Abstract
Chemically defined substrates, which rigorously control protein-surface and cell-surface interactions, can be used to probe the effects of specific biomolecules on cell behavior. Here we combined a chemically-defined, array-based format with automated, time-lapse microscopy to efficiently screen cell-substrate interactions. Self-assembled monolayers (SAMs) of alkanethiolates bearing oligo(ethylene glycol) units and reactive terminal groups were used to present cell adhesion peptides while minimizing non-specific protein interactions. Specifically, we describe rapid fabrication of arrays of 1 mm spots, which present varied densities of the integrin-binding ligand Gly-Arg-Gly-Asp-Ser-Pro (GRGDSP). Results indicate that cell attachment, cell spreading, and proliferation exhibit strong dependencies on GRGDSP density for both human mesenchymal stem cells (hMSCs) and human umbilical vein endothelial cells (HUVECs). Furthermore, relative spreading and proliferation over a broad range of GRGDSP densities are similar for both primary cell types, and detailed comparison between cell behaviors identified a 1:1 correlation between spreading and proliferation for both HUVECs and hMSCs. Finally, time-lapse microscopy of SAM arrays revealed distinct adhesion-dependent migratory behaviors for HUVECs and hMSCs. These results demonstrate the benefits of using an array-based screening platform for investigating cell function. While the proof-of-concept focuses on simple cellular properties, the quantitative similarities observed for hMSCs and HUVECs provides a direct example of how phenomena that would not easily be predicted can be shown to correlate between different cell types.
Keywords: self-assembled monolayers, RGD, mesenchymal stem cells, umbilical vein endothelial cells, array
Introduction
Understanding how cells respond to specific biomolecules present on a cell culture substrate or a surgical implant surface is critical in scenarios ranging from fundamental cell science to biomaterials design. However, mechanistic details associated with how substrate-bound biomolecules influence even basic cell functions, such as adhesion, remain limited by a lack of systematic approaches aimed at probing cell-surface interactions.[1] On a surface, the challenge of deconstructing specific biomolecule influences on cell function is primarily due to rapid and non-specific protein adsorption to most biomaterials surfaces, which occurs within seconds of contact with biological fluids.[2–4] Thus, biological studies conducted on typical cell culture surfaces in which cells interact with adsorbed protein layers that are difficult to characterize and can denature during the adsorption process, do not allow one to draw clear conclusions about cell-material interactions. To address this challenge, substrates designed to limit non-specific protein adsorption and simultaneously present specific biomolecules can be used to chemically define protein-surface and cell-surface interactions.[1–4] Of these types of defined substrates, self-assembled monolayers (SAMs) on gold formed using alkanethiolates bearing oligo(ethylene glycol) units are particularly well-suited to efficiently probe cell-biomolecule interactions, as they allow for strict control over biomolecule presentation and simple functionalization chemistries.[2–9] To date, SAMs have been used to identify unique insights into the effects of specific cell-extracellular matrix interactions[2–9] as well as the effects of surfaces designed to sequester specific soluble molecules such as growth factors and proteoglycans[8, 9]. These previous studies highlight SAMs as unique biological tools.
Here we present a well-defined screening array platform that provides controlled cell-substrate interactions and enables systematic studies of cell behavior. Our approach uses simple elastomeric stencils to form arrays of biomolecules on SAMs formed on gold surfaces (Figure 1), and uses automated, time-lapse microscopy to track cell behavior within array spots. We created arrays containing varying densities of the fibronectin-derived cell adhesion peptide Gly-Arg-Gly-Asp-Ser-Pro (GRGDSP)[10–12] and used them to screen for adhesion-dependent changes in cell behavior. Here, we chose to investigate human umbilical vein endothelial cells (HUVECs) and human mesenchymal stem cells (hMSCs) as they represent two commonly used, but distinct, cell types in regenerative medicine. In particular, HUVECs represent a model cell type that have been used to study endothelial cell roles in blood vessel formation, barrier function, hemostasis, vascular tone, inflammation, leukocyte interactions, and neoangiogenesis.[13–17] Conversely, hMSCs are a multipotent stem cell type present in a variety of tissues that contribute to tissue and immune system homeostasis via mechanisms including differentiation into a range of mature cell types[18–22] as well as secretion of a range of soluble factors[23, 24]. Using SAM arrays to investigate these two cell types, our results provide unique insights into the effects of cell adhesion ligand density on HUVEC and hMSC behaviors, including cell attachment, spreading, proliferation, and migration. Furthermore, direct comparisons between these two cell types identify correlations that could not be obtained without an efficient, array-based approach.
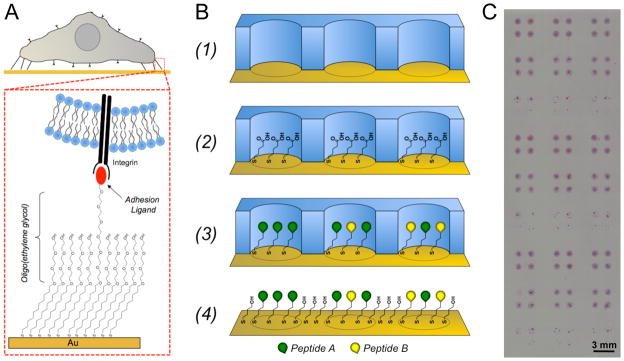
Figure 1.
Generating defined culture substrates using alkanethiolate self-assembled monolayer arrays. (A) SAMs can be designed to present covalently immobilized biomolecules, such the integrin binding ligand Gly-Arg-Gly-Asp-Ser-Pro (GRGDSP), to cells while minimizing the effects of non-specific protein adsorption via oligo(ethylene glycol) moieties that limit protein-surface interaction. (B) Schematic representation of SAM array fabrication: (1) Adhere elastomeric stencil to gold substrate to generate a microwell array superstructure, (2) locally form a SAM in each well with alkanethiolate mixtures containing carboxylic acid-terminated and hydroxyl-terminated oligo(ethylene-glycol) alkanethiolates, (3) covalently conjugate peptides to array spots via carbodiimide condensation of peptide n-terminal primary amine and SAM carboxylic acid terminal moities, and (4) remove mask and backfill with inert SAM. (C) Example of hMSCs on a SAM array containing 1 mm spots (hMSCs were stained using hematoxylin and eosin to visual cells).
Experimental methods
Materials
Carboxylic acid-capped hexa(ethylene glycol) undecanethiol (HS-C11-(O-CH2-CH2)6-O-CH2-COOH) (referred to herein as “HS-C11-EG6-COOH”), was purchased from Prochimia (Sopot, Poland). 11-tri(ethylene glycol)-undecane-1-thiol (HS-C11-(O-CH2-CH2)3-OH) (referred to herein as “HS-C11-EG3-OH”) was synthesized as described elsewhere.[25] Fmoc-protected amino acids and Rink amide MBHA peptide synthesis resin were purchased from NovaBiochem (San Diego, CA). Hydroxybenzotriazol (HOBt) was purchased from Advanced Chemtech (Louisville, KY). Diisopropylcarbodiimide (DIC) was purchased from Anaspec (San Jose, CA). N-hydroxysuccinimide (NHS), n-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), sodium dodecyl sulfate (SDS), trifluoroacetic acid (TFA), diethyl ether, and deionized ultrafiltered water (DIUF H2O) were purchased from Fisher Scientific (Fairlawn, NJ). Triisopropylsilane (TIPS), piperidine, dimethylformamide (DMF), acetone, hexanes, and acetonitrile were purchased from Sigma-Aldrich (St. Louis, MO). Absolute ethanol (EtOH) was purchased from AAPER Alcohol and Chemical Co. (Shelbyville, KY). All purchased items were of analytical grade and used as received. Thin films of 100 Å Au <111>, 20 Å Ti on 1″ × 3″ × 0.040″ glass were purchased from Platypus Technologies, LLC (Madison, WI. Cat. No. AU.0100.ALSI).
Peptide synthesis
Standard solid phase Fmoc-peptide synthesis (Fmoc SPPS) was performed using a 316c automated peptide synthesizer (C S Bio, Menlo Park, CA). Rink amide MBHA resin was used as the solid phase, and HOBt and DIC were used for amino acid activation and coupling. After coupling the final amino acid, a 4-hr incubation in TFA, TIPS, and DIUF H2O (95:2.5:2.5) released the peptide from resin and removed protecting groups. Released peptide was extracted from the TFA/TIPS/DIUF H2O cocktail via precipitation in cold diethyl ether. Lyophilized peptides were analyzed using matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry with a Bruker Reflex II (Billerica, MA). The purity of synthesized peptides was evaluated via HPLC using a C18 analytical column (Shimadzu, Kyoto, Japan) with a gradient of 0–70% H2O + 0.1% TFA/acetonitrile and a flow rate of 0.9 mL/minute. GWGGRGDSP, GWGGRGESP, GWGGIKVAV, and GWGGVKAIV adhesion and mutant peptides were synthesized with tryptophan-bearing spacers to aid in determination of peptide concentration via UV/Vis. Peptide stocks were prepared at 300 μM in PBS at pH 7.4 as determined by absorbance at 280 nm using extinction coefficients outlined by Gill and von Hippel.[26] Fluorescently-labeled GGRGDSPK was synthesized as previously described[7] and peptide concentration was determined by absorbance of the 5(6)-carboxyfluorescein group at 492 nm using an extinction coefficient of 81,000 cm−1M−1
Fabrication of elastomer wells
Elastomeric stencils containing arrays of wells were created using soft lithography.[27, 28] Briefly, master molds containing arrays of 1100 μm diameter posts were fabricated from SU-8 (Microchem, Newton, MA) spin-coated silicon wafers using conventional photolithography techniques. Polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning, Midland, MI) was prepared by mixing a 10:1 ratio of base/curing agent (w/w) followed by degassing for ~30 mins. The degassed mixture was cast over the mold and cured for 4 hrs at 85 °C. Following curing, PDMS stencils were removed from molds and cleaned in hexanes using an overnight Soxhlet extraction.[29] After cleaning, stencils were placed in vacuo to remove residual solvent from the Soxhlet extraction process.
