Abstract
Platelet derived growth factor receptor alpha (PDGFRA)-positive oligodendrocyte progenitor cells (OPC) located within the mature central nervous system may remain quiescent, proliferate, or differentiate into oligodendrocytes. Human glioblastoma multiforme tumors (GBM) often contain rapidly proliferating oligodendrocyte lineage transcription factor 2 (Olig2)-positive cells that resemble OPCs. In this study, we sought to identify candidate pathways that promote OPC differentiation or quiescence rather than proliferation. Gene expression profiling performed in both normal murine OPCs and highly proliferative Olig2-positive glioma cells identified all the transcripts associated with the highly proliferative state of these cells and demonstrated that, among the various cell types found within the brain, Olig2-positive tumor cells are most similar to OPCs. We then subtracted OPC transcripts found in tumor samples from those found in normal brain samples and identified 28 OPC transcripts as candidates for promoting differentiation or quiescence. Systematic analysis of human glioma data revealed that these genes have similar expression profiles in human tumors, and were significantly enriched in genomic deletions, suggesting an anti-proliferative role. Treatment of primary murine glioblastoma cells with agonists of one candidate gene, Gpr17, resulted in a decreased number of neurospheres. Together, our findings demonstrate that comparison of the molecular phenotype of progenitor cells in tumors to the equivalent cells in the normal brain represents a novel approach for the identification of targeted therapies.
Keywords: oliogdendrocyte progenitor, human glioblastoma, translational profiling, molecular targeting, pro-differentiation genes
Introduction
Glioma is the most prevalent type of primary brain tumor in adults and prognosis, especially for the high-grade, stage IV GBM, is dismal. Recently, work at the mRNA (1–3) and protein signaling levels (4), have converged upon similar classification schemes that identify distinct subgroups of high grade glioma. These subgroups show differential gene expression profiles, signaling cascades, and response to treatment. In particular, one subgroup, referred to as ‘proneural’ is characterized by elevated expression of PDGF, amplifications of PDGFRA (4) and expression of Olig2 (1, 2).
In the mature central nervous system the PDGFRA, and the chondroitin sulfate proteoglycan antigen (NG2) have been considered as markers primarily of the OPCs (5, 6). OPCs are sparsely and evenly distributed throughout white and grey matter. The adult OPC was thought to mainly serve as a repository for the generation of new mature oligodendrocytes (6), but may also serve as a neural stem cell (7). Though they are typically quiescent, they are more proliferative than any other population in the brain, with 1–5% actively cycling at any time (8). Regardless of their role, these cells are capable of at least three fates: they may terminally differentiate into oligodendrocytes, they may proliferate to produce additional progenitors, or they may remain quiescent.
We have previously generated a mouse model of glioma in which a Nestin (Nes) positive neural stem cell can be induced to overproduce PDGF-B (9, 10). The PDGFRA is activated by both homo and heterodimers of A and B forms of PDGF (11). These mice generate aggressive gliomas resulting in a tumor mass composed of the highly proliferative cells of origin and cells derived from initially normal progenitors that are recruited to the tumor (9, 12–14). The presence of cells expressing Olig2 and PDGFRA in these tumors suggests that treatment strategies in the 'proneural' subtype, may be informed by knowledge of the signaling pathways that regulate the choice between proliferation, quiescence, and differentiation in these OPCs.
Current glioma treatment, following resection of the tumor, is focused on targeting proliferating cells with Temozolomide and radiotherapy (15). However, as previously noted (16), a complementary strategy would be to promote pathways for maintaining quiescence and/or driving terminal differentiation of the progenitors present in the tumor, as this may also serve to slow tumor growth. Pro-differentiation treatments may be particularly promising; while the tumor environment is likely selecting for mutations in genes that normally suppress cell cycle progression and maintain quiescence, there may not be as much selective pressure on all pro-differentiation pathways.
We undertook a study in this PDGF-B-induced mouse model of glioblastoma to identify transcripts that could be important for the regulation of quiescence and differentiation in OPC like cells. Applying our newly developed methodology for in vivo cell-specific translational profiling (17, 18), we identified all mRNAs specifically enriched in OPCs in normal mouse brain, including those likely to be important for proliferation, quiescence, and differentiation. We then contrasted this to the cell-specific translational profile of Olig2-positive cells in the mouse model of a ‘proneural’ glioma, in which OPC-like cells are committed to proliferation at the expense of differentiation or quiescence. This permitted identification of candidate pathways that may serve as targets for promoting differentiation and quiescence in OPCs in mice. Examination of The Cancer Genome Atlas (TCGA) expression profiles of human gliomas established that analogous pathways are similarly regulated in human ‘proneural’ GBM, suggesting their conservation as targets. Also consistent with an anti-proliferative role, many of these targets show deletions in human GBM. From this combined human and mouse screening, we have identified several candidate pathways for promoting quiescence and differentiation, which may serve as targets for complementary treatments.
Material and Methods
Full materials and methods are available online.
All protocols involving animals were approved by the Rockefeller University and Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee.
New mouse lines
BACs containing genes PDGFRA (RP23-55P22), Cnp1 (RP23-78L12), and Snap25 (RP23-290A18) were modified as described (17) to insert an EGFP-L10a fusion protein into the relevant translation start site.
Histology
Anatomy was performed as described (17, 19). For immunofluorescence samples were incubated with the cell-specific antibodies and quantification was performed on 40× cortical fields imaged with confocal microscopy.
Profiling Tumor Model
Tumors were generated as described (9). Cells producing RCAS-h PDGF-B or RCAS-Cre virus were injected into the progeny of ink4a/arf−/− mice expressing tv-a receptor for RCAS under the Nes promoter, crossed to Olig2-Egfp::L10a mice. TRAP and microarray hybridizations were performed as described (17, 18), except tumors were processed individually.
Microarray Analysis
Microarray data were analyzed with Bionconductor module of the R statistical package, normalized as described (17, 20, 21), and deposited at GEO (GSE30626) (70). To identify messages specific to each cell type Specificity Indices (pSI) were calculated as described (18). Transcripts with pSI<.05 were selected for further analysis.
Heatmaps and hierarchical clustering were performed in R. All color-coded scatterplots show only top 50 transcripts for each cell type, but all statistics were performed on full lists (Supplemental Table 1). Differentiation or Quiescence (DorQ) candidates were selected as those transcripts from the OPC list which were two fold higher in the average of the Olig2-Egfp::L10a normal cortex than the average of all 12 Olig2-Egfp::L10a tumor samples. Statistical comparisons were conducted with LIMMA module of Bioconductor.
For cross species comparisons, human and mouse homologues were mapped by Gene Symbol. For DorQ candidates, mapping was confirmed by Blating mouse protein to human genome (UCSC). Human microarray data were downloaded from TCGA and normalized as described (4).
Gene Ontology Analysis
For each cell type, all gene symbols for messages with a pSI <.05 were analyzed with BINGO (22). 'Biological Process' GO categories were evaluated to identify those with p <.01 using the hypergeometric test and Benjamini-Hochberg correction.
Neurosphere Cell Culture
Primary neurosphere cultures from tumors and wild type mice were generated as described (23). Neurospheres were grown in 20mg/ml EGF and 10mg/ml bFGF, and uridine 5’-diphosphate sodium salt (UDP) (10–50uM), UDP-glucose (100uM), and leukotriene D4 (LTD4) (100 nM) were added on a daily basis. All data are average of cultures from four independent mouse tumors, counted in triplicate in wells of 100–1000 spheres. Counts were normalized within each tumor to the number of spheres in 0 UDP condition.
Results
We recently developed the TRAP strategy, which allows profiling of all mRNAs bound to ribosomes in defined cell populations. This strategy entails using bacterial artificial chromosome (BAC) transgenesis to express EGFP fused to the ribosomal protein L10a under the control of a ‘driver’ gene specific to certain cell types in the brain. Thus, the cells of interest contain ribosomes with an EGFP tag enabling affinity purification of all ribosome-associated mRNA. We have generated and characterized bacTRAP mouse lines for a variety of cell types, including the Olig2-positive oligodendroglia (17).
