Abstract
Background
A few lineages of Group A streptococci (GAS) have been associated with a reemergence of severe invasive streptococcal disease in developed countries. However, the majority of the comparisons between invasive and non-invasive GAS isolates have been performed for collections of reduced genetic diversity or relied on limited typing information to distinguish clones. We characterized by several typing methods and compared a collection of 160 isolates recovered from normally sterile sites with 320 isolates associated with pharyngitis and recovered in the same time period in Portugal.
Results
Although most of the isolates belonged to clones that were equally prevalent in invasive infections and pharyngitis, we identified markers of invasiveness, namely the emm types 1 and 64, and the presence of the speA and speJ genes. In contrast, emm4, emm75, and the ssa and speL/M genes were significantly associated with pharyngitis. There was a strong agreement between the emm type, the superantigen (SAg) genes and the clusters defined by pulsed-field gel electrophoresis (PFGE) profiling. Therefore, combinations of particular emm types and SAg genes frequently co-occurred in the same PFGE cluster, but there was no synergistic or antagonistic interaction between them in determining invasiveness. Only macrolide-susceptible PFGE clones were significantly associated with invasive infections or pharyngitis, while the clones of resistant isolates sharing all other molecular properties analyzed were equally prevalent in the two groups of isolates.
Conclusions
This study confirmed the importance of the widely disseminated emm1-T1-ST28 clone in invasive infections but also identified other clones linked to either invasive infections (emm64-ST164) or pharyngitis (emm4-T4-ST39), which may be more limited in their temporal and geographical spread. Clonal properties like some emm types or SAg genes were associated with disease presentation, highlighting the importance of bacterial genetic factors to the outcome of GAS infections, although other, yet unidentified factors may also play an important role.
Keywords: Streptococcus pyogenes, Streptococcal M protein, Exotoxins, Pharyngitis, Invasive infection
Background
Streptococcus pyogenes (Lancefield group A Streptococcus, GAS) remains one of the most common human pathogens, being responsible for uncomplicated superficial infections of the respiratory tract and skin, such as tonsillo-pharyngitis and impetigo, but also causing severe and rapidly progressing invasive disease such as necrotizing fasciitis, bacteremia, streptococcal toxic shock syndrome (STSS), puerperal sepsis, pneumonia, and meningitis [1]. Although the incidence and severity of GAS infections in industrialized countries decreased for most of the 20th century, a reemerge of GAS invasive disease has been noted since the late 1980s, both in North America and in Europe [2]. The annual incidence of GAS invasive disease has been estimated at 2.45/100 000 for developed countries, with a median case fatality rate of 15% [3].
The increase in the incidence of GAS invasive infections has been frequently associated with specific clones, raising the possibility that the rise of particularly virulent clones was responsible for this reemergence, in particular the M1T1 clone which is dominant among invasive GAS isolates in most developed countries [4,5]. However, a higher representation of a particular clone in invasive infections may be simply due to a high prevalence of that same clone in the general GAS population. To address this question several studies have performed comparisons between the characteristics of the invasive clones and the S. pyogenes isolates associated with carriage or uncomplicated infections in the same time period and geographic region. Some of those studies reported a differential distribution of M/emm types or of T types between invasive and non-invasive isolates and confirmed an association between serotype T1 and M1/emm1 and invasive infection [6-9], but many others found that the major clones responsible for invasive infections had a similar prevalence among non-invasive infections [10-12]. However, most of the studies performing such comparisons were either restricted to small numbers of isolates or were limited in the typing methodologies used, relying essentially on M/emm typing.
Serotyping of GAS based on protein M, a major surface virulence factor, has long been used as the gold standard for the epidemiological surveillance of the infections caused by this pathogen. In recent years it has been widely replaced by an equivalent approach based on sequencing the hypervariable region of the emm gene encoding the M protein. However, recent studies show that emm typing alone is not sufficient to unambiguously identify GAS clones and that it must be complemented with other typing methods such as pulsed-field gel electrophoresis (PFGE) macrorestriction profiling or multilocus sequence typing (MLST) [13]. Streptococcal superantigens (SAgs) secreted by S. pyogenes play an important role in the pathogenesis of the infections caused by this species [14]. The profiling of the eleven SAg genes described so far (speA, speC, speG, speH, speI, speJ, speK, speL, speM, ssa, smeZ) can be used as a typing methodology [15]. Some studies suggested an association between the presence of certain SAg genes or of certain SAg gene profiles and invasive infections [10,16], although others failed to establish such an association, reporting instead a strong link between the SAg profile and the emm type, regardless of the isolation site [12,15].
We have previously characterized a collection of 160 invasive GAS isolates collected throughout Portugal between 2000 and 2005, and found a very high genetic diversity among this collection, but with a dominant clone representing more than 20% of the isolates, which was characterized as emm1-T1-ST28 and carried the gene speA[17]. The aim of the present study was to evaluate if the clone distribution among the invasive GAS isolates in Portugal reflected the clonal structure of the isolates causing pharyngitis, in terms of molecular properties and antimicrobial resistance. In order to do that, 320 non-duplicate isolates collected from pharyngeal exudates associated with tonsillo-pharyngitis in the same time period were studied by emm typing, T typing, SAg profiling, PFGE macrorestriction profiling, and selected isolates were also submitted to MLST analysis. All isolates were also tested for their susceptibility to clinically and epidemiologically relevant antimicrobial agents. The great majority of the clones were found with a similar frequency among invasive infections and pharyngitis. Still, some clones were shown to have a higher invasive disease potential and it was also possible to establish significant associations between some emm types and SAg genes and disease presentation.
Results
Antimicrobial resistance
All isolates were fully susceptible to penicillin, quinupristin/dalfopristin, chloramphenicol, vancomycin, linezolid, and levofloxacin (Table 1). Among the invasive isolates, 19 (12%) were resistant to erythromycin, while the isolates associated with pharyngitis presented significantly higher macrolide resistance – 21% (P = 0.016, two-tailed Fisher’s exact test). Among the invasive macrolide-resistant isolates, 10 (53%) presented the M phenotype and were therefore susceptible to clindamycin, whereas the remaining nine (47%) were also constitutively resistant to clindamycin (cMLSB phenotype). The proportion of the two phenotypes was similar among the pharyngitis isolates, with 37 isolates (55%) presenting the M phenotype and 30 (45%) presenting the MLSB phenotype (one with inducible resistance and the others with constitutive resistance to clindamycin). All the isolates presenting the M phenotype of macrolide resistance carried only the mef(A) variant of the mef determinant. The cMLSB isolates carried only the erm(B) gene, except for one pharyngitis isolate which also harbored mef(A), and the only iMLSB isolate in the collection that presented the erm(A) gene.
Table 1.
