Abstract
Fossorial salamanders typically have elongate and attenuated heads and bodies, diminutive limbs, hands and feet, and extremely elongate tails. Batrachoseps from California, Lineatriton from eastern México, and Oedipina from southern México to Ecuador, all members of the family Plethodontidae, tribe Bolitoglossini, resemble one another in external morphology, which has evolved independently. Whereas Oedipina and Batrachoseps are elongate because there are more trunk vertebrae, a widespread homoplasy (parallelism) in salamanders, the genus Lineatriton is unique in having evolved convergently by an alternate “giraffe-neck” developmental program. Lineatriton has the same number of trunk vertebrae as related, nonelongated taxa, but individual trunk vertebrae are elongated. A robust phylogenetic hypothesis, based on sequences of three mtDNA genes, finds Lineatriton to be deeply nested within a clade characterized by generalized ecology and morphology. Lineatriton lineolus, the only currently recognized taxon in the genus, shows unanticipated genetic diversity. Surprisingly, geographically separated populations of L. lineolus are not monophyletic, but are sister taxa of different species of the morphologically generalized genus Pseudoeurycea. Lineatriton, long thought to be a unique monospecific lineage, is polyphyletic. Accordingly, the specialized morphology of Lineatriton displays homoplasy at two hierarchical levels: (i) with respect to other elongate lineages in the family (convergence), and (ii) within what is currently recognized as a single taxon (parallelism). These evolutionary events are of adaptive significance because to invade the lowland tropics salamanders must be either arboreal or fossorial; the repeated evolution of elongation and attenuation has led to multiple lowland invasions.
Tropical plethodontid salamanders form a monophyletic clade of more than 200 species that display diverse body forms, including extreme elongation accompanied by limb reduction associated with fossorial habits (1). All lowland tropical plethodontids are either fossorial or arboreal, and fossoriality has been an important factor in the invasion of lowlands by these more typically cool-adapted upland animals. The fossorial Oedipina has many species that range south and east of the Isthmus of Tehuantepec, from southern México into northwestern South America; these differ from all other tropical plethodontids in having 18–22 rather than an invariable 14 trunk vertebrae. The other fossorial lineage, the equally attenuate and elongate Lineatriton lineolus, is found only in restricted parts of eastern México, north of the Isthmus of Tehuantepec. Oedipina evolved elongation from a more generalized morphology by increasing the number of body segments, and hence vertebrae, a developmental mechanism that has evolved repeatedly within salamanders as a whole, and even within the clade (Bolitoglossini) to which the tropical species belong. Lineatriton, in contrast, is unique among all salamanders in having evolved an entirely different developmental mechanism. Its trunk vertebrae number only 14, the same as in all other tropical salamanders except for Oedipina, but the individual vertebrae are narrow and elongate. As a consequence, the external form of L. lineolus and its adaptation for fossorial existence are so similar to those of species of Oedipina that it was included in the genus Oedipina until 1950 (2).
We analyzed sequences of mtDNA of many tropical bolitoglossines, including all recognized genera, and determined that Lineatriton and Oedipina are much more closely related to other taxa than to each other (3, 4). Not only was Lineatriton deeply nested within the large, mainly Mexican genus Pseudoeurycea, but populations of Lineatriton from different parts of its geographic range were more closely related to different species of Pseudoeurycea than to each other. Here we analyze molecular data for 1,816 bp of mtDNA derived from three genes, reject the monophyly of Lineatriton, and support an extraordinary case of homoplasy in a putative species that previously has been considered to be extremely specialized, and unique, in both morphology and ecology.