Surface preparation and array fabrication
Gold slides were placed into a 150 mm glass Petri dish, covered with EtOH, and sonicated for ~1 min using an ultrasonic bath (Bransonic 1510, Branson, Danbury, CT). Sonicated gold chips were then rinsed with EtOH and blown dry with N2. SAM arrays were fabricated as follows: An elastomeric stencil containing arrays of 1.1 mm holes was placed on a bare gold surface to form an array of wells on the gold substrate 0Figure 1B, Step 1). Wells were then filled with 1 mM ethanolic alkanethiolate solution and incubated for 10 minutes in a chamber containing a laboratory wipe soaked with ethanol to prevent evaporation during local SAM formation (Figure 1B, Step 2). Alkanethiolate solutions were then aspirated and wells were rinsed with DIUF H2O. Carboxylate groups were then converted to active ester groups by adding a solution of 100 mM NHS and 250 mM EDC in DIUF H2O pH 5.5 to wells and incubated for 10 minutes. After an additional rinse with DIUF H2O, 300 μM solutions of peptide in PBS at pH 7.4 were added to each well and incubated for 1 hr in a humidity controlled chamber to covalently couple peptides to each array spot (Figure 1B, Step 3). After a final rinse with DIUF H2O, regions surrounding array spots were backfilled with HS-C11-EG3-OH. This was achieved by submerging the gold substrate and attached elastomeric stencil in an aqueous 0.1 mM HS-C11-EG3-OH solution (pH 2.0), removing the stencil, and incubating for 10 minutes (Figure 1B, Step 4). Following backfilling, the array was rinsed with 0.1 wt% SDS in DIUF H2O, DIUF H2O, and EtOH and then dried under a stream of N2. Arrays were stored in sterile DIUF H2O at 4 °C and used within 24 hrs.
In this SAM array approach, each spot was designed to contain the same total molar density (mol/cm2) of peptide from spot to spot. Therefore, control over GRGDSP density was achieved by mixing GRGDSP with the mutant GRGESP peptide. In a typical SAM array, SAMs were locally formed within spots using an 1 mM alkanethiolate mixture of 95% HS-C11-EG3-OH and 5% HS-C11-EG6-COOH to create surfaces with a total of 5% carboxylate groups for peptide conjugation. Here, “X%” refers to the mole percent of alkanethiolate present during SAM formation and subsequently the approximate amount of an alkanethiolate present on the surface after SAM formation. Next, to create a spot presenting 5% GRGDSP, a 300 μM peptide solution was used during peptide conjugation. Likewise, to create a spot with 1.6% GRGDSP, a 300 μM peptide solution with 100 μM GRGDSP and 200 μM GRGESP was used during peptide conjugation (Figure 1B, Step 3). In this manner, the amount of GRGDSP peptide could be varied between spots while holding total peptide content constant and a three-fold dilution series of GRGDSP into GRGESP was used to generate arrays with a range of GRGDSP densities such as 5.0, 1.6, 0.6, 0.2, 0.06, 0.02, 0.01, and 0% peptide. Likewise, mixtures of IKVAV and the mutant peptide VKAIV were mixed together to generate arrays with a similar set of IKVAV peptide densities.
Cell Culture
Passage 2 human umbilical vein endothelial cells were expanded at low density (less than 70% confluence) on tissue culture polystyrene to no more than 14 population doublings. During HUVEC expansion and experiments on SAM arrays, HUVECs were cultured in medium 199 (m199, Mediatech, Manassas, VA) containing 1% penicillin/streptomycin (Hyclone, Logan, UT) and supplemented with Clonetics EGM-2 BullitKit (Lonza Walkersville, Inc., Walkersville, MD) containing hydrocortisone, hFGF-B, VEGF, R3-IGF-1, ascorbic acid, heparin, fetal bovine serum, hEGF, GA-1000 growth supplements. Bone marrow-derived human mesenchymal stem cells (Lonza, Walkersville, MD) were expanded at low density on tissue culture polystyrene plates to maintain multipotency as described by Sotiropoulou et al.[30] and used by passage 7. During hMSC expansion and experiments on SAM arrays, hMSCs were cultured in minimum essential medium, alpha (Mediatech, Manassas, VA) containing 10% MSC qualified fetal bovine serum (Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin. For simplicity, throughout the methods section “media” will refer to the respective media used for culture of HUVECs and hMSCs.
Cell Assays on SAM Array
HUVECs or hMSCs were removed from plastic culture plates using a 0.05% trypsin solution, resuspended in media, and seeded onto SAM arrays in sterile polystyrene Petri dishes at 2,500 and 1,500 cells/cm2, respectively, unless specified otherwise. After allowing cells to attach for ~ 1 hr in a humidified incubator at 37 °C and 5% CO2, arrays were dipped in warm media to remove loosely attached cells and then transferred to a rectangular multidish (Thermo Scientific/Nunc, Rochester, NY) with warm media and imaged ~4 hrs later serving as “0 hr.” For the initial time point and subsequent time-lapse imaging, arrays were placed on an incubated stage and each array spot was imaged every 15 mins for 72 hrs. Furthermore, all cell experiments for comparison between HUVECs and hMSCs were run simultaneously, and thus both cell types were exposed to identical environmental conditions and monitored at the same experimental time points.
Immunocytochemistry
At 24 and 72 hr time points during the time-lapse experiment, staining for the actin cytoskeleton and focal adhesion markers was performed as directed by the manufacturer (Catalog No. FAK100, Millipore, Billerica, MA). Briefly, cells on arrays were fixed using 4% formaldehyde in PBS for 15 minutes. Following fixing, arrays were washed and then permeabilized using 0.1 % Triton X-100 (MP Biomedicals, Aurora, OH) in PBS for 5 minutes. After an additional wash and a blocking step using 1% (w/v) bovine serum albumin (Fisher Scientific, Fairlawn, NJ) arrays were exposed to a PBS solution containing an anti-vinculin primary antibody for 1 hr at room temperature. Arrays were then exposed to a PBS solution containing a FITC-conjugated secondary antibody and TRITC-conjugated phalloidin for 60 minutes at room temperature. After a final rinse, arrays were mounted with a cover slip using Prolong Gold Antifade Reagent with DAPI (Invitrogen, Eugene, OR) as indicated by the manufacturer. Integrin staining was performed using a similar protocol except primary antibody staining for β1 (Abcam, Cambridge MA) was performed overnight at 4°C and secondary antibody staining was performed for 2 hrs at room temperature.
Array Imaging
A GE Healthcare Typhoon Trio Variable Mode Imager was used to scan SAM arrays containing fluorescently-labeled peptide and fluorescent intensity was quantified using Image J (ImageJ, Freeware, NIH, Bethesda, MD) imaging software. Cells in culture on SAM arrays were imaged using a Nikon Eclipse Ti inverted microscope equipped with the Perfect Focus System; filters for FITC, TexasRed, and DAPI; and a TIZ Tokai Hit incubated stage that was humidified and maintained at 37 °C and 5% CO2. For phase contrast imaging, a 10X PhL objective was used to capture 4 images of each spot, which were automatically stitched together using the Nikon NIS Elements software. Immunofluorescence images of each array spot were acquired at 30X and stitched together from 20 images. For immunofluorescence imaging, exposure times for each channel were kept constant from array to array.
Analysis, Quantification, and Statistics
Relative cell attachment at 0 hrs was quantified by counting the number of attached cells per spot and normalizing cell numbers to maximal attachment conditions. Normalized cell number was calculated by dividing the number of cells per spot at 72 hrs (C72) to the cell number observed on the same spot at 0 hrs (C0). Relative proliferation was determined by dividing normalized cell numbers at 72 hrs by the maximum average normalized cell number observed across GRGDSP density conditions. Similarly, relative spreading was determined by dividing projected cell areas to the maximum average projected cell area observed across GRGDSP density conditions. Analysis of projected cell area was achieved using Nikon NIS Elements Software (Melville, NY). Briefly, stacked images of array spots were thresholded and then automated measurements of area and counts were tabulated. For each spot, average cell projected area was calculated by dividing the thresholded actin staining (red channel) by the total number of nuclei in the same spot. For cell tracking, the cell tracking module in NIS Elements was used to monitor single cell migration in time lapse images over 6 hrs starting at ~12 hrs into cell culture experiments. During quantification, migration was classified as a single cell that migrated more than an average distance of 2 nuclei and did not divide or interact with other cells. Additionally, migration speed was only reported for conditions in which more than 15% of single cells were migrating. To clearly display phase contrast and fluorescence images in the manuscript, adjustments to brightness and contrast were performed in accordance to guidelines outlined by the Journal of Cell Biology.[31] Statistical analysis of all data sets was performed using a two-tailed Student’s t-test, where p < 0.05 is used to denote statistical significance.