PFGE clusters presenting antimicrobial resistant isolates collected from invasive infections and pharyngitis in Portugal
| PFGE cluster a | Antimicrobial resistance b |
No. of resistant isolates |
|
|---|---|---|---|
| Invasive | Pharyngitis | ||
| C38 |
Tet |
|
1 |
| D36 |
MLSB |
|
1 |
| M |
|
1 |
|
| G27 |
M |
6 |
19 |
| M,Tet |
1 |
|
|
| H26 |
MLSB,Bac |
6 |
17 |
| Tet |
|
1 |
|
| I24 |
MLSB,Tet |
|
1 |
| J16 |
Tet |
12 |
1 |
| K14 |
M |
|
1 |
| L13 |
MLSB,Tet |
1 |
6 |
| Tet |
2 |
|
|
| M11 |
MLSB,Tet |
1 |
|
| N10 |
Tet |
1 |
1 |
| MLSB,Tet |
|
1 |
|
| O9 |
M |
4 |
5 |
| R6 |
M |
|
3 |
| S6 | M | 1 | |
a Clusters are designated by capital letters and a subscript number indicating the number of isolates in each cluster;
b The antibiotics tested were penicillin quinupristin/dalfopristin, chloramphenicol, vancomycin, linezolid, levofloxacin, erythromycin, clindamycin, tetracycline, and bacitracin. M, presenting the M phenotype of macrolide resistance; MLSB, presenting the MLSB phenotype of macrolide resistance; Tet, non-susceptibility to tetracycline; M,Tet, presenting the M phenotype of macrolide resistance and resistance to tetracycline; MLSB,Tet, presenting the MLSB phenotype of macrolide resistance and resistance to tetracycline; MLSB,Bac, presenting the MLSB phenotype of macrolide resistance and resistance to bacitracin.
In contrast to erythromycin, tetracycline resistance was much lower among the pharyngitis isolates when compared with the invasive group (6% vs 17%, P < 0.001). One invasive isolate presented intermediate resistance to tetracycline (MIC = 3μg/ml). All the resistant strains carried the tet(M) gene, except one pharyngitis isolate for which no PCR product was obtained for any of the screened tetracycline-resistance genes. The tet(L) gene was detected in only one pharyngitis isolate, which also harbored tet(M), while the genes tet(K) and tet(O) were not amplified in any of the studied isolates. Overall there was a positive association between the genes tet(M) and erm(B) (P < 0.001), but that association was not observed among the subset of invasive isolates, since only three of the 27 tet(M)-positive invasive isolates also carried the erm(B) gene (P = 0.178).
Bacitracin resistance was detected in a total of 23 isolates (5%), with no significant differences among the two types of infection considered. All these isolates expressed the cMLSB phenotype of macrolide resistance and were tetracycline-susceptible.
Characterization of GAS clones
Globally, among the 480 isolates there were 36 emm types, 17 T types, and 49 SAg profiles (the genes included in each SAg profile are presented in Additional file 1). In the subset of 170 isolates (100 from pharyngitis and 70 from invasive infections) selected for MLST analysis, 49 different STs were identified. Nineteen PFGE clusters (groups of > 5 isolates presenting ≥ 80% similarity on the PFGE profile) were obtained including 268 pharyngitis isolates and 143 invasive isolates (86% of all isolates) (Table 2 and Table 3). Except for R6, isolates grouped into PFGE clusters presented some variability in their emm type, ST, T type, or SAg profile, with most variability found in the later two properties. Still, in most PFGE clusters the majority of the isolates were characterized by a single profile of dominant properties. The emm diversity among the PFGE clusters differed significantly (Table 4). Within each PFGE cluster, different emm types were associated with distinct SAg profiles (Table 2 and Table 3), although globally the emm and PFGE had a similar predictive power over the SAg profile (data not shown).
Table 2.
Properties of the PFGE clusters with >15 GAS isolates collected from invasive infections and tonsillo-pharyngitis in Portugal
| PFGE cluster a | emm type |
No. of isolates (% of total) |
T type b (no. of isolates) | SAg genes profile (no. of isolates) | ST c (no. of isolates) | |
|---|---|---|---|---|---|---|
| Invasive | Pharyngitis | |||||
| A51 |
3 |
15 (9.4) |
36 (11.25) |
3 (22), NT (14), 3/13 (13), 1 (2) |
8 (48), 37 (2), 2 (1) |
406 (8), 15 (4), 315 (2) |
| B49 |
1 |
28 (17.5) |
20 (6.3) |
1 (46), NT (2) |
10 (47), 3 (1) |
28 (10) |
| stIL103 |
1 (0.6) |
0 |
1 (1) |
10 (1) |
28 (1) |
|
| C38 |
89 |
12 (7.5) |
25 (7.8) |
B3264 (37) |
27 (21), 29 (8), 46 (5), 43 (2), 40 (1) |
408 (5), 553 (1), 101 (2) |
| 75 |
0 |
1 (0.3) |
25 (1) |
42 (1) |
150 (1) |
|
| D36 |
12 |
10 (6.3) |
25 (7.8) |
12 (29), NT (6) |
33 (29), 16 (5), 46 (1) |
36 (13), 551 (2) |
| 94 |
1 (0.6) |
0 |
B3264 (1) |
35 (1) |
89 (1) |
|
| E30 |
6 |
11 (6.9) |
19 (5.9) |
6 (27), NT (2), 2(1) |
2 (28), 5 (1), 9 (1) |
382 (6), 411 (3) |
| F29 |
4 |
1 (0.6) |
28 (8.8) |
4 (29) |
23 (27), 22 (2) |
39 (5) |
| G27 |
4 |
8 (5.0) |
19 (5.9) |
4 (23), B3264 (2), 2/27/44 (1), 2/4 (1) |
23 (23), 30 (2), 40 (1), 41 (1) |
39 (8), 561 (1) |
| H26 |
28 |
7 (4.4) |
17 (5.3) |
28 (23), NT (1) |
27 (13), 24 (10), 15 (1) |
52 (10) |
| 22 |
0 |
1 (0.3) |
12 (1) |
3 (1) |
nd |
|
| 75 |
0 |
1 (0.3) |
NT (1) |
7 (1) |
481 (1) |
|
| I24 |
44/61 |
6 (3.8) |
16 (5.0) |
2/27/44 (19), NT (2), 12 (1) |
32 (16), 12 (6) |
25 (5), 554 (1) |
| 75 |
0 |
1 (0.3) |
25 (1) |
36 (1) |
150 (1) |
|
| 89 |
0 |
1 (0.3) |
5/27/44 (1) |
6 (1) |
555 (1) |
|
| J16 | 64 |
11 (6.9) |
0 |
3/13 (5), NT (4), 1 (2) |
46 (10), 43 (1) |
164 (4), 124 (1) |
| 53 |
2 (1.3) |
0 |
NT (2) |
26 (2) |
11 (1) |
|
| 74 |
0 |
1 (0.3) |
B3264 (1) |
11 (1) |
120 (1) |
|
| 87 |
0 |
1 (0.3) |
28 (1) |
38 (1) |
62 (1) |
|
| 89 | 0 | 1 (0.3) | B3264 (1) | 43 (1) | 568 (1) | |
a Clusters are designated by capital letters and a subscript number indicating the number of isolates in each cluster; b NT, non-typeable; c nd, not determined.
Table 3.