Materials and Methods
We collected specimens of all populations reported herein and preserved either frozen tissues or placed small pieces of tail tissue in 95% ethanol. The in-group includes eight samples of L. lineolus from central western Veracruz, México, and four samples from the Los Tuxtlas area of coastal southeastern Veracruz. The first of sequential out-groups includes samples of 15 species of Pseudoeurycea from México. More distant out-groups were selected from other members (Batrachoseps and Thorius) of the plethodontid tribe Bolitoglossini. Because of the surprising results, several populations of the in-group and Pseudoeurycea were collected on repeated field trips and sequenced independently. DNA was extracted following standard techniques (4). We obtained 544 bp of the large 16S subunit ribosomal mtDNA (5), 569 bp of the cytochrome b gene (6), and 709 bp of the ND4 gene (7). Amplification was done via the PCR (8) using the primers 16Sar and 16Sbr (9) for 16S, MVZ15 and MVZ18 (10) for cytochrome b, and ND4 (7) and MVZ112 (3) for the ND4 gene. Following standard techniques (4), cycle-sequencing products were purified by using ethanol precipitation and separated by electrophoresis on a 6% polyacrylamide gel using an ABI 377 DNA sequencer (Applied Biosystems). Sequences were read from both strands and aligned to each other by eye in the program sequence navigator version 1.0.1 (Applied Biosystems). Sequence divergences were estimated by using the Kimura two-parameter distance (11). Phylogenetic analyses were performed by using PAUP 4.0b2a (D. Swofford, Smithsonian Institution, Washington, DC). Parsimony analysis used heuristic searches by stepwise random addition of taxa, with 20 replications and tree bisection and reconnection branch swapping with the mulpars option in effect. Zero-length branches were collapsed. Both the consistency index (12), and retention index (13) were calculated. We used two character weighting schemes: equal weighting for all codon positions, and differential weighting in which we downweighted third codon position transitions to a ratio of 1:0.16 transversion/transition. The transversion to transition ratio was estimated by maximum likelihood analysis. Neighbor-joining reconstructions (14) used Kimura two-parameter distances. Maximum likelihood analysis used the heuristic algorithm, empirical base frequencies, and the Hasegawa-Kishino-Yano (15) model of character evolution. Decay indices and bootstrap values in excess of 50% are reported (1,000 pseudoreplicates for parsimony analysis with heuristic search and simple addition of taxa).
Morphological comparisons are based on study of proportions of preserved individuals, cleared and stained skeletal preparations, and radiographs of preserved specimens.
Results
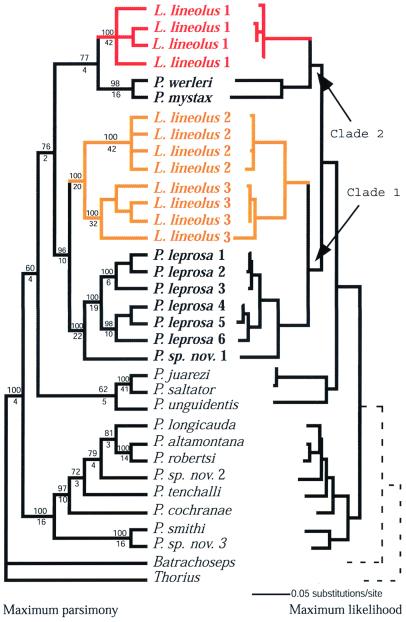
The three genes studied each display extensive differentiation among populations of L. lineolus, with Kimura two-parameter distances as great as 18% (cytochrome b), 16% (ND4), and 5% (16S). These levels are much greater than typically found within other species of bolitoglossine salamanders (e.g., ref. 4), or other taxa (16). Independent and combined data analyses are consistent in finding mitochondrial para- or polyphyly of Lineatriton. Mitochondrial haplotypes do not form a monophyletic group but rather fall out in different parts of the mtDNA gene tree. A maximum parsimony analysis of a combined data set using equal weighting produced 80 equally parsimonious trees (L = 2,183 steps; 545 characters were parsimony informative; consistency index = 0.478, retention index = 0.763). In this analysis (Fig. 1), populations of L. lineolus are deeply nested within Pseudoeurycea, and samples of Lineatriton are not monophyletic. Samples of L. lineolus from southeast Veracruz (population 1, Los Tuxtlas) form a clade that is sister to the closely related Oaxacan species Pseudoeurycea werleri and Pseudoeurycea mystax. A well supported clade consisting mainly of samples of Pseudoeurycea leprosa is the sister taxon of a clade that includes samples of L. lineolus from central western Veracruz (populations 2 and 3, Cerro Chicahuaxtla and Barranca de San Miguel). We recover two well supported clades (clade 1, bootstrap value 96%, decay index 10, and clade 2, bootstrap value 77%, decay index 4; Fig. 1), each containing Lineatriton, and these clades are deeply nested within Pseudoeurycea. When all samples of L. lineolus are forced to be monophyletic, the most parsimonious tree is 17 steps longer than the most parsimonious tree overall. A Wilcoxon signed-ranks test (17) indicates that these two trees differ significantly (n = 58, P = 0.0356), thus monophyly of L. lineolus is rejected. Independent and combined data sets with different weighting schemes resulted in topologies identical to that obtained in the equally weighted analysis, with minor differences in the bootstrap support for clades. The topology obtained in the maximum likelihood analysis is nearly identical to the parsimony analysis topology (Fig. 1, −Ln = 11,928.72), the only difference being that this analysis provides more structure for relationships at the base of the tree, within Pseudoeurycea.