Results and Discussion
Control Over Peptide Identity and Density in Cell Culture
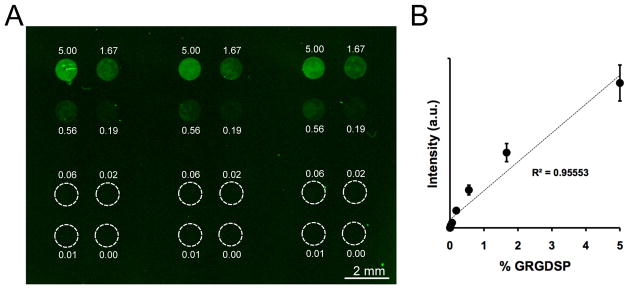
An elastomeric stencil approach was used to generate SAM arrays with precise spot-to-spot control over peptide composition (Figure 1). The local density of GRGDSP was controlled by mixing GRGDSP and GRGESP (a non-bioactive mutant peptide) together during peptide coupling, maintaining a constant total peptide density of 5%. To visualize differences in peptide density on a typical array, 5(6)-carboxyfluorescein-labeled GRGDSPK was mixed with GRGESP at varied ratios and coupled to SAM array spots presenting 5% total HS-C11-EG6-COOH (Figure 2A). 5% HS-C11-EG6-COOH density was chosen for this work as previous results have demonstrated good control over peptide density while limiting non-specific interactions with background functional groups.[6, 7] On SAM arrays, fluorescence was confined to the array spots and the fluorescence intensity in each spot correlated with the amount of labeled peptide included in the coupling reaction (Figure 2A). Furthermore, this trend exhibited a linear relationship between fluorescent intensity and peptide density (Figure 2B), indicating control over peptide density that is similar to what we have reported in a previous SAM array approach.[7] Taken together, fluorescent visualization of peptide incorporation demonstrated control over peptide density between spots and the ability to generate a range of peptide conditions on a single SAM array.
Figure 2.
SAM arrays presenting varied GRGDSP density. (A) Fluorescent surface scan of a SAM array presenting varied densities of fluorescently-labeled peptide. (B) Quantification of fluorescence intensity in SAM array spots (Error bars represent standard error of the mean).
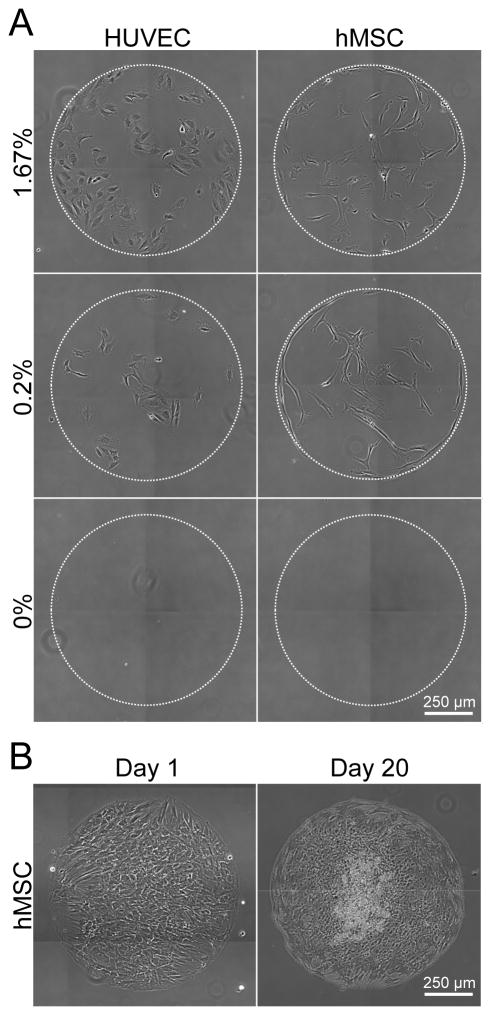
SAM arrays provide a robust and controllable platform for primary, human cell culture (Figure 3). HUVECs and hMSCs were seeded onto identical SAM arrays presenting varied densities of GRGDSP, and imaged using time-lapse microscopy over 72 hrs (See Supplement Movies 1A–D). Over the course of an experiment, HUVEC and hMSC attachment and growth was limited to SAM regions presenting GRGDSP and there was no cell attachment observed on background oligo (ethylene glycol) regions or on spots containing only GRGESP and no GRGDSP (Figure 3A). In long term studies in which hMSCs were grown for 20 days with repeated media exchange, cells remained confined within GRGDSP-presenting array spots (Figure 3B). Thus, these results demonstrate that SAM arrays can be used to probe for ligand specific effects on cell behavior and suggest that arrays can be used for long-term studies such as those aimed at understanding stem cell differentiation.
Figure 3.
SAM arrays for assaying cell behavior. (A) HUVECs and hMSCs were seeded on array spots presenting varied densities of GRGDSP: 1.6%, 0.2%, and 0% GRGDSP shown at t=48 hrs. (Time-lapse videos of GRGDSP spots over 48–72 hrs included in Supplement Movies 1A–D). (B) hMSCs were seeded on SAM arrays at 50,000 cells/cm2 and imaged at 1 and 20 days.
The elastomeric stencil approach used here has several advantages over previously described SAM array approaches developed by us[7] and others[32]. In a previous study[7], we varied the density of HS-C11-EG6-COOH between spots prior to peptide coupling, resulting in variation in total peptide density across the array. In our current approach we instead varied biologically active peptide density while holding the total peptide density constant by including an inactive (scrambled) peptide. These two approaches both yield SAM arrays with control over peptide density, similar to the approach described by Orner et al.[32]. However, by maintaining constant total peptide density, the current approach limits chemical variation from spot to spot and thus provides a more robust strategy for probing the effects of a specific peptide sequence. From a practical standpoint, an additional advantage of the current approach is that functionalization does not require localized SAM removal via UV irradiation[32] or borohydride chemistry[7] and complete cell-seeded arrays can be generated in less than 2 hrs in a few steps with a standard pipette and PDMS stencil (Figure 1B). Additionally, arrays generated on substrates the size of a standard microscope slide (3″ × 1″) can contain over 100 spots, each with a different cell-interactive substrate (Figure 1C). Taken together, the approach described here reduces the logistical complexity required to rapidly generate SAM arrays with a wide range of peptide densities, enabling systematic investigation of chemically-defined cell culture substrates without a need for specific expertise in organic synthesis or SAMs chemistry.
Cell Attachment
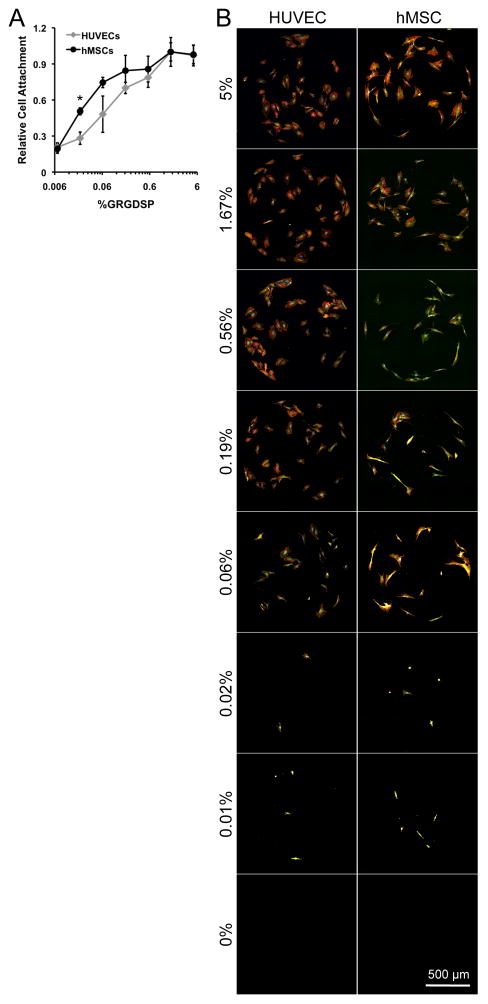
HUVEC and hMSC attachment were each GRGDSP-dependent and both cell types showed similar cell attachment trends across conditions (Figure 4), suggesting similar response to changes in adhesion ligand density. HUVEC and hMSC attachment was similar over 3 orders of magnitude of GRGDSP density, and minimal required GRGDSP densities for cell attachment were also similar (~0.006% GRGDSP) (Figure 4A). The similarities in hMSC and HUVEC attachment to GRGDSP functionalized SAMs is interesting in the context of previous studies, which have reported attachment for bovine aortic endothelial cells (BAECs) [33] and hMSCs [6, 7] on SAMs functionalized with cell adhesion ligand. BAEC attachment was insensitive to adhesion ligand density above the minimum GRGD density required for adhesion at 0.001%[33] while minimum GRGDSP density required for hMSC attachment has been reported in several studies[5–7] with values ranging from 0.1%[6] to 0.0005%[7] GRGDSP. It is unclear whether discrepancies between previous and current results for minimal cell attachment conditions are due to subtle differences in cell culture protocols, the differing cell adhesion ligand identity (RGD vs. RGDSP), or the different endothelial cell types used (bovine vs. human derived endothelial cells). However, the difficulty in directly comparing these studies highlights the utility of standardized platforms that can probe similarities and differences in cell behavior while minimizing experimental errors or differences in experimental conditions. Here, we were able to probe a wide range of GRGDSP densities using identical substrate conditions, and thus identify previously unreported similarities in the mechanisms governing the initial attachment of these primary cell types to GRGDSP-terminated surfaces.
