Abstract
FtsY and Ffh are structurally similar prokaryotic Signal Recognition Particle GTPases that play an essential role in the Signal Recognition Particle (SRP)-mediated cotranslational targeting of proteins to the membrane. The two GTPases assemble in a GTP-dependent manner to form a heterodimeric SRP targeting complex. We report here the 2.1 Å X-ray structure of FtsY from T. aquaticus bound to GDP. The structure of the monomeric protein reveals, unexpectedly, canonical binding interactions for GDP. A comparison of the structures of the monomeric and complexed FtsY NG GTPase domain suggests that it undergoes a conformational change similar to that of Ffh NG during the assembly of the symmetric heterodimeric complex. However, in contrast to Ffh, in which the C-terminal helix shifts independently of the other subdomains, the C-terminal helix and N domain of T. aquaticus FtsY together behave as a rigid body during assembly, suggesting distinct mechanisms by which the interactions of the NG domain “module” are regulated in the context of the two SRP GTPases.
Keywords: SRP, Ffh, FtsY, SR, GTPase, GDP, X-ray crystallography
INTRODUCTION
FtsY, the GTPase receptor for the Signal Recognition Particle (SRP) in prokaryotes,1,2 forms a GTP-dependent targeting complex with Ffh, the structurally similar GTPase of the SRP, that plays an essential role in co-translational targeting of proteins to the membrane.2–4 The SRP is a highly conserved ribonucleoprotein, and homologs of FtsY and Ffh in higher organisms are known as SRα and SRP54, respectively.1,5 The GTPase domains of FtsY and Ffh comprise two structural subdomains, termed N and G (or “NG”).6 The NG GTPase is a modular functional unit that occurs at the N-terminus in the sequence of Ffh, and at the C-terminus of FtsY.7,8 In some prokaryotes, such as T. aquaticus, however, FtsY comprises only the NG GTPase,9 and the FtsY NG GTPase alone has been shown to be sufficient for function. 10–12
Previous X-ray structures of the SRP NG GTPases from Ffh and FtsY6,13 have revealed a common threedimensional fold [Fig. 1(A)], similar to that of other members of the GTPase superfamily.15,16 Interestingly, the N- and C-termini of the NG domains are closely apposed in their three-dimensional structures; therefore, although the position of the modular GTPase domain in the primary structure of each protein is different, its spatial relationship to the auxiliary domains of each protein—the N-terminal A-domain of FtsY and the C-terminal M domain of Ffh—is similar. In Ffh, the M domain, C-terminal to the GTPase domain, mediates interaction with the signal sequence of the nascent polypeptide and also binds to the RNA component of SRP.17–19 However, little is known about its interaction with NG—a dynamic relationship between the M- and NG-domains has been demonstrated by fluorescence resonance energy transfer, 20 susceptibility to proteolysis of the linker polypeptide between the M and NG domains,17 and disorder between the two domains in the crystal structure of the intact T. aquaticus Ffh (“NG-M”),21 but modification of one domain of Ffh (e.g. the NG GTPase) affects the function of the other (e.g. the M domain),22 and there are specific interactions between the two domains in the presence and the absence of RNA.23,24 The relationship between the FtsY NG GTPase domain and its N-terminal A-domain is also poorly understood.11,25–27 The A domain may function in interaction with the plasma membrane, 26,27 and proteolysis between the A and NG domains, possibly coupled to the targeting mechanism, has been reported.12 However, the A domain of E. coli FtsY is not essential for SRP-mediated targeting,11 and the A domain is absent in T. aquaticus.9 In eukaryotic SRα the role of the N-terminal “SRX” domain is more clearly defined, as it provides the specific interaction between SRα and its membrane anchor SRβ.28,29
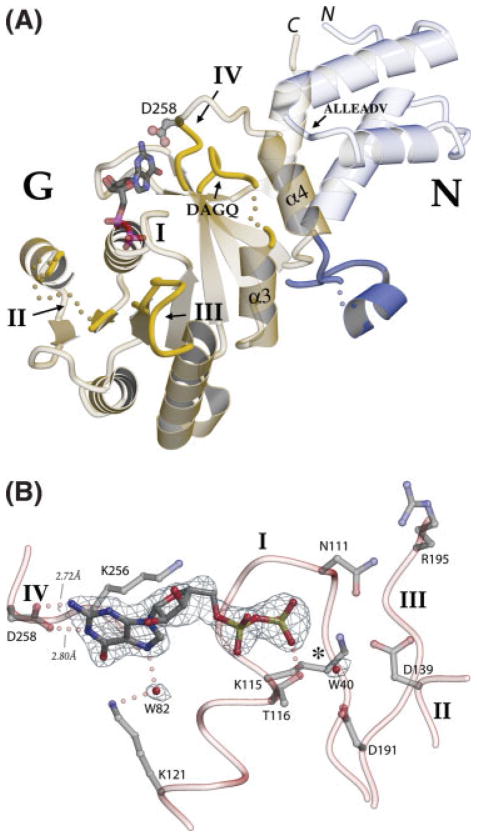
Fig. 1.
