are some of the major genetic alterations found in low-grade gliomas as well as secondary GBMs. When the low-grade tumors progress toward the high-grade secondary GBMs, additional changes such as CDK-dependent kinase inhibitor (CDKN) 2A/CDKN2B deletion are acquired (Figure 1). In the primary GBMs, however, a distinct set of genetic changes are observed, such as epidermal growth factor receptor (EGFR) amplification/mutation, phosphatase and tensin homolog (PTEN) mutations/deletion and Mdm2 p53 binding protein homolog (MDM2) overexpression (Figure 1), suggesting a different cell-of-origin of these tumors is responsible for generating primary GBMs[2],[4],[6]–[8].
In this review, we briefly discuss PDGF signaling with an emphasis on PDGFRα signaling and its role in normal glial cell development. We then summarize the evidence obtained from various studies of human glioma tissues and animal models of PDGF-induced tumorigenesis. Lastly, we provide insights into how new treatments targeting PDGF signaling will benefit patients with this subset of brain tumors.
PDGFRα Signaling
PDGF was purified as a molecule released by platelets into the whole blood serum, stimulating the proliferation of various mesenchymal[9]–[11] and glial cells[12],[13]. The mitogenic effect of this growth factor requires that target cells express the receptors for PDGF (PDGFR)[14]. The PDGF family consists of four ligands, PDGF-A, -B, -C, and -D, and two receptors, PDGFRα and PDGFRβ. The structures of PDGF-A and -B are largely similar, consisting of a PDGF/VEGF core domain with conserved cysteine residues called the cysteine knot motif and an N-terminal propeptide region that is removed intracellularly for activation prior to secretion[15]. Additionally, in PDGF-B and a membrane-bound, long alternative splice form of PDGF-A, there is a C-terminal basic “retention motif” that can interact with haparan sulfate proteoglycans of the extracellular matrix (ECM)[16]. The retention motif needs to be cleaved prior to secretion of the ligand. Expression of long- or short-form PDGF-A results from alternative splicing of exon 6 of PDGFA and is cell-type specific. Human glioma cells produce mainly the long form[17], whereas normal human endothelial cells express the short form of PDGF-A. Interestingly, the differential alternative splicing of PDGF-A might determine its mitogenic capacity in these cells[17]. The two newly found PDGFs, PDGF-C and PDGF-D, contain a distinct N-terminal domain called the CUB domain. It is cleaved extracellularly after secretion of the growth factors and this cleavage is important for their activity[18]. PDGFRs share a common structure including five extracellular immunoglobulin (Ig) loops and a split intracellular tyrosine kinase (TK) domain separated by a kinase insert region[15],[19]. Similar structures can also be found in other receptor tyrosine kinases (RTKs) such as vascular endothelial growth factor receptors (VEGFRs), c-Kit, c-Fms and fms-related tyrosine kinase 3 (Flt-3)[19].
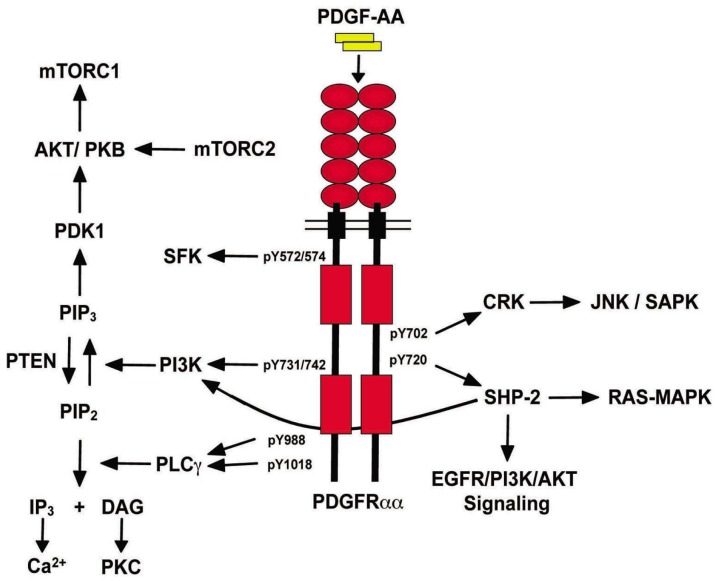
PDGF ligands function as disulfide-linked homo- or hetero-dimers, PDGF-AA, PDGF-AB, PDGF-BB, PDGF-CC and PDGF-DD[18]. These dimeric isoforms are capable of binding to two structurally related RTKs with different specificities (PDGFRα and PDGFRβ)[18],[20]. The binding of PDGF ligands induces dimerization of PDGFRs and juxtaposition of their intracellular tyrosine kinase domains leading to trans-autophosphorylation of multiple tyrosine residue sites[21]. The subsequent association between different SH2 domain–containing signaling molecules and phosphorylated tyrosine residues engages various downstream signaling cascades (Figure 2), gene transcription events and various cellular behaviors, such as cell proliferation, apoptosis, actin reorganization and chemotaxis[15],[22]. The SH2 domain–containing signaling effectors include phosphoinositide-3-kinase (PI3K), phospholipase C (PLC)–γ, Src family kinase (SFK), protein tyrosine phosphatase SHP-2, GTPase-activating protein (GAP) for Ras, signal-transducer and activator of transcription proteins (STATs), as well as adaptor proteins such as growth factor receptor-bound protein (Grb)2, Grb7, SHC-adaptor protein (Shc), SH2/SH3 adaptor protein (Nck) and v-crk sarcoma virus CT10 Oncogene homolog (Crk)[22]. PDGFRα and PDGFRβ bind to distinct but overlapping sets of these signaling molecules upon ligand stimulation. An allelic series of mutant PDGFRs have been generated and it appears that in comparison to PDGFRβ, PDGFRα relies more on the activation of specific signaling pathways to function properly in specific stages and organs during animal development[23],[24]. For example, PI3K signaling is indispensable for PDGFRα during early development, whereas for PDGFRβ, disruption of PI3K alone has minimal effect on normal development[19]. Cytoplasmic domain swapping experiments further reveal that in contrast to PDGFRβ, it is not the intrinsic properties of the receptor but the ability of PDGFRα to engage specific signaling molecules that determines the activity and function of receptor signaling[25],[26]. In this review, we focus on the downstream SH2 domain–containing signaling effectors of PDGFRα (Figure 2).
Figure 2. PDGFRα/PDGF-A signaling.
PDGF-AA homodimers bind and induce the dimerization and autophosphorylation of the PDGFRαα homodimer receptor. Subsequently, SH2 domain-containing signaling effectors are recruited to the receptor by binding to specific phosphorylated tyrosine residues, initiating downstream signaling cascades.
Members of the PI3K family consist of a regulatory p85 subunit and a catalytic p110 subunit. Upon association with the phosphorylated tyrosine residues 731 and 742 (Tyr-731/42) of the PDGFRα intracellular domain[15], PI3K is activated and produces phosphatidylinositol 3,4,5-triphosphate [PI (3,4,5)P3 or PIP3] in the plasma membrane. PIP3 then recruits several downstream effectors such as Akt/PKB[15],[22]. Activation of the PI3K pathway leads to several cellular effects including cell growth, survival, chemotaxis and actin reorganization[15],[22] (Figure 2). PLC-γ shares the same substrate as PI3K. Upon association to Tyr-988 and -1018, it phosphorylates PIP2 to generate inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG), which then mobilize intracellular Ca2+ and activates the PKC family, respectively[15],[22] (Figure 2). PDGF-induced PLC-γ activation is responsible for cellular effects such as cell motility and growth. The full activation of PLC-γ, however, is dependent on PIP3 generated by PI3K, since the recruitment of PLC-γ requires association of its pleckstrin homology (PH) domain with PIP3 in the plasma membrane[15],[22]. The SFK represents a family of tyrosine kinases including Src. Their binding sites on PDGFRα autophosphorylated tyrosine sites are Tyr-572 and -574. PDGFRα-mediated Src activation appears to be dispensable for mitogenic responses in some cell types[15],[22] (Figure 2). SHP-2 is an SH2-containing protein tyrosine phosphatase that binds to phosphorylated Tyr-720 of activated PDGFRα. Its phosphatase activity can potentially dephosphorylate the RTK and downstream effectors, leading to inactivation of these proteins. However, it has also been shown that SHP-2 is involved in the up-regulation of several effectors such as the Ras/MAPK and Src pathways, possibly through its role as an adaptor[15],[22] (Figure 2). Crk is an adaptor that binds to phosphorylated Typ-702 of PDGFRα. Upon binding, it then activates the nucleotide exchange protein C3G and the subsequent activation of the JNK/SAPK pathway[15],[22] (Figure 2).