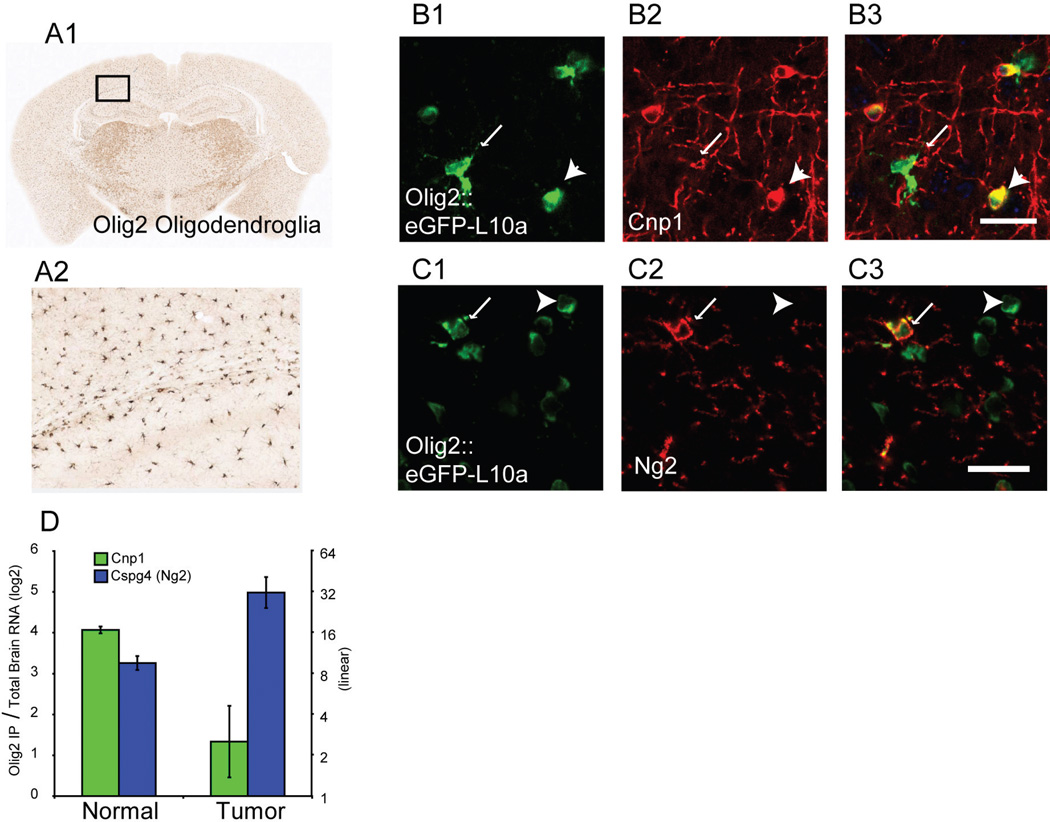
Because Olig2 is frequently overexpressed in human glioma (25) and high levels of expression may particularly characterize the 'proneural' subtype (2). We recently examined the expression of neural lineage markers, proliferation markers and translational profile of Olig2::Egfp-L10a in our mouse model of glioma (12). This work revealed a severe perturbation of the translational profile of Olig2-positive cells. In a normal Olig2::Egfp-L10a mouse, both NG2-positive OPCs and 2',3'-cyclic nucleotide 3'-phosphodiesterase 1 (Cnp1)-positive mature oligodendrocytes express EGFP-L10a (Figure 1A–C) (17). The tumor translational profiles for the Olig2::Egfp-L10a cells reveal a strong increase in translation for Cspg4 (the RNA corresponding to NG2) and decrease in Cnp1, relative to normal Olig2::Egfp-L10a cells (Figure 1D). While the data come from only two markers, it suggests the preponderance of Olig2::Egfp-L10a cells are in an immature state within the tumor. To investigate this thoroughly, we decided to identify the translational profile of normal adult OPCs. With this data for comparison, we would then be able to determine if Olig2::Egfp-L10a are in a more OPC-like state in the tumor, and if so, we could then examine how this state may differ from normal OPCs, as these differences may regulate cell fate choices and represent potential treatment strategies.
Figure 1. The Olig2::Egfp-L10a mouse permits study of oligodendroglia.
A1, A2: DAB Immunohistochemistry (IHC) for EGFP-L10a Olig2::Egfp-L10a mouse reveals a pattern consistent with expression in both OPCs and mature oligodendrocytes. B1–B3: Confocal immunofluorescence (IF) for EGFP-L10a and Cnp1 confirms localization in mature oligodendrocytes (white arrowheads). C1–C3: Confocal immunofluorescence for EGFP-L10a and NG2 confirms localization in OPCs (white arrows). D: In normal brain, Olig2::Egfp-L10a translational profiles show enrichment of both Cnp1 and Cspg4 (Ng2), compared to total brain RNA. In glioma, Olig2::Egfp-L10a cells show a relative increase of Cspg4 and loss of Cnp1 RNA. All scale bars 20 microns.
Identification of the translational profile of the normal oligodendrocyte progenitors
Identification of transcripts that are specific to OPCs requires not only knowledge of their profile, but also the profiles of the other cell types. The brain contains four major classes of cells: neurons, astrocytes, and two types of oligodendroglia; mature oligodendrocytes, and OPCs. We have previously generated and thoroughly characterized mouse lines that allow translational profiling from astrocytes (Aldh1L1::Egfp-L10a), mature oligodendrocytes (Cnp1::Egfp-L10a), and the pan-oligodendroglial line (Olig2::Egfp-L10a) (17). However, to identify those mRNAs that are enriched in each of the major cell classes we generated additional bacTRAP mouse lines targeting all neurons (Snap25::Egfp-L10a), as well as Pdgfra expressing cells (Pdgfra::Egfp-L10a).
To target neurons, we selected two genes as putative pan neuronal drivers, complexin 1 (Cplx1) and synaptosomal-associated protein 25 (Snap25). Examination of multiple EGFP-L10a lines for both constructs revealed that Snap25 was the brighter and more reliable construct and thus selected for further analysis.
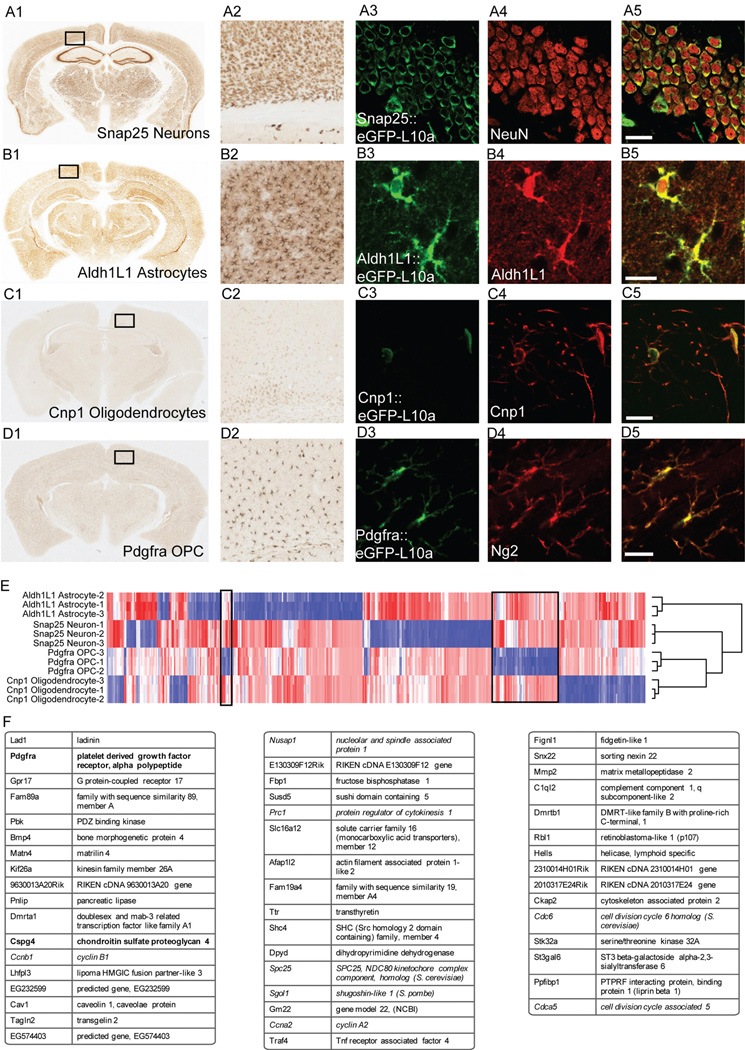
To target PDGF-responsive OPCs, we tested a BAC covering the Pdgfra gene. Multiple lines showed similar but not identical patterns of expression, targeting NG2-positive cells with varying intensity, but also subpopulations of neurons in some lines, as well as robust expression in the choroid plexus. For this study, we selected the line that targeted cortical NG2-positive OPCs most specifically. For each of the four cell classes, the lines utilized for further experiments were carefully characterized with confocal immunofluorescence to confirm accuracy of transgene expression, and identity of each cell type (Figure 2A–D).