Figure 4.
Screening for cell attachment using SAM arrays. HUVECs and hMSCs were seeded onto SAM arrays presenting varied GRGDSP densities for 1 hr and then rinsed to remove unattached cells. (A) Cell attachment per spot was scaled to maximal attachment conditions to observe relative cell attachment. (B) Focal adhesion complexes and cytoskeletal organization (vinculin:green, actin:red, and nuclei:blue) of HUVECs and hMSCs on SAM arrays at 24 hrs. (Error bars indicate standard error of the mean. Asterisks indicates significant increase in hMSC attachment compared to HUVECs at the same GRGDSP density, p < 0.05.)
HUVEC and hMSC attachment to arrays presenting varied densities of the well-known laminin-derived cell adhesion ligand Ile-Lys-Val-Ala-Val, IKVAV (Supplement Figure 1) was minimal (less than 5 cells per spot) and equivalent to non-bioactive control or PEG-only background. This result may contrast with previous studies have demonstrated that IKVAV adsorbed onto plastic cell culture plates can support HUVEC attachment. [34, 35] However, several other studies have demonstrated that HUVEC attachment to IKVAV is minimal and significantly less than RGD-functionalized materials [36] [37]. Similarly, previous studies have demonstrated that hMSC spreading on polystyrene-block-poly(ethylene oxide)-copolymer surfaces presenting IKVAV is minimal when compared to RGD.[38] Taken together, these previous results demonstrate that HUVEC and hMSC attachment to IKVAV is significantly lower that RGD and our current results suggest that when presented in a chemically well-defined context, neither cell type adheres to IKVAV. It is noteworthy that our array-based platform allowed us to efficiently explore 8 distinct IKVAV densities that span 3 orders of magnitude, which strongly indicates that these cell types do not adhere to IKVAV.
Expression of Cytoskeletal and Adhesion Components
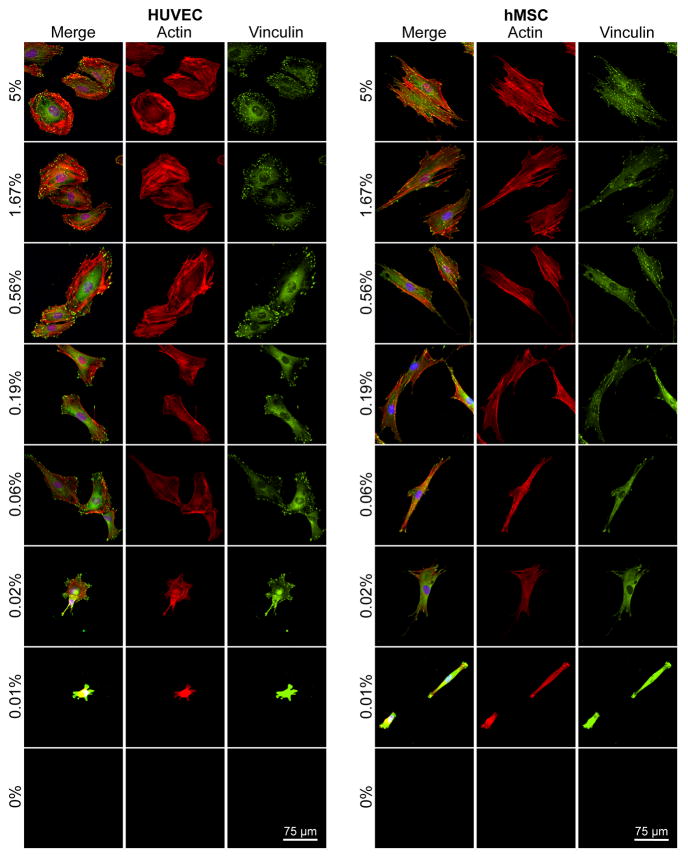
Immunofluorescence was used to visualize f-actin (red, for cytoskeleton) and vinculin (green, a component of focal adhesions) to gain insight about how adhesion ligand density induced changes in cell structure (Figure 5). Both cell types had vinculin-containing focal adhesions [39] and well-organized f-actin structure at high GRGDSP densities (5.00% and 1.67%). However, HUVECs and hMSCs had distinctly different cytoskeletal structure and focal adhesion organization at each GRGDSP density. Specifically, at high GRGDSP densities HUVECs expressed thick f-actin stress fibers terminated by focal adhesions prominently around the cell perimeter, while hMSCs were more elongated and fibroblastic, with f-actin stress fibers running longitudinally (parallel to the long axis of the cell) through the cell and terminated by focal adhesions. As GRGDSP density was decreased, HUVECs became more elongated (Figure 5, 0.19%, 0.06% GRGDSP) with stress fibers taking on a more longitudinal orientation and focal adhesions occurring on the ends of the elongated cell body. Changes for hMSCs were subtler over the intermediate GRGDSP densities, with hMSCs maintaining an elongated shape and longitudinal stress fiber orientation but adopting a more spindle-shaped morphology. At the lowest GRGDSP density (Figure 5, 0.01% GRGDSP), HUVECs and hMSCs both became morphologically compact with highly condensed f-actin and vinculin staining throughout the cell that made structure difficult to distinguish. Quantified focal adhesion density, size, and average staining intensity (Supplemental Figure 2) also exhibited GRGDSP dependent changes in focal adhesion expression for both cell types, although HUVECs exhibited significantly higher focal adhesion density compared to hMSCs for several GRGDSP densities. These changes in hMSC morphology with respect to GRGDSP density are consistent with those previously observed in our work using SAM substrates.[5–7] Additionally, while there has been little previous work done to characterize HUVEC response using RGD presenting SAM substrates, the gradual changes in cytoskeletal morphology observed in this work are similar to results observed using other systems to vary the density of RGD on HUVEC culture surfaces.[40] It is interesting to note that at intermediate GRGDSP densities we could identify similarities in morphology and structure between hMSCs and HUVECs (e.g. HUVECs at 0.19% and hMSCs at 0.56%), which is in stark contrast to observations from high adhesion ligand density conditions (5%) or standard culture of these cell types.
Figure 5.
Screening for cell morphology using SAM arrays presenting varied GRGDSP densities. HUVECs and hMSCs cultured for 24 hrs on arrays were stained for focal adhesion complexes and cytoskeletal organization (vinculin:green, actin:red, and nuclei:blue).
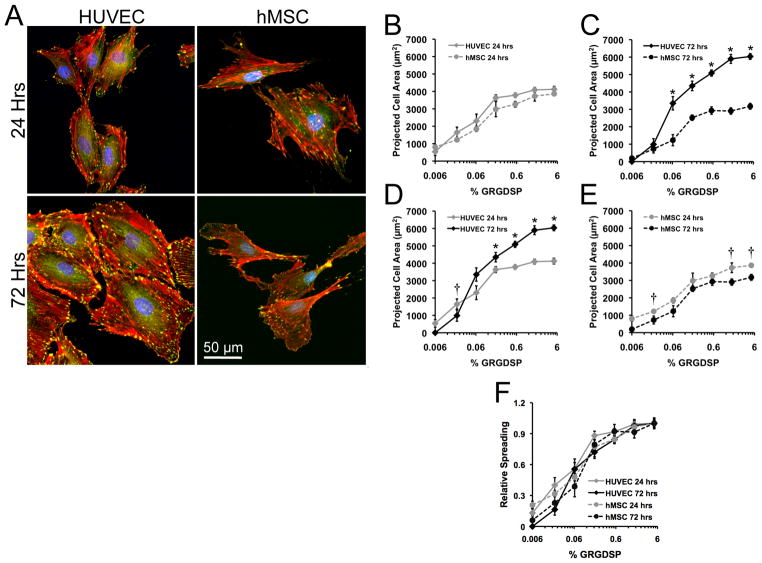
Projected cell area was not significantly different for HUVECs and hMSCs at 24 hrs for any of the conditions investigated, with maximal projected cell areas of greater than 4000 μm2 occurring at 1.6% and 5% GRGDSP (Figure 6A-B). These maximal projected cell areas at 24 hrs agree with previous reports of HUVEC spreading on glass substrates[41–43] and hMSC spreading on GRGDSP presenting SAMs.[5–7] Furthermore, both cell types exhibited GRGDSP density-dependent changes in spreading. In the case of hMSCs spreading with respect to ligand density, results agree well with results from our previously described SAM array system.[7] Taken together, these spreading results and the initial attachment results (Figure 4A) highlight that, despite differences in cytoskeletal and adhesion component structures (Figure 5 and Supplemental Figure 2), the two different cell types exhibited remarkably similar attachment and spreading.
Figure 6.