An allelic series of PDGFRA tyrosine-to-phenylalanine mutations disrupting association between the RTK with different downstream effectors and signaling pathways have been generated[27]. This study demonstrated that PDGF-AA–induced DNA synthesis was strictly dependent on PI3K signaling only, whereas Src, PI3K and PLC-γ are required for PDGF-mediated chemotaxis with Src being the most important effector for cell motility[27]. Surprisingly, all PDGFRα mutants in the “add-back panel” that can only initiate one of the five downstream pathways were not able to restore the PDGF-induced chemotaxis to the level achieved by WT PDGFRα, indicating that activation of multiple effectors is required for full activity of the receptor to mediate certain cellular responses[27]. Using a similar strategy, PI3K and PLC-γ were shown to play predominant roles in preventing apoptosis of mesoderm cells during early Xenopus embryo development[28]. These studies suggest that distinct downstream effectors of PDGFR may be required for different cell types, species and stages of development. To further dissect the contribution of signaling pathways emanating from PDGFRα, Klinghoffer et al.[23] further generated knock-in mice that harbored one of the three mutants, PDGFRα-F7, PDGFRα-F731/42 or PDGFRα-F572/74. Interestingly, mice that homozygously harbored PDGFRα-F731/42, as well as PDGFRα-F7, displayed phenotypes comparable to PDGF-A– or PDGFRα-null animals, including growth retardation, skeletal and lung development abnormalities[23]. However, Src association with PDGFRα was only required for oligodendrocyte development, possibly due to the role of Src in promoting progenitor cell migration[23]. These experiments suggest that PI3K is the major effector of PDGFRα signaling during embryogenesis.
Role of PDGFRα and PDGF-A during Development of the CNS and Injury
The developmental functions of PDGFRs and PDGFs have predominantly been unveiled through genetic studies in mice [19],[29]. The role of the closely related PDGF/VEGF superfamily in neural and glial development is evolutionarily conserved: in Caenorhabditis elegans, VERs, the VEGF family which are structurally similar to vertebrate VEGFRs, are expressed in neurons and glial cells[30]; in Drosophila melanogaster, the PDGFRA/EGFR homologue PDGFR/VEGFR-related receptor (PVR) and its ligands, PDGF- and VEGF-related factor (PVF), are required for ventral midline glia cell survival[31]. In the mammalian CNS, messenger RNA (mRNA) and protein expression of PDGF ligands and receptors are found throughout various brain regions[32]. In rodents, the PDGFRα-positive oligodendrocyte progenitor cells (OPCs)[33] originate at the ventricular surface near the floor plate of spinal cord in embryonic day 12.5 (E12.5) of mice (E14 for rats)[34]. Neurons and astrocytes throughout the CNS produce PDGF-A, which acts as a mitogen for OPCs[35]–[38]. Early studies showed PDGF-A–knockout mice develop tremor caused by severe hypomyelination on neuronal projections throughout the CNS[39],[40]; PDGFRα-null mice die prenatally and the OPC isolated from the premortem animals shows defects in proliferation and differentiation[41]. Subsequent studies using PDGFRα-mutant–knock-in mice showed that downstream SFK and PI3K are important for normal myelination of the CNS[23]. Further analyses revealed that PDGF-A, but not PDGF-B, is required for proliferation, migration and normal differentiation of PDGFRα-positive OPCs[36],[37],[42],[43], and that over-production of PDGF-A in the CNS neurons or astrocytes induces hyper-proliferation of the PDGFRα-positive O-2A progenitor cells in a paracrine manner[40],[44]. The mitogenic effect of PDGF-A appears to rely on various environmental cues endowed by the ECM, which activate various integrins[45] and chondroitin sulfate proteoglycan 4 (CSPG4)/nerve/glial antigen (NG2)[46]. The proliferation rate and the amount of PDGF-A available are proportional and the OPC population continues to grow until the concentration of PDGF-A becomes limiting; that is, the rate of PDGF-A consumption exceeds the rate of its production due to the increased number of OPCs[47]. The OPCs may also be able to sense the amount of PDGF-A by the “rheostat-like” PDGFRα function, switching downstream signaling from the PI3K pathway to PLC–γ under higher PDGF-A concentration[41]. Similar mitogenic function of PDGFRα/PDGF-A signaling has also been demonstrated during CNS injury. After experimental demyelination, the ensuing remyelination often involves increased proliferation of PDGFRα- and NG2-positive OPCs[48]. A decrease in genomic PDGFRα dosage, as shown in heterozygous PDGFRα+/− mice[49], resulted in impaired remyelination after myelin damage to the CNS[50].
In addition to its role as a mitogen for OPCs, PDGF-A acts as a lineage specification factor, directing the differentiation of embryonic neural progenitor cells into oligodendrocytes[51],[52]. In cultured O-2A progenitor cells isolated from rat optic nerve, PDGF-A and basic fibroblast growth factor (bFGF) are also capable of maintaining proliferation and self-renewal potential, as well as preventing terminal differentiation into oligodendrocytes[53]. Interestingly, the effect of these growth factors on OPC proliferation and differentiation appear to be reserved in the adult CNS in that following demyelination, PDGF-A and bFGF levels are increased in order to promote OPC proliferation and subsequent differentiation into mature oligodendrocytes for remyelination[54]. The observation that PDGF and bFGF convert adult rat O-2A cells to their perinatal counterparts after injury to the adult brain[55] implies their capacity to reprogram OPCs to their progenitors[56].
In addition to the role of PDGF-A in glial-restricted progenitor cell development, PDGF-A is capable of stimulating the proliferation of PDGFRα-positive neural stem cells (NSCs) residing in the subventrical zone (SVZ) of the adult murine brain, which are capable of differentiating into neurons and oligodendrocytes[57]. The presence of this common progenitor has been long appreciated, and identified in embryonic mouse spinal cord by expression of the basic helix-loop-helix transcription factor Oligs[58].Through observations employing Olig2-null animals, one of the Olig genes, Olig2, was found to be important for oligodendrocyte development[58]. In situ hybridization experiments further revealed that expression of Olig2 precedes that of PDGFRα in mouse embryonic oligodendrocyte precursors[58],[59], suggesting a possible link between the genes during glial development. Indeed, ectopic expression of Olig2 in vivo induces expression of the transcription factor Sox10, which together with Sox9, controls transcription of the gene encoding PDGFRα[60],[61]. Intriguingly, in mouse embryos, Olig expression is regulated by Shh[59] that is also required for self-renewal of the embryonic forebrain progenitors induced by PDGF-A and bFGF[62], evincing a complex interaction of Shh, Sox, Olig and PDGF in oligodendrocyte development. Surprisingly, normal astrocytic development does not seem to be affected in Olig2-null animals[58]. Whether OPCs are derived from glial-restricted O-2A cells or neuron/oligodendrocyte common progenitor cells is still an open question[62]. However, these findings suggest the existence of distinct PDGF-responsive neural progenitor cells, each of which is Spatiotemporally responsible for generating various neural cells during CNS development. These observations in normal CNS development not only helped to elucidate the identity of the long pursued cells-of-origin of the malignant gliomas, but also raised questions such as whether these intrinsic developmental and physiological properties and functions are preserved after cell transformation.
PDGF-B, -C, and -D during Development of the CNS
While several studies have shown that PDGF-B was able to potently induce glioma development from resident glial (progenitor) cells[63], its role in normal glial cell development is still elusive. Postmitotic CNS neurons are the major cells that express PDGF-B ligand. However, unlike PDGF-A, neuron-specific PDGF-B knockout did not impact normal CNS development or astroglial and angiogenic responses to CNS injury[64]. The role of neuron-derived PDGF-B in normal CNS development and injury is thus obscure.