Figure 2. Four bacTRAP lines accurately target EGFP-L10a expression to the major cell classes of the CNS, and allow for identification of transcripts specific to each population.
A: A BAC for Snap25 was tested as a pan-neuronal driver for EGFP-L10a transgene. A1, A2: DAB IHC for EGFP-L10a reveals a pattern consistent with expression in all neurons and only neurons. A3–A5: Confocal IF for EGFP-L10a and the neuronal nuclear marker NeuN confirm pan-neuronal expression. Dentate gyrus shown here. There was no overlap with markers for the other cell types below (not shown). B: Aldh1L1 BAC targets astrocytes. B1, B2: DAB IHC reveals a pattern consistent with expression in all astrocytes. B3–B5: IF reveals concordance between expression of EGFP-L10a and pan-astroglial Aldh1L1 protein. Cortex shown here. EGFP-L10 population includes all GFAP+ astrocytes, but not markers for other glial cell types (NG2 & Cnp1, not shown). C: Cnp1 BAC targets mature oligodendrocytes. C1, C2: DAB IHC reveals a pattern consistent with expression in oligodendrocytes, with densest labeling in white matter tracts. C3–C5: IF reveals concordance between expression of EGFP-L10a and oligodendrocyte protein Cnp1 in the cytoplasm. Superficial cortex shown here. Note that Cnp1 protein (red) is also found abundantly in fine processes of oligodendrocytes while ribosome bound EGFP-L10a is not. Egfp-L10 does not overlap with OPCs (NG2, not shown). D: A BAC for PDGFRA was tested as a driver for OPCs. D1, D2: For this line (JD340), DAB IHC reveals a pattern consistent with expression in OPCs of adult brain, in small cells with few processes evenly spaced through white and gray matter. D3–D5: IF reveals concordance of expression between EGFP-L10a and OPC marker NG2. In all lines, EGFP-L10a is found predominantly in the cytoplasm, consistent with inclusion in ribosomes. All scale bars 20 microns. E: Hierarchical clustering and heatmap for microarrays of TRAP data. Replicate arrays on independent samples for show consistent patterns of ribosome bound RNA for each cell type. F: Top 50 transcripts, ranked by specificity index, for OPCs. List includes the known markers for these cells (bold), as well as a variety of cell cycle transcripts (italics). Complete lists for all cell types are in Supplemental Table 1.
For the newly generated lines, we quantified, from randomly collected confocal images of mouse cortex, the overlap of GFP with DAPI and with each of two cell specific markers. For Snap25, we labeled with pan-neuronal maker NeuN (Figure 2A), or Tbr1 (not shown), a marker of deep layer neurons. In cortex, of 663 DAPI positive cells counted, 57.6% were positive for GFP, consistent with the large proportion of neurons in this tissue. 96.8% of GFP positive cells were positive for NeuN, and 98.6% of NeuN positive cells were positive for GFP. For Tbr1, of 592 Tbr1 positive cells counted, 99.8% were labeled with GFP. For Pdgfra, of 3015 DAPI positive cells counted, 8.2% were positive for GFP, consistent with the relative rarity of the OPC cells. 88.8% of GFP positive cells were labeled with Ng2, and though receptors are often localized on processes far from the GFP labeled cell bodies, obfuscating counting, at least 51.4% of GFP positive cells were clearly labeled with Pdgfra antibodies. Most of these cells distinctly had the morphology of OPCs, though both Ng2 and Pdgfra label pericytes as well. A subset of Pdgfra::eGFP-L10a cells also labeled with the proliferative marker Ki-67 (<2%), consistent with expectation for these cells (8).
For each new line, TRAP was conducted in triplicate on pooled cortices from 2–3 mice, and cell-specific polysomal mRNAs were queried with microarrays. Replicate arrays showed high reproducibility (minimum correlation =0.978), though each cell type had a distinct profile (Figure 2E). We used the specificity index statistic (26) to select for those transcripts significantly enriched in each cell population (Snap25, Ald1L1, Cnp1, Pdgfra) relative to the other three. These lists included known markers for each cell type (Supplemental Table 1, and Figure 2F). The non-OPC cell-types had significant overrepresentation of Gene Ontology (GO) categories consistent with their known roles in synaptic transmission (Neurons, Supplemental Figure 2A), myelination (Oligodendrocytes, Supplemental Figure 2B), or a role in lipid metabolism (Astrocytes, Supplemental Figure 2C) (27). As the OPC population is the most proliferative population in the cortex (8), the transcripts specific to OPCs showed a highly significant over-representation of categories related to the cell cycle (p<1E-7, hypergeometric test with B-H correction) (Supplemental Figure 3), as well as in categories related to protein phosphorylation and biopolymer modification (p<1E-3). For all cell types there were also a variety of transcripts not currently annotated in known pathways. Overall, this evidence, along with presence of all known markers for OPCs on the list, suggests we have identified an OPC specific profile, as well as generated a useful tool for assessment of these cells in vivo.
Characterization of Olig2-positive cells in mouse model of glioblastoma
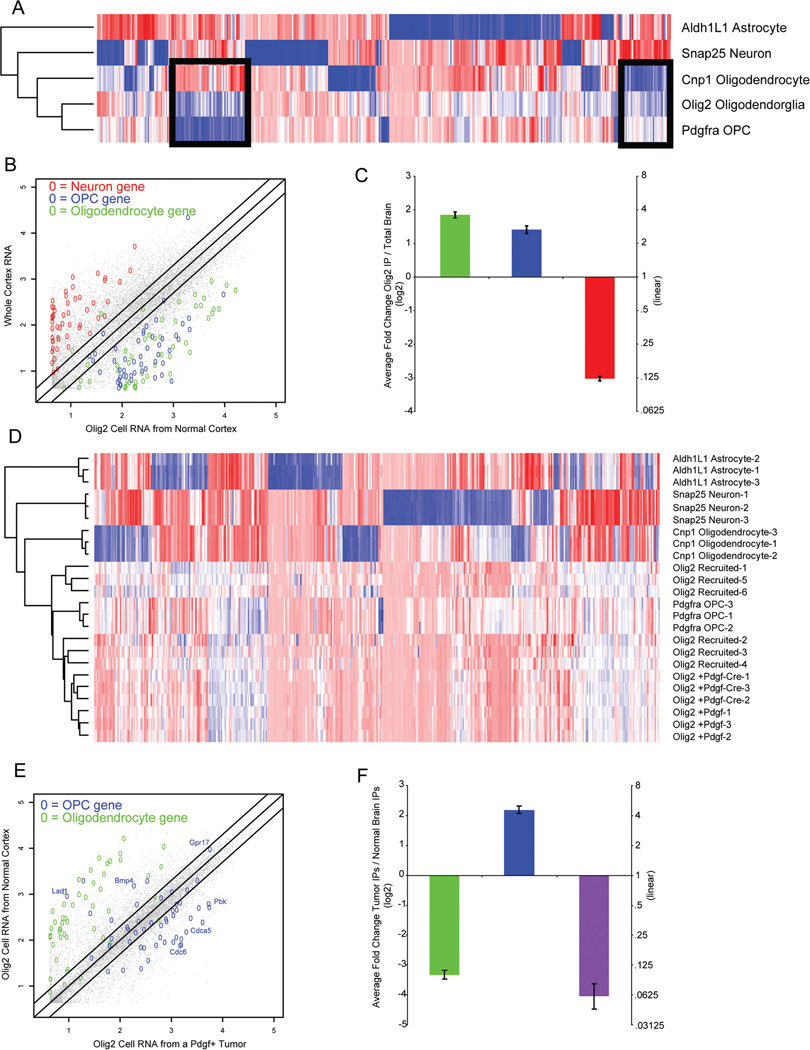
We next applied our tools to characterize the oligodendroglial lineage cells in the context of a mouse model of GBM. Murine PDGF-driven GBM show high expression levels of Olig2. In a normal brain, the transgenic line Olig2::Egfp-L10a labels all oligodendroglial cells: both the NG2-positive OPC, and Cnp1-positive myelinating oligodendrocytes (Figure 1A–C and (17)). Hierarchical clustering of TRAP microarray data positions the profile of these cells between that of mature oligodendrocytes and PDGFRA-positive OPCs (Figure 3A). Comparison of the Olig2::Egfp-L10a profile to whole cortex reveals that it is enriched in both the OPC and myelinating oligodendrocyte transcripts (Figure 3B), consistent with it covering the entire lineage.
