Abstract
The GTPases Ffh and FtsY are components of the prokaryotic signal recognition particle protein-targeting pathway. The two proteins interact in a GTP-dependent manner, forming a complex that can be stabilized by use of the non-hydrolyzable GTP analog GMPPCP. Crystals of the complex of the NG GTPase domains of the two proteins have been obtained from ammonium sulfate solutions. Crystals grow with several different morphologies, predominately as poorly diffracting plates and needle clusters, but occasionally as well diffracting rods. It has been demonstrated that all forms of the crystals observed contain an intact complex. Diffraction data to 2.0 Å resolution have been measured.
1. Introduction
Ffh and FtsY are the GTPase components of the signal recognition particle (SRP) pathway for co-translational targeting of proteins to the membrane (Keenan et al., 2001). Ffh, the homolog of the eukaryotic SRP54 (Bernstein et al., 1989), is a component of the SRP that comprises an M domain, which mediates interaction with signal peptide and the RNA component, and an NG domain GTPase that mediates the GTP-dependent interaction of SRP with its receptor, FtsY (Lu et al., 2001; Miller et al., 1994; Zopf et al., 1993). The NG domain may also interact with the signal sequence (Cleverley & Gierasch, 2002). FtsY is itself a three-domain protein comprising an A domain implicated in membrane association (de Leeuw et al., 1997; Millman & Andrews, 1999; Zelazny et al., 1997) and an NG GTPase domain structurally homologous to that of Ffh (Montoya et al., 1997). The NG ‘modules’ in this system are linked to their respective M and A domains via relatively unstructured linker peptides that are sensitive to proteolysis (Millman & Andrews, 1999; Zopf et al., 1990) and it has been shown that the NG domains of both Ffh and FtsY function in protein targeting even when placed in a non-native context (Macao et al., 1997; Zelazny et al., 1997).
We have previously found conditions under which a complex of the NG domains of Thermus aquaticus Ffh and FtsY could be stabilized and purified in the presence of the non-hydrolyzable GTP analog β-γ-methyleneguanosine 5′-triphosphate (GMPPCP; Shepotinovskaya & Freymann, 2002). Our strategy took advantage of the observation that association of the Ffh and FtsY in the absence of the cognate SRP RNA is slow (Peluso et al., 2000) and the kinetics of formation seemed consistent with this. However, we found that complex formation was associated with the presence of an unidentified protease activity which cleaved the FtsY NG domain at two sites, one of which was accessible only on formation of the heterodimeric complex (Shepotinovskaya & Freymann, 2002). Nevertheless, the purified complex was shown to comprise both the Ffh and FtsY NG domains and to behave in a manner consistent with a dependence on both magnesium and the GTP analog for their interaction (Shepotinovskaya & Freymann, 2002).
The determination of the three-dimensional structure of the GTPase ‘core’ of the SRP targeting complex would provide insight into a key event in the co-translational targeting pathway and would provide a structural basis for understanding the novel reciprocal GTPase-activation activity of the two proteins (Powers & Walter, 1995). Here, we report crystallization conditions for the purified complex that yield a number of different crystal morphologies, most of which are twinned and diffract to only low resolution and which in some cases interconvert over the month-long period of a crystallization experiment. However, we have been able to demonstrate that crystals of all morphologies contain the expected complex and we have found well formed crystals that diffract to ~2 Å resolution.