A GDP-bound structure of T. aquaticus FtsY. (A) A ribbon diagram of the structure of the FtsY:GDP complex. The NG GTPase domain comprises the “N” subdomain (right), an α-helical bundle, and the “G” subdomain (left), which exhibits the fold, characterized by four conserved sequence motifs (I–IV), common to all members of the GTPase superfamily. Motif II occurs as part of an β-α-β “IBD” subdomain unique to the SRP GTPases (at left). The loops of the T. aquaticus FtsY G domain motifs II, III, IV, and DAGQ are highlighted in yellow; disordered regions of the polypeptide are shown as dots (yellow in the G domain, blue at the N-G linker). The polypeptide that links the N and G domains is highlighted in dark blue; it packs against a neighboring molecule across a crystal contact [Fig. 3(B)]. Note the N- and C-termini of the FtsY NG domain (“N,” “C,” top) are closely apposed in space. (B) A 2Fo-Fc electron density map for the bound GDP molecule and two water molecules within the FtsY active site contoured at 1.0σ. The positions of GTPase sequence motifs I–IV are indicated. The paired hydrogen bonds (2.72 and 2.80 Å) between motif IV Asp258 and the N1 and 2-amino nitrogens of guanine are canonical specificity-determining interactions, seen in all GTPases. A hydrogen bond with a buried water (W82) mediates interaction between the guanine N7 and Lys121, as in the structures of GDP-bound Ffh.14 There is no evidence for Mg2+ ion binding (the predicted position for Mg2+ ion is indicated by an asterisk).
Coordinate structural changes of the FtsY and Ffh NG GTPase domains occur during the assembly of their cotranslational targeting complex30,31 that may act to signal assembly of the complex30 and provide a mechanism for regulating its assembly by recruitment of other elements of the cotranslational targeting machinery (i.e. the ribosome, the nascent polypeptide, and the translocon 32–35). In the GMPPCP-stabilized heterodimeric complex between Ffh and FtsY these changes generate a “latched” configuration of the two GTPase domains that exhibits a striking two-fold pseudo-symmetry.30,31 In the complex, the C-terminal helices of both proteins pack symmetrically at the interface of their respective N and G subdomains, displacing their N-termini such that, in Ffh NG, the first five residues become disordered,30,31 and in FtsY NG, the N-terminal ~20 residues become susceptible to proteolysis.9 That the linker peptides to the N-terminus (in FtsY) and the C-terminus (in Ffh) of the NG domain are located similarly adjacent to the N/G interface means that their interactions may modulate common structural features of the two GTPases. Neither linker is visualized in structures of the NG GTPase domain alone. But the modular structure of the paired NG GTPase domains, their two different primary sequence contexts, and their symmetric reorientation on formation of the targeting complex,30 suggests that structural rearrangements common to the architecture of both the Ffh and FtsY GTPases may be exploited in different ways by the two proteins to signal their distinct partners.