Figure 3. Analysis of Olig2::Egfp-L10a translational profiles in normal brains and tumors identifies candidates for differentiation and quiescence of OPCs.
A: Hierarchical clustering of Olig2::Egfp-L10a bacTRAP data from normal cortex reveals a profile intermediate to oligodendrocytes and OPCs, consistent with transgene expression in both populations. B: Scatterplot of Olig2 bacTRAP microarray data (x-axis) compared to whole cortex microarray data (y-axis) for all transcripts. Olig2 shows clear enrichment of OPC (blue) and mature oligodendrocyte transcripts (green), and no enrichment of neuronal transcripts (red). C: Average ratios (log2 scale) between Olig2::Egfp-L10a translational profiles in normal brain and whole cortex RNA for all transcripts of mature oligodendrocytes (green), OPCs (blue), and neuronal (red) lists. D: Heatmap and hierarchical clustering of Olig2::Egfp-L10a translational profiles for three variations of the tumor model, PDGF+, PDGF-Cre, and Recruited, show most similarity to normal OPC cells, though there are transcripts that distinguish them from OPCs (arrow). E: Scatterplot of a representative Olig2::Egfp-L10a bacTRAP data in a PDGF+ tumor (x-axis) compared to normal brain y-axis), shows depletion of mature oligodendrocyte transcripts (green) and a clear enrichment of most OPC transcripts in tumors (blue), including those involved in cell cycle (eg. Pbk, Cdca5, Cdc6). However, there are a subset of OPC transcripts (eg. Lad1, Bmp4, and Gpr17), enriched in normal brain. These are designated as DorQ candidates. F: Average ratios (log2 scale) between Olig2::Egfp-L10a translational profiles in tumors and in normal brain for all transcripts of mature oligodendrocytes (green), OPCs (blue, excluding DorQ candidates), and DorQ candidate (purple) lists. All Error bars SEM.
First, we translationally profiled Ntv-a/ink4a/arf+/− Olig2::EgfpL10a cells in primary tumors from our model (PDGF-B tumors). Hierarchical clustering revealed that these cells are most similar to OPCs of the normal brain. To test this observation, we repeated the analysis in two additional variations of our tumor model: primary tumors from this model with additional deletion of Pten (PDGF-Cre tumors), and in tumors induced by transplanting EGFP-L10a negative tumor cells from PDGF-driven primary murine tumors into Ntv-a/ink4a/arf+/− Olig2::Egfp-L10a mice (Recruited). While this increases the variability amongst the tumor samples, all samples cluster most closely with OPC cells (Figure 3D). This provides systematic evidence that Olig2::Egfp-L10a cells are in a more OPC-like state in glioma.
The most parsimonious explanation of these findings is that the excess of PDGF-B ligand produced by the Nes-positive cell has driven the recruitment and expansion of the proliferating PDGFRA-positive cells, at the expense of quiescence or differentiation. To examine this, we plotted the translational profile of Olig2::Egfp-L10a cells in a normal mouse cortex versus Olig2::Egfp-L10a cells in tumors, and labeled those transcripts that are specific to either OPCs or mature oligodendrocytes. On average, the tumor contains a clear enrichment of transcripts specific to OPCs, and depletion of transcripts found in mature oligodendrocytes (Figure 3E). Not surprisingly, the subset of OPC transcripts identified by GO as involved in cell cycle/cell proliferation, such as Melk (28) and Pbk (29), are highly enriched in the tumor samples, consistent with the Olig2:Egfp-L10a population being in a highly proliferative state inside of the tumor. However, surprisingly, there is also a subset of OPC transcripts that seem enriched in the normal brain.
Identification of Differentiation or Quiescence Candidate transcripts in OPCs
In a normal brain, OPCs may proliferate, remain quiescent, or differentiate, but in the context of a tumor they seem to proliferate at the expense of differentiation or quiescence. Comparison of the transcriptional profiles of Olig2::Egfp-L10a cells from normal brain and tumor pointed to a subset of 28 transcripts translated greater than two fold higher in normal brain (Figure 3F, Table 1). We hypothesized that this subset contains those transcripts employed in either differentiation or quiescence in oligodendroglia.
Table 1. List of DorQ candidates.
OPC genes with expression > 2 fold higher in Olig2 cells from normal cortex than tumors. Includes average fold changes (linear scale), and LIMMA p-value. More detail can be found in Supplemental Table 3.
| Mouse Symbol | Entrez | Fold Change |
p value | w/FDR | Name |
|---|---|---|---|---|---|
| 1810041L15Rik | 72301 | 4.29 | 0.0004 | 0.0126 | RIKEN cDNA 1810041L15 gene |
| 2310031A18Rik | 69627 | 3.51 | 0.0120 | 0.1103 | RIKEN cDNA 2310031A18 gene |
| 9030409G11Rik | 71529 | 2.26 | 0.0013 | 0.0278 | RIKEN cDNA 9030409G11 gene |
| 9630013A20Rik | 319903 | 20.05 | 0.0156 | 0.1305 | RIKEN cDNA 9630013A20 gene |
| BC057371 | 194237 | 5.03 | 0.0003 | 0.0098 | |
| Bmp4 | 12159 | 9.96 | 0.0002 | 0.0077 | bone morphogenetic protein 4 |
| C1ql1 | 23829 | 3.69 | 0.1154 | 0.4434 | complement component 1, q subcomponent -like 1 |
| C1qtnf2 | 69183 | 3.51 | 0.0043 | 0.0581 | C1q and tumor necrosis factor related protein 2 |
| Calb2 | 12308 | 4.32 | 0.0015 | 0.0305 | calbindin 2 |
| Caskin2 | 140721 | 2.91 | 0.0001 | 0.0061 | CASK-interacting protein 2 |
| Cdkn1c | 12577 | 2.78 | 0.0588 | 0.2918 | cyclin -dependent kinase inhibitor 1C (P57) |
| Dct | 13190 | 3.48 | 0.0425 | 0.2401 | dopachrome tautomerase |
| Gpr17 | 574402 | 2.80 | 0.0283 | 0.1888 | G protein -copled receptor 17 |
| Kank1 | 107351 | 2.02 | 0.0035 | 0.0508 | KN motif and ankyrin repeat domains 1 |
| Kif26a | 668303 | 2.66 | 0.0566 | 0.2854 | kinesin family member 26A |
| Lad1 | 16763 | 43.22 | 0.0002 | 0.0097 | ladinin |
| Lims2 | 225341 | 3.30 | 0.0016 | 0.0309 | LIM and senescent cell antigen like domains 2 |
| Npm3-ps1 | 108176 | 3.35 | 0.0965 | 0.3969 | Pseudogene |
| Rab32 | 67844 | 3.32 | 0.0111 | 0.1044 | RAB32, member RAS oncogene family |
| Rasl12 | 70784 | 3.33 | 0.0014 | 0.0286 | RAS-like, family 12 |
| Rassf10 | 78748 | 7.10 | 0.0055 | 0.0664 | Ras association domain family member 10 |
| Rlbp1 | 19771 | 2.29 | 0.0160 | 0.1331 | retinaldehyde binding protein 1 |
| Rnf43 | 207742 | 2.19 | 0.0008 | 0.0194 | ring finger protein 43 |
| Rxrg | 20183 | 2.71 | 0.0158 | 0.1318 | retinoid X receptor gamma |
| Sema5a | 20356 | 2.11 | 0.0993 | 0.4039 | semaphorin 5A |
| Snx22 | 382083 | 2.17 | 0.0932 | 0.3873 | sorting nexin 22 |
| Susd5 | 382111 | 3.59 | 0.2071 | 0.6356 | sushi domain containing 5 |
| Ttr | 22139 | 8.69 | 0.0664 | 0.3124 | transthyretin |
| Ust | 338362 | 2.84 | 0.0621 | 0.3019 | uronyl -2-sulfotransferase |
Careful examination of current literature supports this hypothesis. For example Cdkn1c is required for the regulation of cell cycle and specification of OPCs (30), while Sema5a, an oligodendrocyte specific semaphorin, plays a role in axon growth inhibition (31). G-coupled protein receptor Gpr17 has the role in myelination during development (20). Other DorQ candidate transcripts are known to be expressed in oligodendroglia although their role is unclear. They include Rlbp (32) and C1qtnf2, whose mRNA expression pattern is consistent in part with the oligodendrocyte expression (33). Additional screening of the expression patterns of DorQ candidate transcripts in Allen Brain Atlas and Gene Expression Nervous System Atlas (GENSAT) confirmed that Gpr17, Bmp4, Rlbp1, Rnf43, 90304409g11Rik, Rxrg, Caskin2, Rasl12, 2310031a18Rik expression patterns are consistent with OPC localization. The presence of these transcripts in OPCs suggests that the DorQ candidates may have some relevance as complementary targets for treatment, if they are conserved in humans.