To a similar extent, the roles of the two new members of the PDGF family, PDGF-C and PDGF-D, in CNS development remain relatively elusive. PDGF-C was first discovered in the spinal cord of chick embryo. Thus, it was originally named spinal cord-derived growth factor (SCDGF)[65]. In rats, PDGF-C is expressed predominantly in embryonic brain and spinal cord, while PDGF-D expression is evident in adults rather than in embryos[66]. In addition, PDGF-C was detected by immunohistochemistry (IHC) in developing brain and spinal cord in mouse embryo[67]. Interestingly, PDGF-C expression was also found in the transient external granular layer cells of the cerebellum in mouse E13 embryo[68]. The requirement of PDGF-C for normal spinal cord development was further demonstrated by studies of PDGF-C knockout mice[69]. Mice deficient in PDGF-C displayed a range of abnormalities and died perinatally due to difficulties in breathing and eating. Examination of the mouse embryos revealed the development of spina bifida occulta, an incomplete closure and deformation of the vertebrae[69]. PDGF-D knockout animals have not been reported yet and studies on its role in normal CNS development still fall behind. However, PDGF-D has been shown to stimulate blood vessel formation and wound healing processes[70], suggesting that PDGF-D might have an impact on tumor angiogenesis.
PDGFRα Signaling in Human Glioma
The first implication that PDGF autocrine signaling can contribute to cell transformation comes from the discoveries of amino acid sequence similarity between simian sarcoma virus (SSV) Oncogene v-sis and PDGF-B chain gene[71],[72]. In human glioma, a glioma cell line was shown to secrete a growth-promoting factor[73]–[75] that was later found to be three disulfide-linked dimers, PDGF-AA, PDGF-AB and PDGF-BB[76]–[78]. Each of these dimers had distinct binding affinities to receptors α and β[20]. Subsequent studies revealed that mRNA for PDGF ligands and receptors were expressed in a series of established glioma cell lines[79],[80]. In situ hybridization and IHC analyses further demonstrated that in human glioma tissue, tumor cells express PDGF-A, -B, and PDGFRα, whereas surrounding hyperplastic endothelial cells express PDGF-B and PDGFRα, suggesting the presence of autocrine and paracrine PDGF stimulation in glioma development[81]–[84]. As described in the previous section during development of the CNS, the OPC number increases in response to the level of paracrine PDGF-A expression. Whereas in an autocrine situation, in which PDGF ligand is over-expressed in PDGFRα-positive progenitor cells, the rate of PDGF-A production increases together with the number of the progenitor cells, theoretically rendering these cells capable of proliferating indefinitely[29]. Thus in an autocrine situation, the growth factor sensing mechanism of OPCs might be lost, potentially leading to tumor formation.
Overexpression and gene amplification of PDGFRα occurred mostly in lower-grade gliomas as well as secondary GBMs[2],[4],[85]–[87], representing a distinct subtype of GBMs from those with EGFR overexpression (Figure 1)[88]. In a clinical study of IHC staining on 103 grade II astrocytomas, PDGFR expression was found to be an independent prognostic factor for these patients with low-grade gliomas[89]. Overexpression of PDGF ligands, however, varies among gliomas of different grades, with PDGF-A being expressed in all grades and PDGF-B only in high-grade GBMs[90], suggesting PDGF-B may be involved in the conversion of low- to high-grade gliomas. In fact, it has been shown that in human oligodendroglioma tissue[91] and in a mouse glioma model[92], the level of PDGF signaling may predict grade and malignancy.
High-throughput approaches such as comparative genomic hybridization (CGH) and microarray have been established, enabling glioma profiling in a whole-genome scale[93]–[96]. The most comprehensive and reliable analysis of genomic alteration in primary GBM tissue has been conducted by The Cancer Genome Atlas Research Network (TCGA)[97], a collaborative project sponsored by the National Cancer Institute (NCI), National Institutes of Health in USA[97]. This multi-institutional effort provides invaluable resources for gene expression and mutation, DNA copy number alterations (CNA), microRNA expression and DNA methylation data for GBMs. Most of the previously appreciated genetic aberrations such as alterations in the RB, TP53 and RTK pathways were re-captured in the initial TCGA studies. However, an unexpected higher frequency (∼13% ) of focal amplifications of the 4q12 locus harboring PDGFRA was reported for primary GBMs in the TCGA collection than that published previously [97],[98]. Further classification of these GBMs by gene expression signatures revealed that PDGFRA overexpression occurs together with TP53 and IDH1 mutations in the Proneural subtype of GBMs, which also express oligodendrocyte lineage genes such as OLIG2 and SOX [99]. Interestingly, within this subtype of GBMs, PDGFRA amplifications and PIK3CA/PIK3R1 mutations (leading to constitutively active PI3K subunits p110 and p85, respectively) mostly occur in a mutually exclusive manner[99], signifying an overlapping functionality between these two alterations in gliomagenesis. Indeed, analyses in different cohorts of gliomas of different grades using other methods such as single nucleotide polymorphism array (SNP-Chip)[100] or a Western blot– based proteomic analysis[101] confirmed the importance of PI3K/AKT/mTOR pathway activation in PDGFRA amplified gliomas.
The Proneural subtype of GBMs harboring PDGFRA amplification from the TCGA dataset share similar gene expression profiles with the previously reported pro-neural (PN) subclass[94]. Nearly all WHO grade III tumors including astrocytomas, oligodendrogliomas and mixed oligoastrocytomas have been classified as a PN subclass, as well as secondary GBMs[94],[99]. Patients with PN tumors are younger and survive longer. Analyses of signature gene expression in PN tumors also revealed that these tumors resemble neuroblast/glial progenitor cells in the fetal and adult brain indicating a possible cell of origin of this subclass of glioma[94]. Indeed, putative NSCs residing in the adult mouse SVZ are PDGFRα-positive and respond to PDGF-A stimulation by forming a glioma-like lesion[57], suggesting susceptibility of adult NSCs to oncogenic transformation by PDGFRA alteration.
PDGF-C and PDGF-D in Human Glioma
The two novel PDGF ligands, PDGF-C and PDGF-D, were first implicated in the development of glioma by a study that examined the expression of PDGF ligands and receptors in glioma cell lines and primary glioma tissues[102],[103]. Lokker and colleagues found that PDGF-C and -D were expressed at high levels in tumor tissues as compared with those in the normal brain. Additionally, autocrine loops involving PDGF-C/PDGFRα and PDGF-D/PDGFRβ exist in all tumors examined. However, the pathologic consequences of PDGF-C and -D overexpression and whether they compensate PDGF-A and -B functions or merely initiate redundant signaling cascades remains elusive. Interestingly, in a subcutaneous tumor model, PDGF-C was associated with tumor resistance to anti-VEGF therapies. The resistant tumors tend to up-regulate the level of PDGF-C after anti-VEGF treatment, possibly compensating the function of the inhibited VEGF in stimulating angiogenesis[104]. Indeed, in patients with recurrent GBMs after anti-VEGF therapy, PDGF-C together with c-Met was expressed at high levels, especially in the center of the tumor mass[105]. The mechanism of resistance to anti-VEGF therapy may involve the capacity of PDGF-C to alter the structure of blood vessels by recruiting perivascular cells[106], thus stabilizing the vessels and rendering them insensitive to anti-VEGF treatment. The function of PDGF-D/PDGFRα autocrine signaling remains unknown despite its presence in several GBM cell lines and primary tissues[102],[103]. However, it has been shown that PDGF-D was responsible for inducing chemotactic tropism of PDGFR-positive stem cells toward glioma cells in the mouse brain[107]. This observation is likely important for the development of therapeutic strategies using stem cells as a drug carrier.
PDGF-induced Brain Tumors in Animal Models
Despite intensive research, the prognosis for patients diagnosed with high-grade glioma remains dismal. A major hurdle to progress is the lack of animal models accurately recapitulating the behavior and neuropathological features of human tumors[108]. Several genetically engineered mouse (GEM) models of spontaneous glioma formation have been developed by generating mice with astrocyte-specific transgenic expression of v-src[109] or H-ras[101] and with the concomitant loss of the neurofibromatosis 1 (NF1) and p53 tumor suppressors[111],[112]. Recently, transgenic mice that harbor astrocyte-specific expression of PDGF-B (hGFAPpPDGFB mice) were generated and assessed for spontaneous glioma formation[113] (Table 1). The loss of p53 or the overexpression of PDGF-B alone was unable to induce brain tumor growth. However, when Hede and colleagues crossed hGFAPpPDGFB mice with p53-null mice, the resultant hGFAPpPDGFB/p53-null mice developed an aggressive GBM-like tumor with characteristics frequently found in human GBMs[113]. The researchers also observed the increased expression of PDGFRα in tumor cells, while PDGFRβ was expressed in the tumor vasculature, and that the tumor expressed a series of lineage markers indicating the presence of various cell types including neurons, astrocytes and oligodendrocytes, especially in large tumors.