Dynamic cell spreading. (A) High magnification images of HUVECs and hMSCs on 5% RGD at 24 and 72 hrs (vinculin:green, actin:red, and nuclei:blue). Quantification of projected cell areas between cell types at (B) 24 and (C) 72 hrs as well as comparison within (D) HUVECs and (E) hMSCs at both time points. (F) Relative spreading was determined by scaling projected cell area to maximum projected areas for each cell type and time point. (Error bars indicate standard error of the mean. Asterisk indicates significant increase and cross indicates significant decrease in projected cell area at the indicated GRGDSP density, p < 0.05.)
While spreading at 24 hrs was similar for HUVECs and hMSCs, changes in spreading differed markedly for the two cell types from 24 to 72 hrs in culture (Figure 6C), with HUVECs increasing their projected cell area for all GRGDSP densities above 0.06% (Figure 6D), and hMSCs projected area either decreasing or remaining constant with increasing adhesion ligand density (Figure 6E). Previous studies of cell spreading have typically examined projected cell area at a static 24 hr time point or for changing time over the first few hrs to 24 hrs. For example, HUVECs on glass substrates have previously been documented to continually increase their projected cell area until maximal spreading is achieved between 3 and 24 hrs.[41–43] However, at 72 hours we observe maximal HUVEC spreading reaching 6000 μm2 on 1.67% and 5% GRGDSP as well as a general increase in spreading across several adhesion ligand densities. Conversely, we observed slight decreases in hMSC projected cell areas from 24 to 72 hrs across several GRGDSP densities (Figure 6E). These data suggest that it may be desirable to track cell adhesion over greater than 24 hrs to understand dynamics of cell phenotype, as cell spreading has been clearly correlated to changes in differentiated function of both HUVECs and hMSCs.[44–51] Interestingly, even with temporal changes in spreading, the relative spreading is almost identical for both cell types at both 24 and 72 hrs (Figure 6F). Therefore, while these results indicate that temporal changes extend beyond timeframes typically used to characterize cell spreading, they also suggest that cell behavior maintains a similar dependence on adhesion ligand density over time.
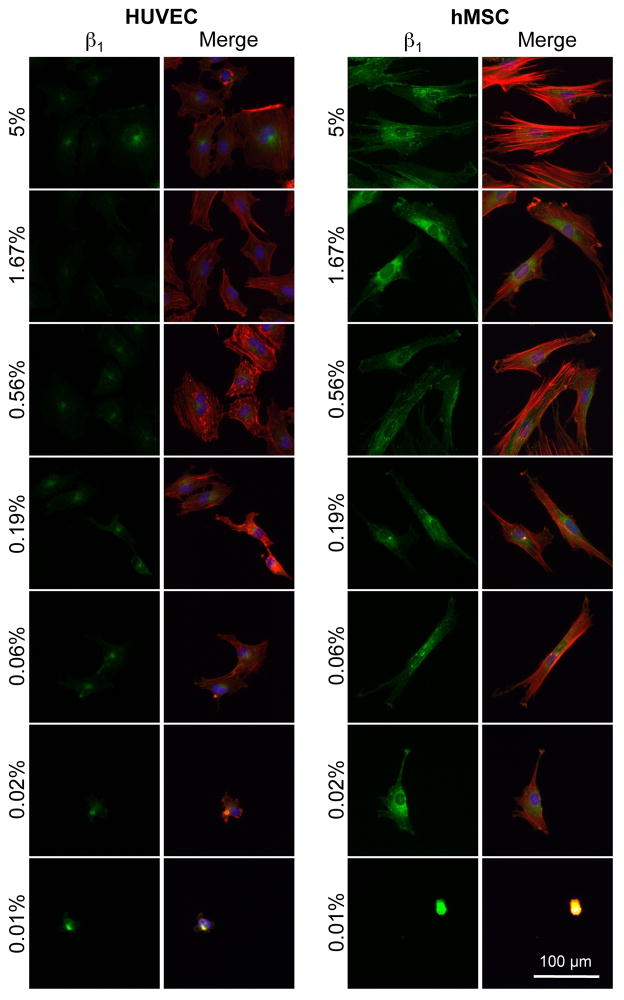
Antibody staining revealed differences in HUVEC and hMSC β1 integrin expression (Figure 7). HUVECs and hMSCs have the capacity to attach through a wide range of integrins, with each expressing a variety of different α and β subunits [52, 53] [54]. However, β1-integrins are known to facilitate a range of cellular functions such as attachment, spreading, and migration, and several integrin receptors with β1 subunits are known to bind RGD [10–12, 55–57]. Therefore, we compared expression of β1-integrin for HUVECs and hMSCs using immunohistochemistry (Figure 7). After 24 hours on SAM arrays presenting GRGDSP, hMSCs exhibited increased levels of staining for β1 integrins compared to HUVECs across all RGD densities that promoted cell attachment (Figure 7). The expression of β1-integrin at an elevated level for hMSCs is interesting, as we also observed elevated focal adhesion expression for HUVECs (Supplement Figure 2). Importantly, these observations indicate that despite global similarities in cell attachment and spreading at 24 hours, the adhesion mechanisms that facilitate these processes may be quite different in terms of the involvement of β1 integrin subunits and the density and size of focal adhesions.
Figure 7.
Staining for integrin expression using SAM arrays presenting varied GRGDSP densities. HUVECs and hMSCs cultured for 24 hrs on arrays were stained for β1 integrin expression. (β1 integrin: green, actin:red, and nuclei:blue).
Cell Proliferation
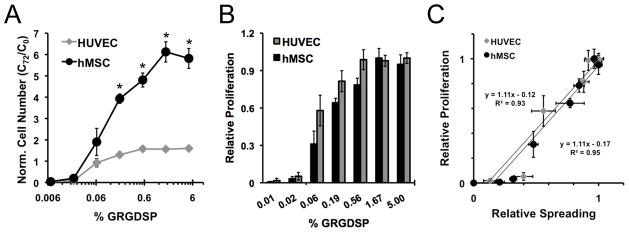
To further characterize cell adhesion-dependent influences on cell behavior using SAM arrays, we investigated how systematic changes in adhesion ligand density influenced proliferation for HUVECs compared to hMSCs (Figure 8). Proliferation was quantified by normalizing the cell number at 72 hrs (C72) to the number of cells at 0 hr (C0). Based on this notation, normalized cell numbers greater than one (C72/C0 > 1) indicate proliferation and values less than one (C72/C0 < 1) indicate cell death or detachment. Cell types exhibited different proliferation rates with HUVECs achieving a maximum normalized cell number of 1.6 and hMSCs achieving a maximum of 6.1 (Figure 8A). Despite these differences in maximum proliferation, proliferation as a function of ligand density was similar for both HUVECs and hMSCs with increasing proliferation over GRGDSP densities ≥ 0.06% and no proliferation below 0.06% GRGDSP (Figures 8A). Furthermore, calculation of relative proliferation depicts similar proliferative responses to adhesion ligand density (Figure 8B). Here we see that both cell types exhibit similar adhesion ligand dependencies for proliferation. This positive correlation adhesion and proliferation coincides well with what other investigations have reported in a range of cell types.[58–61] However, an important difference here is that through a well-controlled comparison, we found that adhesion dependence on proliferation is identical for two distinct cell types over a wide range of adhesion ligand density.
Figure 8.
Screening for proliferation using SAM arrays. Arrays seeded with HUVECs and hMSCs were imaged at 0 and 72 hrs in culture and quantification of proliferation was achieved by normalizing cell number at 72 hrs (C72) to 0 hrs (C0) for (A) HUVECs and hMSCs. (B) Relative proliferation over 72 hours for each cell type was determined by scaling proliferation at each condition to maximum proliferation and was also compared to (C) relative spreading at 24 hrs. (Error bars indicate standard error of the mean. Asterisk indicates significant difference between HUVECs and hMSCs at a specific GRGDSP density, p < 0.05.)