Here, we report the structure of the GDP-bound T. aquaticus FtsY, allowing direct comparison with its structure in the heterodimeric SRP GTPase targeting complex.30,31 In contrast to T. aquaticus Ffh,6 the surface loops of FtsY that contribute to assembly of the heterodimeric interface are, in general, poorly ordered in the monomeric protein. Furthermore, GDP is bound such that the specificity-determining hydrogen bonds with motif IV are fully formed, in contradiction to the proposal that they might be absent prior to assembly.36 Comparison of the conformational rearrangements that occur in Ffh and FtsY during assembly into their heterodimeric complex suggests that while they adopt symmetric configurations in the complex, the conformational path taken by each protein is different. These data support the hypothesis that the C-terminal helix, which is positioned distinctly in the SRP subfamily relative to its position in other small GTPases,15,16 plays a specific role in regulating the structural state of the targeting complex.
MATERIALS AND METHODS
Expression and Purification of FtsY NGd20
The vector pJGS3 expressing the protein FtsY NGd20, in which the N-terminal residues 2–20 of T. aquaticus FtsY are deleted, was constructed by PCR-based site-directed mutagenesis of pTP88.37,38 Initial expression trials in the BL21(DE3)/RP E. coli expression strain (Stratagene) resulted in low levels of protein expression, and the construct was further modified to optimize the sequence and position of the Shine-Dalgarno sequence. All sequences were verified by sequencing at the Northwestern University Biotechnology Laboratory. The FtsY NGd20 deletion of the T. aquaticus FtsY NG domain (comprising residues 21–304) was expressed from pJGS3 using the Rosetta-2(DE3)/pLysS strain of E. coli. (Novagen). Cells were pelleted by centrifugation and resuspended in 20 mL of buffer containing 20 mM EPPS, pH 8.0, 2 mM EDTA, 250 mM NaCl, 10% glycerol, 1 mM DTT, and 0.1 mg/mL lysozyme. Following three freeze/thaw cycles, the cell paste was disrupted by sonication and then incubated at 70°C for 20 min. The resultant supernatant was filtered through 0.8 and 0.2 μm cellulose acetate membranes and passed over a 75 mL DE53 column equilibrated with 50 mM Tris, pH 8.0, 250 mM NaCl, and 1.0 mM DTT, then dialyzed against 10 mM HEPES, pH 7.5, and 1.0 mM DTT overnight at 4°C. The FtsY NGd20 protein was purified over a HiTrap 5 mL Blue column eluted using a linear gradient to 1.0M NaCl, desalted and then passed over a HiTrap 5 mL SP column (Amersham Pharmacia) equilibrated with 10 mM HEPES, pH 7.5, 1.0 mM DTT, and eluted using a linear gradient to 1.0M NaCl. The desalted fraction was, finally, passed over HiTrap 5 mL Q Sepharose HP column (Amersham Pharmacia) equilibrated with 50 mM Tris, pH 8.0, 1.0 mM DTT, and the protein eluted using a linear gradient to 1.0M NaCl. Approximately 12 mg of protein were obtained from 2 L of culture.
Crystallization and Data Collection
Crystals of the FtsY:GDP complex were obtained during screens for crystallization of the GDP:AlF4-stabilized Ffh:FtsY heterodimer. The proteins were each at ~5.5 mg/mL in 2 mM AlCl3, 20 mM NaF, 1 mM GDP, 50 mM Hepes, 30 mM Tris, 250 mM NaCl, and 2 mM MgCl2, pH 7.5. Crystals grew from 0.2M ammonium acetate and 2.2M ammonium sulfate, and were frozen using a nylon loop from a cryo-mother liquor supplemented with 10% ethylene glycol.39 The crystal used for data collection was annealed by thawing in cryo-mother liquor three times, during which the diffraction limit increased from ~3.0 to ~2.0 Å. Data were measured at APS Bio-CARS beamline 14BMC using a wavelength of 0.900 Å and an exposure time of 10 s for 0.7° rotation over a total rotation angle of 196°. Data were integrated with MOSFLM,40 and scaled using the CCP4 suite.41 The diffraction symmetry indicated space group I4 (or I41) and assuming one NG domain in the asymmetric unit, the crystal contained 45.8% solvent. Molecular replacement using the NG domains of both Ffh and FtsY (1OKK.pdb) as probes in PHASER42 yielded clearly distinguishable results (LLGs of −5 and 522 for Ffh and FtsY, respectively).