Properties of the PFGE clusters with <15 GAS isolates collected from invasive infections and tonsillo-pharyngitis in Portugal
| PFGE cluster a | emm type |
No. of isolates (% of total) |
T type b (no. of isolates) | SAg genes profile (no. of isolates) | ST(no. of isolates) | |
|---|---|---|---|---|---|---|
| Invasive | Pharyngitis | |||||
| K14 |
2 |
1 (0.6) |
13 (4.1) |
2 (13), 4 (1) |
31 (12), 48 (2) |
55 (5) |
| L13 |
22 |
1 (0.6) |
7 (2.2) |
12 (8) |
21 (6), 13 (1), 19 (1) |
46 (2), 389 (1) |
| 9 |
1 (0.6) |
1 (0.3) |
9 (1), NT (1) |
46 (2) |
75 (2) |
|
| 2 |
0 |
1 (0.3) |
2 (1) |
31 (1) |
55 (1) |
|
| 74 |
1 (0.6) |
0 |
9 (1) |
5 (1) |
120 (1) |
|
| st106M |
1 (0.6) |
0 |
4 (1) |
49 (1) |
53 (1) |
|
| M11 |
28 |
8 (5.0) |
3 (0.9) |
28 (11) |
24 (7), 27 (3), 15 (1) |
52 (5) |
| N10 |
87 |
2 (1.3) |
7 (2.2) |
28 (8), 6 (1) |
20 (3), 27 (3), 2 (1), 18 (1), 44 (1) |
62(2) |
| 22 |
0 |
1 (0.3) |
12 (1) |
21 (1) |
46 (1) |
|
| O9 |
1 |
4 (2.5) |
5 (1.6) |
1 (8), 13 (1) |
10 (9) |
28 (4) |
| P8 |
78 |
4 (2.5) |
4 (1.3) |
11 (7), 3/13 (1) |
29 (8) |
409 (3) |
| Q8 |
43 |
4 (2.5) |
0 |
3/13 (2), NT (2) |
11 (4) |
3 (2) |
| 58 |
2 (1.3) |
2 (0.6) |
NT (4) |
17 (3), 14 (1) |
410 (3), 176 (1) |
|
| R6 |
75 |
0 |
6 (1.9) |
25 (6) |
39 (6) |
150 (2) |
| S6 | 9 |
1 (0.6) |
4 (1.3) |
9 (5) |
40 (5) |
75 (2) |
| 12 | 0 | 1 (0.3) | 12 (1) | 33 (1) | 36 (1) | |
a Clusters are designated by capital letters and a subscript number indicating the number of isolates in each cluster; b NT, non-typeable.
Table 4.
Simpson’s index of diversity and 95% Confidence intervals (CI95%) of emm types for each PFGE cluster
| PFGE cluster a | No. emm types | SID [CI 95% ] |
|---|---|---|
| B49 |
2 |
0.041 [0–0.118] |
| C38 |
2 |
0.053 [0–0.151] |
| D36 |
2 |
0.056 [0–0.159] |
| H26 |
3 |
0.151 [0–0.336] |
| I24 |
3 |
0.163 [0–0.361] |
| J16 |
5 |
0.533 [0.255-0.812] |
| L13 |
5 |
0.628 [0.353-0.903] |
| N10 |
2 |
0.200 [0–0.504] |
| Q8 |
2 |
0.571 [0.571-0.571] |
| S6 | 2 | 0.333 [0–0.739] |
a PFGE clusters A51, E30, F29, G27, K14, M11, O9, P8, and R6 include only one emm type (SID=0).
Unrelated STs within the same PFGE clusters were associated with isolates of different emm types, while isolates of the same emm type presented the same ST or single-locus variants (SLVs) (Table 2 and Table 3). The only exceptions were ST39 and ST561 that were both associated with cluster G27 and emm4, but were double-locus variants (DLVs) of each other. In clone I24, four distinct STs were found. While ST25 and ST554 were SLVs and were both associated with emm44/61, ST150 belonged to a different clonal complex, but was also associated with a different emm type (emm75). Finally, ST555 despite being associated with an isolate of a different emm type (emm89) is a SLV of ST25, which may explain why this isolate was clustered in I24 and not in the major PFGE cluster associated with this emm type (C38).
Isolates expressing the M phenotype of macrolide resistance belonged mostly to clusters G27, O9, and R6, while the majority of MLSB isolates were clustered in H26 and L13 (Table 1). Isolates belonging to emm types 1, 4, and 28 were separated into different PFGE clusters according to macrolide resistance (B49 and O9 for emm1; F29 and G27 for emm4; H26 and M11 for emm28, respectively), while the remaining characteristics (T type, ST, and SAg profile) of each pair of PFGE clusters were the same.
Bacitracin-resistant isolates were all clustered in PFGE H26 and were characterized as emm28-T28, except for one isolate that was emm22-T12. However, this cluster was not restricted to bacitracin-resistant isolates, since it also included three bacitracin susceptible isolates, two of which were also emm28-T28, while the other was emm75 but T non-typable.
Surface antigen differences between invasive and pharyngitis isolates
The invasive isolates were significantly less diverse than the pharyngitis isolates by T typing and SAg profiling (Table 5). However, while the emm type distribution varied between the invasive and pharyngitis isolates (P < 0.001) no differences were noted in the T types. Sixteen emm types occurred only in invasive infection or pharyngitis, but in most cases the small number of isolates associated with these emm types prevented the differences from reaching statistical significance (Figure 1). In contrast, the overrepresentation of emm types 1, 4, 64, and 75 in one of the groups was statistically supported.
Table 5.
Simpson’s index of diversity and 95% Confidence intervals (CI 95% ) of the typing methods used in the analysis of the 160 invasive isolates and 320 pharyngitis isolates
| Typing method |
Invasive |
Pharyngitis |
||
|---|---|---|---|---|
| No. types | SID [CI 95% ] | No. types | SID [CI 95% ] | |
| T typing |
13 |
0.882 [0.859-0.904] |
17 |
0.915 [0.907-0.923] |
|
emm typing |
30 |
0.920 [0.900-0.940] |
26 |
0.921 [0.911-0.931] |
| PFGE profiling |
30 |
0.930 [0.912-0.947] |
44 |
0.947 [0.939-0.954] |
| SAg profiling | 27 | 0.911 [0.891-0.931] | 46 | 0.941 [0.932-0.951] |
Figure 1.
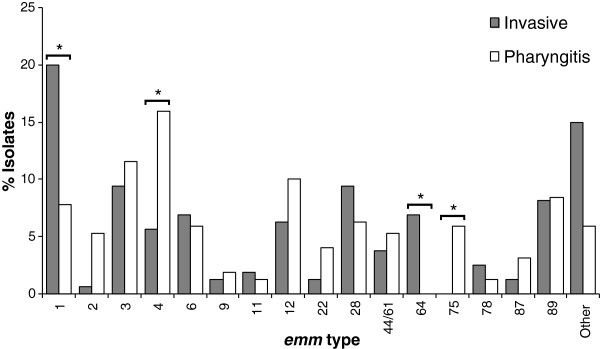
Distribution of emm types among 160 invasive isolates and 320 pharyngitis isolates. Other includes emm types identified in ≤ 5 isolates - emm18 (n=4), 25 (n=1), 29 (n=2), 30 (n=1), 43 (n=4), 48 (n=1), 53 (n=3), 58 (n=5), 74 (n=2), 77 (n=4), 90 (n=1), 94 (n=3), 102 (n=3), 103 (n=1), 113 (n=3), 114 (n=1), 118 (n=1), st106M (n=1), stG1750 (n=1), stIL103 (n=1). The asterisk indicates significant differences (P<0.001).