Figure 1.
(Left) Consensus tree of maximum parsimony analysis, combined data set with equal weighting. (Right) Maximum likelihood phylogram of combined data set. For the parsimony tree bootstrap values (above lines) and decay indices (below lines) for relevant nodes. Shown are four individuals each for three populations currently assigned to L. lineolus: L. lineolus 1 (red) from the slopes of Volcán San Martín, Los Tuxtlas, Veracruz, México; L. lineolus 2 (yellow) from Barranca de San Miguel, Fortín de las Flores, Veracruz, México; and L. lineolus 3 (yellow) from slopes of Cerro Chicahuaxtla, Cuautlápan, Veracruz, México. Other samples used in analysis include (all from México except an outgroup, Batrachoseps, from California): P. werleri, Volcán San Martín, Los Tuxtlas, Veracruz; P. mystax, near Ayutla, Oaxaca; P. leprosa 1, Xometa, Veracruz; P. leprosa 2, east of Santa Marta, México; P. leprosa 3, Las Vigas, Veracruz; P. leprosa 4, Tres Mogotes, Puebla; P. leprosa 5, La Malinche, Puebla; P. leprosa 6, Tres Mogotes; P. sp. nov. 1, near Chiconquiaco, Veracruz; Pseudoeurycea juarezi, Cerro Pelón, Oaxaca; Pseudoeurycea saltator, La Esperanza, Oaxaca; Pseudoeurycea unguidentis, Cerro Machín, Oaxaca; Pseudoeurycea longicauda, Lengua de Vaca, México; Pseudoeurycea altamontana, east of Santa Marta, México; Pseudoeurycea robertsi, Raíces, México; P. sp. nov. 2, Tlaxiaco, México; Pseudoeurycea tenchalli, Filo de Caballo, Guerrero; Pseudoeurycea cochranae, San Pedro el Alto, Oaxaca; P. smithi, Cerro San Felipe, Oaxaca; P. sp. nov. 3, Cerro Pelón, Oaxaca; Thorius troglodytes, Puerto del Aire, Veracruz.
Discussion
The phylogenetic hypotheses derived from the mitochondrial data unambiguously reject the monophyly not only of Lineatriton but also of L. lineolus. This is extraordinary because the morphology (Figs. 2 and 3) of the single recognized species is extreme in its degree of specialization (miniaturization, elongation, and attenuation of body, tail and limbs), and has been considered to be unique in the combination of characters (especially with respect to elongation of the vertebrae) within the order Caudata (1). The evolutionary implication of our phylogenetic hypothesis is that the extreme morphology of the different populations of L. lineolus has been acquired independently from ancestors with generalized morphology.