Refinement
The structure was built and refined using Arp/wARP43 and REFMAC,44 and the model inspected and manually corrected using O.45 Both the N- and C-terminal residues (Ala21 and Asp304) are clearly located in the electron density map. Residues 80–82 and 95–96 of the linker peptide between the N and G domains cannot be located, but an intervening segment (83–94) is well ordered across a crystal packing contact (see text). In the G domain, residues 141–146 of motif II and residues 235–236 of the DAGQ motif cannot be placed in electron density. The two adjacent loops, residues 165–167 of the IBD and residues 195–201 of motif III, are both poorly ordered (B factors of 60–68 Å2) but can be located in the map. Electron density for the bound GDP is well defined, but there is some evidence for partial occupancy of the nucleotide in the site—stronger density at the β-phosphate than at the α-phosphate is consistent with full occupancy at that position either by GDP or by SO4 from the crystallization mother liquor. Following refinement to consistent B factors, we estimate that the occupancy of GDP is ~65%.
Analysis
Coordinates were superimposed using LSQMAN.46 Superposition over the nucleotide was carried out using all GDP atoms, yielding an rmsd over 28 atoms of 0.85 and 0.37 Å, respectively, when compared with structures of the Mg2+GDP bound Ffh, 1NG1, and 1O87.14,47 The RMS deviations of the residues that pack against the nucleotide—the motif I loop (FtsY residues 109–118, Ffh residues 105–114), motif IV loop (FtsY residues 255–258, Ffh 245–248), and the closing loop (FtsY 281–286, Ffh 271–276)—were evaluated using this superposition [Fig. 2(A)]. The rmsd is 0.80 Å for all 134 atoms, and 0.54 Å for the 21 Cα atoms only. Also, the T. aquaticus FtsY G domain was superimposed over 173 Cα atoms of the G domain of T. maritima FtsY with a 0.97 Å rmsd, E. coli FtsY (165 atoms with rmsd 1.26 Å), and M. mycoides FtsY (165 atoms, rmsd of 1.63 Å).
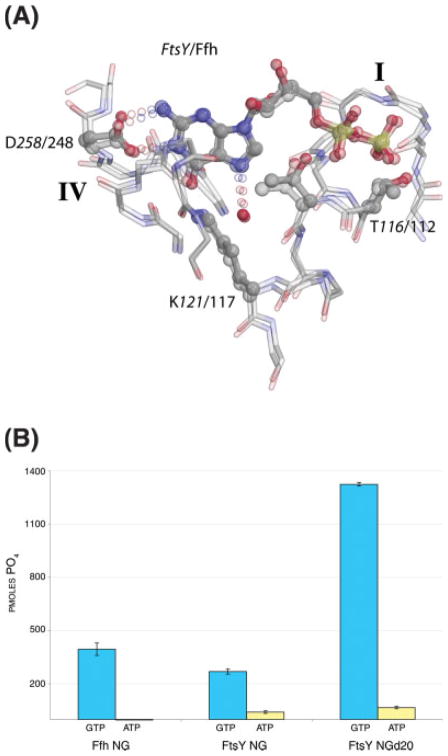
Fig. 2.
T. aquaticus FtsY binds GDP and hydrolyzes GTP with specificity. (A) Comparison of the GDP-binding interactions of the FtsY:GDP complex with the Ffh:Mg2+:GDP complex (1O87).47 The structures were superimposed over the nucleotide—shown are the residues of motifs I and IV and the closing loop (left foreground) that mediate all packing interactions with the nucleotide in the two proteins. The relative positions of the specificity determining residue Asp258 (Asp248 in Ffh) (left) and the motif I P-loop (right) are identical in the two structures. (B) GTP hydrolysis activity of T. aquaticus FtsY wt and FtsY NGd20 construct. Hydrolysis of GTP (pmoles phosphate released) is shown in blue and ATP in yellow. Under these experimental conditions (which are saturating for GTP, but not for ATP, see Methods) the activity with respect to ATP is less than 5–15% that for GTP. The activity of T. aquaticus Ffh NG under the same conditions is shown for comparison.