SAg differences between invasive and pharyngitis isolates
A detailed analysis of the SAg profiling results of the isolates is performed elsewhere [18]. Briefly, the chromosomally encoded SAg genes smeZ and speG were the most frequent among the 480 isolates (n = 461 and 417, respectively), followed by speC (n = 247), ssa (n = 170), speJ (n = 157), speA (n = 154), speK (n = 118), speH (n = 82), speI (n = 73), and speL and speM, which were always detected together (n = 44). The association of individual SAg genes with disease presentation was tested. In the analysis of these results, the SAg genes speG and smeZ were not considered because they were present in nearly all isolates, and the genes speL and speM were considered as a single entity, since they always co-occurred. Individually, the genes speA and speJ were both associated with invasive isolates (P < 0.001). As expected, strains carrying both these genes were also associated with invasive infections (P < 0.001), but no synergistic effect between the two genes was observed, since the presence of one did not significantly increase the representation of the other among invasive isolates. In contrast, speC (P = 0.002), ssa (P < 0.001), and speL/M (P < 0.001) were individually associated with pharyngitis. The combinations speC+speL/M and ssa+speL/M were both associated with pharyngitis (P = 0.004 and 0.012, respectively), but there was also no synergistic effect relative to the presence of a single gene. However, the association of speC with pharyngitis isolates can be explained by a high frequency of co-occurrence of this gene with ssa, since the isolates harboring speC without ssa were not significantly associated with any of the groups. An interesting situation occurred when analyzing the interaction between speJ (associated with invasive infections) and ssa (associated with pharyngitis). Among isolates carrying speJ, the group that also carried ssa was no longer associated with invasive infections, while the association of isolates carrying ssa with pharyngitis was not significantly altered by the presence of speJ. This argues for a dominant effect of the presence of ssa over that of speJ in determining the invasive capacity of individual isolates. The association of SAg profiles with disease presentation was also tested. Two SAg profiles presented a significant association with invasive isolates, namely SAg10 (speA+speG+speJ+smeZ+) and SAg46 (speG+smeZ+) (P < 0.001). The remaining profiles were not significantly associated with any of the two groups of isolates.
When the same kind of analysis was performed for emm types and individual SAg genes, three combinations with statistical significance emerged: the association of isolates presenting emm1 and speA, and emm1 and speJ with invasive infections (P < 0.001), and the association of isolates carrying emm75 and speL/M with pharyngitis (P = 0.001). In all cases, no synergistic or antagonistic interaction was detected between emm type and SAg gene, since the emm type did not alter the association of the SAg gene with a particular group of isolates.
Differences between the PFGE clusters found among invasive infection and pharyngitis
The associations described above can be correlated with the PFGE clusters which were also different between the invasive and pharyngitis groups of isolates (P < 0.001), in agreement with the differences found in emm types (Figure 1 and Figure 2). All the 19 major PFGE clusters occurred in both invasive and pharyngitis isolates, except for R6 (emm75-T25-ST150-SAg39), which was present only among pharyngeal isolates, but the difference did not reach statistical significance due to the small number of isolates in this cluster. PFGE distinguished several groups of isolates belonging to emm types 1 and 4. The difference in the distribution between pharyngitis and invasive infection was not found for all PFGE clusters containing isolates carrying each of these two emm types, but only for those including macrolide-susceptible isolates (B49 associated with invasive infections and F29 associated with pharyngitis, respectively, P < 0.001). The emm1 and emm4 isolates expressing macrolide resistance (M phenotype) were grouped into PFGE clusters O9 and G27, respectively, which presented a similar prevalence among invasive infections and pharyngitis (Figure 2). PFGE J16, which included all emm64 isolates, was also associated with invasive infections (P < 0.001). The emm75 association with pharyngitis was not translated into an association of a specific PFGE cluster, since the 19 emm75 strains were scattered into various PFGE clusters (Table 2 and Table 3).
Figure 2.
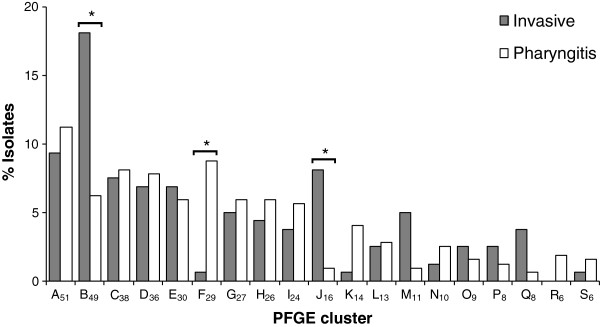
PFGE clusters found among 160 invasive isolates and 320 pharyngitis isolates. Approximately 11% of invasive and 16% of non-invasive isolates were included in PFGE clusters of ≤ 5 isolates that are not represented. The asterisk indicates significant differences (P<0.001).
Not surprisingly, three emm-PFGE cluster combinations showed significant associations with infection type: emm1-B49 and emm64-J16 were associated with invasive infections, while emm4-F29 was associated with pharyngitis (P < 0.001). It was not possible to detect any synergistic or antagonistic interaction between PFGE and emm type in modulating the association of the isolates with either group. The same was true for the statistically significant combinations between PFGE clusters and individual SAg genes, namely the combination of B49 with speA and with speJ (both associated with invasive infections, P < 0.001), and the combination of F29 with speC and with ssa (both associated with pharyngitis, P < 0.001).
Discussion
Several studies yielding conflicting results have attempted to compare the clonal composition of GAS populations causing invasive and non-invasive infections in order to identify particularly virulent clones or properties that may be used as epidemiological markers of invasiveness [7,8,11,12,16]. However, many of those studies were limited in the size and diversity of the GAS collections studied or in the typing methodologies used, with most of them relying essentially on emm typing, which has been shown to be insufficient for the complete identification of GAS clones [13]. In this work, we used several different typing methods to compare a collection of genetically diverse GAS isolates recovered from normally sterile sites during a period of six years in Portugal [17] with isolates recovered from pharyngeal exudates of patients presenting with tonsillo-pharyngitis, during the same time period and in the same geographical region. The nasopharyngeal mucosa has been suggested to be the main reservoir for GAS isolates associated with invasive infections [19,20].
The differences among the GAS clones associated with invasive infections and pharyngitis were reflected in antimicrobial resistance, with the invasive group of isolates presenting lower macrolide resistance and higher tetracycline resistance, when compared with the pharyngitis group. Among isolates belonging to the same emm types, namely emm1 and emm4, only the macrolide-susceptible clones were associated with either invasive infections or pharyngitis. The macrolide-resistant clones of these emm types are reflected in invasive infections according to their prevalence in pharyngitis, suggesting that these are translating more the antibiotic selective pressure than the invasive capacity of the clones. Tetracycline is not currently used in the treatment of GAS infections but resistance to this antimicrobial in S. pyogenes is usually acquired by horizontal transfer, since the resistance genes are frequently encoded in mobile genetic elements with a wide host range [21]. These elements often carry macrolide resistance genes as well, and in S. pyogenes a significant association between the presence of the genes erm(B) and tet(M) has been reported and it has been suggested that tetracycline use could contribute to the selection of macrolide-resistant GAS isolates [21,22]. In our study, the association between the presence of the genes erm(B) and tet(M) was observed globally, but not among the invasive isolates, suggesting that the genetic elements carrying tetracycline resistance conferring genes may be different between the two bacterial populations.
Bacitracin susceptibility is routinely used for the presumptive identification of GAS, although resistant clones have been identified in several countries [23-25]. In our GAS collection, all the bacitracin-resistant isolates (5%), regardless of the type of infection, were clustered in the same PFGE clone (H26) and belonged to ST52, although one was emm22-T12 while the others were all emm28-T28. Isolates with such characteristics had been previously reported in Portugal associated with tonsillo-pharyngitis, skin infections and asymptomatic carriage [26,27], but not with invasive infections. Bacitracin resistance among invasive isolates has been previously reported only among isolates recovered in France and in San Francisco [24,25].