Differentiation and Quiescence Candidate transcripts show similar expression pattern in human glioma
As human ‘proneural’ tumors are characterized by an abundance of PDGF-B and activation of PDGFRA signaling pathways, we sought to determine if they contained a preponderance of OPC transcripts. The results, summarized in Supplemental Table 2, Supplemental Figure 2 and Supplemental Figure 4, showed that human ‘proneural’ specific gene list has a highly significant enrichment of OPC transcripts. We performed hierarchical clustering of the human gliomas using the OPC transcripts (excluding those categorized by GO as involved in cell cycle or DNA replication as enhanced cell cycle is common to all glioblastoma subtypes). Using just these transcripts, most of the ‘proneural’ tumors clustered into a single large branch, suggesting these OPC transcripts are sufficient to distinguish the ‘proneural’ subtype (Supplemental Figure 4B). On average, the non-cell cycle OPC transcripts are expressed at a higher relative level in ‘proneural’ tumors (Supplemental Figure 4C), though they are present at detectable levels in all subtypes. This suggests that part of what distinguishes the ‘proneural’ subtype from the other types may be the relative abundance of OPC-like cells.
While the analysis above demonstrated that OPC transcripts in general have potential relevance to human glioma, we next examined the DorQ candidate subset of OPC transcripts specifically. We first analyzed the human TCGA dataset to determine if these transcripts demonstrate the same pattern of expression that is present in the mouse tumors. Since the DorQ candidates were identified by using translational profiling data specifically for Olig2::Egfp-L10a cells, which is unavailable in the human samples, to generate an analysis analogous to human samples we examined the expression of the DorQ candidates in total RNA profiles from mouse gliomas and normal mice (Supplemental Figure 5A). With this method all OPC transcripts, including the DorQ candidates, are somewhat enriched in tumors compared to normal brain. However, the DorQ candidate transcripts are significantly less increased in tumors than the other OPC transcripts are (Supplemental Figure 5D, T-test p < 1E-6.). We repeated this same analysis in human ‘proneural’ tumors compared to normal human cortex, using the human homologues of these DorQ transcripts (Supplemental Figure 5B). We find that, as with the mouse, the DorQ candidate transcripts are enriched, but significantly less so than the other OPC transcripts (p < .05) (Supplemental Figure 5D). Overall, the log2 fold change values of glioma/normal brain for all OPC transcripts is correlated at .62 (Pearsons correlation) across mouse and human comparisons, suggesting that the general pattern of expression of these pathways is conserved across mouse and human glioma (Supplemental Figure 5C).
Differentiation and Quiescence Candidates are frequently deleted in human glioma
We next sought to determine if the DorQ candidates were mutated in human glioma. Common source of genetic variation in the cancer genome are copy number variations (CNVs), often resulting in deletions of tumor suppressor genes, and amplifications of particular oncogenes (34). However, there are also a large number of apparently neutral CNVs that occur in glioma. Also, even if a certain gene is very commonly deleted in glioma, it does not necessarily implicate that gene in glioma biology; it could merely be adjacent to an important tumor suppressor. Nonetheless, if we consider these transcripts as a group, then the DorQ transcripts, particularly those that have a role in quiescence, or autocrine pro-differentiation pathways, should have a higher prevalence of deletions compared to duplications.
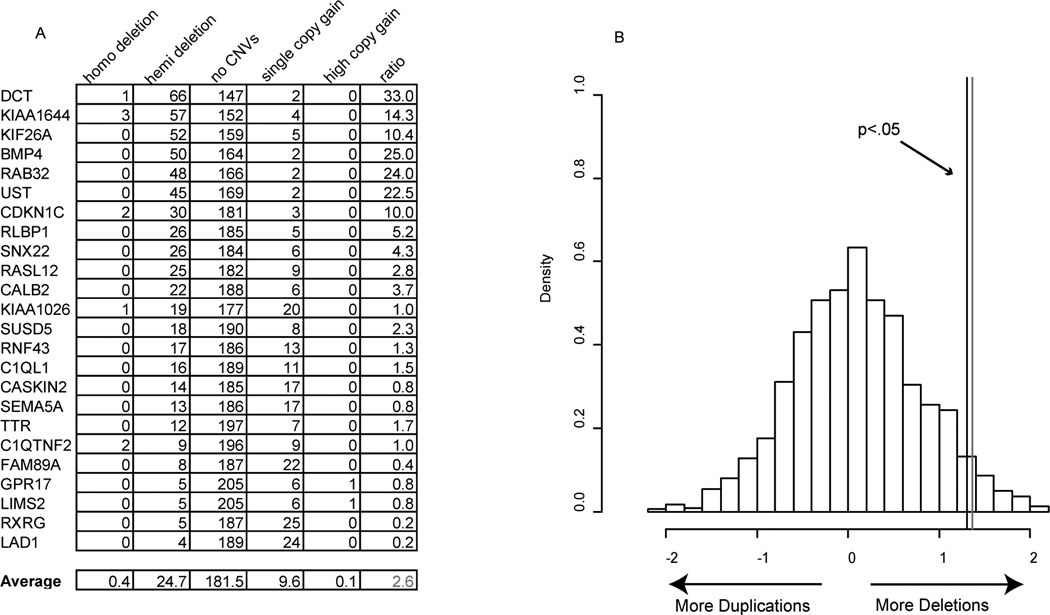
Thus we examined the CNVs for the DorQ candidates using the RAE algorithm for the set of human gliomas analyzed here (Figure 4A) (1, 35). Half of the DorQ genes showed a preponderance of deletions relative to duplications and six were deleted in nearly one quarter of GBM samples. On average, the 24 transcripts showed a 2.6 to 1 preponderance of deletions to duplications in human glioma, a result unlikely to be due to chance (p<.05, permutation testing, Figure 4B), and consistent with the role of some of the DorQ candidates as tumor suppressors in human glioma through promotion of quiescence or differentiation.
Figure 4. Differentiation or Quiescence candidates have a preponderance of deletions in human glioma.