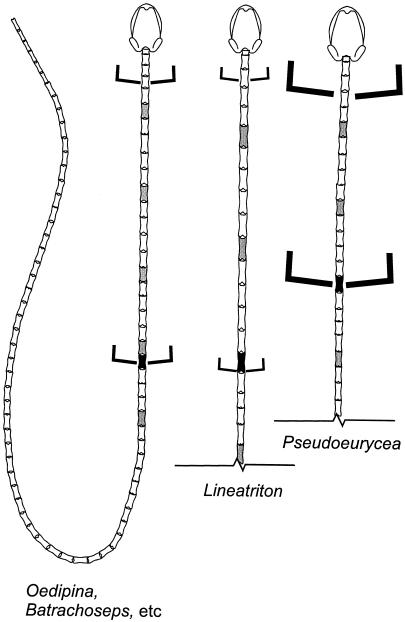
Figure 2.
Ideograms showing the relationship between attenuation and vertebral proportions for individuals of three species of plethodontid salamanders. Three specimens displaying different morphologies are shown. Each was radiographed to show the skeleton. (Left) Oedipina stenopodia from Guatemala. (Center) L. lineolus from México. (Right) P. leprosa from Mexico. The two elongate species have the same head + body length, and the major difference between them is that the vertebrae of Lineatriton are longer than those of Oedipina, which are about the same length as those of Pseudoeurycea, a shorter-bodied form with a head slightly larger than the other two individuals illustrated. Every fifth vertebra is shaded, to better display the effect of vertebral lengthening. Each species has a single cervical vertebra (the atlas) and a single sacral vertebra; there are 20 trunk vertebrae in Oedipina but only 14 in Lineatriton and Pseudoeurycea. The transverse processes of the sacral vertebra of Lineatriton are located in an extreme posterior position, and they are directed posteriorly, effectively adding to the relative interlimb distance as compared with the other species. The morphology displayed by Oedipina is a common homoplasy in plethodondontid salamanders, seen for example in the distantly related bolitoglossine genus Batrachoseps from California.
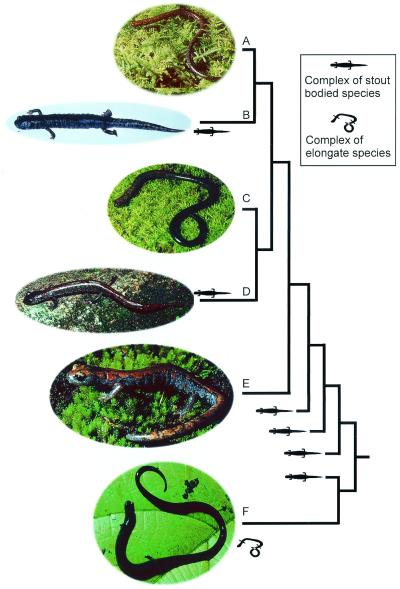
Figure 3.
Photographs of species studied together with simplified tree, showing body form and two levels of homoplasy with respect to elongation, attenuation, and fossorial specialization in tropical salamanders. The first is convergence for elongation related to fossorial life shown in comparisons of Lineatriton and Oedipina, while the second is parallelism with respect both to mode of development for elongation and adaptation for fossorial living shown for the paraphyletic taxon Lineatriton. (A) L. lineolus from Los Tuxtlas, Veracruz, México. (B) P. mystax from Ayutla, Oaxaca, Mexico. (C) L. lineolus from Cerro Chicahuaxtla, Veracruz, México. (D) P. leprosa from slopes of Pico de Orizaba, Veracruz, México. (E) P. robertsi from near Raices, México, México. (F) Oedipina uniformis from slopes of Volcán Irazú, San José, Costa Rica.
Alternative explanations for our findings include: (i) copies of the sequenced genes have been transposed to the nucleus and thus our comparison was inappropriate (18), (ii) ancestral mitochondrial polymorphisms exist (19), and (iii) local hybridization and subsequent introgressive gene flow has occurred (20). We consider these in order.