Although 74% of the invasive isolates in our collection belonged to clones which were equally frequent among pharyngitis, suggesting that a significant part of the invasive GAS population mirrors the clonal structure of the circulating GAS isolates, the remaining isolates represented clones that had an enhanced capacity to cause invasive disease. We also found significant associations between individual properties or pairwise combinations of properties and disease presentation. Since in most cases these were also characteristics of the more invasive clones, we cannot exclude that the associations of individual properties or pairwise combinations of properties can reflect, at least partially, the distribution of genetic lineages in the two GAS populations analyzed.
Individually, emm types 1 and 64 were associated with invasive infections. Isolates belonging to these emm types presented the only two SAg profiles significantly associated with invasive infections. Surprisingly, only one of these SAg profiles includes a phage-encoded SAg gene (speA). In agreement with our observation, a previous study found that within the same PFGE-emm group, the SAg profiles significantly associated with invasive infections had a smaller number of SAg genes than the dominant profiles in pharyngitis [28]. These results suggest that although some SAg genes may significantly contribute to the virulence of S. pyogenes, the rise and success of highly virulent GAS clones may not hinge upon the acquisition of phage-encoded SAg genes. Still, in our study, the SAg genes speA and speJ were both significantly more prevalent among invasive isolates. This association was not substantially affected by emm type, PFGE clone, nor by the presence of other SAg genes, suggesting that speA and speJ can be regarded by themselves as markers for invasiveness. Although such association has not been previously noted for speJ, the speA gene has been frequently associated with invasive infections [6,8,16] and the production of SpeA by GAS isolates has been linked to streptococcal toxic shock syndrome [29].
On the other hand, we identified an association of pharyngitis isolates with emm types 4 and 75, and with the SAg genes speC, ssa, and speL/M. The association of speC with non-invasive infections has been previously reported [6,16,30], but in our collection this association could be explained simply by a high frequency of co-occurrence of this gene with ssa which was strongly associated with pharyngitis, as was also noted in a recent study [16]. The presence of the genes speL and speM was not previously associated with non-invasive infections.
Since there is a strong correlation of the SAg profile with emm type and of both these properties with PFGE type, some of these individual factors frequently co-occurred in the same clones. Therefore, combinations of these characteristics were also significantly associated with disease presentation. However, we could not detect any synergistic or antagonistic interactions between most of these characteristics, meaning that their co-occurrence in a particular isolate does not make it more invasive than isolates sharing only one of these characteristics.
Two PFGE clusters were significantly more prevalent among isolates associated with invasive disease than among those causing tonsillo-pharyngitis. One of these was a cluster of macrolide-susceptible isolates characterized as emm1-T1-ST28 and by the presence of the SAg genes speA, speG, speJ, and smeZ (B49), which accounted for 18% of the invasive isolates. M1T1 isolates have been frequently associated with severe invasive GAS disease, and the acquisition of prophage-encoded virulence genes, as well as horizontal gene transfer events by homologous recombination were implicated in the increased virulence of these isolates [31,32]. However, many epidemiological studies found a similar prevalence of this clone among invasive and non-invasive isolates [10-12], questioning its enhanced invasive capacity. In contrast to these findings, but similarly to those of others [6-9], we found an association between this clone and invasive GAS disease in Portugal, although it can also frequently cause milder infections such as pharyngitis (it accounted for 6% of the pharyngitis isolates analyzed in this study).
The other cluster significantly associated with invasive infections in Portugal was J16, which was dominated by isolates belonging to emm64-ST164 and carrying the SAg genes speG and smeZ. A clone with these characteristics has not been previously associated with invasive disease and emm64 has been infrequently reported among invasive GAS isolates [4,33,34]. The higher invasive capacity of this clone cannot be attributed to its SAg repertoire, since these isolates do not harbor any of the SAg genes associated with invasive infection. Other, still unidentified, characteristics may be responsible for the properties of this clone.
In contrast to these PFGE clones, the F29 clone of macrolide-susceptible isolates characterized by emm4-T4-ST39 and harboring the genes speC, ssa and smeZ was associated with pharyngitis, suggesting that this clone may have a reduced ability to cause invasive disease, in agreement with the negative association between emm4 and invasive infection that has been suggested elsewhere [16]. The association of emm75 with pharyngitis has not been previously reported and was not translated into particular PFGE clusters due to the high diversity of emm75 isolates.
Our data confirms that the widely dispersed M1T1 clone has enhanced invasiveness but we also identified clones with increased or decreased invasive capacity that may have emerged locally and that have a more limited temporal and geographical spread. The emm alleles and the SAg genes characteristic of these clones were associated with particular disease presentations. Other individual emm alleles and SAg genes were also associated with a higher propensity to cause invasive infections or pharyngitis indicating the importance of these characteristics in determining an isolate’s invasive capacity.
Other factors that were not evaluated in this study may contribute to a different distribution of GAS clones in less severe and more severe infections. These include bacterial factors, such as the occurrence of mutations in transcriptional regulators controlling the expression of virulence factors, which seems to play an important role in the pathogenesis of some GAS isolates [35]. For other clones, the ability to cause invasive infections may be more dependent on exploiting host factors, like the HLA class II haplotype [36], which may vary in frequency in different human populations.
Conclusions
This study established links between particular genetic lineages and the type of infection, indicating that genetic characteristics of the bacteria play an important role in determining the outcome of their interaction with the human host. The different distribution of clones in the two types of infection supports the relevance of PFGE as a typing methodology for GAS [13]. This was further evidenced by the fact that the macrolide-resistant emm1 and emm4 PFGE clones were not associated with any particular disease presentation, contrary to the susceptible clones carrying the same emm types that were associated with invasive infections and pharyngitis, respectively. Moreover, in contrast to other reports [12,15] we found associations between particular emm alleles and SAg genes and disease presentation. In this study, we identified emm4, emm75, ssa and speL/M as independent markers for pharyngitis and emm1, emm64, speA, and speJ as independent markers for invasiveness. Our data re-enforces the multi-factorial nature of GAS invasive capacity and highlighted lineages and characteristics, in addition to the well known M1T1 lineage, that are associated with particular disease presentations and that may further increase in importance.
Methods
Bacterial isolates
The invasive isolates (n = 160) were collected from normally sterile sites, and their partial characterization was previously reported [17]. A total of 320 non-duplicate GAS isolates were randomly selected among a collection of 1604 isolates recovered from pharyngeal exudates of patients presenting with tonsillo-pharyngitis in 32 laboratories distributed throughout Portugal, between 2000 and 2005, in the proportion of 1:2 (invasive:pharyngitis) for each studied year. These isolates were recovered from pediatric patients (<18 yrs) and showed a balanced distribution by gender. The subset of macrolide-resistant pharyngeal isolates had been partially characterized [27,37]. Strains were identified by the submitting laboratories and confirmed in our laboratory by colony morphology, β-hemolysis and the presence of the characteristic group antigen (Slidex Strepto A, BioMérieux, Marcy l’Etoile, France).