Migration
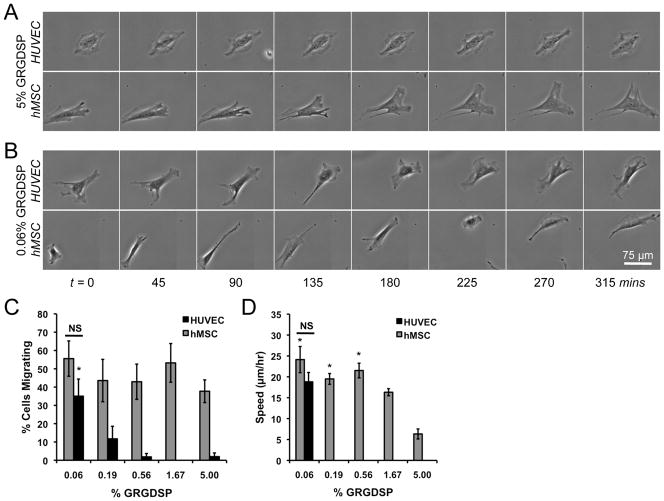
Although HUVEC and hMSC spreading and proliferation trends over 72 hrs were similar, we observed stark differences in migration morphology and relative number of migrating cells as a function of cell adhesion (Figure 9). The fraction of migrating hMSCs was insensitive to adhesion ligand densities with greater than 37% of the cell migrating for each condition (Figure 9C). In contrast, HUVEC migration was highly dependent on adhesion ligand density with virtually no cells migrating on high GRGDSP densities but over 30% of the cells migrating at lower GRGDSP densities. The morphology of migrating cells was also a function of adhesion ligand density (Figures 9A, B) and more clearly observed in time-lapse movies (Supplemental Movies 2A–D). HUVECs exhibited static behavior on 5% GRGDSP in which the cell body continuously oscillated, but little net movement was observed. In contrast, hMSCs migrating on high GRGDSP densities exhibited migratory phenotypes characterized by polarized membranes with leading edges driven by active membrane protrusions such as lammellipodia and filopodia, consistent with phenotypes of migrating fibroblasts.[62] At lower densities of adhesion ligand, both cell types exhibit migratory phenotypes that were similar to hMSC behavior on 5% GRGDSP (polarized cell body with leading edge protrusions) but had subtle differences. hMSCs on high densities of adhesion ligand concertedly progressed through the following steps of migration: i) protrusion of the leading edge, ii) attachment to the surface, iii) contraction of the cytoplasm, and iv) release and retraction of the rear end.[62–64] hMSC and HUVEC migration on low densities of adhesion ligand also followed these steps of migration, but each step was more clearly defined giving the cells a more sporadic migration appearance (Figure 9B). These results suggest a potential adhesion-dependent role for protrusion dynamics, with more fan or sheet-like protrusions (lammellipodia) at high RGD densities, but more pronounced spike-like protrusions (filopodia) at low density. Changes in migratory phenotypes also corresponded to changes in migration speed, with hMSCs exhibiting maximal migrations speeds below 1.67 % GRGDSP (Figure 9D). Interestingly, HUVECs on 0.06% GRGDSP (the only condition with a significant number of cells to track for speed) exhibited migration speeds similar to those observed for hMSCs. This change in migration morphology and speed is similar to recent work in which fish keratocytes, a model migratory cell type, exhibited two distinct migration regimes when cultured on surfaces with varied densities of RGD.[65] Here, we see qualitative evidence of differences in migration phenotypes in two primary human cell types, however a more thorough investigation is required to decipher potential differences in migratory mechanisms with respect to adhesion ligand density.
Figure 9.
Screening for cell migration using SAM arrays. Arrays seeded with HUVECs and hMSCs were imaged every 15 mins using automated microscopy and tracked over 12–18hrs. Example sequences of cells on (A) 5% GRGDSP and (B) 0.06% GRGDSP are shown and single cell tracking was used to quantify the (C) percentage of cells migrating and (D) migration speed on each GRGDSP condition. (Error bars indicate standard error of the mean. Asterisk indicates significant increase compared to 5% GRGDSP conditions and “NS” indicates no significant difference between cell types at a single GRGDSP density, p < 0.05. Note: Migration speed is only reported for conditions in which more than 15% of single cells were migrating.) Time-lapse videos of these cells are included in Supplement Movies 2A–D.
Adaptability of SAM arrays to various human cell types
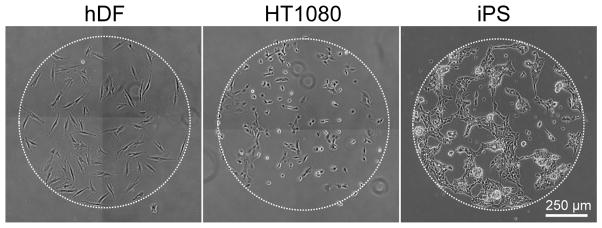
The SAM array platform described here affords enhanced throughput experimental methodologies that not only enable screening a large set of experimental variables, but also direct comparison of multiple cell types. In addition to the carbodiimide chemistries used in this work for peptide immobilization, the SAM array fabrication process described here is also amenable to other immobilization chemistries such as copper catalyzed azide-alkyne cycloadditions previously used in our lab[5, 6] (data not shown) offering several strategies for immobilization of a range of different biomolecules (i.e. peptides, nucleic acids, polysaccharides). To date, we have successfully used GRGDSP SAM arrays with primary, immortalized, and pluripotent cell types (Figure 10). Taken together, we demonstrate that SAM arrays provide a flexible strategy for biomolecule immobilization that is compatible with several cell types and has the potential to probe a wide variety of fundamental biological questions aimed at deconstructing the influence of the microenvironment on cell function.
Figure 10.
SAM array versatility. Arrays presenting GRGDSP can be used to culture a range of cell types including human dermal fibroblasts (hDFs), human fibrosarcoma cells (HT-1080s), and induced pluripotent stem cells, iPS(IMR90)-4 (iPS).
Implications of similarities between cell types
Numerous studies have linked attachment, spreading, proliferation, and migration to changes in adhesion and have identified strong correlations between them for a given cell type[2, 4, 40, 50, 58–61, 66–70], yet comparisons between cell types as a function of systematically varied adhesion[53] ligand density have not typically been investigated. Direct comparison between hMSC and HUVEC attachment and spreading using SAM arrays exposed several similarities between these cell types that would not necessarily be easily predicted. For example, initial attachment and spreading at 24 hrs were nearly identical for both cell types, despite substantial differences in cytoskeletal structure (Figures 4–6) and expression of focal adhesions (Supplement Figure 2) and β1-integrin (Figure 7). One possible reason for these similarities may be that both cell types recover from trypsinization at similar rates. Previous work has shown that buffer conditions and duration of trypsinization significantly impact HUVEC spreading and attachment dynamics over 24 hrs and that cell attachment was dependent on synthesis of new integrins.[43] Therefore, although we observed differences in β1 integrin expression (Figure 7), the rates at which other integrins are synthesized following trypsinization may be similar leading to similar attachment and projected cell areas at 24 hrs. We speculate that after this initial recovery phase, cell behavior may be regulated by additional, perhaps more cell type-dependent, mechanisms that promote the observed differences in spreading behavior at 72 hrs. However, the relative dependence of spreading on ligand density at 72 hrs remains similar to that observed at 24 hrs.
Analogous to trends observed for attachment and spreading, proliferation for both cell types exhibited similar dependencies on RGDSP (Figure 8A). Further, when relative proliferation is compared to relative spreading, both HUVECs and hMSCs exhibit a 1:1 correlation between spreading and proliferation (Figure 8C), suggesting a similar fundamental relationship between these properties across two distinct cell types. In a landmark paper, Folkman and colleagues used poly(2-hydroxyethyl methacrylate) coatings to control the extent to which bovine aortic endothelial cells (BAECs) spread on culture surfaces and found that proliferation exhibited a positive correlation with cell spreading.[71] Additionally, Ingber and co-workers observed a similar positive correlation between bovine capillary endothelial cell spreading and proliferation when fibronectin, an extracellular matrix protein that contains the GRGDSP motif, was adsorbed onto polystyrene surfaces at varied densities.[40] Since these studies, a range of other studies have investigated this relationship [58–61], including approaches that explicitly defined endothelial cell spreading using microscale adhesive islands, and implicated cell spreading is the dominating factor influencing proliferation.[67–70] The observations in the current manuscript are consistent with these previous observations of endothelial cell behavior. While there are several studies investigating the relationship between endothelial cell spreading and proliferation, there has been little investigation of this effect in hMSCs. Instead, prior studies have focused on correlating changes in cell shape and projected cell area to hMSC differentiation down osteogenic, adipogenic, myogenic, chondrogenic, and neurogenic lineages.[45–51] Here we observe clear correlations between spreading and proliferation indicating that changes in hMSC proliferation in response to a given cell culture substrate may also need to be considered. In addition to identification of this relationship in hMSCs, we believe this is the first example in which the influence of cell spreading on proliferation for two distinct primary human cell types has been shown to follow identical trends, a result that suggests a similar fundamental adhesion-dependent mechanism.
While attachment and spreading follow similar trends for HUVECs and hMSCs, migration was significantly different for several RGD densities. Specifically, at RGD densities above 0.06%, a higher fraction of hMSCs were motile and had increased migration speed compared to HUVECs (Figure 9C–D), a result that may be correlated to differences in expression of integrins (Figure 7) and focal adhesions (Suppliment Figure 2). Focal adhesions play an important role for migration[72], but maturation leads to a more stationary phenotype [73–76]. Many previous studies have implicated β1 integrin-containing heterodimers as regulators of cell migration [77, 78]. Additionally, while the increased β1 integrin expression observed for hMSCs may play a role in the increased migratory phenotype compared to HUVECs, there are several integrins that could facilitate motility and thus further investigation will be required to better understand the specific role for integrins on RGD-SAMs.
The clear change in HUVEC behavior from static to migratory phenotype at 0.06% GRGDSP (Figure 9C) coincides with several other observations in this work. In particular, the observed induction of migration at 0.06% GRGDSP corresponds with changes in cytoskeletal and focal adhesion organization observed in HUVECs at low adhesion ligand densities where cells take on a more elongated phenotype (Figure 5). Furthermore, 0.06% GRGDSP also appears to be a transition point for HUVEC proliferation; above 0.06% HUVECs proliferate to varied extents and below 0.06% HUVECs detach and/or die (Figure 8). Therefore, this suggests that HUVECs change between migratory and proliferative phenotypes when cultured on GRGDSP presenting surfaces, while hMSCs may not exhibit the phenomenon. The ECM-dependent migration data may have implications for function of these cell types in vivo. hMSCs and endothelial cells can each migrate into sites of injury, and the changing ECM conditions during wound healing may significantly influence this type of cell infiltration into a wound site.[79, 80] In addition, recent discoveries that hMSCs express markers that are similar to pericytes suggest that these two cell types can migrate to similar locations during tissue development and maintenance.[81–83] Further analyses of ECM-dependent migration of these cell types in 3-D and further insights into the ontogeny of hMSCs in future studies will help to illuminate the importance of their ECM-dependent migration. Regardless, these results further implicate the adhesive state of a cell as a major determinant of cell behavior and highlight the utility of the SAM arrays as a means to interrogate these dependencies.