Table 1. Summary of animal models of PDGF-induced spontaneous brain tumor formation.
| Method | Vector | PDGF | Strain | Age | Target Cell | Histology | Reference(s) |
| Virus Sup I.C. Inj | MoMuLV | PDGF-B | C57BL/6 | Neonatal | Non-specific | GBM / PNET | [115] |
| Virus Sup I.C. Inj | MoMuLV | PDGF-B | p53−/ | Neonatal | Non-specific | GBM / PNET | [123] |
| Virus Sup I.C. Inj | MoMuLV | PDGF-B | Ink4a/Arf−/ | Neonatal | Non-specific | GBM / PNET | [123] |
| Packaging Cell I.C. Inj | RCAS | PDGF-B | Ntv-a | Neonatal | Nestin+ cells | Low-grade Oligo | [116] |
| Packaging Cell I.C. Inj | RCAS | PDGF-B | Ntv-a, Ink4a/Arf−/− | Neonatal | Nestin+ cells | Anaplastic Oligo | [116] |
| Packaging Cell I.C. Inj | RCAS | PDGF-B | Gtv-a | Neonatal | GFAP+ cells | Low-grade Oligo/Oligoastro | [116] |
| Packaging Cell I.C. Inj | RCAS | PDGF-B | Gtv-a, Ink4a/Arf−/− | Neonatal | GFAP+ cells | Anaplastic Oligo/Oligoastro | [116] |
| Packaging Cell I.C. Inj | RCAS | PDGF-B | Gtv-a, p53−/ | Neonatal | GFAP+ cells | Low-grade Oligo/Oligoastro | [116] |
| Packaging Cell I.C. Inj | RCAS | ACC-PDGF B-HA | Ntv-a | Neonatal | Nestin+ cells | High-grade Oligo | [92] |
| Virus Sup C.C. Inj | pQ (Retroviral) | PDGF-B | Sprague Dawley Rat | Adult | Non-specific | GBM | [117] |
| Virus Sup LV, Inj | pQ(Retroviral) | PDGF-B | Sprague Dawley | Neonatal | SVZ Cells | GBM | [117] |
| Packaging Cell LV. Inj | pCEG (Retroviral) | PDGF-B | Rat C57BL/6 | E14 | SVZ Cells | High-grade Oligo | [52], [118] |
| Protein L.V. Infusion | NA | PDGF-A | CD-1 | Adult | SVZ Cells | Low-grade Glioma | [57] |
| Zygote Pronuclear Inj | IRESβGEO | PDGF-B | Transgenic hGFAPpPDGFB, p53−/ | E0 | GFAP+ cells | GBM / High-grade Oligo | [113] |
| Zygote Pronuclear Inj | IRESβGEO | Long-form PDGF-A | Transgenic hGFAPpPDGFA | E0 | GFAP+ cells | Grade III Oligoastro | [114] |
Spontaneous gliomas have been found to develop in transgenic mice overexpressing the long isoform of PDGF-A, PDGF-AL, under the control of a GFAP promoter[114] (Table 1). Unlike hGFAPpPDGFB mice, hGFAPpPDGFAL mice did not require additional genetic aberrations such as p53 deletion in order to develop spontaneous tumors. These mice were also found to harbor neoplastic cells positive for PDGFRα, Olig2 and NG2 in the SVZ, corpus callosum, hippocampus and cerebellum [114], suggesting that the resident OPCs in these areas of the brain were likely the target of transformation. Alternatively, GFAP-positive NSCs could also be the cell of origin for PDGF-induced gliomas in the hGFAPpPDGFAL mice, since PDGF-A has been reported to be capable of biasing the fate of stem cells residing in the SVZ toward OPC-like cells[57]. Histologically, spontaneous gliomas formed in hGFAPpPDGFAL mice showed characteristics of both astrocytes and oligodendrocytes and were determined to be WHO grade III oligoastrocytomas (Table 1). Although it is difficult to assess the tumor-promoting potential of PDGF-B, short-and long-form PDGF-A by comparing the transgenic mice generated by overexpression of these genes, due to the differences in promoter strength and transgenic copy numbers among these mice, these studies have suggested that glioma-specific long-form PDGF-A may exert tumorigenic effects through distinct signaling pathways than those stimulated by PDGF-B and short-form PDGF-A[114].
Other GEM models of PDGF-induced glioma development have also been described, most of which involve a single intracranial injection of retroviruses expressing PDGF-B ligand [115]–[119] (Table 1) targeted nonspecifically, or to astrocytes (GFAP-positive) or neural progenitor cells (nestin-positive) using cell type-specific promoters. In these studies, PDGF-B overexpression induces glioma growth from various cell types including glial progenitor cells, astrocytes and neural progenitor cells, and the resulting tumors frequently exhibited characteristics of oligodendrogliomas or mixed oligoastrocytomas (Table 1). These two models, termed germ-line transgenic or retrovirus induction, differ in many ways such as the incidence of tumorigenesis and histological features. Discrepancy may be due to different times of stimulation during animal development, different cells being targeted or retroviral-mediated insertional mutagenesis in genes that cooperate with PDGF to induce tumorigenesis[120]. Additionally, the local injection of various agents including cells or viruses into the brain of mice may induce the proliferation of VEGF-expressing Olig2-positive glial cells and a local increase in angiogenesis[121],[122]. These factors may contribute to the differential tumor induction potentials observed in these models where PDGF-B alone was targeted to GFAP-positive cells in the brain. Despite the differences between the transgenic and the virus-mediated models, an increase in tumor malignancy by concomitantly introducing a loss of p53 function or the Ink4a/Arf locus was evident in all studies[113],[116],[119],[123] (Table 1), demonstrating a cooperative effect of PDGF overexpression and the disruption of a p53 or RB pathway in glioma tumorigenesis. These observations in the animal models thus provide an experimental basis for TCGA analyses, demonstrating that the three core signaling pathways (RTK, RB, and P53) are important for GBM viability.
PDGFRα-positive progenitor cells that respond to PDGF stimulation can be found in various parts of the adult brain [57],[113],[117]. Exogenous PDGF-A infusion into the adult SVZ induces PDGFRα-positive neural progenitor cells to generate glioma-like lesions in the mouse brain[57]. To investigate how PDGF-A–mediated PDGFRα activation contributes to glioma formation, we utilized an orthotopic transplantation model, in which we implanted various PDGF receptor or ligand overexpressing human or mouse cells into the brains of mice to assess their tumorigenic potentials[124]. In order to determine which signaling pathway(s) downstream of PDGFRα are necessary for its oncogenic capacity, we chose PDGF-A instead of PDGF-B since PDGF-A only stimulates PDGFRα but not PDGFRβ. When the tyrosine phosphorylation sites for PI3K binding were mutated to phenylalanine, PDGFRα lost the capacity to transform mouse astrocytes, both in vitro in soft agar and in vivo in the brain of mice. Unexpectedly, the association between PDGFRα and a downstream effecter (SHP-2) is required for maximal tumorigenic potential observed in cells overexpressing the wild-type receptor. This is significant, since a recent automated signaling network-based analysis on the TCGA genomic data identified SHP-2/PTPN11 as one of six linker genes that are not altered in GBMs but are statistically enriched for interactions with the commonly altered genes in GBMs[125], including EGFR, PDGFRA, PIK3R1, KIT and KDR. Our observation thus provides functional validation of the role of SHP-2/PTPN11 in mediating PDGFRα signaling in gliomagenesis.