GTPase assay
GTP hydrolysis was measured using a colorimetric assay under saturating nucleotide conditions.48–50 Protein was at a final concentration of 2 μM in 50 mM Hepes, pH 7.5, 10 mM MgCl2, and 50 mM NaCl; the reaction was started by adding 20 μL of a 1 mM GTP (or ATP) solution in ddH2O to a 20 μL premix at 2× protein and buffer concentration. Reactions were incubated at 42°C for 60 min and stopped by placing in liquid nitrogen. The samples were thawed in a water bath and assayed by addition of 80 μL of freshly prepared filtered Malachite Green reagent.49 After 10 min, the colorimetric reaction was stopped by adding 5.2 μL of 34% w/v NaCitrate. Samples (100 μL) were assayed in 96-well plates using a Molecular Devices Spectramax absorbance reader at 620 nm. A standard curve generated by twofold dilution was linear over the range 50–1600 pmol free phosphate. Each measurement is the average of duplicates.
Coordinates
Coordinates and structure factors have been submitted to the Protein Data Bank and assigned codes 2iyl and r2iylsf, respectively.
RESULTS
We describe here the structure of T. aquaticus FtsY from a crystal obtained during a screen for the heterodimeric complex of T. aquaticus Ffh and FtsY that was found to contain only the FtsY monomer, with GDP bound. The crystallization screen was carried out using an FtsY protein construct in which the N-terminal 20 amino acids were truncated, mimicking the proteolysis observed on assembly of the heterodimeric SRP GTPase complex.9,30,37 The 2.1 Å resolution X-ray structure was determined by molecular replacement and refined to an Rcryst of 0.21 and Rfree 0.29 (Table I). The model comprises the FtsY NGd20 monomer (residues 21–304), one bound GDP, one sulfate ion, and 144 water molecules [Fig. 1(A)]. The electron density map clearly defines the position of the bound nucleotide [Fig. 1(B)]. Although magnesium was present in the crystallization buffer, there is no evidence for Mg2+ ion in the active site.
TABLE I.
Crystallographic Statistics (FtsY:GDP)
| Spacegroup/cell | I4/105.20, 105.20, 54.50 Å |
| Resolution range (Å) | 18.92–2.06 (2.17–2.06) |
| Rsym | 0.080 (0.440) |
| I/σ | 20.1 (3.9) |
| Completeness (%) | 99.4 (100.0) |
| Redundancy | 7.4 (7.7) |
| Rcryst | 0.208 (0.225) |
| Rfree | 0.291 (0.305) |
| No. of atoms | 2251 |
| <Temperature factor> | 45.18 Å2 |
| RMS bonds/angles | 0.014 Å/1.527° |
Rsym = ΣhklΣi |Io − 〈Io〉|/ΣhklΣi Io.
Rcryst = Σhkl |Fo − Fc|/ΣhklFo.
Values in parenthesis calculated for the high resolution bin.
Rfree calculated for 5% reflections omitted from the refinement.
FtsY Binds GDP in a Canonical Configuration
Two solvent exposed loops, comprising much of GTPase motif II (residues 141–146)51 and the “DAGQ” motif (235–236)30 adjacent to the binding site, cannot be located in the electron density map, and the adjacent loops of GTPase motif III (196–201) and the connecting loop between β2a and α2b of the IBD (165–166) are poorly ordered [Fig. 1(A)]. The disorder of these regions contrasts with the well-ordered conformations adopted in structures of the T. aquaticus Ffh,6 which are stabilized by a network of interactions that couple them to each other.6 Only motif II exhibits substantial disorder14 or conformational variability47 in structures of the GDP complexes of T. aquaticus Ffh. Motifs II and III correspond approximately to the GTPase “switch 1” and “switch 2” loops that are often disordered in the GDP-bound state of other GTPases,16,52 and three of the motifs—II, III, and DAGQ—undergo concerted rearrangement on formation of the heterodimeric SRP GTPase targeting complex to generate an intricate set of hydrogen bonding and van der Waals packing interactions across the interface.30 That the residues of these loops are disordered presumably reflects their propensity to adopt different structural configurations in the monomeric and complexed states; however, why more flexibility is seen in the context of FtsY than in the structurally similar Ffh (35% identity, 52% similarity over the “NG” GTPase domain) is unclear.
