Abstract
We introduce clustering with overlapping neighborhood expansion (ClusterONE), a method for detecting potentially overlapping protein complexes from protein-protein interaction data. ClusterONE-derived complexes for several yeast data sets showed better correspondence with reference complexes in the Munich Information Center for Protein Sequence (MIPS) catalog and complexes derived from the Saccharomyces Genome Database (SGD) than the results of seven popular methods. The results also showed a high extent of functional homogeneity.
Recent developments in experimental procedures have resulted in the publication of many high-quality, large-scale protein-protein interaction (PPI) data sets for different organisms. These data can be represented as undirected graphs, in which nodes represent proteins and edges represent interactions between pairs of proteins. Often an estimation of the reliability of such interactions is available and is included as edge labels (weights). One can formulate the problem of identifying protein complexes from PPI data as that of detecting dense regions containing many connections in PPI networks (or regions with large weights in weighted networks).
Densely connected regions in graphs are most frequently identified by some unsupervised clustering method1,2. However, standard clustering is not ideal for PPI networks: proteins may have multiple functions, and therefore the corresponding nodes may belong to more than one cluster; for example, 207 of 1,628 proteins in the CYC2008 hand-curated yeast complex data set3 participate in more than one complex. Such nodes present a challenge to classical graph clustering algorithms that assign each node of the graph to just one of the clusters. Recently, algorithms have been proposed that detect overlapping clusters, but in many cases they are limited to unweighted PPI data4,5 and can be applied to weighted networks only after ‘binarizing’ them by removing edges with weights below a given threshold. Although it is difficult to assess the reliability of a single edge weight, we found that taking into account network weights can greatly improve the detection of protein complexes (Supplementary Discussion and Supplementary Figs. 1 and 2); therefore, when available, weights should be used.
Intuitively, a subgraph representing a protein complex should satisfy two simple structural properties: it should contain many reliable interactions between its subunits, and it should be well-separated from the rest of the network. We formalized these two properties in a quality measure that we called cohesiveness and developed an algorithm that detects possibly overlapping protein complexes from weighted networks, using cohesiveness to guide the search.
Our algorithm consists of three major steps (Online Methods). First, starting from a single seed vertex, a greedy procedure adds or removes vertices to find groups with high cohesiveness. The growth process is repeated from different seeds to form multiple, possibly overlapping groups. Although some overlaps are likely to have biological importance, groups overlapping to a very high extent in comparison to their sizes should likely be merged. In the second step, we quantify the extent of overlap between each pair of groups and merge those for which the overlap score4 is above a specified threshold. In the third step, we discard complex candidates that contain less than three proteins or whose density is below a given threshold. Note that our method potentially can be used to recognize not only partial overlaps but also cases in which a complex is completely contained in another complex.
We tested ClusterONE on five large scale yeast PPI data sets (Supplementary Data 1), four weighted6–8 and one unweighted9, and compared ClusterONE to a representative set of other approaches: Markov cluster (MCL)1, molecular complex detection (MCODE)4, affinity propagation10, restricted neighborhood search clustering algorithm (RNSC)2, CFinder11, clustering based on maximal cliques (CMC)5 and repeated random walks (RRW)12 (Supplementary Discussion). We compared predicted complexes to two reference complex sets: the first derived from the MIPS catalog of protein complexes13 and the second from Gene Ontology–based complex annotations in the SGD (Supplementary Data 2). We assessed the quality of the predicted complexes by three scores: the fraction of protein complexes matched by at least one predicted complex; the geometric accuracy measure14; and the maximum matching ratio, a score that we propose here based on a maximal one-to-one mapping between predicted and reference complexes (Online Methods).