There are high levels of Akt phosphorylation in our model of PDGFRα/PDGF-A–overexpressing tumors[124]. However, using a RCAS/tv-a system, Dai and colleagues have shown that combined activation of Akt and K-Ras generated astrocytomas while PDGF-B overexpression led mostly to oligodendrogliomas[126]. They further showed that the blockade target of rapamycin (mTOR) converted PDGF-B–induced astrocytomas to oligodendroglioma-like tumors[127]. Thus it appears that in the RCAS/tv-a system, PDGF-B and Ras/Akt/mTOR signaling pathways promote the formation of distinct histological types of animal brain tumors. However, in human glioma, as shown by a proteomic study, the activation of mTOR and Ras/Erk pathways frequently co-occurred in the PDGF-B–enriched subgroup[101] and two of four oligodendrogliomas in this study were of the PDGF-B/mTOR/Ras activation subtype, suggesting a more complex connection between PDGF signaling and glioma cell lineage specification may exist in humans than found in mice. Additionally, since there was no concomitant overexpression of PDGFRα in tumors generated from RCAS/tv-a–mediated PDGF-B expression, it will be interesting to determine whether targeted co-overexpression of PDGFRα and PDGF-B in the mouse brain will generate gliomas with similar histopathological characteristics as does PDGF-B alone.
Prospects for PDGF-targeted Therapy in Glioma
Although PDGF targeted therapies using various tyrosine kinase inhibitors have been extensively studied over the past decade, the results were generally disappointing and have yet to shift the standard clinical treatment of GBM, which encompasses maximal surgical resection, followed by TMZ and radiation[128]. In order to design therapeutic strategies that can benefit patients with glioma driven and maintained by PDGF signaling, more thorough pre-clinical studies are warranted. Various animal models that are candidates for this purpose have been proposed[129]. Additionally, the identification of new potential molecular targets in tumors induced by PDGF signaling is also important. The application of systems biology to the genomic data obtained from patient samples is expected to aid in this process[130]. To generate a network of genes that share a regulatory program, the commonly altered genes in GBM were grouped into several functional “modules.” The interactions between each cancer-altered gene and other non-altered “linkers” can be identified within and among different modules[101]. SHP-2/PTPN11 was identified in this network approach as one of the six linkers and was also shown to be important in the PDGF-driven gliomagenesis in our model[124]. Molecules such as SHP-2/PTPN11, which are not commonly overexpressed or mutated in GBM, are often understudied. However, several signaling pathways may converge on these molecules to exert downstream function. Besides its involvement in regulating the PDGFRα/PI3K/Akt pathway in gliomas[124], SHP-2 has also been known to mediate the EGFR/PI3K/Akt [131] and the Ras/MAPK signaling pathways[132] (Figure 2). Thus, it represents a molecule whose loss may lead to the disruption of multiple signaling cascades involved in tumorigenesis. As tumors are considered to be “network-addicted” rather than “oncogene-addicted” [133], strategies targeting such a molecule in monotherapy or in combination with other cytotoxic drugs may prove more efficient than targeting multiple upstream RTKs. Recent advances in large-scale whole-genome profiling of GBMs should yield to the discovery of more promising targets similar to these. However, preclinical studies of targeted drugs require a more stringent selection of animal models that accurately emulate human tumors. In addition, personalized medicine should be administered to each individual, employing the data gained from genomic profiling and systems biology.
Acknowledgments
This work was supported in part by grants from NIH CA130966 and the Pennsylvania Department of Health and Innovative Research Scholar Awards of the Hillman Foundation to Shi-Yuan Cheng and Bo Hu, and a James S McDonnell Foundation Researching Award in Brain Cancers to Bo Hu.
Footnotes
Conflict of Interest Statement: The authors have declared that no conflict of interest exists.
References
- 1.Central Brain Tumor Register of the United States 2010 CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2004–2006 [R] 2010. http://www.cbtrus.org/2010-NPCR-SEER/CBTRUS-WEBREPORT-Final-3-2-10.pdf.
- 2.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment [J] Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumors of the central nervous system [J] Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen, PY, Kesari, S Malignant gliomas in adults [J] N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma [J] N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma [J] Am J Pathol. 2007;170(5):1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas [J] Cancer Sci. 2009;100(12):2235–2241. doi: 10.1111/j.1349-7006.2009.01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keisner SV, Shah SR. Pazopanib: the newest tyrosine kinase inhibitor for the treatment of advanced or metastatic renal cell carcinoma [J] Drugs. 2011;71(4):443–454. doi: 10.2165/11588960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Ross R, Glomset J, Kariya B, et al. A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro [J] Proc Natl Acad Sci USA. 1974;71(4):1207–1210. doi: 10.1073/pnas.71.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohler N, Lipton A. Platelets as a source of fibroblast growth-promoting activity [J] Exp Cell Res. 1974;87(2):297–301. doi: 10.1016/0014-4827(74)90484-4. [DOI] [PubMed] [Google Scholar]
- 11.Ross R, Vogel A. The platelet-derived growth factor [J] Cell. 1978;14(2):203–210. doi: 10.1016/0092-8674(78)90107-1. [DOI] [PubMed] [Google Scholar]
- 12.Westermark B, Wasteson A. A platelet factor stimulating human normal glial cells [J] Exp Cell Res. 1976;98(1):170–174. doi: 10.1016/0014-4827(76)90476-6. [DOI] [PubMed] [Google Scholar]
- 13.Heldin CH, Westermark B, Wasteson A. Platelet-derived growth factor: purification and partial characterization [J] Proc Natl Acad Sci USA. 1979;76(8):3722–3726. doi: 10.1073/pnas.76.8.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heldin CH, Westermark B, Wasteson A. Specific receptors for platelet-derived growth factor on cells derived from connective tissue and glia [J] Proc Natl Acad Sci USA. 1981;78(6):3664–3668. doi: 10.1073/pnas.78.6.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor [J] Physiol Rev. 1999;79(4):1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 16.Betsholtz C, Johnsson A, Heldin CH, et al. cDNA sequence and chromosomal localization of human platelet-derived growth factor A-chain and its expression in tumour cell lines [J] Nature. 1986;320(6064):695–699. doi: 10.1038/320695a0. [DOI] [PubMed] [Google Scholar]
- 17.Collins T, Bonthron DT, Orkin SH. Alternative RNA splicing affects function of encoded platelet-derived growth factor A chain [J] Nature. 1987;328(6131):621–624. doi: 10.1038/328621a0. [DOI] [PubMed] [Google Scholar]
- 18.Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms [J] Cytokine Growth Factor Rev. 2004;15(4):197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine [J] Genes Dev. 2008;22(10):1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heldin CH, Backstrom G, Ostman A, et al. Binding of different dimeric forms of PDGF to human fibroblasts: evidence for two separate receptor types [J] Embo J. 1988;7(5):1387–1393. doi: 10.1002/j.1460-2075.1988.tb02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly JD, Haldeman BA, Grant FJ, et al. Platelet-derived growth factor (PDGF) stimulates PDGF receptor subunit dimerization and intersubunit trans-phosphorylation [J] J Biol Chem. 1991;266(14):8987–8992. [PubMed] [Google Scholar]
- 22.Heldin CH, Ostman A, Ronnstrand L. Signal transduction via platelet-derived growth factor receptors [J] Biochim Biophys Acta. 1998;1378(1):F79–F113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 23.Klinghoffer RA, Hamilton TG, Hoch R, et al. An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development [J] Dev Cell. 2002;2(1):103–113. doi: 10.1016/s1534-5807(01)00103-4. [DOI] [PubMed] [Google Scholar]