Antimicrobial susceptibility testing
Susceptibility tests were performed by disk diffusion on Mueller-Hinton agar supplemented with 5% defibrinated sheep blood, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) using the following antibiotic disks (Oxoid, Basingstoke, UK): penicillin, vancomycin, erythromycin, tetracycline, levofloxacin, chloramphenicol, clindamycin, quinupristin/dalfopristin, and linezolid. Whenever isolates with intermediate susceptibility were identified, the results were confirmed by MIC determination using E-test strips (BioMérieux, Marcy l’Etoile, France). The macrolide resistance phenotype was determined as previously described [38]. Susceptibility to bacitracin was determined for all isolates using disks containing 0.05 U of bacitracin (Oxoid, Basingstoke, UK), as described elsewhere [27].
Detection of antimicrobial resistance genetic determinants
Bacterial DNA was prepared according to the protocols of the Centers for Disease Control and Prevention (CDC) [39]. Determination of the macrolide resistance genotype was performed for strains presenting either the M or the MLSB macrolide resistance phenotype, by a multiplex PCR reaction with primers to detect the erm(B), erm(A) and mef genes, as previously described [40]. Isolates carrying the mef gene were subjected to a second PCR reaction in order to discriminate between mef(A) and mef(E) [37]. Tetracycline resistant isolates were PCR-screened for the presence of the genes tet(K), tet(L), tet(M), and tet(O) as previously described [41]. Strains harboring each of the resistance genes were used as positive controls for the PCR reactions.
T-typing
Strains were cultured in Todd-Hewitt broth (Oxoid, Basingstoke, UK) at 30°C overnight and treated with swine pancreatic extract, using the Auxiliary Reagents for Hemolytic Streptococcus Typing (Denka Seiken, Tokyo, Japan), and following the manufacturer’s instructions. T serotypes were determined by slide agglutination with 5 polyvalent and 19 monovalent sera (Hemolytic Streptococcus Group-A Typing Sera, Denka Seiken).
emm-typing and SAg gene profiling
The emm-typing of all isolates was performed according to the protocols and recommendations of the CDC, and the first 240 bases of each sequence were searched against the emm CDC database [39]. Identity of ≥ 95% with previously described sequences over the 150 bases considered allowed the assignment of an emm type. The presence of the SAg genes speA, speC, speG, speH, speI, speJ, speK, speL, speM, smeZ, and ssa, and of the chromosomally encoded exotoxin genes speB and speF (used as positive control fragments) was assessed in all 160 invasive and 320 non-invasive GAS isolates by two multiplex PCR reactions as described elsewhere [18].
PFGE macrorestriction profiling and MLST
Agarose plugs of bacterial DNA were prepared as previously described [27]. After digestion with SmaI or Cfr9I (Fermentas, Vilnius, Lithuania), the fragments were resolved by PFGE [27]. The isoschizomer Cfr9I was used only for the isolates with the M phenotype, which were not digested by SmaI [13,27]. The macrorestriction patterns generated were compared using the Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) to create UPGMA (unweighted pair group method with arithmetic mean) dendrograms. The Dice similarity coefficient was used, with optimization and position tolerance settings of 1.0 and 1.5, respectively. PFGE clones were defined as groups of >5 isolates presenting profiles with ≥ 80% relatedness on the dendrogram [13]. MLST analysis was performed as described elsewhere [42] for representatives of each PFGE cluster (a total of 100 non-invasive and 70 invasive isolates). When more than one emm or T-type was present in the same PFGE cluster, isolates expressing different surface antigens were selected. Allele and sequence type (ST) identification was performed using the S. pyogenes MLST database [43]. Whenever new alleles were identified, chromatograms of both strands were submitted to the database curator for approval and an allele number was assigned. Clonal complexes were determined using the goeBURST algorithm implemented in PHYLOViZ [44].
Statistical analysis
The diversities of the different PFGE clusters were compared using the Simpson’s index of diversity (SID) with corresponding 95% confidence intervals (CI95%) [13]. Differences in antibiotic resistance between the invasive and non-invasive groups of isolates were evaluated using Fisher’s exact test. P values < 0.05 were considered to indicate statistical significance. SAg genes, emm types and PFGE types were screened for associations with the invasive group by computing an odds-ratio and an associated Fisher’s exact test. Additionally, pairs of individual SAg genes with each other or with emm types or PFGE types were similarly tested for the association of each pairs’ co-occurrence with the invasive group of isolates. For the pairs where at least one of the types individually or their co-occurrence were associated (either positively or negatively) with the invasive group, two more tests were done, to investigate if the association of one of the types individually was modified by the co-occurrence of the other type in the pair (synergism or antagonism). Considering a pair of types A and B, this test compares the proportion of invasive isolates among the ones that have A type but not B with the same proportion among isolates that have both A and B types. If the proportions are statistically different, according to a Fisher’s exact test, we can conclude that type B modifies the association of type A with the invasive group of isolates. Conversely, if the proportion of invasive isolates among the ones that have the B type but not A differs from the same proportion among isolates that have both A and B types, type A modifies the association of type B with the invasive group. If the isolates that are simultaneously of the A and B type show a significantly stronger association with invasive infection than the one observed for isolates having either the A or B type, the types are said to be synergistic. If, on the other hand, isolates that are simultaneously of the A and B type show a significantly weaker association with invasive infection than the one observed for isolates having either the A or B type, the types are said to be antagonistic. All the p-values obtained in each step of the screening procedure were corrected for multiple testing through the False Discovery Rate (FDR) linear procedure [45].
Competing interests
Dr José Melo-Cristino has received research grants administered through his university and received honoraria for consulting and serving on the speakers bureaus of Pfizer, Bial, GlaxoSmithKline and Novartis. Dr Mário Ramirez has received honoraria for consulting and serving on speakers bureau of Pfizer. The other authors declare no conflict of interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This work was partially supported by Fundação para a Ciência e Tecnologia, Portugal (PTDC/SAU-ESA/72321/2006), Fundação Calouste Gulbenkian and unrestricted research grant from Glaxo SmithKline.
Authors’ contributions
AF, CSC performed the majority of the experiments. AF, MR and JMC have made substantial contributions to conception and design. AF, FRP and MR analysed and interpreted the data. All authors have been involved in drafting the manuscript and revising it critically for important intellectual content. All authors read and approved the final manuscript.
Supplementary Material
SAg genes profiles identified in GAS isolates in Portugal.
Contributor Information
Ana Friães, Email: afriaes@fm.ul.pt.
Francisco R Pinto, Email: frpinto@fc.ul.pt.
Catarina Silva-Costa, Email: anacosta@fm.ul.pt.
Mario Ramirez, Email: ramirez@fm.ul.pt.
José Melo-Cristino, Email: melo_cristino@fm.ul.pt.