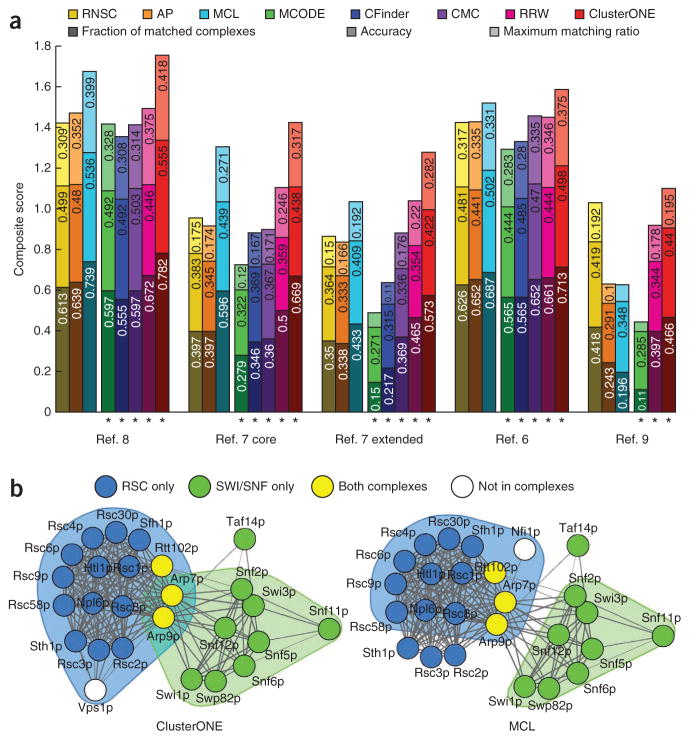
We present the three quality scores obtained using the MIPS reference set (Fig. 1a); larger scores are better, and the sum of the three scores is a composite score. Our benchmarks showed that ClusterONE outperformed the other approaches both on weighted and unweighted PPI networks, matching more complexes with a higher accuracy and providing a better one-to-one mapping with reference complexes in almost all the data sets. MCL yielded the closest score to ClusterONE. But MCL cannot handle overlaps. We also provide results using the reference set derived from the Gene Ontology annotations in the SGD and the expected values of each score for randomized predicted complex sets (Supplementary Discussion and Supplementary Fig. 3).
Figure 1.
Benchmark results. (a) Results using MIPS data sets. Shades of the same color denote individual quality scores; the total height of each bar is the value of the composite score. Numbers are the values for each score. The first four data sets are weighted, BioGRID is unweighted. Asterisks mark algorithms that could handle overlaps. AP, affinity propagation. (b) Subunits of RSC and SWI/SNF chromatin-remodeling complexes in the reference 8 data set, as detected by ClusterONE and MCL. Shaded areas denote detected complexes.
To examine the biological relevance of predicted complexes we calculated the ‘co-localization’ score of the entire predicted complex set and conducted over-representation analysis of Gene Ontology annotations for each predicted complex. As MCL yielded the closest score to ClusterONE in the MIPS benchmarks, we present these scores for ClusterONE and MCL (Supplementary Tables 1 and 2). Results for ClusterONE indicated that predicted complexes tended to consist of proteins in the same cellular component and these proteins are likely to have similar functions and/or participate in the same biological process. Comparison of co-localization and overrepresentation scores of ClusterONE and MCL revealed that ClusterONE complexes had higher scores on almost all data sets. We present an example of how a pair of complexes with a known overlap was detected by ClusterONE and MCL (Fig. 1b); we also present results obtained using the other algorithms and another example (Supplementary Discussion and Supplementary Figs. 4–6).
ClusterONE could find applications in other areas such as the study of social networks. A fast, free implementation of ClusterONE is available at http://www.paccanarolab.org/cluster-one/.
ONLINE METHODS
The ClusterONE algorithm
The algorithm we outlined builds on the concept of the cohesiveness score and uses a greedy growth process to find groups in a protein-protein interaction network that are likely to correspond to protein complexes. Cohesiveness measures how likely it is for a group of proteins to form a protein complex, and it was defined as follows. Let win(V) denote the total weight of edges contained entirely by a group of proteins V, and let wbound(V) denote the total weight of edges that connect the group with the rest of the network. The cohesiveness of V is then given by
| (1) |
where p|V| is a penalty term whose purpose is to model the uncertainty in the data by assuming the existence of yet undiscovered interactions in the protein interaction network. Letting p > 0 offsets the boundary weight wbound(V) by p|V|, practically assuming that every protein in V has p additional boundary connections that we could not identify owing to limitations in the experimental procedure. This definition could be extended to use different values of p for different proteins based on biological assumptions; thus a well-studied protein may have a lower p value because it is less likely to have undiscovered interactions.
Cohesiveness provides an easy and efficient way to assess how well a given subgraph fits the two above-mentioned structural properties: a subgraph with many reliable edges has a high win, and a well-separated subgraph has a low wbound, both having the effect of increasing f(V). f(V) > 1/3 also implies that vertices of the subgraph have more internal weight than external weight on average, satisfying the conditions of being a community in the weak sense15.