2. Crystallization
The NG GTPase domains of T. aquaticus FtsY and Ffh were expressed and purified as described previously (Freymann et al., 1997; Shepotinovskaya & Freymann, 2002). The expressed proteins have a molecular weight of ~33 kDa, comprise residues 2–304 of FtsY and residues 1–294 or 1–297 of Ffh, and correspond to the NG domains of T. aquaticus FtsY and Ffh as defined by previous biochemical and structural studies (Keenan et al., 1998; Montoya et al., 1997; Ramirez et al., 2002; Shepotinovskaya & Freymann, 2002; Zelazny et al., 1997). The different C-terminal truncations of the Ffh NG were constructed using PCR (QuickChange) to introduce stop codons 3′ to Met294 and Val297, residues that occur at the C-terminus of the C-terminal α-helix of the NG domain and at the site of elastase cleavage of its C-terminal linker peptide, respectively (Freymann et al., 1997; Keenan et al., 1998) (D. M. Freymann, unpublished data). The complex between the two NG domains was formed by incubation of 15 µM FtsY NG and 10 µM Ffh NG in 50 mM HEPES pH 7.5, 2 mM MgCl2, 50 mM NaCl and 1 mM β-γ-methyleneguanosine 5′-triphosphate (Sigma) in a 5 ml reaction mixture for 4 d at 310 K and the complex was purified by ion-exchange chromatography as described previously (Shepotinovskaya & Freymann, 2002). The presence of the intact complex was confirmed by gelfiltration chromatography. Complex formation was associated with proteolysis at two sites in the FtsY NG domain, as shown previously (Shepotinovskaya & Freymann, 2002) and verified by SDS–PAGE. Aliquots were stored in 50 mM Tris pH 8, 2 mM MgCl2, 0.2 M NaCl and 1 mM GMPPCP at 253 K. Prior to crystallization trials, aliquots were thawed at 293 K in a water bath and concentrated using a Centricon 30 (Amicon) to 4–20 mg ml−1. Protein concentration was determined with the Bradford assay (Biorad) using conversions for T. aquaticus Ffh and FtsY NG domains derived from quantitative amino-acid analysis.
We assayed the stability of the purified complex under room-temperature crystallization solution conditions by gel-filtration HPLC. These tests, which demonstrated that the complex could be concentrated to >7 mg ml−1, that it could be stabilized for at least a month in the presence of the nucleotide analog at room temperature and that it remained intact in the presence of high-salt solutions, provided a foundation for proceeding with crystallization trials. GMPPCP was also found to be stable under the complex-formation and incubation conditions as assayed using ion-exchange HPLC (data not shown).
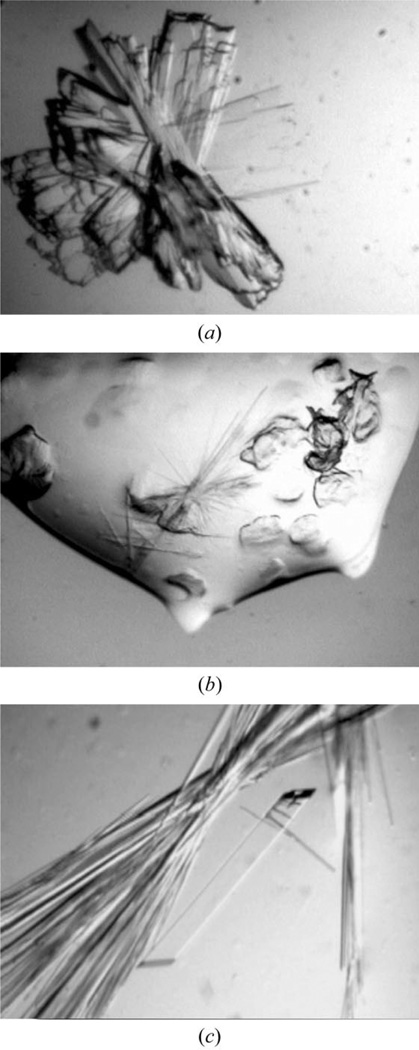
Crystallization trials were carried out using sitting-drop vapor diffusion at room temperature. Initial crystallization screens were carried out with Crystal Screens I and II (Hampton Research) using 1 µl protein solution plus 1 µl well solution equilibrated over 0.5 ml wells. After 3 d, clusters of twinned plate-like ‘sheaves’ or needle-like crystals were found under three related conditions at 1.8–2.0 M ammonium sulfate, 0.1 M Tris pH 8.5, 2.0 M ammonium sulfate (Crystal Screen I, No. 4), 0.1 M HEPES pH 7.5, 2% PEG 400, 2.0 M ammonium sulfate (Crystal Screen I, No. 39) and 0.1 M MES pH 6.5, 10 mM CoCl2, 1.8 M ammonium sulfate (Crystal Screen II, No. 25). These conditions were used as starting points for optimization experiments using selected reagents from Additive Screens (Hampton Research) and varying the buffer, temperature and protein concentration. Despite some improvement, it was difficult to reliably obtain single crystals and crystal morphology was, in general, very poor (Fig. 1). Interestingly, under several conditions we observed that two crystal morphologies appeared in the same crystallization drop or that one crystal form (generally sheaves) appeared over several days only to dissolve and transform into the second crystal form (generally long needles) over the course of a month (Fig. 1b). We do not yet know the difference between the two crystal forms and in particular do not know whether it represents further chemical modification of the protein in the complex (in addition to the known proteolysis) over time. However, we have been able to show that all crystal forms tested (sheaves, needles and rods) contained the Ffh/FtsY NG complex.