Acknowledgements
Members of the Portuguese Group for the Study of Streptococcal Infections are: Teresa Vaz, Marília Gião, Rui Ferreira, Iryna Klyeshtorna (Centro Hospitalar do Barlavento Algarvio), Ana Buschy Fonseca (Hospital de Cascais), Henrique Oliveira (Centro Hospitalar de Coimbra), Ana Cristina Silva, Hermínia Costa, Maria Fátima Silva, Maria Amélia Afonso (Centro Hospitalar de Entre Douro e Vouga), Margarida Pinto, Odete Chantre, João Marques, Isabel Peres, Isabel Daniel, Cristina Marcelo (Centro Hospitalar de Lisboa Central), Lurdes Monteiro, Luís Marques Lito (Centro Hospitalar Lisboa Norte), Teresa Marques, Maria Ana Pessanha, Elsa Gonçalves (Centro Hospitalar Lisboa Ocidental), Paulo Lopes, Luísa Felício, Angelina Lameirão (Centro Hospitalar de Vila Nova de Gaia / Espinho), Ana Paula Mota Vieira, Margarida Tomaz (Centro Hospitalar do Alto Ave), Rosa Bento (Centro Hospitalar do Baixo Alentejo), Maria Helena Ramos, Ana Apula Castro (Centro Hospitalar do Porto), Fernando Fonseca (Centro Hospitalar da Póvoa do Varzim / Vila do Conde), Ana Paula Castro (Centro Hospitalar Trás-os-Montes e Alto Douro), Graça Ribeiro, Luísa Boaventura, Catarina Chaves, Teresa Reis (Hospitais da Universidade de Coimbra), Nuno Canhoto, Teresa Afonso (Hospital Central do Funchal), Teresa Pina, Helena Peres (Hospital Curry Cabral, Lisboa), Ilse Fontes, Paulo Martinho (Hospital de Santa Luzia, Elvas), Ana Domingos, Gina Marrão (Hospital de Santo André, Leiria), Manuela Ribeiro, Helena Gonçalves (Hospital de São João, Porto), Maria Alberta Faustino , Maria Carmen Iglesias, Adelaide Alves (Hospital de Braga), Maria Paula Pinheiro, R. Semedo (Hospital Dr. José Maria Grande, Portalegre), Adriana Coutinho (Hospital do Espírito Santo, Évora), Luísa Cabral, Olga Neto (Hospital dos SAMS, Lisboa), Luísa Sancho (Hospital Dr. Fernando da Fonseca, Amadora / Sintra), José Diogo, Ana Rodrigues, Isabel Nascimento (Hospital Garcia de Orta, Almada), Elmano Ramalheira, Raquel Diaz (Hospital Infante D. Pedro, Aveiro), José Miguel Ribeiro, Isabel Vale, Ana Carvalho (Hospital de São Teotónio,Viseu) , Maria Antónia Read, Margarida Monteiro, Valquíria Alves (Hospital Pedro Hispano, Matosinhos), Engrácia Raposo, Maria Lurdes Magalhães, Helena Rochas, Anabela Silva (Instituto Nacional de Saúde Ricardo Jorge, Porto), Margarida Rodrigues (Hospital Reynaldo dos Santos, Vila Franca de Xira), Eulália Carvalho, Karine Hyde (Hospital do Divino Espírito Santo, Ponta Delgada), Clotilde Roldão (Hospital Distrital de Abrantes).
Filipa Vaz and Paulo Lopes are gratefully thanked for technical support.
References
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/CMR.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiou A. Group A streptococci in the 1990s. J Antimicrob Chemother. 2000;45:3–12. doi: 10.1093/jac/45.suppl_1.3. [DOI] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Luca-Harari B, Darenberg J, Neal S, Siljander T, Strakova L, Tanna A, Creti R, Ekelund K, Koliou M, Tassios PT, van der Linden M, Straut M, Vuopio-Varkila J, Bouvet A, Efstratiou A, Schalén C, Henriques-Normark B, Jasir A. Clinical and microbiological characteristics of severe Streptococcus pyogenes disease in Europe. J Clin Microbiol. 2009;47:1155–1165. doi: 10.1128/JCM.02155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, Albanese BA, Farley MM, Barrett NL, Spina NL, Beall B, Harrison LH, Reingold A, Van Beneden C. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin Infect Dis. 2007;45:853–862. doi: 10.1086/521264. [DOI] [PubMed] [Google Scholar]
- Creti R, Gherardi G, Imperi M, von Hunolstein C, Baldassarri L, Pataracchia M, Alfarone G, Cardona F, Dicuonzo G, Orefici G. Association of group A streptococcal emm types with virulence traits and macrolide-resistance genes is independent of the source of isolation. J Med Microbiol. 2005;54:913–917. doi: 10.1099/jmm.0.46035-0. [DOI] [PubMed] [Google Scholar]
- Ekelund K, Darenberg J, Norrby-Teglund A, Hoffmann S, Bang D, Skinhøj P, Konradsen HB. Variations in emm type among group A streptococcal isolates causing invasive or noninvasive infections in a nationwide study. J Clin Microbiol. 2005;43:3101–3109. doi: 10.1128/JCM.43.7.3101-3109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes M, Ardanuy C, Tamayo E, Domènech A, Liñares J, Pérez-Trallero E. Epidemiological and molecular analysis of Streptococcus pyogenes isolates causing invasive disease in Spain (1998–2009): comparison with non-invasive isolates. Eur J Clin Microbiol Infect Dis. 2011;30:1295–1302. doi: 10.1007/s10096-011-1226-x. [DOI] [PubMed] [Google Scholar]
- Wajima T, Murayama SY, Sunaoshi K, Nakayama E, Sunakawa K, Ubukata K. Distribution of emm type and antibiotic susceptibility of group A streptococci causing invasive and noninvasive disease. J Med Microbiol. 2008;57:1383–1388. doi: 10.1099/jmm.0.2008/002642-0. [DOI] [PubMed] [Google Scholar]
- Descheemaeker P, Van Loock F, Hauchecorne M, Vandamme P, Goossens H. Molecular characterisation of group A streptococci from invasive and non-invasive disease episodes in Belgium during 1993–1994. J Med Microbiol. 2000;49:467–471. doi: 10.1099/0022-1317-49-5-467. [DOI] [PubMed] [Google Scholar]
- Rivera A, Rebollo M, Miró E, Mateo M, Navarro F, Gurguí M, Mirelis B, Coll P. Superantigen gene profile, emm type and antibiotic resistance genes among group A streptococcal isolates from Barcelona, Spain. J Med Microbiol. 2006;55:1115–1123. doi: 10.1099/jmm.0.46481-0. [DOI] [PubMed] [Google Scholar]
- Rogers S, Commons R, Danchin MH, Selvaraj G, Kelpie L, Curtis N, Robins-Browne R, Carapetis JR. Strain prevalence, rather than innate virulence potential, is the major factor responsible for an increase in serious group A streptococcus infections. J Infect Dis. 2007;195:1625–1633. doi: 10.1086/513875. [DOI] [PubMed] [Google Scholar]
- Carriço JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol. 2006;44:2524–2532. doi: 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriskandan S, Faulkner L, Hopkins P. Streptococcus pyogenes: Insight into the function of the streptococcal superantigens. Int J Biochem Cell Biol. 2007;39:12–19. doi: 10.1016/j.biocel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Schmitz F-J, Beyer A, Charpentier E, Normark BH, Schade M, Fluit AC, Hafner D, Novak R. Toxin-gene profile heterogeneity among endemic invasive European group A streptococcal isolates. J Infect Dis. 2003;188:1578–1586. doi: 10.1086/379230. [DOI] [PubMed] [Google Scholar]
- Lintges M, van der Linden M, Hilgers R-D, Arlt S, Al-Lahham A, Reinert RR, Plücken S, Rink L. Superantigen genes are more important than the emm type for the invasiveness of group A Streptococcus infection. J Infect Dis. 2010;202:20–28. doi: 10.1086/653082. [DOI] [PubMed] [Google Scholar]