Our algorithm consists of three steps. In the first step, the algorithm grows groups with high cohesiveness from selected seed proteins. Initially, it selects the protein with the largest number of connections (highest degree) as the first seed, and grows a cohesive group from it using a greedy procedure. Whenever the growth process finishes, the algorithm selects the next seed by considering all the proteins that have not been included in any of the protein complexes found so far and taking the one with the highest degree again. The entire procedure terminates when there are no proteins remaining to consider.
A step-by-step description of the greedy growth process starting from v0 is as follows. Step 1: let V0 = {v0}. Set the step number t = 0. Step 2: calculate the cohesiveness of Vt and let Vt + 1 = Vt. Step 3: for every external vertex v incident on at least one boundary edge, calculate the cohesiveness of V′ = Vt ∪ {v}. If f(V′) > f(Vt + 1), let Vt + 1 = V′. Step 4: for every internal vertex v incident on at least one boundary edge, calculate the cohesiveness of V″ = Vt \ {v}. If f(V″) > f(Vt + 1), let Vt + 1 = V″. Step 5: if Vt ≠ Vt + 1, increase t and return to step 2. Otherwise, declare Vt a locally optimal cohesive group.
The growth process allows the removal of any vertex from the cohesive group being grown, including the original seed vertex. If the original seed vertex is not included in the final cohesive group, the seed vertex is considered an outlier and it will not be included in any of the clusters, except in the case when another cohesive group grown from a different seed vertex absorbs it.
To illustrate the above procedure, let us consider an example graph with 11 nodes, seven of which are marked by letters A to G (Supplementary Fig. 7). Assuming p = 0, the cohesiveness of the marked set is 10/15. In steps 3 and 4, the algorithm can either extend the current set by adding C, F or G, or contract the set by removing A, B, D or E. The best choice is to add C to the set, because it converts three boundary edges to internal ones and does not bring in any new boundary edges. After adding C, the cohesiveness increases to 13/15 and the group becomes locally optimal, as adding F would result in a cohesiveness of 14/17 and adding G would yield 14/18.
In the second step of the algorithm, highly overlapping pairs of locally optimal cohesive groups are merged. In our benchmarks, we have merged pairs of groups with an overlap score ω larger than 0.8, where the overlap score of two protein sets A and B is defined as follows4:
| (2) |
Such merges may be performed one after another (in which case the overlap scores have to be recalculated after each merge) or concurrently; the reference implementation of ClusterONE uses the latter approach. More precisely, given a set of cohesive groups, ClusterONE first calculates the overlap scores for each pair of groups and constructs an overlap graph in which each vertex represents a cohesive group, and two groups are connected by an edge if they overlap substantially. Groups that are connected to each other (either directly by an edge or indirectly by a path of edges) are then merged into protein complex candidates. If a group has no connection to other groups, it is promoted to a protein complex candidate without any additional merging. In the third and final step of the algorithm, we discard complex candidates that contain less than three proteins or whose density is below a given threshold δ (where the density of a complex with n proteins is defined as the total weight of its internal edges, divided by n(n - 1)/2).
Comparing predicted complexes with a gold standard: the maximum matching ratio
To assess the performance of ClusterONE, we needed to compare an arbitrary set of predicted complexes with a predefined gold standard complex set (Supplementary Data 2). The comparison was made difficult by the fact that a match between a predicted complex and a gold standard one was often only partial. Moreover, a gold standard complex can have a (partial) match with more than one predicted complex and vice versa.
Here we propose a measure called the maximum matching ratio (MMR) to evaluate a complex detection algorithm. The MMR builds on maximal matching in a bipartite graph, in which the two sets of nodes represent the reference and predicted complexes, respectively, and an edge connecting a reference complex with a predicted one is weighted by the overlap score between the two (equation (2) and Supplementary Fig. 8). We selected the maximum weighted bipartite matching on this graph; that is, we chose a subset of edges such that each predicted and reference complex was incident on at most one selected edge and the sum of the weights of such edges was maximal. The chosen edges then represent an optimal assignment between reference and predicted complexes such that no reference complex is assigned to more than one predicted complex and vice versa. The MMR between the predicted and the reference complex set is then given by the total weight of the selected edges, divided by the number of reference complexes. This ratio is a measure of how accurately the predicted complexes represent the reference complexes. MMR offers a natural, intuitive way to compare predicted complexes with a gold standard and it explicitly penalizes cases when a reference complex is split into two or more parts in the predicted set, as only one of its parts is allowed to match the correct reference complex.