A: Output of RAE analysis on 204 human glioma samples analyzed for the 24 DorQ candidates with clear human homologues. 12/24 of the genes have at least two times more deletions than duplications, and 6/24 are deleted in more than a quarter of the GBM samples. On average, these genes have a ratio of single deletions to duplications of 2.6 (black line). B: Distribution of deletion/duplication ratios (in log2) for 1500 random sets of 24 human genes from the same analysis reveals a ratio of 2.6 (black line) occurs in less than 5% of permuted cases (gray line).
Agonists of DorQ candidate Gpr17 can decrease clonogenicity of glioblastoma cells in vitro
Previously, others have tested perturbations of Bmp4 (36) and Rxrg (37) signaling for treatment of glioma and neuroblastoma, respectively. We sought to determine if any of the other DorQ candidates identified here might also drive the glioma OPCs towards differentiation or quiescence. We decided to first test Gpr17 for four reasons. First, receptors make attractive targets for eventual treatment strategies, and in this case ligands have been recently characterized: Gpr17 is a G-Protein coupled receptor that responds to both the uracil nucleotides and cysteinyl-leukotrienes (38). Second, our survey of translational profiles across a variety of neuronal and glial cell types suggested that Gpr17 is restricted to cells of the oligodendroglial lineage in the mature CNS (17), consistent with the Allen Brain Atlas, and Gpr17 knock in mice (20), suggesting treatments targeting this receptor will have few off target effects, at least in the brain. Third, there is reasonably strong support for Gpr17 regulating the differentiation in oligodendrocytes, though there are unresolved differences in the direction of this regulation between the genetic and pharmacological studies (20, 39). Finally, Gpr17 only rarely shows deletions in the human GBM samples (Figure 4A), suggesting that this pathway may be intact but inactive in the tumor environment.
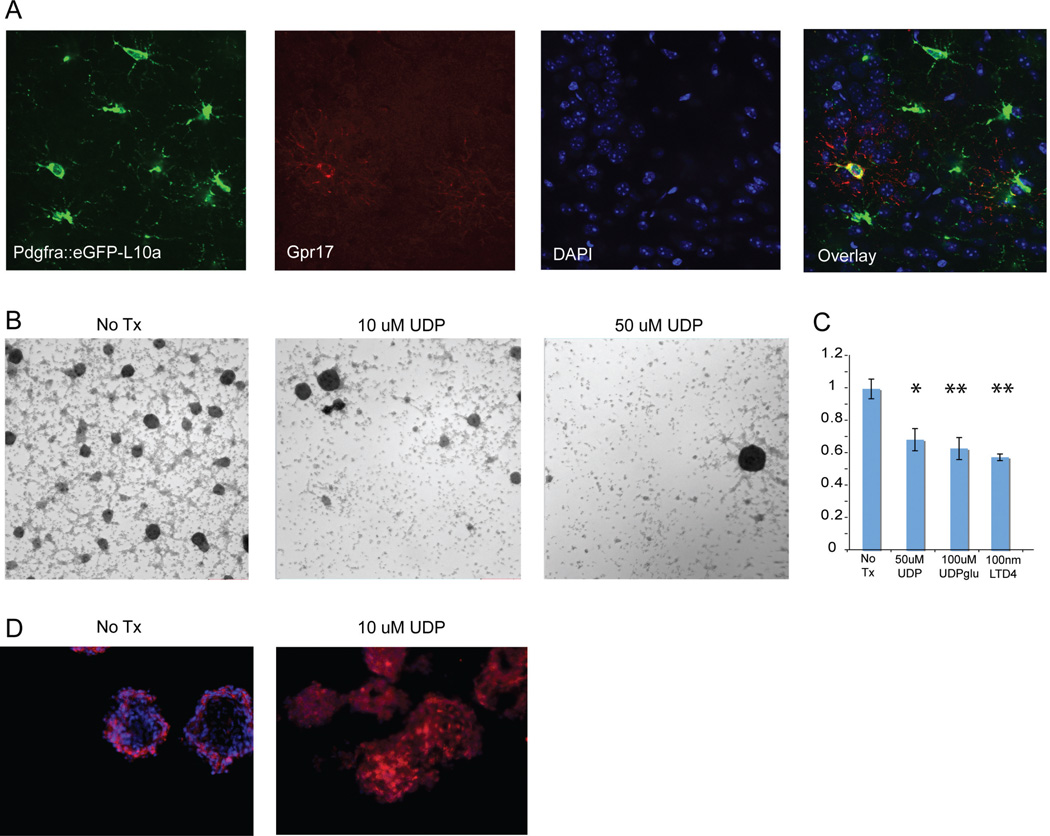
First, to confirm that Gpr17 has a pattern consistent with a DorQ candidate, we examined the Gpr17 protein expression in the cortex. DorQ candidates should overlap with at least a subset of Pdgfra cells. Figure 5A shows that a subset (11.5%, n =148) of Pdgfra::eGFP-L10a expressed GPR17 protein. Next, to address the effects of Gpr17 activation on glioma cells, we counted formation of primary neurospheres grown from fresh murine glioma cells in EGF/bFGF containing media, with or without the Gpr17 agonist UDP (Figure 5B). At 2–3 days after plating, the number of cells in treated and untreated cultures was similar, indicating that UDP treatment did not have apparent toxicity at the concentrations used. However, treatment with 10uM or 50uM UDP resulted in reduced formation of glioma spheres, suggesting that activation of Gpr17 can affect both the proliferation and self-renewal of murine glioma cells (Figure 5B, C). We further expanded our findings to include the effects of additional agonists of both classes for Gpr17; UDP-glucose (100uM) and leukotriene D4 (LTD4) (100 nM) on primary glioma sphere formation (Figure 5B, C). Although the magnitude of the effects of these agents were variable across tumors, with a decrease ranging anywhere from 15 to 60% in primary neurosphere formation, each of the above compounds had a significant effect, consistent with activation of Gpr17 decreasing the proliferation and/or changing the potential of the sphere forming (stem cell-like) glioma cells. In contrast, neither UDP nor LTD4 had significant effects on the number of spheres formed from wild type mouse cortices, when they were passaged into agonist containing media (data not shown).
Figure 5. GPR17 agonists changed cellular properties and decreased clonogenicity in primary glioma cells.
A: Confocal immunofluorescence for EGFP-L10a and Gpr17 confirms localization in a subset of OPCs. B: Phase contrast images of primary murine glioma spheres treated with 0uM, 10uM or 50uM UDP. UDP treatment reduced sphere formation C: Counts of primary neurosphere formation is consistently decreased by Gpr17 agonists UDP, UDP-glu, LTD4. *p<.05, ** p<.01, paired T-Test. D: Olig2 staining of untreated or 10uM UDP-treated sectioned murine glioma neurospheres. Red – Olig2, Blue - DAPI.
Intriguingly, exposure to UDP also results in a dramatic increase in the relative number of Olig2 expressing cells in neurospheres (red, Figure 5D). This suggests that activation of Gpr17 favored the selective survival and/or expansion of Olig2 cells in vitro, and altered the overall self-renewal and proliferation capacity of the glioma cells.
Discussion
Human gliomas show remarkable heterogeneity in cell composition, activated pathways and gene copy number variations. This heterogeneity poses an enormous challenge in finding effective treatments for this highly fatal tumor. An increasing number of mouse models help elucidate the basic processes of tumor initiation, growth and maintenance, as well as serve as preclinical trials for selective glioma treatments (40). In particular, there is a previously proposed diagnostic concept of ‘glioblastoma multiforme with oligodendroglial component’ (41), for the subset of GBM displaying some oligodendroglial or oligodendroglioma-like component by histology. The most likely transcriptomic correspondent to this histological definition is the ‘proneural’ GBM; in addition to the shared cellular features, both have been shown to be more responsive to treatment then GBM in general (1, 2, 42). The features of the mouse model used in this study are most similar to those of this human ‘proneural’ type of glioma, such as overexpression of PDGF-B, loss of ink4a/arf/pten tumor suppressor genes, and it displays a similar histological phenotype to human tumors (12). In this model, the tumor-initiating cell is a PDGF-B producing Nestin-positive stem cell that can recruit OPCs into the tumor environment, OPCs which can then develop sufficient mutations to be independently tumorgenic (12). A different mouse model has shown that OPC cells can also generate a tumor by the cell autonomous expression of a constitutively active Egf receptor (43), though TCGA data suggest that human proneural tumors are not generally characterized by increased Egf signaling (1, 2). Nonetheless, this does suggest that both cell autonomous and non-cell autonomous mechanisms are capable of initiating tumors derived largely of OPC like cells, which may respond to pro-differentiation OPC signals.