The binding configuration of the GDP molecule and its interactions with the conserved GTPase motifs I–IV15 are similar to that for other GTPases16 and for the binding of GDP to T. aquaticus Ffh14 [Fig. 1(A)]. These interactions include the specificity-determining H-bonding interaction, provided by the aspartate sidechain (Asp258) of motif IV, that would have to be displaced if a noncognate nucleotide (i.e. ATP, XTP) were to be accommodated within the site [Fig. 1(B)]. Indeed, when the magnesium- free binding configuration for GDP in the T. aquaticus FtsY structure is superimposed over the nucleotide alone with the GDP configuration in the Mg2+GDP complex of T. aquaticus Ffh, the relative positions of the three polypeptide segments that mediate all interactions with the nucleotide in the two structures—motif I, motif IV, and the closing loop—are essentially identical [Fig. 2(A)]. These observations appear to be inconsistent with a requirement for assembly into its targeting complex with Ffh prior to acquisition of nucleotide specificity by the T. aquaticus FtsY,36 and are consistent with other biochemical studies of FtsY from different species.53,54 However, in solution, both the full length T. aquaticus FtsY (which comprises the NG domain) and the FtsY NGd20 construct used in the crystal structure determination retain some activity towards the noncognate nucleotide, ATP [Fig. 2(B)].
A Common Arrangement of the N and C Terminus Prior to Assembly
In addition to the structure of the T. aquaticus FtsY described here, there are three other published crystal structures of the NG domain of FtsY, all in the apo state—from E. coli (1FTS),13 T. maritima (1VMA),55 and M. mycoides (1ZU4, 1ZU5).56 Their different sequences are ~35% identical,56 and the G domains share a common core structure that allows them to be directly superimposed with an rmsd of 1.0–1.6 Å [Fig. 3(A)]. The relative orientations of the N- and G-domains of the proteins vary, as has been noted previously for different structures of Ffh14; when superimposed over the 18 atoms of the G domain motif I P-loop (rmsd ranges from 0.28 to 0.46 Å for the three pairs), the N domains of E. coli FtsY and T. maritima FtsY are superimposable in a configuration that is similar in orientation, but not in detail (see below), to that found when FtsY assembles with Ffh in its GTP-dependent targeting complex,30,31 and the N domains of T. aquaticus and M. mycoides FtsY are rotated away from this position to different extents around the interface helix α4.57 However, all four structures share two commonalities—the α4 interface helix, which couples the N and G domains, is superimposable in a position ~2.5 Å away from that adopted by the helix in the T. aquaticus Ffh:FtsY complex, and despite the disparate disposition of each N domain, in each case the C-terminal helix that packs against it adopts a common orientation that is similar between structures and that is distinct from that adopted in the Ffh:FtsY complex [Fig. 3(A)].
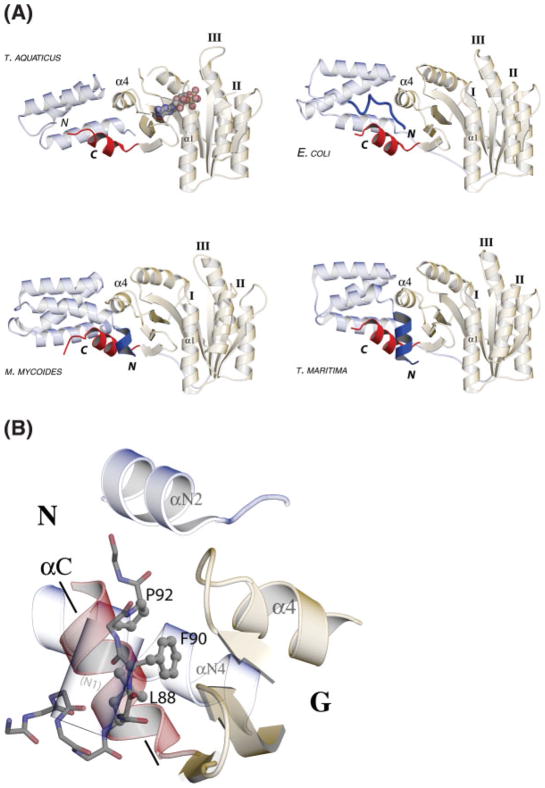
Fig. 3.