Besides the MMR, several other measures were also considered to compare predicted complexes to a gold standard. We used the number of matched complexes with an overlap score ω (equation (2)) larger than 0.25, the clustering-wise sensitivity14 (Sn), the positive predictive value14 (PPV) and the geometric accuracy14 (Supplementary Discussion).
Co-localization and overrepresentation score of a predicted complex set
Owing to the fact that the gold standard sets are incomplete16, a predicted complex that does not match any of the reference complexes may belong to a valid but still uncharacterized complex. A possible way to quantify the quality of such unmatched complexes is by recognizing that a protein complex can be formed only when its constituents are to be found in the same cellular compartment17, and that it is more likely for proteins of similar function to form a protein complex. We used the ‘co-localization score’18 using localization annotations of yeast proteins19 and a standard overrepresentation analysis of biological process, molecular function and cellular component terms from the Gene Ontology to assess the biological relevance of predicted complexes. The significance levels of the overrepresentation analysis were adjusted according to the Benjamini-Hochberg method20 to keep the overall significance level of the test at 0.05.
Data sources of interacting protein pairs
We used two experimental yeast PPI data sets6,7, a combined computational interaction map8 and the entire set of physical protein-protein interactions in yeast from BioGRID9 (Supplementary Table 3). Here we refer to these as the Gavin6, Krogan7, Collins8 and BioGRID9 data sets. The Gavin data set was obtained by considering all PPIs with a socio-affinity index larger than five6. The Krogan data set7 was used in two variants: the core data set (referred to as Krogan core) contained only highly reliable interactions (probability > 0.273), and the extended data set (referred to as Krogan extended) contained more interactions with less overall reliability (probability > 0.101). The socio-affinity and probability cutoffs we used have been proposed by the original authors. In the Collins data set, we used the top 9,074 interactions according to their purification enrichment score8, as suggested in the original paper. When applying algorithms that cannot handle weights (MCODE, CMC, RNSC and CFinder) to the above networks, weights were ignored. The BioGRID data set was downloaded from version 3.1.77 and contained all physical interactions that involve yeast proteins only. Self-interactions and isolated proteins were filtered from all the data sets. As BioGRID provides weights for only 18.05% of the interactions, we treated the entire BioGRID network as unweighted, keeping the weighted interactions but disregarding their weights.
Gold standard protein complexes
The most recent version of the MIPS catalog of protein complexes13 (18 May 2006) and the Gene Ontology (GO)-based protein complex annotations from SGD21 (11 Aug 2010) were used as gold standards (Supplementary Table 4).
The MIPS catalog is organized hierarchically: complexes may consist of subcomplexes extending to at most five hierarchy levels deep. An example of such a deeply embedded complex is the SAGA complex (MIPS identifier 510.190.10.20.10), a multifunctional coactivator that regulates transcription by RNA polymerase II. However, some MIPS categories do not correspond to complexes but rather to a set of related complexes; for instance, the category 510.180 corresponds to all ‘DNA-repair complexes’. To avoid selection bias, we decided to consider all MIPS categories containing at least three and at most 100 proteins as protein complexes. We also excluded MIPS category 550 and all its descendants, as these categories correspond to unconfirmed protein complexes that were predicted by computational methods.
The SGD maintains GO annotations for all yeast proteins. These annotations formed the basis of the SGD complex set that we used as a gold standard; a similar approach has been used before12. To create this data set, we first downloaded the mapping of yeast genes and proteins to GO terms21, and the most recent version of the GO structure22. We then ran an inference engine on the cellular component aspect of GO using the standard GO inference rules to find all terms that are descendants of the GO term GO:0043234 (protein complex) using “is_a” relations only. (Code of the inference engine is available as a forked version of Biopython at http://github.com/ntamas/biopython.) These GO terms were then treated as protein complex annotations. Finally, we retrieved all the yeast proteins that are assigned to the GO terms selected in the previous step and are supported by at least one non–inferred from electronic annotation (IEA) evidence code, and grouped them into protein complexes based on their GO annotations. Annotations with modifiers such as ‘not’ or ‘colocalizes_with’ were ignored.