Figure 1.
Crystals of the Ffh/FtsY NG complex. Crystals grew with a number of different morphologies. (a) Sheaves. Crystals were grown from Ffh/FtsY NG complex at 20 mg ml−1 in 50 mM Tris pH 8.0, 200 mM NaCl, 2 mM MgCl2, 1 mM GMPPCP equilibrated against 1.51 M ammonium sulfate, 100 mM Tris pH 8, 100 mM NaCl, 10 mM MgCl2 at room temperature. (b) Needles and sheaves. Crystals were grown from the protein used in (a) equilibrated against 1.91 M ammonium sulfate, 100 mM bis-tris pH 6.7, 100 mM NaCl, 20 mM MgCl2, 4% PEG 400. Sheaves appeared within 1–2 d; however, after 2.5–3 weeks needle clusters appeared, coexisting with the sheaves, which then dissolved such that after a month only needle clusters were present in the drop. (c) A large well formed rod growing among needle clusters. Crystals were grown from protein solution at 9.9 mg ml−1, equilibrated against 1.91 M ammonium sulfate, 100 mM bis-tris pH 6.7, 100 mM NaCl, 4% PEG 400. While the sheaf and needle morphologies were readily reproducible, well formed rods appeared in only a small subset of the crystallization drops.
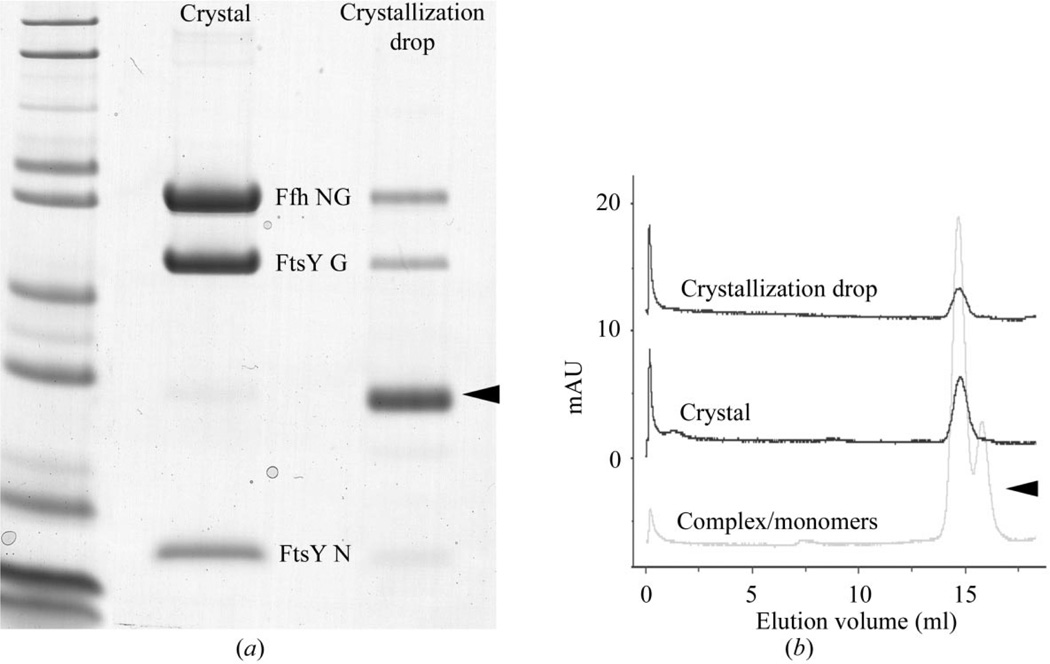
Because both the formation and purification of the complex and its crystallization behavior were considered problematic, we were particularly careful at this point to establish that the crystals in fact comprised the Ffh/FtsY NG domain complex. Therefore, crystals exhibiting a number of different morphologies were harvested and examined by SDS–PAGE, and crystals mounted for characterization of X-ray diffraction were recovered from the mounting loop and were also analyzed by SDS–PAGE. In some experiments, we added an equal volume of lysozyme at 5 mg ml−1 in the crystallization buffer as a wash control to the crystallization drop prior to harvesting (Fig. 2). Crystals were moved using a 100 µm nylon loop, passed through a 4 µl drop containing protein-free mother liquor and then dissolved in water before adding loading buffer for SDS–PAGE (Novagen). In each case, we found that the crystal contained both the Ffh NG and proteolyzed FtsY NG domains, as expected for the purified complex (Fig. 2a). In addition, in two cases an aliquot of the washed and dissolved crystal was assayed by gel-filtration HPLC and found to elute at the position expected for the intact complex (Fig. 2b).