- Friães A, Ramirez M, Melo-Cristino J. the Portuguese Group for the Study of Streptococcal Infections. Nonoutbreak surveillance of group A streptococci causing invasive disease in Portugal identified internationally disseminated clones among members of a genetically heterogeneous population. J Clin Microbiol. 2007;45:2044–2047. doi: 10.1128/JCM.00496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friães A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J. Superantigen gene complement of Streptococcus pyogenes-relationship with other typing methods and short-term stability. Eur J Clin Microbiol Infect Dis. 2012. In press. ( http://dx.doi.org/10.1007/s10096-012-1726-3) [DOI] [PubMed]
- Cockerill FR, MacDonald KL, Thompson RL, Roberson F, Kohner PC, Besser-Wiek J, Manahan JM, Musser JM, Schlievert PM, Talbot J, Frankfort B, Steckelberg JM, Wilson WR, Osterholm MT. An outbreak of invasive group A streptococcal disease associated with high carriage rates of the invasive clone among school-aged children. JAMA. 1997;277:38–43. doi: 10.1001/jama.1997.03540250046030. [DOI] [PubMed] [Google Scholar]
- Fiorentino TR, Beall B, Mshar P, Bessen DE. A genetic-based evaluation of the principal tissue reservoir for group A streptococci isolated from normally sterile sites. J Infect Dis. 1997;176:177–182. doi: 10.1086/514020. [DOI] [PubMed] [Google Scholar]
- Ayer V, Tewodros W, Manoharan A, Skariah S, Luo F, Bessen DE. Tetracycline resistance in group A streptococci: emergence on a global scale and influence on multiple-drug resistance. Antimicrob Agents Chemother. 2007;51:1865–1868. doi: 10.1128/AAC.01341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HUK, Hammerum AM, Ekelund K, Bang D, Pallesen LV, Frimodt-Møller N. Tetracycline and macrolide co-resistance in Streptococcus pyogenes: co-selection as a reason for increase in macrolide-resistant S. pyogenes? Microb Drug Resist. 2004;10:231–238. doi: 10.1089/mdr.2004.10.231. [DOI] [PubMed] [Google Scholar]
- Malhotra-Kumar S, Wang S, Lammens C, Chapelle S, Goossens H. Bacitracin-resistant clone of Streptococcus pyogenes isolated from pharyngitis patients in Belgium. J Clin Microbiol. 2003;41:5282–5284. doi: 10.1128/JCM.41.11.5282-5284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaila-Amrouche L, Bouvet A, Loubinoux J. Clonal spread of emm type 28 isolates of Streptococcus pyogenes that are multiresistant to antibiotics. J Clin Microbiol. 2004;42:3844–3846. doi: 10.1128/JCM.42.8.3844-3846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York MK, Gibbs L, Perdreau-Remington F, Brooks GF. Characterization of antimicrobial resistance in Streptococcus pyogenes isolates from the San Francisco Bay area of northern California. J Clin Microbiol. 1999;37:1727–1731. doi: 10.1128/jcm.37.6.1727-1731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires R, Rolo D, Mato R, Feio de Almeida J, Johansson C, Henriques-Normark B, Morais A, Brito-Avô A, Gonçalo-Marques J, Santos-Sanches I. Resistance to bacitracin in Streptococcus pyogenes from oropharyngeal colonization and noninvasive infections in Portugal was caused by two clones of distinct virulence genotypes. FEMS Microbiol Lett. 2009;296:235–240. doi: 10.1111/j.1574-6968.2009.01642.x. [DOI] [PubMed] [Google Scholar]
- Silva-Costa C, Ramirez M, Melo-Cristino J. Identification of macrolide-resistant clones of Streptococcus pyogenes in Portugal. Clin Microbiol Infect. 2006;12:513–518. doi: 10.1111/j.1469-0691.2006.01408.x. [DOI] [PubMed] [Google Scholar]
- Darenberg J, Luca-Harari B, Jasir A, Sandgren A, Pettersson H, Schalén C, Norgren M, Romanus V, Norrby-Teglund A, Normark BH. Molecular and clinical characteristics of invasive group A streptococcal infection in Sweden. Clin Infect Dis. 2007;45:450–458. doi: 10.1086/519936. [DOI] [PubMed] [Google Scholar]
- Proft T, Sriskandan S, Yang L, Fraser JD. Superantigens and streptococcal toxic shock syndrome. Emerging Infect Dis. 2003;9:1211–1218. doi: 10.3201/eid0910.030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haukness HA, Tanz RR, Thomson RB, Pierry DK, Kaplan EL, Beall B, Johnson D, Hoe NP, Musser JM, Shulman ST. The heterogeneity of endemic community pediatric group A streptococcal pharyngeal isolates and their relationship to invasive isolates. J Infect Dis. 2002;185:915–920. doi: 10.1086/339407. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Edwards RA, Taylor WW, Low DE, McGeer A, Kotb M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J Bacteriol. 2005;187:3311–3318. doi: 10.1128/JB.187.10.3311-3318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- Nir-Paz R, Korenman Z, Ron M, Michael-Gayego A, Cohen-Poradosu R, Valinsky L, Beall B, Moses AE. Streptococcus pyogenes emm and T types within a decade, 1996–2005: implications for epidemiology and future vaccines. Epidemiol Infect. 2010;138:53–60. doi: 10.1017/S0950268809002805. [DOI] [PubMed] [Google Scholar]
- Szczypa K, Sadowy E, Izdebski R, Strakova L, Hryniewicz W. Group A streptococci from invasive-disease episodes in Poland are remarkably divergent at the molecular level. J Clin Microbiol. 2006;44:3975–3979. doi: 10.1128/JCM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 2010;6:e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotb M, Norrby-Teglund A, McGeer A, El-Sherbini H, Dorak MT, Khurshid A, Green K, Peeples J, Wade J, Thomson G, Schwartz B, Low DE. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–1404. doi: 10.1038/nm1202-800. [DOI] [PubMed] [Google Scholar]
- Silva-Costa C, Pinto FR, Ramirez M, Melo-Cristino J. Portuguese Suveillance Group for the Study of Respiratory Pathogens. Decrease in macrolide resistance and clonal instability among Streptococcus pyogenes in Portugal. Clin Microbiol Infect. 2008;14:1152–1159. doi: 10.1111/j.1469-0691.2008.02104.x. [DOI] [PubMed] [Google Scholar]
- Melo-Cristino J, Fernandes ML. Portuguese Surveillance Group for the Study of Respiratory Pathogens. Streptococcus pyogenes isolated in Portugal: macrolide resistance phenotypes and correlation with T types. Microb Drug Resist. 1999;5:219–225. doi: 10.1089/mdr.1999.5.219. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention - CDC Streptococcus Laboratory. [ http://www.cdc.gov/ncidod/biotech/strep/strepindex.htm]
- Figueira-Coelho J, Ramirez M, Salgado MJ, Melo-Cristino J. Streptococcus agalactiae in a large Portuguese teaching hospital: antimicrobial susceptibility, serotype distribution, and clonal analysis of macrolide-resistant isolates. Microb Drug Resist. 2004;10:31–36. doi: 10.1089/107662904323047772. [DOI] [PubMed] [Google Scholar]
- Trzcinski K, Cooper BS, Hryniewicz W, Dowson CG. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2000;45:763–770. doi: 10.1093/jac/45.6.763. [DOI] [PubMed] [Google Scholar]
- Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun. 2001;69:2416–2427. doi: 10.1128/IAI.69.4.2416-2427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MLST - Multilocus Sequence Typing - Streptococcus pyogenes. [ http://spyogenes.mlst.net/]
- Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carriço JA. PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinforma. 2012;13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approch to multiple testing. J R Stat Soc Ser B Statistical Methodology. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SAg genes profiles identified in GAS isolates in Portugal.