- 24.Tallquist M, Kazlauskas A. PDGF signaling in cells and mice [J] Cytokine Growth Factor Rev. 2004;15(4):205–213. doi: 10.1016/j.cytogfr.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Klinghoffer RA, Mueting-Nelsen PF, Faerman A, et al. The two PDGF receptors maintain conserved signaling in vivo despite divergent embryological functions [J] Mol Cell. 2001;7(2):343–354. doi: 10.1016/s1097-2765(01)00182-4. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton TG, Klinghoffer RA, Corrin PD, et al. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms [J] Mol Cell Biol. 2003;23(11):4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenkranz S, DeMali KA, Gelderloos JA, et al. Identification of the receptor-associated signaling enzymes that are required for platelet-derived growth factor-AA-dependent chemotaxis and DNA synthesis [J] J Biol Chem. 1999;274(40):28335–28343. doi: 10.1074/jbc.274.40.28335. [DOI] [PubMed] [Google Scholar]
- 28.Van Stry M, Kazlauskas A, Schreiber SL, et al. Distinct effectors of platelet-derived growth factor receptor-alpha signaling are required for cell survival during embryogenesis [J] Proc Natl Acad Sci USA. 2005;102(23):8233–8238. doi: 10.1073/pnas.0502885102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betsholtz C. Insight into the physiological functions of PDGF through genetic studies in mice [J] Cytokine Growth Factor Rev. 2004;15(4):215–228. doi: 10.1016/j.cytogfr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Popovici C, Isnardon D, Birnbaum D, et al. Caenorhabditis elegans receptors related to mammalian vascular endothelial growth factor receptors are expressed in neural cells [J] Neurosci Lett. 2002;329(1):116–120. doi: 10.1016/s0304-3940(02)00595-5. [DOI] [PubMed] [Google Scholar]
- 31.Learte AR, Forero MG, Hidalgo A. Gliatrophic and gliatropic roles of PVF/PVR signaling during axon guidance [J] Glia. 2008;56(2):164–176. doi: 10.1002/glia.20601. [DOI] [PubMed] [Google Scholar]
- 32.Valenzuela CF, Kazlauskas A, Weiner JL. Roles of platelet-derived growth factor in the developing and mature nervous systems [J] Brain Res Brain Res Rev. 1997;24(1):77–89. doi: 10.1016/s0165-0173(97)00012-x. [DOI] [PubMed] [Google Scholar]
- 33.Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium [J] Nature. 1983;303(5916):390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- 34.Pringle NP, Richardson WD. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage [J] Development. 1993;117(2):525–533. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- 35.Richardson WD, Pringle N, Mosley MJ, et al. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system [J] Cell. 1988;53(2):309–319. doi: 10.1016/0092-8674(88)90392-3. [DOI] [PubMed] [Google Scholar]
- 36.Noble M, Murray K, Stroobant P, et al. Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell [J] Nature. 1988;333(6173):560–562. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- 37.Raff MC, Lillien LE, Richardson WD, et al. Platelet-derived growth factor from astrocytes drives the clock that times oligodendrocyte development in culture [J] Nature. 1988;333(6173):562–565. doi: 10.1038/333562a0. [DOI] [PubMed] [Google Scholar]
- 38.Yeh HJ, Ruit KG, Wang YX, et al. PDGF A-chain gene is expressed by mammalian neurons during development and in maturity [J] Cell. 1991;64(1):209–216. doi: 10.1016/0092-8674(91)90222-k. [DOI] [PubMed] [Google Scholar]
- 39.Fruttiger M, Karlsson L, Hall AC, et al. Defective oligodendrocyte development and severe hypomyelination in PDGF-A knockout mice [J] Development. 1999;126(3):457–467. doi: 10.1242/dev.126.3.457. [DOI] [PubMed] [Google Scholar]
- 40.Calver AR, Hall AC, Yu WP, et al. Oligodendrocyte population dynamics and the role of PDGF in vivo [J] Neuron. 1998;20(5):869–882. doi: 10.1016/s0896-6273(00)80469-9. [DOI] [PubMed] [Google Scholar]
- 41.McKinnon RD, Waldron S, Kiel ME. PDGF alpha-receptor signal strength controls an RTK rheostat that integrates phosphoinositol 3′-kinase and phospholipase Cgamma pathways during oligodendrocyte maturation [J] J Neurosci. 2005;25(14):3499–3508. doi: 10.1523/JNEUROSCI.5049-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armstrong RC, Harvath L, Dubois-Dalcq ME. Type 1 astrocytes and oligodendrocyte-type 2 astrocyte glial progenitors migrate toward distinct molecules [J] J Neurosci Res. 1990;27(3):400–407. doi: 10.1002/jnr.490270319. [DOI] [PubMed] [Google Scholar]
- 43.Barres BA, Hart IK, Coles HS, et al. Cell death and control of cell survival in the oligodendrocyte lineage [J] Cell. 1992;70(1):31–46. doi: 10.1016/0092-8674(92)90531-g. [DOI] [PubMed] [Google Scholar]
- 44.Woodruff RH, Fruttiger M, Richardson WD, et al. Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination [J] Mol Cell Neurosci. 2004;25(2):252–262. doi: 10.1016/j.mcn.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Baron W, Shattil SJ, ffrench-Constant C. The oligodendrocyte precursor mitogen PDGF stimulates proliferation by activation of alpha(v)beta3 integrins [J] EMBO J. 2002;21(8):1957–1966. doi: 10.1093/emboj/21.8.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishiyama A, Lin XH, Giese N, et al. Interaction between NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells is required for optimal response to PDGF [J] J Neurosci Res. 1996;43(3):315–330. doi: 10.1002/(SICI)1097-4547(19960201)43:3<315::AID-JNR6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 47.van Heyningen P, Calver AR, Richardson WD. Control of progenitor cell number by mitogen supply and demand [J] Curr Biol. 2001;11(4):232–241. doi: 10.1016/s0960-9822(01)00075-6. [DOI] [PubMed] [Google Scholar]
- 48.Redwine JM, Armstrong RC. In vivo proliferation of oligodendrocyte progenitors expressing PDGFalphaR during early remyelination [J] J Neurobiol. 1998;37(3):413–428. doi: 10.1002/(sici)1097-4695(19981115)37:3<413::aid-neu7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 49.Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites [J] Development. 1997;124(14):2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 50.Murtie JC, Zhou YX, Le TQ, et al. PDGF and FGF2 pathways regulate distinct oligodendrocyte lineage responses in experimental demyelination with spontaneous remyelination [J] Neurobiol Dis. 2005;19(1–2):171–182. doi: 10.1016/j.nbd.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 51.Hu JG, Fu SL, Wang YX, et al. Platelet-derived growth factor-AA mediates oligodendrocyte lineage differentiation through activation of extracellular signal-regulated kinase signaling pathway [J] Neuroscience. 2008;151(1):138–147. doi: 10.1016/j.neuroscience.2007.10.050. [DOI] [PubMed] [Google Scholar]
- 52.Appolloni I, Calzolari F, Tutucci E, et al. PDGF-B induces a homogeneous class of oligodendrogliomas from embryonic neural progenitors [J] Int J Cancer. 2009;124(10):2251–2259. doi: 10.1002/ijc.24206. [DOI] [PubMed] [Google Scholar]
- 53.Bogler O, Wren D, Barnett SC, et al. Cooperation between two growth factors promotes extended self-renewal and inhibits differentiation of oligodendrocyte-type-2 astrocyte (O-2A) progenitor cells [J] Proc Natl Acad Sci USA. 1990;87(16):6368–6372. doi: 10.1073/pnas.87.16.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frost EE, Nielsen JA, Le TQ, et al. PDGF and FGF2 regulate oligodendrocyte progenitor responses to demyelination [J] J Neurobiol. 2003;54(3):457–472. doi: 10.1002/neu.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolswijk G, Noble M. Cooperation between PDGF and FGF converts slowly dividing O-2Aadult progenitor cells to rapidly dividing cells with characteristics of O-2Aperinatal progenitor cells [J] J Cell Biol. 1992;118(4):889–900. doi: 10.1083/jcb.118.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondo T, Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells [J] Science. 2000;289(5485):1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- 57.Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling [J] Neuron. 2006;51(2):187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 58.Lu QR, Sun T, Zhu Z, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection [J] Cell. 2002;109(1):75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 59.Lu QR, Yuk D, Alberta JA, et al. Sonic hedgehog—regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system [J] Neuron. 2000;25(2):317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 60.Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors [J] Neuron. 2000;25(2):331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 61.Finzsch M, Stolt CC, Lommes P, et al. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression [J] Development. 2008;135(4):637–646. doi: 10.1242/dev.010454. [DOI] [PubMed] [Google Scholar]