In summary, here we used a chemically-defined array-based platform to report the influence of cell adhesion ligand density on primary human cell behavior. Our results demonstrated that multiple fundamental cellular properties (attachment, spreading, and proliferation) for two distinct primary human cell types (HUVECs and hMSCs) followed virtually identical trends across a wide range of cell adhesion conditions. This result was unexpected, as one would generally predict that distinct cell lineages would display very different responses to the extracellular environment. However, a series of recent studies suggest an important link between endothelial cells and mesenchymal stem cells. Recent in vivo studies suggest that hMSCs may represent pericytes that surround the endothelium in blood vessels. [81–83] In addition, prior in vitro studies have demonstrated that hMSCs can differentiate into an endothelial cell phenotype[84], and HUVECs can transform into an hMSC-like phenotype.[85, 86] While the goal here was not to investigate these phenotypic transitions, emerging evidence suggests their important role in several diseases including fibrosis and heterotopic ossification[83, 85, 86]. These prior studies, coupled with the quantitative similarities observed in our current study, suggest interesting phenotypic similarities between these distinct cell lineages, and motivate further discovery-based screening to characterize phenotypic similarities and ECM dependencies.
Supplementary Material
Insight, Innovation, Integration.
Insight
Our results indicate that human umbilical vein endothelial cells and human mesenchymal stem cells exhibit previously unrealized similarities in attachment, spreading, and proliferation as a function of cell adhesion ligand density, yet stark differences in migratory behavior.
Innovation
These studies combined a chemically-defined self-assembled monolayer array platform with automated time-lapse microscopy to screen for the effects of cell adhesion ligand density (over several orders of magnitude) on several different cell behaviors.
Integration
Investigation of identical sets of conditions for both cell types using a synthetic, chemically defined array allowed us to (i) rigorously compare behaviors within a single cell type and (ii) compare behaviors between both cell types to identify links between cell adhesion and critical cell behaviors.
Acknowledgments
The authors would like to acknowledge funding from the National Institutes of Health (R01HL093282 and the Biotechnology Training Program NIGMS 5 T32GM08349) and the National Science Foundation (DMR 0906123). Fluorescent scans were obtained using a GE Healthcare Typhoon Trio Variable Mode Imager at the Scientific Instrumentation Facility of the UW Carbone Cancer Center, Madison, WI.
References
- 1.Dubiel EA, Martin Y, Vermette P. Bridging the Gap Between Physicochemistry and Interpretation Prevalent in Cell-Surface Interactions. Chemical Reviews. 2011;111(4):2900–2936. doi: 10.1021/cr9002598. [DOI] [PubMed] [Google Scholar]
- 2.Mrksich M. Using self-assembled monolayers to model the extracellular matrix. Acta Biomaterialia. 2009;5(3):832–841. doi: 10.1016/j.actbio.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudalla GA, Murphy WL. Chemically well-defined self-assembled monolayers for cell culture: toward mimicking the natural ECM. Soft Matter. 2011;7(20):9561–9571. doi: 10.1039/C1SM05596H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mrksich M. A surface chemistry approach to studying cell adhesion. Chemical Society Reviews. 2000;29(4):267–273. [Google Scholar]
- 5.Hudalla GA, Murphy WL. Immobilization of peptides with distinct biological activities onto stem cell culture substrates using orthogonal chemistries. Langmuir. 2010;26(9):6449–56. doi: 10.1021/la1008208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudalla GA, Murphy WL. Using “click” chemistry to prepare SAM substrates to study stem cell adhesion. Langmuir. 2009;25(10):5737–46. doi: 10.1021/la804077t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koepsel JT, Murphy WL. Patterning discrete stem cell culture environments via localized self-assembled monolayer replacement. Langmuir. 2009;25(21):12825–34. doi: 10.1021/la901938e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudalla GA, Koepsel JT, Murphy WL. Surfaces that sequester serum-borne heparin amplify growth factor activity. Adv Mater. 2011;23(45):5415–8. doi: 10.1002/adma.201103046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudalla GA, et al. Harnessing endogenous growth factor activity modulates stem cell behavior. Integr Biol (Camb) 2011;3(8):832–42. doi: 10.1039/c1ib00021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierschbacher MD, Ruoslahti E. Cell Attachment Activity of Fibronectin Can Be Duplicated by Small Synthetic Fragments of the Molecule. Nature. 1984;309(5963):30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 11.Ruoslahti E. RGD and other recognition sequences for integrins. Annual Review of Cell and Developmental Biology. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 12.Ruoslahti E, Pierschbacher MD. New Perspectives in Cell-Adhesion - Rgd and Integrins. Science. 1987;238(4826):491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 13.Aird WC. Spatial and temporal dynamics of the endothelium. J Thromb Haemost. 2005;3(7):1392–406. doi: 10.1111/j.1538-7836.2005.01328.x. [DOI] [PubMed] [Google Scholar]
- 14.Bachetti T, Morbidelli L. Endothelial cells in culture: a model for studying vascular functions. Pharmacol Res. 2000;42(1):9–19. doi: 10.1006/phrs.1999.0655. [DOI] [PubMed] [Google Scholar]
- 15.Cines DB, et al. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91(10):3527–61. [PubMed] [Google Scholar]
- 16.Krishnaswamy G, et al. Human endothelium as a source of multifunctional cytokines: molecular regulation and possible role in human disease. J Interferon Cytokine Res. 1999;19(2):91–104. doi: 10.1089/107999099314234. [DOI] [PubMed] [Google Scholar]
- 17.Michiels C. Endothelial cell functions. J Cell Physiol. 2003;196(3):430–43. doi: 10.1002/jcp.10333. [DOI] [PubMed] [Google Scholar]
- 18.Caplan AI, Bruder SP. Mesenchymal stem cells: building blocks for molecular medicine in the 21st century. Trends Mol Med. 2001;7(6):259–64. doi: 10.1016/s1471-4914(01)02016-0. [DOI] [PubMed] [Google Scholar]
- 19.Deans RJ, Moseley AB. Mesenchymal stem cells: biology and potential clinical uses. Exp Hematol. 2000;28(8):875–84. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 20.Godara P, Nordon RE, McFarland CD. Mesenchymal stem cells in tissue engineering. Journal of Chemical Technology and Biotechnology. 2008;83(4):397–407. [Google Scholar]
- 21.Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9(1):204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krampera M, et al. Mesenchymal stem cells for bone, cartilage, tendon and skeletal muscle repair. Bone. 2006;39(4):678–683. doi: 10.1016/j.bone.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Krampera M, et al. Regenerative and immunomodulatory potential of mesenchymal stem cells. Current Opinion in Pharmacology. 2006;6(4):435–441. doi: 10.1016/j.coph.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Caplan AI. What’s in a name? Tissue Eng Part A. 2010;16(8):2415–7. doi: 10.1089/ten.TEA.2010.0216. [DOI] [PubMed] [Google Scholar]
- 25.Prime KL, Whitesides GM. Adsorption of Proteins onto Surfaces Containing End-Attached Oligo(Ethylene Oxide) - a Model System Using Self-Assembled Monolayers. Journal of the American Chemical Society. 1993;115(23):10714–10721. [Google Scholar]
- 26.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Analytical Biochemistry. 1989;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 27.Walker GM, Beebe DJ. A passive pumping method for microfluidic devices. Lab on a Chip. 2002;2(3):131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 28.Jo BH, et al. Three-dimensional micro-channel fabrication in polydimethylsiloxane (PDMS) elastomer. Journal of Microelectromechanical Systems. 2000;9(1):76–81. [Google Scholar]
- 29.Thibault C, et al. Poly(dimethylsiloxane) Contamination in Microcontact Printing and Its Influence on Patterning Oligonucleotides. Langmuir. 2007;23(21):10706–10714. doi: 10.1021/la701841j. [DOI] [PubMed] [Google Scholar]
- 30.Sotiropoulou PA, et al. Characterization of the optimal culture conditions for clinical scale production of human mesenchymal stem cells. Stem Cells. 2006;24(2):462–71. doi: 10.1634/stemcells.2004-0331. [DOI] [PubMed] [Google Scholar]
- 31.Rossner M, Yamada KM. What’s in a picture? The temptation of image manipulation. J Cell Biol. 2004;166(1):11–5. doi: 10.1083/jcb.200406019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orner BP, et al. Arrays for the combinatorial exploration of cell adhesion. J Am Chem Soc. 2004;126(35):10808–9. doi: 10.1021/ja0474291. [DOI] [PubMed] [Google Scholar]
- 33.Roberts C, et al. Using Mixed Self-Assembled Monolayers Presenting RGD and (EG)3OH Groups To Characterize Long-Term Attachment of Bovine Capillary Endothelial Cells to Surfaces. Journal of the American Chemical Society. 1998;120(26):6548–6555. [Google Scholar]
- 34.Malinda KM, et al. Identification of laminin alpha1 and beta1 chain peptides active for endothelial cell adhesion, tube formation, and aortic sprouting. FASEB J. 1999;13(1):53–62. [PubMed] [Google Scholar]
- 35.Ponce ML, et al. Identification of endothelial cell binding sites on the laminin gamma 1 chain. Circ Res. 1999;84(6):688–94. doi: 10.1161/01.res.84.6.688. [DOI] [PubMed] [Google Scholar]