The results from independent analyses of each of the three genes are consistent with the analyses of all data combined in revealing the mitochondrial paraphyly of Lineatriton. Such congruence between the three genes would be unlikely if paralogous copies of either gene were sequenced (18). Nuclear copies are under different mutational constraints and are expected to show different evolutionary patterns compared with the authentic mitochondrial genes. Nucleotide substitutions occur more randomly in nuclear genes, and nuclear insertions show less marked transition and codon position biases than is the case for mitochondrial genes (18). Analysis of molecular evolution of the mitochondrial genes shows that the genes sequenced have a typical “mitochondrial” behavior, e.g., most variable sites are in the third codon position as is typical for protein coding regions, so it is unlikely that the paraphyly of the two populations of Lineatriton can be explained by the presence of nuclear copies of the mtDNA genes.
Retention of polymorphisms (19) is highly unlikely given the long period since the splitting of the relevant lineages. Using a cytochrome b calibration (20, 21) for North American salamandrids, the estimated divergence time between the populations of Lineatriton from central Veracruz and P. leprosa group (clade 1) is about 20–25 million years, and between Lineatriton from Oaxaca and P. werleri (clade 2) is about 16–18 million years.
Hybridization with sympatric forms is unlikely, given that Lineatriton is a highly specialized burrower. The mechanics of courtship (22) argue against mating between species with extremely different morphology. Furthermore, hybridization has been demonstrated only once within the entire tropical bolitoglossine clade, and that occurred between two close relatives with different ecologies after habitat modification by humans (23).
High bootstrap support and high decay indices in critical parts of the tree show that our phylogenetic hypothesis is well resolved. Accordingly, the phylogeny recovered from our analysis of mtDNA probably reflects the organismal phylogeny.
Nonsister lineages, at present considered to be populations of L. lineolus, share the derived morphological features previously thought to have been uniquely derived, but which we believe evolved twice independently (Fig. 2). We infer, on grounds of parsimony, that the ancestral morphology of the clade that contains the two subsets of L. lineolus, as well as P. leprosa, P. werleri and P. mystax, is that of nonfossorial Pseudoeurycea (the three terrestrial species mentioned appear typical of the modal morphology in the genus). The possibility that the ancestral morphology of this clade is that found in Lineatriton would require that the morphology found in P. leprosa, P. werleri, and P. mystax be secondarily derived, i.e., an evolutionary reversal to the condition typical of distantly related species of Pseudoeurycea, which would require multiple morphological reversions. Examples of unusual morphological specialization include the ribs of Lineatriton, which are greatly reduced in size (24), the rib heads and rib bearers, which are specialized in structure (2, 24), and the transverse processes of the sacral vertebrae, which are located in an unusual position near the end of the centrum (Fig. 2) and are oriented more posteriorly than laterally. Although it is difficult to imagine a restoration of the ancestral conditions from these extremes, it is also difficult to understand how these several features could have evolved in parallel.
The populations we studied that currently are referred to L. lineolus belong to at least two species that are very similar morphologically. The type locality in the original description is vague (Mexican Tableland, ref. 25), but is more appropriate to the populations from central western Veracruz than from the Los Tuxtlas area. Complicating species determination is the fact that in central western Veracruz (populations 2 and 3) divergences in the three genes sequenced are so great (greatest Kimura two-parameter distances for the three genes in this area are 13% for cytochrome b, 9% for ND4, and 1.6% for 16S) that we suspect that several additional species eventually will be recognized in this limited area alone.
The adaptive significance of the “Lineatriton” phenomenon appears to be local and limited. Given the fact that only one clade, Oedipina, of the large number of tropical bolitoglossine clades has evolved an escape from the constraint of a fixed number of trunk vertebrae, it might seem reasonable to expect the adaptive radiation of tropical salamanders to have exploited the mode of evolution used by Lineatriton. However, only in Veracruz has this option been used, although it appears to have evolved twice. In contrast, the developmental option used by Oedipina, apparently difficult to achieve in the large tropical bolitoglossine clade because it has only evolved once (Oedipina is a well supported monophyletic clade, ref. 4), has been more successful, as evidenced by the wide geographic range (from Chiapas throughout Central America to Colombia and Ecuador), great elevational range (from sea level to over 2,500 m), and large number of species (more than 20). Although apparently adaptive advantages followed the evolution of increased numbers of trunk vertebrae in ancestral Oedipina, no adaptive radiation or burst of species formation followed vertebral elongation, and the salamanders displaying it can only be considered terminal twigs in the bolitoglossine radiation, which encompasses about half of the living species of salamanders.