- 62.Chojnacki, A, and Weiss, S Isolation of a novel platelet-derived growth factor-responsive precursor from the embryonic ventral forebrain [J] J Neurosci. 2004;24(48):10888–10899. doi: 10.1523/JNEUROSCI.3302-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calzolari F, Malatesta P. Recent insights into PDGF-induced gliomagenesis [J] Brain Pathol. 2010;20(3):527–538. doi: 10.1111/j.1750-3639.2009.00335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Enge M, Wilhelmsson U, Abramsson A, et al. Neuron-specific ablation of PDGF-B is compatible with normal central nervous system development and astroglial response to injury [J] Neurochem Res. 2003;28(2):271–279. doi: 10.1023/a:1022421001288. [DOI] [PubMed] [Google Scholar]
- 65.Hamada T, Ui-Tei K, Miyata Y. A novel gene derived from developing spinal cords, SCDGF, is a unique member of the PDGF/VEGF family [J] FEBS Lett. 2000;475(2):97–102. doi: 10.1016/s0014-5793(00)01640-9. [DOI] [PubMed] [Google Scholar]
- 66.Hamada T, Ui-Tei K, Imaki J, et al. The expression of SCDGF/PDGF-C/fallotein and SCDGF-B/PDGF-D in the rat central nervous system [J] Mech Dev. 2002;112(1–2):161–164. doi: 10.1016/s0925-4773(01)00625-6. [DOI] [PubMed] [Google Scholar]
- 67.Aase K, Abramsson A, Karlsson L, et al. Expression analysis of PDGF-C in adult and developing mouse tissues [J] Mech Dev. 2002;110(1–2):187–191. doi: 10.1016/s0925-4773(01)00560-3. [DOI] [PubMed] [Google Scholar]
- 68.Ding H, Wu X, Kim I, et al. The mouse Pdgfc gene: dynamic expression in embryonic tissues during organogenesis [J] Mech Dev. 2000;96(2):209–213. doi: 10.1016/s0925-4773(00)00425-1. [DOI] [PubMed] [Google Scholar]
- 69.Ding H, Wu X, Bostrom H, et al. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling [J] Nat Genet. 2004;36(10):1111–1116. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- 70.Reigstad LJ, Varhaug JE, Lillehaug JR. Structural and functional specificities of PDGF-C and PDGF-D, the novel members of the platelet-derived growth factors family [J] Febs J. 2005;272(22):5723–5741. doi: 10.1111/j.1742-4658.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 71.Waterfield MD, Scrace GT, Whittle N, et al. Platelet-derived growth factor is structurally related to the putative transforming protein p28sis of simian sarcoma virus [J] Nature. 1983;304(5921):35–39. doi: 10.1038/304035a0. [DOI] [PubMed] [Google Scholar]
- 72.Doolittle RF, Hunkapiller MW, Hood LE, et al. Simian sarcoma virus onc gene, v-sis, is derived from the gene (or genes) encoding a platelet-derived growth factor [J] Science. 1983;221(4607):275–277. doi: 10.1126/science.6304883. [DOI] [PubMed] [Google Scholar]
- 73.Nister M, Heldin CH, Wasteson A, et al. A platelet-derived growth factor analog produced by a human clonal glioma cell line [J] Ann N Y Acad Sci. 1982;397:25–33. doi: 10.1111/j.1749-6632.1982.tb43414.x. [DOI] [PubMed] [Google Scholar]
- 74.Betsholtz C, Heldin CH, Nister M, et al. Synthesis of a PDGF-like growth factor in human glioma and sarcoma cells suggests the expression of the cellular homologue to the transforming protein of simian sarcoma virus [J] Biochem Biophys Res Commun. 1983;117(1):176–182. doi: 10.1016/0006-291x(83)91557-7. [DOI] [PubMed] [Google Scholar]
- 75.Nister M, Heldin CH, Wasteson A, et al. A glioma-derived analog to platelet-derived growth factor: demonstration of receptor competing activity and immunological crossreactivity [J] Proc Natl Acad Sci USA. 1984;81(3):926–930. doi: 10.1073/pnas.81.3.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heldin CH, Johnsson A, Wennergren S, et al. A human osteosarcoma cell line secretes a growth factor structurally related to a homodimer of PDGF A-chains [J] Nature. 1986;319(6053):511–514. doi: 10.1038/319511a0. [DOI] [PubMed] [Google Scholar]
- 77.Hammacher A, Nister M, Westermark B, et al. A human glioma cell line secretes three structurally and functionally different dimeric forms of platelet-derived growth factor [J] Eur J Biochem. 1988;176(1):179–186. doi: 10.1111/j.1432-1033.1988.tb14266.x. [DOI] [PubMed] [Google Scholar]
- 78.Nister M, Hammacher A, Mellstrom K, et al. A glioma-derived PDGF A chain homodimer has different functional activities from a PDGF AB heterodimer purified from human platelets [J] Cell. 1988;52(6):791–799. doi: 10.1016/0092-8674(88)90421-7. [DOI] [PubMed] [Google Scholar]
- 79.Nister M, Libermann TA, Betsholtz C, et al. Expression of messenger RNAs for platelet-derived growth factor and transforming growth factor-alpha and their receptors in human malignant glioma cell lines [J] Cancer Res. 1988;48(14):3910–3918. [PubMed] [Google Scholar]
- 80.Harsh GR, Keating MT, Escobedo JA, et al. Platelet derived growth factor (PDGF) autocrine components in human tumor cell lines [J] J Neurooncol. 1990;8(1):1–12. doi: 10.1007/BF00182081. [DOI] [PubMed] [Google Scholar]
- 81.Hermansson M, Nister M, Betsholtz C, et al. Endothelial cell hyperplasia in human glioblastoma: coexpression of mRNA for platelet-derived growth factor (PDGF) B chain and PDGF receptor suggests autocrine growth stimulation [J] Proc Natl Acad Sci USA. 1988;85(20):7748–7752. doi: 10.1073/pnas.85.20.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hermanson M, Funa K, Hartman M, et al. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops [J] Cancer Res. 1992;52(11):3213–3219. [PubMed] [Google Scholar]
- 83.Plate KH, Breier G, Farrell CL, et al. Platelet-derived growth factor receptor-beta is induced during tumor development and upregulated during tumor progression in endothelial cells in human gliomas [J] Lab Invest. 1992;67(4):529–534. [PubMed] [Google Scholar]
- 84.Maxwell M, Naber SP, Wolfe HJ, et al. Coexpression of platelet-derived growth factor (PDGF) and PDGF-receptor genes by primary human astrocytomas may contribute to their development and maintenance [J] J Clin Invest. 1990;86(1):131–140. doi: 10.1172/JCI114675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martinho O, Longatto-Filho A, Lambros MB, et al. Expression, mutation and copy number analysis of platelet-derived growth factor receptor A (PDGFRA) and its ligand PDGFA in gliomas [J] Br J Cancer. 2009;101(6):973–982. doi: 10.1038/sj.bjc.6605225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hermanson M, Funa K, Koopmann J, et al. Association of loss of heterozygosity on chromosome 17p with high platelet-derived growth factor alpha receptor expression in human malignant gliomas [J] Cancer Res. 1996;56(1):164–171. [PubMed] [Google Scholar]
- 87.Smith JS, Wang XY, Qian J, et al. Amplification of the platelet-derived growth factor receptor-A (PDGFRA) gene occurs in oligodendrogliomas with grade IV anaplastic features [J] J Neuropathol Exp Neurol. 2000;59(6):495–503. doi: 10.1093/jnen/59.6.495. [DOI] [PubMed] [Google Scholar]
- 88.Fleming TP, Saxena A, Clark WC, et al. Amplification and/or overexpression of platelet-derived growth factor receptors and epidermal growth factor receptor in human glial tumors [J] Cancer Res. 1992;52(16):4550–4553. [PubMed] [Google Scholar]
- 89.Varela M, Ranuncolo SM, Morand A, et al. EGF-R and PDGF-R, but not bcl-2, overexpression predict overall survival in patients with low-grade astrocytomas [J] J Surg Oncol. 2004;86(1):34–40. doi: 10.1002/jso.20036. [DOI] [PubMed] [Google Scholar]
- 90.Mapstone T, McMichael M, Goldthwait D. Expression of platelet-derived growth factors, transforming growth factors, and the ros gene in a variety of primary human brain tumors [J] Neurosurgery. 1991;28(2):216–222. doi: 10.1097/00006123-199102000-00007. [DOI] [PubMed] [Google Scholar]