- 36.Jung JP, et al. Co-assembling peptides as defined matrices for endothelial cells. Biomaterials. 2009;30(12):2400–10. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant DS, et al. Interaction of endothelial cells with a laminin A chain peptide (SIKVAV) in vitro and induction of angiogenic behavior in vivo. J Cell Physiol. 1992;153(3):614–25. doi: 10.1002/jcp.1041530324. [DOI] [PubMed] [Google Scholar]
- 38.Frith JE, et al. Tailored integrin-extracellular matrix interactions to direct human mesenchymal stem cell differentiation. Stem Cells Dev. 2012;21(13):2442–56. doi: 10.1089/scd.2011.0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geiger B, et al. Vinculin, an intracellular protein localized at specialized sites where microfilament bundles terminate at cell membranes. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1980;77(7):4127–4131. doi: 10.1073/pnas.77.7.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingber DE, et al. Cell shape, cytoskeletal mechanics, and cell cycle control in angiogenesis. Journal of Biomechanics. 1995;28(12):1471–1484. doi: 10.1016/0021-9290(95)00095-x. [DOI] [PubMed] [Google Scholar]
- 41.Kopp PM, et al. Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1. European Journal of Cell Biology. 2010;89(9):661–673. doi: 10.1016/j.ejcb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stroka KM, Aranda-Espinoza H. Effects of Morphology vs. Cell-Cell Interactions on Endothelial Cell Stiffness. Cell Mol Bioeng. 2011;4(1):9–27. doi: 10.1007/s12195-010-0142-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown MA, et al. The use of mild trypsinization conditions in the detachment of endothelial cells to promote subsequent endothelialization on synthetic surfaces. Biomaterials. 2007;28(27):3928–3935. doi: 10.1016/j.biomaterials.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bischoff J. Cell adhesion and angiogenesis. J Clin Invest. 1997;100(11 Suppl):S37–9. [PubMed] [Google Scholar]
- 45.McBeath R, et al. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–95. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 46.Engler AJ, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 47.Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am J Physiol Cell Physiol. 2008;295(4):C1037–44. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 48.Gao L, McBeath R, Chen CS. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells. 2010;28(3):564–72. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kilian KA, et al. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 2010;107(11):4872–7. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frith JE, Mills RJ, Cooper-White JJ. Lateral spacing of adhesion peptides influences human mesenchymal stem cell behaviour. J Cell Sci. 2012 doi: 10.1242/jcs.087916. [DOI] [PubMed] [Google Scholar]
- 51.Kilian KA, Mrksich M. Directing Stem Cell Fate by Controlling the Affinity and Density of Ligand-Receptor Interactions at the Biomaterials Interface. Angew Chem Int Ed Engl. 2012 doi: 10.1002/anie.201108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Short SM, Talbott GA, Juliano RL. Integrin-mediated signaling events in human endothelial cells. Mol Biol Cell. 1998;9(8):1969–80. doi: 10.1091/mbc.9.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baranska P, et al. Expression of Integrins and Adhesive Properties of Human Endothelial Cell Line EA. hy 926. Cancer Genomics - Proteomics. 2005;2(5):265–269. [PubMed] [Google Scholar]
- 54.Prowse AB, et al. Stem cell integrins: implications for ex-vivo culture and cellular therapies. Stem Cell Res. 2010;6(1):1–12. doi: 10.1016/j.scr.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 55.Danen EHJ, Yamada KM. Fibronectin, integrins, and growth control. Journal of Cellular Physiology. 2001;189(1):1–13. doi: 10.1002/jcp.1137. [DOI] [PubMed] [Google Scholar]
- 56.Ruoslahti E. Integrins. Journal of Clinical Investigation. 1991;87(1):1–5. doi: 10.1172/JCI114957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruoslahti E. Fibronectin and Its Receptors. Annual Review of Biochemistry. 1988;57:375–413. doi: 10.1146/annurev.bi.57.070188.002111. [DOI] [PubMed] [Google Scholar]
- 58.Aplin AE, Howe AK, Juliano RL. Cell adhesion molecules, signal transduction and cell growth. Curr Opin Cell Biol. 1999;11(6):737–44. doi: 10.1016/s0955-0674(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 59.Stupack DG, Cheresh DA. Get a ligand, get a life: integrins, signaling and cell survival. J Cell Sci. 2002;115(Pt 19):3729–38. doi: 10.1242/jcs.00071. [DOI] [PubMed] [Google Scholar]
- 60.Schwartz MA, Assoian RK. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J Cell Sci. 2001;114(Pt 14):2553–60. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]
- 61.Bacakova L, et al. Modulation of cell adhesion, proliferation and differentiation on materials designed for body implants. Biotechnol Adv. 2011;29(6):739–67. doi: 10.1016/j.biotechadv.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Mogilner A, Keren K. The shape of motile cells. Curr Biol. 2009;19(17):R762–71. doi: 10.1016/j.cub.2009.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lauffenburger DA, Horwitz AF. Cell migration: A physically integrated molecular process. Cell. 1996;84(3):359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 64.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 65.Barnhart EL, et al. An adhesion-dependent switch between mechanisms that determine motile cell shape. PLoS Biol. 2011;9(5):e1001059. doi: 10.1371/journal.pbio.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mrksich M. Using self-assembled monolayers to understand the biomaterials interface. Current Opinion in Colloid & Interface Science. 1997;2(1):83–88. [Google Scholar]
- 67.Huang S, Chen CS, Ingber DE. Control of cyclin D1, p27(Kip1), and cell cycle progression in human capillary endothelial cells by cell shape and cytoskeletal tension. Molecular Biology of the Cell. 1998;9(11):3179–3193. doi: 10.1091/mbc.9.11.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang S, Ingber DE. Shape-dependent control of cell growth, differentiation, and apoptosis: Switching between attractors in cell regulatory networks. Experimental Cell Research. 2000;261(1):91–103. doi: 10.1006/excr.2000.5044. [DOI] [PubMed] [Google Scholar]
- 69.Thomas CH, et al. Surfaces designed to control the projected area and shape of individual cells. Journal of Biomechanical Engineering-Transactions of the Asme. 1999;121(1):40–48. doi: 10.1115/1.2798041. [DOI] [PubMed] [Google Scholar]
- 70.Chen CS, et al. Geometric control of cell life and death. Science. 1997;276(5317):1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 71.Folkman J, Moscona A. Role of Cell-Shape in Growth-Control. Nature. 1978;273(5661):345–349. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 72.Burridge K, et al. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- 73.Couchman JR, Rees DA. The behaviour of fibroblasts migrating from chick heart explants: changes in adhesion, locomotion and growth, and in the distribution of actomyosin and fibronectin. Journal of Cell Science. 1979 Oct;39:149–165. doi: 10.1242/jcs.39.1.149. [DOI] [PubMed] [Google Scholar]
- 74.Herman IM, Crisona NJ, Pollard TD. Relation between cell activity and the distribution of cytoplasmic actin and myosin. Journal of Cell Biology. 1981;90(1):84–91. doi: 10.1083/jcb.90.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Couchman JR, et al. Fibronectin has a dual role in locomotion and anchorage of primary chick fibroblasts and can promote entry into the division cycle. Journal of Cell Biology. 1982;93(2):402–410. doi: 10.1083/jcb.93.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolega J, et al. Rapid cellular translocation is related to close contacts formed between various cultured cells and their substrata. Journal of Cell Science. 1982 Apr;54:23–34. doi: 10.1242/jcs.54.1.23. [DOI] [PubMed] [Google Scholar]
- 77.Huttenlocher A, Horwitz AR. Integrins in Cell Migration. Cold Spring Harbor Perspectives in Biology. 2011;3(9) doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harb Perspect Biol. 2011;3(9):a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karp JM, Leng Teo GS. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell. 2009;4(3):206–16. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 80.Lamalice L, Le Boeuf F, Huot J. Endothelial cell migration during angiogenesis. Circ Res. 2007;100(6):782–94. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- 81.Crisan M, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3(3):301–13. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 82.Nombela-Arrieta C, Ritz J, Silberstein LE. The elusive nature and function of mesenchymal stem cells. Nat Rev Mol Cell Biol. 2011;12(2):126–31. doi: 10.1038/nrm3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bautch VL. Stem cells and the vasculature. Nat Med. 2011;17(11):1437–43. doi: 10.1038/nm.2539. [DOI] [PubMed] [Google Scholar]
- 84.Oswald J, et al. Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells. 2004;22(3):377–84. doi: 10.1634/stemcells.22-3-377. [DOI] [PubMed] [Google Scholar]
- 85.Medici D, et al. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16(12):1400–6. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Piera-Velazquez S, Li Z, Jimenez SA. Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am J Pathol. 2011;179(3):1074–80. doi: 10.1016/j.ajpath.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.