Here we showed that translational profile of Olig2-positive cells in our model closely resembles the profile of OPC cells, and that OPC transcripts are sufficient to distinguish the human ‘proneural’ glioma subtype. This confirmed the results from a study that used similar glioblastoma model with the pten/p53 deletion (44). Our data also confirmed the expression of stem and progenitor cell markers, as well as high expression of genes involved in cell cycle, in Olig2-positive cells in glioblastoma, consistent with numerous studies of human tumors and mouse models (25, 45–50). Our study went a step further generating the list of candidate transcripts, deleted often in human gliomas that may serve to promote the differentiation or quiescence of Olig2 cells in the tumor. Of course, future work will need to be done to establish which of these candidates are bona fide pro-differentiation or quiescence factors.
Gene expression and transcriptome analysis could provide valuable information about potential cell-specific therapy targets, and the expression profile of glioma cells has been extensively studied (reviewed in (51)). However, these analyses were performed either on glioma cell lines or using the total mRNA from the tumor mass. The pitfall of this approach is that glioma cell lines do not possess the physiological characteristics of the tumor cells in vivo and analysis based on the total mRNA extracted from the tumor does not reflect the differences in expression profile of specific cell subpopulations. In addition, this approach does not identify the actively translated pool of mRNAs. It has been shown that translational regulation plays an important role in tumorigenesis (52, 53). Thus, this study is the first that used the bacTRAP methodology to analyze the actively translated mRNAs in a cell specific manner within the tumor.
The prevalence of hemideletions for DorQ genes in many human glioma samples suggested that they might serve as targets for glioma therapy. Bone morphogenetic factor 4 (Bmp4) and four other DorQ genes (Kank1, Lims2, Rab32, and Rassf10) are frequently epigenetically silenced or deleted in other cancer types, and/or are suggested to function as tumor suppressors (54–57). Several DorQ candidates were already tested either in vivo or in vitro (36, 58, 59). When we tested the activation of Gpr17 signaling in the murine GBM cells in vitro our results suggested that Gpr17 agonists may drive the differentiation of highly proliferative, uncommitted tumor cells towards oligodendoglial fates. Similarly, Bmp4 has been tested in animal model of glioma and human glioblastoma cells (36) as well as in meduloblastoma (60). Cells isolated from human glioblastomas treated with Bmp4 showed decreased proliferation and increased differentiation in vitro, while in vivo treatment with Bmp4 blocked the tumor growth. Finally, agonists of the retinoid receptor gamma Rxrg promote differentiation in human neuroblastoma (37) and glioblastoma (58) cell lines and have already shown some efficacy as part of combination treatments for thyroid cancer (59). In addition, many of the candidate DorQ transcripts have been detected in other types of tumors (54–57, 60–66), suggesting these candidates may have some relevance beyond GBM.
Human glioma tumors contain an abundance of cells expressing markers of glial progenitors and stem cells, suggesting that tumorigenesis involves expansion of a glial progenitor-like cell (67). The comparison of the translational profiles of tumors to OPCs and normal mature cells of the neuronal, oligodendroglial and astrocyte lineage supports this hypothesis. The analysis confirmed the similarities of tumor cells with the immature PDGFRA-positive cells from the normal brain. It was also suggested that OPCs might serve as source of gliomas (43, 44, 67–69). While this study could not provide the information about cell-of-origin for gliomas, the approach would be useful if normal immature glial and stem-like cell profiles are compared with glioma cells at different time points during tumorigenesis. The mouse line generated for this study (Pdgfra::Egfp-L10a) will provide reproducible access to OPCs for future translational profiling. However, beyond applications to glioma, this line should permit reproducible in vivo access for researchers interested in studies of the basic biology of these cells both in the normal mouse brain, and experimental models of OPC-relevant diseases such as multiple sclerosis or white matter stroke.
Supplementary Material
Acknowledgments
We thank Victoria Isakova and Daviana Martinez Osorio for their technical assistance, Elizabeth Griggs for image processing, and N. Ramanan and members of the N. Heintz laboratory for their advice and support. We also thank the RU Genomics Resource Center and the MSKCC Flow Cytometry Resource Center.
This work was supported by the Tri-Institutional Stem Cell Initiative award to A. Milosevic and E.C. Holland, the NIH (4R00NS067239-03 to J. Dougherty) and the Adelson Medical Research Foundation.
Footnotes
Authors declare no conflict of interest.
References
- 1.TCGARN. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishiyama A, Chang A, Trapp BD. NG2+ glial cells: a novel glial cell population in the adult brain. J Neuropathol Exp Neurol. 1999;58:1113–1124. doi: 10.1097/00005072-199911000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Dawson MR, Levine JM, Reynolds R. NG2-expressing cells in the central nervous system: are they oligodendroglial progenitors? J Neurosci Res. 2000;61:471–479. doi: 10.1002/1097-4547(20000901)61:5<471::AID-JNR1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Nishiyama A, Watanabe M, Yang Z, Bu J. Identity, distribution, and development of polydendrocytes: NG2-expressing glial cells. J Neurocytol. 2002;31:437–455. doi: 10.1023/a:1025783412651. [DOI] [PubMed] [Google Scholar]
- 9.Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, Holland EC. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res. 2004;64:4783–4789. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- 10.Uhrbom L, Dai C, Celestino JC, Rosenblum MK, Fuller GN, Holland EC. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 2002;62:5551–5558. [PubMed] [Google Scholar]
- 11.Tallquist M, Kazlauskas A. PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 2004;15:205–213. doi: 10.1016/j.cytogfr.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Fomchenko EI, Dougherty JD, Helmy KY, Katz AM, Pietras A, Brennan C, et al. Recruited cells can become transformed and overtake PDGF-induced murine gliomas in vivo during tumor progression. PLoS One. 2011;6:e20605. doi: 10.1371/journal.pone.0020605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26:6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 16.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 17.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleau AM, Howard BM, Taylor LA, Gursel D, Greenfield JP, Lim Tung HY, et al. New strategy for the analysis of phenotypic marker antigens in brain tumor-derived neurospheres in mice and humans. Neurosurg Focus. 2008;24:E28. doi: 10.3171/FOC/2008/24/3-4/E27. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Wu H, Wang S, Koito H, Li J, Ye F, et al. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat Neurosci. 2009;12:1398–1406. doi: 10.1038/nn.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Research. 2010;38:4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 24.Charles N, Ozawa T, Squatrito M, Bleau AM, Brennan CW, Hambardzumyan D, et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6:141–152. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 26.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profilling data for the identification of genes enriched in specific cells. 2010 doi: 10.1093/nar/gkq130. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano I, Paucar AA, Bajpai R, Dougherty JD, Zewail A, Kelly TK, et al. Maternal embryonic leucine zipper kinase (MELK) regulates multipotent neural progenitor proliferation. J Cell Biol. 2005;170:413–427. doi: 10.1083/jcb.200412115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dougherty JD, Garcia AD, Nakano I, Livingstone M, Norris B, Polakiewicz R, et al. PBK/TOPK, a proliferating neural progenitor-specific mitogen-activated protein kinase kinase. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:10773–10785. doi: 10.1523/JNEUROSCI.3207-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dugas JC, Ibrahim A, Barres BA. A crucial role for p57 (Kip2) in the intracellular timer that controls oligodendrocyte differentiation. J Neurosci. 2007;27:6185–6196. doi: 10.1523/JNEUROSCI.0628-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldberg JL, Vargas ME, Wang JT, Mandemakers W, Oster SF, Sretavan DW, et al. An oligodendrocyte lineage-specific semaphorin, Sema5A, inhibits axon growth by retinal ganglion cells. J Neurosci. 2004;24:4989–4999. doi: 10.1523/JNEUROSCI.4390-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saari JC, Huang J, Possin DE, Fariss RN, Leonard J, Garwin GG, et al. Cellular retinaldehyde-binding protein is expressed by oligodendrocytes in optic nerve and brain. Glia. 1997;21:259–268. [PubMed] [Google Scholar]