Figure 2.
Crystals contain intact NG-domain complex. (a) SDS–PAGE demonstrates that both proteins are present in the crystal. The crystallization drop was supplemented with mother liquor containing 5 mg ml−1 lysozyme as a wash control (arrow). A crystal was removed using a 100 µm nylon loop, passed through successive lysozyme-free mother-liquor washes and dissolved in water. The dissolved crystal and a sample of the crystallization mother liquor were analyzed by SDS–PAGE. The expected products are the intact Ffh NG domain (~33 kDa) and the proteolysis products of FtsY NG, migrating at ~22 and ~6 kDa. The crystal used in the experiment shown had a ‘sheaf’ morphology and was grown at 7 mg ml−1 under 1.7 M ammonium sulfate, 0.1 M MES pH 6.5, 1 mM SrCl2 crystallization conditions. Similar experiments with other crystals (e.g. having the ‘needle’ morphology) gave the same result. (b) Gel filtration demonstrates that the proteins are associated in a bona fide complex. The crystals were grown using protein at 7 mg ml−1 and 1.7 M ammonium sulfate, 0.1 M MES pH 6.5 crystallization conditions. The mother liquor was removed from the drop and the crystals were first washed with protein-free mother liquor and then dissolved in 2 µl of a mobile phase buffer, 50 mM HEPES, 50 mM NaCl, 2 mM MgCl2, supplemented with 1 mM GMPPCP. The samples were then diluted to 80 µl and injected onto a Superdex 200 column. Both the mother-liquor sample and the washed crystals are shown. The bottom trace indicates the elution positions of the complex (large peak) and the two monomeric proteins (which migrate at the same size, arrow) (Shepotinovskaya & Freymann, 2002).
3. Data collection
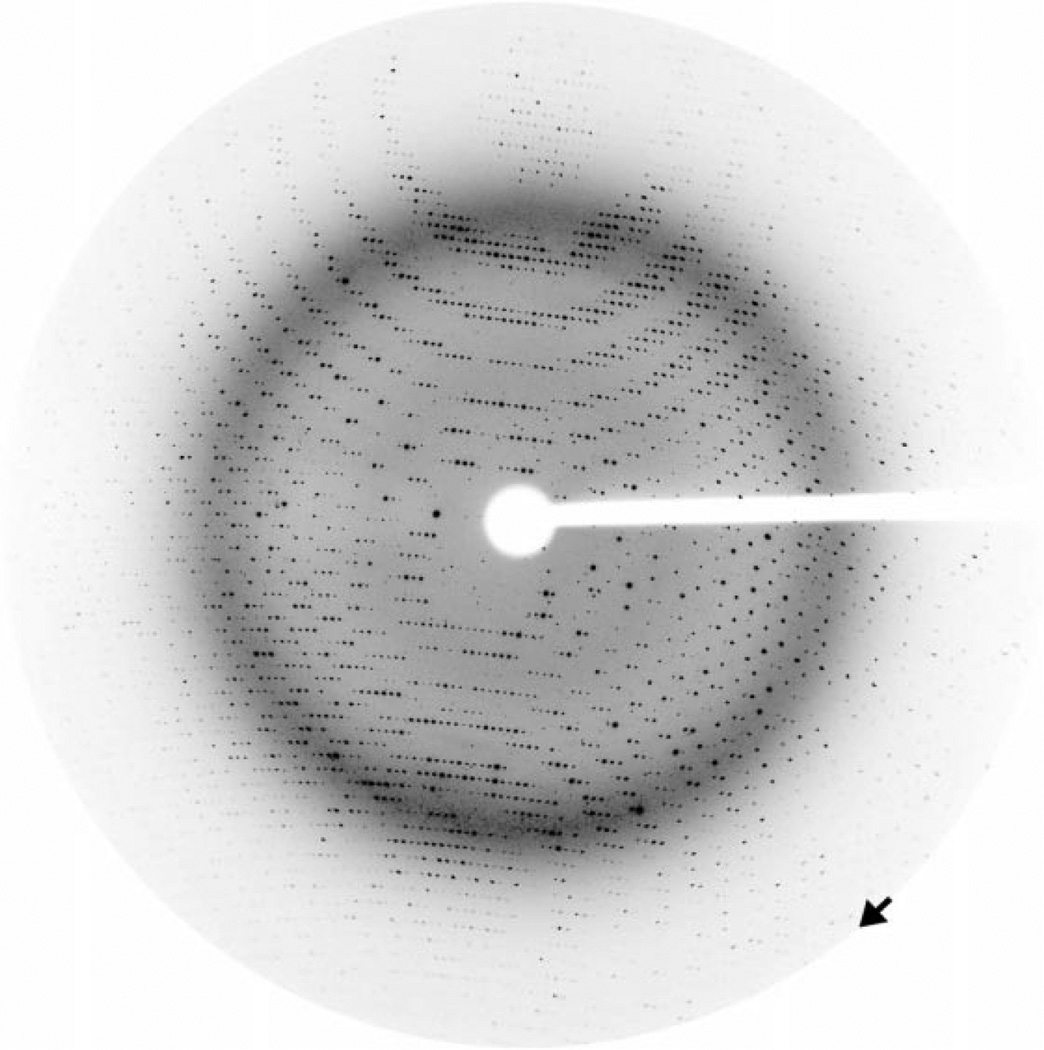