Data sources of functional classifications and annotations
Subcellular localizations were obtained from a previously published dataset19, which assigns 4,155 yeast proteins to one or more of 21 cellular components. The functional classification of yeast proteins was obtained from the Gene Ontology annotations in the SGD21–23 on 11 August 2010. The GO classification is hierarchical, that is, proteins annotated by a given term are also annotated by all the ancestors of that term in the ontology. For the GO, annotations with ‘inferred from electronic annotation’, ‘no biological data available’ and ‘nontraceable author statement’ evidence codes (IEA, ND and NAS, respectively) were ignored. We also evaluated the case when the ‘inferred from protein interactions’ (IPI) evidence code was ignored (Supplementary Discussion).
An efficient and user-friendly implementation of ClusterONE
To make our method easily accessible for the scientific community, we developed a reference implementation of ClusterONE, which can be downloaded for free from http://www.paccanarolab.org/cluster-one/. The implementation consists of a single Java archive file that can be operated in standalone mode from the command line or as a plugin from the popular Cytoscape24 and ProCope25 platforms. The command line version is suitable for integrating ClusterONE into high-throughput data processing pipelines. The website includes a comprehensive manual for both the command line and the plugin interface, and a 1-min guide that explains the basic use-cases of ClusterONE. The source code of the application is also made available under the conditions of the GNU General Public License.
Besides clustering an entire network, both the Cytoscape plugin and the command line interface allow the user to initiate the protein complex detection process from a preselected set of seed proteins. This can be used to detect protein complexes that involve a particular protein or set of proteins. The Cytoscape plugin can also be operated in an exploratory analysis mode, where the user can select proteins manually from the network and calculate the total weight of the internal edges of the selection, the total weight of the edges connecting the selection to the rest of the network, the density and the cohesiveness score. This mode can also be used to fine-tune the obtained complexes as the application readily gives feedback about the quality of the complex when the user tries to extend it by adding new proteins or contract it by removing proteins that are not likely to be part of the complex based on expert biological knowledge.
Supplementary Material
Acknowledgments
T.N. was supported by the Newton International Fellowship Scheme of the Royal Society grant NF080750. A.P. was supported by the Biotechnology and Biological Sciences Research Council New Investigator grant BB/F00964X/1. H.Y. was supported by US National Institute of General Medical Sciences grant R01 GM097358.
Footnotes
AUTHOR CONTRIBUTIONS
T.N. and A.P. conceived the study. T.N. devised and implemented the algorithm and conducted benchmarks. H.Y. evaluated the biological relevance of the results. A.P. supervised the project. H.Y., T.N. and A.P. discussed the results and implications. A.P. and T.N. wrote the manuscript.
Note: Supplementary information is available on the Nature Methods website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Enright AJ, van Dongen S, Ouzounis CA. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King A, Pržulj N, Jurisica I. Bioinformatics. 2004;20:3013–3020. doi: 10.1093/bioinformatics/bth351. [DOI] [PubMed] [Google Scholar]
- 3.Pu S, Wong J, Turner B, Cho E, Wodak S. Nucleic Acids Res. 2009;37:825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bader GD, Hogue CW. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G, Wong L, Chua HN. Bioinformatics. 2009;25:1891–1897. doi: 10.1093/bioinformatics/btp311. [DOI] [PubMed] [Google Scholar]
- 6.Gavin A, et al. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 7.Krogan N, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 8.Collins SR, et al. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Stark C, et al. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey BJ, Dueck D. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 11.Palla G, Derényi I, Farkas I, Vicsek T. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 12.Macropol K, Can T, Singh A. BMC Bioinformatics. 2009;10:283. doi: 10.1186/1471-2105-10-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mewes HW, et al. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brohée S, van Helden J. BMC Bioinformatics. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radicchi F, Castellano C, Cecconi F, Loreto V, Parisi D. Proc Natl Acad Sci USA. 2004;101:2658–2663. doi: 10.1073/pnas.0400054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen R, Gerstein M. Curr Opin Microbiol. 2004;7:535–545. doi: 10.1016/j.mib.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Jansen R, et al. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 18.Friedel CC, Krumsiek J, Zimmer R. J Comput Biol. 2009;16:971–987. doi: 10.1089/cmb.2009.0023. [DOI] [PubMed] [Google Scholar]
- 19.Huh W-KK, et al. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 21.Hong E, et al. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dwight S, et al. Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, et al. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krumsiek J, Friedel CC, Zimmer R. Bioinformatics. 2008;24:2115–2116. doi: 10.1093/bioinformatics/btn376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.