Acknowledgments
We thank M. García-París, J. Hanken, F. Pedroche, M. López, M. Cerón, C. Cerón, and L. F. García for assistance in the field, herpetological collections at Universidad Autonoma de México (L. Canseco-Marquez, O. Flores-Villela) for samples, Secretaría de Medio Ambiente y Recursos Naturales for providing collecting permits, and M. García-París, J. Hanken, J. Patton, M. Slatkin, J. Rodríquez-Robles, S. Kuchta, and M. Wake for discussion. A. Summers provided valuable advice and created Figs. 2 and 3. Studies were financed in part by grants from the U.S. National Science Foundation and the National Geographic Society. G.P.-O. was sponsored by a fellowship from Consejo Nacional de Ciencia y Tecnología.
Footnotes
References
- 1.Wake D B. Am Nat. 1991;138:543–567. [Google Scholar]
- 2.Tanner W. Great Basin Nat. 1950;10:27–44. [Google Scholar]
- 3.Parra-Olea G. Ph.D. Thesis. Berkeley: Univ. of California; 1999. [Google Scholar]
- 4.García-París M, Wake D B. Copeia. 2000;2000:42–70. [Google Scholar]
- 5.Anderson S, Bunkier A T, Barrell B G, Debruijn M H L, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 6.Roe B A, Ma D P, Wilson R K, Wong J F. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- 7.Arévalo E, Davis S K, Sites J W. Syst Biol. 1994;43:387–418. [Google Scholar]
- 8.Saiki R K, Delfand D H, Stooffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 9.Palumbi S R, Martin A P, Romano S, McMillan W O, Stice L, Grabowski G. Special Publication, Department of Zoology. Honolulu: Univ. of Hawaii; 1991. [Google Scholar]
- 10.Moritz C, Schneider C J, Wake D B. Syst Biol. 1992;41:273–291. [Google Scholar]
- 11.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 12.Kluge A G, Farris J S. Syst Zool. 1969;18:1–32. [Google Scholar]
- 13.Farris J S. Syst Zool. 1989;38:406–407. [Google Scholar]
- 14.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 16.Johns G C, Avise J C. Mol Biol Evol. 1998;15:1481–1490. doi: 10.1093/oxfordjournals.molbev.a025875. [DOI] [PubMed] [Google Scholar]
- 17.Templeton A. Evolution. 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang D-X, Hewitt G M. Trends Ecol Evol. 1996;11:247–251. doi: 10.1016/0169-5347(96)10031-8. [DOI] [PubMed] [Google Scholar]
- 19.Pamilo P, Nei M. Mol Biol Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- 20.Funk D J. Mol Biol Evol. 1999;16:67–82. doi: 10.1093/oxfordjournals.molbev.a026039. [DOI] [PubMed] [Google Scholar]
- 21.Tan A-M, Wake D B. Mol Phyl Evol. 1995;4:383–394. doi: 10.1006/mpev.1995.1036. [DOI] [PubMed] [Google Scholar]
- 22.Arnold S J. In: The Reproductive Biology of Amphibians. Taylor D H, Guttman S I, editors. New York: Plenum; 1977. pp. 141–183. [Google Scholar]
- 23.Wake D B, Yang S Y, Papenfuss T J. Herpetologica. 1980;36:335–345. [Google Scholar]
- 24.Wake D B. Memoirs So Calif Acad Sci. 1966;4:1–111. [Google Scholar]
- 25.Cope E D. Proc Acad Nat Sci Phil. 1865;17:185–198. [Google Scholar]