Crystals were mounted on nylon loops for data collection, first transferring them to a cryoprotectant comprising mother liquor supplemented with 10% ethylene glycol and then flash-cooling by immersion in liquid nitrogen. Diffraction experiments were carried out at APS DND-CAT BL 5-ID-B with a MAR CCD detector, at BioCARS BL 14-ID-B with a Quantum4 CCD detector and at SSRL BL 9-2 with a Quantum315 CCD detector. The crystals were maintained at 100 K using an Oxford Cryostream. In general, the crystals diffracted poorly, with the plate-like sheaf and the needle morphologies yielding diffraction to 3.5 Å resolution at best. However, the well formed rod-like crystals which were occasionally found (see Fig. 1c) diffracted to at least 2.0 Å resolution (Fig. 3) and a 99% complete data set to 2.05 Å resolution has been measured from one crystal at the DND-CAT beamline. The unit-cell parameters were found to be a = b = 98.9, c = 130.7 Å, α = β = γ = 90°. A similar unit cell is found on indexing low-resolution diffraction data from ‘sheaves’ and ‘needles’, suggesting a close relationship between the space groups of the different crystal forms. The space-group assignment is P41212 and the overall Rsym for the data set is 6.5% (Table 1).
Figure 3.
Diffraction from rod-shaped crystals to 2.0 Å resolution. A diffraction image from a rod-shaped crystal obtained at DND-CAT beamline 5-ID-B at the APS shows diffraction to 2.05 Å (arrow; note the strong water ring at ~3.5 Å). The exposure for the 0.5° oscillation image was 5 s at a wavelength of 1.0 Å and a detector distance of 150 mm.
Table 1.
Cystallographic data statistics
| Resolution range (Å) | 25.0−2.05 (2.10−2.05) |
| Unique reflections | 41332 |
| Redundancy | 10.9 (10.4) |
| Completeness | 99.9 (100.0) |
| I/σ(I) | 33.4 (16.2) |
| Rsym | 0.065 (0.158) |
Values in parentheses are for the last resolution shell.