- 91.Majumdar K, Radotra BD, Vasishta RK, et al. Platelet-derived growth factor expression correlates with tumor grade and proliferative activity in human oligodendrogliomas [J] Surg Neurol. 2009;72(1):54–60. doi: 10.1016/j.surneu.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 92.Shih AH, Dai C, Hu X, et al. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis [J] Cancer Res. 2004;64(14):4783–4789. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- 93.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme [J] Proc Natl Acad Sci USA. 2005;102(16):5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis [J] Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 95.Kotliarov Y, Steed ME, Christopher N, et al. High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic imbalances [J] Cancer Res. 2006;66(19):9428–9436. doi: 10.1158/0008-5472.CAN-06-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme [J] Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cancer Genome Altas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways [J] Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maher EA, Brennan C, Wen PY, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities [J] Cancer Res. 2006;66(23):11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- 99.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1 [J] Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yin D, Ogawa S, Kawamata N, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray [J] Mol Cancer Res. 2009;7(5):665–677. doi: 10.1158/1541-7786.MCR-08-0270. [DOI] [PubMed] [Google Scholar]
- 101.Brennan C, Momota H, Hambardzumyan D, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations [J] PLoS One. 2009;4(11):e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lokker NA, Sullivan CM, Hollenbach SJ, et al. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors [J] Cancer Res. 2002;62(13):3729–3735. [PubMed] [Google Scholar]
- 103.LaRochelle WJ, Jeffers M, Corvalan JR, et al. Platelet-derived growth factor D: tumorigenicity in mice and dysregulated expression in human cancer [J] Cancer Res. 2002;62(9):2468–2473. [PubMed] [Google Scholar]
- 104.Crawford Y, Kasman I, Yu L, et al. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment [J] Cancer Cell. 2009;15(1):21–34. doi: 10.1016/j.ccr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 105.di Tomaso E, Snuderl M, Kamoun WS, et al. Glioblastoma recurrence after Cediranib therapy in patients: lack of “rebound” revascularization as mode of escape [J] Cancer Res. 2011;71(1):19–28. doi: 10.1158/0008-5472.CAN-10-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.di Tomaso E, London N, Fuja D, et al. PDGF-C induces maturation of blood vessels in a model of glioblastoma and attenuates the response to anti-VEGF treatment [J] PLoS One. 2009;4(4):e5123. doi: 10.1371/journal.pone.0005123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gondi CS, Veeravalli KK, Gorantla B, et al. Human umbilical cord blood stem cells show PDGF-D-dependent glioma cell tropism in vitro and in vivo [J] Neuro Oncol. 2010;12(5):453–465. doi: 10.1093/neuonc/nop049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weiss WA, Israel M, Cobbs C, et al. Neuropathology of genetically engineered mice: consensus report and recommendations from an international forum [J] Oncogene. 2002;21(49):7453–7463. doi: 10.1038/sj.onc.1205936. [DOI] [PubMed] [Google Scholar]
- 109.Weissenberger J, Steinbach JP, Malin G, et al. Development and malignant progression of astrocytomas in GFAP-v-src transgenic mice [J] Oncogene. 1997;14(17):2005–2013. doi: 10.1038/sj.onc.1201168. [DOI] [PubMed] [Google Scholar]
- 110.Ding H, Roncari L, Shannon P, et al. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas [J] Cancer Res. 2001;61(9):3826–3836. [PubMed] [Google Scholar]
- 111.Reilly KM, Loisel DA, Branson RT, et al. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects [J] Nat Genet. 2000;26(1):109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- 112.Zhu Y, Guignard F, Zhao D, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma [J] Cancer Cell. 2005;8(2):119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hede SM, Hansson I, Afink GB, et al. GFAP promoter driven transgenic expression of PDGFB in the mouse brain leads to glioblastoma in a Trp53 null background [J] Glia. 2009;57(11):1143–1153. doi: 10.1002/glia.20837. [DOI] [PubMed] [Google Scholar]
- 114.Nazarenko I, Hedren A, Sjodin H, et al. Brain abnormalities and glioma-like lesions in mice overexpressing the long isoform of PDGF-A in astrocytic cells [J] PLoS One. 2011;6(4):e18303. doi: 10.1371/journal.pone.0018303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Uhrbom L, Hesselager G, Nister M, et al. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus [J] Cancer Res. 1998;58(23):5275–5279. [PubMed] [Google Scholar]
- 116.Dai C, Celestino JC, Okada Y, et al. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo [J] Genes Dev. 2001;15(15):1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Assanah M, Lochhead R, Ogden A, Bet al. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses [J] J Neurosci. 2006;26(25):6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Calzolari F, Appolloni I, Tutucci E, et al. Tumor progression and Oncogene addiction in a PDGF-B-induced model of gliomagenesis [J] Neoplasia. 2008;10(12):1373–1382. doi: 10.1593/neo.08814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tchougounova E, Kastemar M, Brasater D, et al. Loss of Arf causes tumor progression of PDGFB-induced oligodendroglioma [J] Oncogene. 2007;26(43):6289–6296. doi: 10.1038/sj.onc.1210455. [DOI] [PubMed] [Google Scholar]
- 120.Johansson FK, Brodd J, Eklof C, et al. Identification of candidate cancer-causing genes in mouse brain tumors by retroviral tagging [J] Proc Natl Acad Sci USA. 2004;101(31):11334–11337. doi: 10.1073/pnas.0402716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krum JM, Rosenstein JM. VEGF mRNA and its receptor flt-1 are expressed in reactive astrocytes following neural grafting and tumor cell implantation in the adult CNS [J] Exp Neurol. 1998;154(1):57–65. doi: 10.1006/exnr.1998.6930. [DOI] [PubMed] [Google Scholar]
- 122.Chen Y, Miles DK, Hoang T, et al. The basic helix-loop-helix transcription factor olig2 is critical for reactive astrocyte proliferation after cortical injury [J] J Neurosci. 2008;28(43):10983–10989. doi: 10.1523/JNEUROSCI.3545-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hesselager G, Uhrbom L, Westermark B, et al. Complementary effects of platelet-derived growth factor autocrine stimulation and p53 or Ink4a-Arf deletion in a mouse glioma model [J] Cancer Res. 2003;63(15):4305–4309. [PubMed] [Google Scholar]
- 124.Liu KW, Feng H, Bachoo R, et al. SHP-2/PTPN11 mediates gliomagenesis driven by PDGFRA and INK4A/ARF aberrations in mice and humans [J] J Clin Invest. 2011;121(3):905–917. doi: 10.1172/JCI43690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cerami E, Demir E, Schultz N, et al. Automated network analysis identifies core pathways in glioblastoma [J] PLoS One. 2010;5(2):e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dai C, Lyustikman Y, Shih A, et al. The characteristics of astrocytomas and oligodendrogliomas are caused by two distinct and interchangeable signaling formats [J] Neoplasia. 2005;7(4):397–406. doi: 10.1593/neo.04691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hu X, Pandolfi PP, Li Y, et al. mTOR promotes survival and astrocytic characteristics induced by Pten/AKT signaling in glioblastoma [J] Neoplasia. 2005;7(4):356–368. doi: 10.1593/neo.04595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.De Witt Hamer PC. Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies [J] Neuro Oncol. 2010;12(3):304–316. doi: 10.1093/neuonc/nop068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hu X, Holland EC. Applications of mouse glioma models in preclinical trials [J] Mutat Res. 2005;576(1–2):54–65. doi: 10.1016/j.mrfmmm.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 130.Suzuki K, Momota H, Tonooka A, et al. Glioblastoma simultaneously present with adjacent meningioma: case report and review of the literature [J] J Neurooncol. 2010;99(1):147–153. doi: 10.1007/s11060-009-0109-9. [DOI] [PubMed] [Google Scholar]
- 131.Zhan Y, Counelis GJ, O'Rourke DM. The protein tyrosine phosphatase SHP-2 is required for EGFRvlll oncogenic transformation in human glioblastoma cells [J] Exp Cell Res. 2009;315(14):2343–2357. doi: 10.1016/j.yexcr.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chan G, Kalaitzidis D, Neel BG. The tyrosine phosphatase Shp2 (PTPN11) in cancer [J] Cancer Metastasis Rev. 2008;27(2):179–192. doi: 10.1007/s10555-008-9126-y. [DOI] [PubMed] [Google Scholar]
- 133.Tonon G. From Oncogene to network addiction: the new frontier of cancer genomics and therapeutics [J] Future Oncol. 2008;4(4):569–577. doi: 10.2217/14796694.4.4.569. [DOI] [PubMed] [Google Scholar]