- 33.Iijima T, Miura E, Watanabe M, Yuzaki M. Distinct expression of C1q-like family mRNAs in mouse brain and biochemical characterization of their encoded proteins. Eur J Neurosci. 2010;31:1606–1615. doi: 10.1111/j.1460-9568.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 34.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor BS, Barretina J, Socci ND, Decarolis P, Ladanyi M, Meyerson M, et al. Functional copy-number alterations in cancer. PLoS One. 2008;3:e3179. doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 37.Das A, Banik NL, Ray SK. Retinoids induce differentiation and downregulate telomerase activity and N-Myc to increase sensitivity to flavonoids for apoptosis in human malignant neuroblastoma SH-SY5Y cells. Int J Oncol. 2009;34:757–765. doi: 10.3892/ijo_00000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciana P, Fumagalli M, Trincavelli ML, Verderio C, Rosa P, Lecca D, et al. The orphan receptor GPR17 identified as a new dual uracil nucleotides/cysteinyl-leukotrienes receptor. Embo J. 2006;25:4615–4627. doi: 10.1038/sj.emboj.7601341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecca D, Trincavelli ML, Gelosa P, Sironi L, Ciana P, Fumagalli M, et al. The recently identified P2Y-like receptor GPR17 is a sensor of brain damage and a new target for brain repair. PLoS One. 2008;3:e3579. doi: 10.1371/journal.pone.0003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huse JT, Holland EC. Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol. 2009;19:132–143. doi: 10.1111/j.1750-3639.2008.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuller GN, Scheithauer BW. The 2007 Revised World Health Organization (WHO) Classification of Tumours of the Central Nervous System: newly codified entities. Brain Pathol. 2007;17:304–307. doi: 10.1111/j.1750-3639.2007.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vordermark D, Ruprecht K, Rieckmann P, Roggendorf W, Vince GH, Warmuth-Metz M, et al. Glioblastoma multiforme with oligodendroglial component (GBMO): favorable outcome after post-operative radiotherapy and chemotherapy with nimustine (ACNU) and teniposide (VM26) BMC Cancer. 2006;6:247. doi: 10.1186/1471-2407-6-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson AI, Petritsch C, Swartling FJ, Itsara M, Sim FJ, Auvergne R, et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lei L, Sonabend AM, Guarnieri P, Soderquist C, Ludwig T, Rosenfeld S, et al. Glioblastoma Models Reveal the Connection between Adult Glial Progenitors and the Proneural Phenotype. PLoS One. 2011;6:e20041. doi: 10.1371/journal.pone.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu QR, Park JK, Noll E, Chan JA, Alberta J, Yuk D, et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci U S A. 2001;98:10851–10856. doi: 10.1073/pnas.181340798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhee W, Ray S, Yokoo H, Hoane ME, Lee CC, Mikheev AM, et al. Quantitative analysis of mitotic Olig2 cells in adult human brain and gliomas: implications for glioma histogenesis and biology. Glia. 2009;57:510–523. doi: 10.1002/glia.20780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellis JA, Castelli M, Bruce JN, Canoll P, Ogden AT. Retroviral delivery of platelet-derived growth factor to spinal cord progenitor cells drives the formation of intramedullary gliomas. Neurosurgery. 2012;70:198–204. doi: 10.1227/NEU.0b013e31822ce963. discussion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivkovic S, Canoll P, Goldman JE. Constitutive EGFR signaling in oligodendrocyte progenitors leads to diffuse hyperplasia in postnatal white matter. J Neurosci. 2008;28:914–922. doi: 10.1523/JNEUROSCI.4327-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mischel PS, Cloughesy TF, Nelson SF. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat Rev Neurosci. 2004;5:782–792. doi: 10.1038/nrn1518. [DOI] [PubMed] [Google Scholar]
- 52.Bilanges B, Stokoe D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene. 2007;26:5973–5990. doi: 10.1038/sj.onc.1210431. [DOI] [PubMed] [Google Scholar]
- 53.Parsa AT, Holland EC. Cooperative translational control of gene expression by Ras and Akt in cancer. Trends Mol Med. 2004;10:607–613. doi: 10.1016/j.molmed.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, Chiaramonte R, et al. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42. doi: 10.1186/1476-4598-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shibata D, Mori Y, Cai K, Zhang L, Yin J, Elahi A, et al. RAB32 hypermethylation and microsatellite instability in gastric and endometrial adenocarcinomas. Int J Cancer. 2006;119:801–806. doi: 10.1002/ijc.21912. [DOI] [PubMed] [Google Scholar]
- 56.Kim SK, Jang HR, Kim JH, Noh SM, Song KS, Kim MR, et al. The epigenetic silencing of LIMS2 in gastric cancer and its inhibitory effect on cell migration. Biochem Biophys Res Commun. 2006;349:1032–1040. doi: 10.1016/j.bbrc.2006.08.128. [DOI] [PubMed] [Google Scholar]
- 57.Sarkar S, Roy BC, Hatano N, Aoyagi T, Gohji K, Kiyama R. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J Biol Chem. 2002;277:36585–36591. doi: 10.1074/jbc.M204244200. [DOI] [PubMed] [Google Scholar]
- 58.Das A, Banik NL, Ray SK. Retinoids induced astrocytic differentiation with down regulation of telomerase activity and enhanced sensitivity to taxol for apoptosis in human glioblastoma T98G and U87MG cells. J Neurooncol. 2008;87:9–22. doi: 10.1007/s11060-007-9485-1. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Jia S, Liu Y, Li B, Wang Z, Lu H, et al. A clinical study of all-trans-retinoid-induced differentiation therapy of advanced thyroid cancer. Nucl Med Commun. 2007;28:251–255. doi: 10.1097/MNM.0b013e3280708ebf. [DOI] [PubMed] [Google Scholar]
- 60.Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008;22:722–727. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lubbe SJ, Pittman AM, Matijssen C, Twiss P, Olver B, Lloyd A, et al. Evaluation of germline BMP4 mutation as a cause of colorectal cancer. Hum Mutat. 2010 doi: 10.1002/humu.21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci. 2010;67:3535–3548. doi: 10.1007/s00018-010-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyamoto K, Sakurai H, Sugiura T. Proteomic identification of a PSF/p54nrb heterodimer as RNF43 oncoprotein-interacting proteins. Proteomics. 2008;8:2907–2910. doi: 10.1002/pmic.200800083. [DOI] [PubMed] [Google Scholar]
- 64.Sendoel A, Kohler I, Fellmann C, Lowe SW, Hengartner MO. HIF-1 antagonizes p53-mediated apoptosis through a secreted neuronal tyrosinase. Nature. 2010;465:577–583. doi: 10.1038/nature09141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vicent S, Luis-Ravelo D, Anton I, Garcia-Tunon I, Borras-Cuesta F, Dotor J, et al. A novel lung cancer signature mediates metastatic bone colonization by a dual mechanism. Cancer Res. 2008;68:2275–2285. doi: 10.1158/0008-5472.CAN-07-6493. [DOI] [PubMed] [Google Scholar]
- 66.Aalto Y, El-Rifa W, Vilpo L, Ollila J, Nagy B, Vihinen M, et al. Distinct gene expression profiling in chronic lymphocytic leukemia with 11q23 deletion. Leukemia. 2001;15:1721–1728. doi: 10.1038/sj.leu.2402282. [DOI] [PubMed] [Google Scholar]
- 67.Fomchenko EI, Holland EC. Stem cells and brain cancer. Exp Cell Res. 2005;306:323–329. doi: 10.1016/j.yexcr.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 68.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 69.Lindberg N, Kastemar M, Olofsson T, Smits A, Uhrbom L. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene. 2009;28:2266–2275. doi: 10.1038/onc.2009.76. [DOI] [PubMed] [Google Scholar]
- 70. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.