A solution to the structure has been found by molecular replacement with the program BEAST (Read, 2001) using a probe derived from the G domain of a GDP complex of the T. aquaticus Ffh NG (i.e. residues 99–283) with all ligands removed (Freymann et al., 1999). Ffh and FtsY share 34% sequence identity over their NG domains. Two distinct rotation solutions, one corresponding to the G domain of Ffh and the other to the structurally similar G domain of FtsY, could be distinguished readily. The log-likelihood gain (LLG) scores, which measure the significance of the solutions (Read, 2001), were 33.0 (7.1σ above the mean) and 30.4 (6.5σ), respectively. Determination of the relative translations of the two solutions yielded a final LLG of 1050.5. There is one heterodimeric complex in the asymmetric unit. The molecular-replacement model, which comprised two identical copies of the Ffh G domain, was refined using REFMAC (Murshudov et al., 1997), yielding crystallographic R and Rfree values of 0.43 and 0.48, respectively. Subsequent density modification using the ‘prime-and-switch’ protocol of RESOLVE (Terwilliger, 2000) yielded a figure of merit of 0.48, a bias ratio of 1.19 and confirmed the solution by clearly indicating the position of two bound GMPPCP molecules in the heterodimer. The unusual characteristics of complex formation (e.g. the uncharacterized proteolysis) may have contributed to the unpredictable and heterogeneous behavior during crystallization trials. It is likely that determination of the crystal structure, as well as ongoing biochemical and mass-spectrometric studies, will allow us to better understand these phenomena.
Acknowledgments
This work was supported by grant GM58500 from the NIH and by support from the R. H. Lurie Cancer Center provided to the Structural Biology Facility at Northwestern University. The DuPont–Northwestern–Dow Collaborative Access Team (DND-CAT) at the Advanced Photon Source is supported by the US National Science Foundation through Grant DMR-9304725 and the State of Illinois through the Department of Commerce and the Board of Higher Education Grant IBHE HECA NWU 96. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources under grant No. RR07707. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Energy Research under Contract No. W-31-102-Eng-38. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and the National Institute of General Medical Sciences.
References
- Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Nature (London) 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Cleverley RM, Gierasch LM. J. Biol. Chem. 2002;277:46763–46768. doi: 10.1074/jbc.M207427200. [DOI] [PubMed] [Google Scholar]
- Freymann DM, Keenan RJ, Stroud RM, Walter P. Nature (London) 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- Freymann DM, Keenan RJ, Stroud RM, Walter P. Nature Struct. Biol. 1999;6:793–801. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Walter P, Stroud RM. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- Leeuw E de, Poland D, Mol O, Sinning I, ten Hagen-Jongman CM, Oudega B, Luirink J. FEBS Lett. 1997;416:225–229. doi: 10.1016/s0014-5793(97)01238-6. [DOI] [PubMed] [Google Scholar]
- Lu Y, Qi HY, Hyndman JB, Ulbrandt ND, Teplyakov A, Tomasevic N, Bernstein HD. EMBO J. 2001;20:6724–6734. doi: 10.1093/emboj/20.23.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macao B, Luirink J, Samuelsson T. Mol. Microbiol. 1997;24:523–534. doi: 10.1046/j.1365-2958.1997.3551729.x. [DOI] [PubMed] [Google Scholar]
- Miller JD, Bernstein HD, Walter P. Nature (London) 1994;367:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- Millman JS, Andrews DW. J. Biol. Chem. 1999;274:33227–33234. doi: 10.1074/jbc.274.47.33227. [DOI] [PubMed] [Google Scholar]
- Montoya G, Svensson C, Luirink J, Sinning I. Nature (London) 1997;385:365–369. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Peluso P, Herschlag D, Nock S, Freymann DM, Johnson AE, Walter P. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- Ramirez UD, Minasov G, Focia PJ, Stroud RM, Walter P, Kuhn P, Freymann DM. J. Mol. Biol. 2002;320:783–799. doi: 10.1016/s0022-2836(02)00476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ. Acta Cryst. 2001;D57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Shepotinovskaya IV, Freymann DM. Biochim. Biophys. Acta. 2002;1597:107–114. doi: 10.1016/s0167-4838(02)00287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Acta Cryst. 2000;D56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny A, Seluanov A, Cooper A, Bibi E. Proc. Natl Acad. Sci. USA. 1997;94:6025–6029. doi: 10.1073/pnas.94.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D, Bernstein HD, Johnson AE, Walter P. EMBO J. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D, Bernstein HD, Walter P. J. Cell. Biol. 1993;120:1113–1121. doi: 10.1083/jcb.120.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]