Common features of the structures of FtsY from different species. (A) Structures of FtsY from T. aquaticus, E. coli, M. mycoides, and T. maritima are aligned using a superimposition based on the motif I P-loop; the α1 helix that follows the motif is vertical at the center of each image. The α4 helix (labeled) defines the interface between the N and G subdomains. The C-terminal helix (highlighted in red) adopts a similar configuration in each of the structures, despite the slightly different orientation of each N domain. In the structures of the apo M. mycoides and T. maritima FtsY an N-terminal helix extension (“N1”, highlighted in blue) packs between the C-terminal helix and the N/G interface. In the E. coli structure the N-terminal polypeptide is poorly structured (blue coil) but appears to play a similar role. The GDP in the T. aquaticus FtsY complex is indicated with a CPK model. In each structure of the apo or GDP-bound state of FtsY shown here, the motif II and motif III loops (labeled) adopt distinct (or disordered) conformations. (B) The N/G linker peptide packs across a crystal contact, occupying space that would accommodate the “N1” N-terminal helix seen in other structures of FtsY. The position of the N1 helix is indicated by a ghosted cylinder. The backbone atoms of the linker splayed across the crystal contact are shown as sticks; the hydrophobic sidechains of Leu88, Phe90, and Pro92 (ball-and-stick) pack into a conserved hydrophobic pocket at the interface. The orientation of the C-terminal helix, αC, is indicated by an axis line. The ribbon representation includes helices αN2 and αN4 of the N domain, and the α4 helix, the motif IV/DARGG loop that precedes it, and the closing loop (bottom) of the G domain, which together contribute to the hydrophobic pocket at the N/G interface.
The configuration of the C-terminal helix in each of the structures is stabilized by an N-terminal interposition that packs between the C-terminal helix and the adjacent motif IV of the G domain.13,56 In the M. mycoides and T. maritima FtsY crystal structures,55,56 a short α-helix at the N-terminus of the N domain helical bundle (“N1”) packs against the C-terminus [Fig. 3(A)], and in the crystal structure of the E. coli FtsY NG domain a cleaved and relatively unstructured N-terminus extends around the C-terminal helix.13 In the former cases the helix packing is mediated by a highly conserved heptad repeat at the N-terminus of FtsY NG domain sequences56 that nestles into a conserved hydrophobic surface at the N/G domain interface.58 The structural and functional implications of these interactions have recently been discussed56—in the intact FtsY they may couple the putative targeting function of the N-terminal A-domain (or perhaps, the N1 helix itself) to the NG GTPase domain.12,26,59
That a similar orientation of the C-terminal helix is maintained in the structure of T. aquaticus FtsY, despite the first 20 amino acids of the NG domain being absent (see above), is explained by a crystal packing interaction that compensates—although much of the linker peptide between the N- and G-domains is disordered, residues 83–94 can be located unambiguously in the electron density map, splayed across a crystal contact to pack at the N/G interface of an adjacent monomer [Fig. 3(B)]. The linker adopts an extended conformation that allows three hydrophobic sidechains (Leu88, Phe90, and Pro92) to pack between the C-terminal helix and the motif IV/DARGG loop. The packing interactions of the linker peptide in this structure closely mimic the hydrophobic interactions of the N-terminal helix (in T. maritima and M. mycoides FtsY) or unstructured peptide (in E. coli FtsY) against the hydrophobic face of the N/G interface [Fig. 3(A)].
Specific Structural Transitions Between the Free and Complexed FtsY
The structural changes that occur on assembly of the heterodimer can be inferred from comparison of the monomeric FtsY:GDP and assembled FtsY:GMPPCP complex structures. The G domain of T. aquaticus FtsY in its GDP-bound state can be superimposed on its structure in the GMPPCP-stabilized heterodimer complex with an rmsd of only 0.97 Å for 171 Cαs, as the conformational changes occurring within the G domain (but not between the N and G domains) are largely limited to the loops at the interface. GTP binding itself does not functionally activate SRP receptor prior to its interaction with SRP,60,61 and a noncanonical binding mode is observed in the T. aquaticus Ffh NG:GMPPNP complex, 62 suggesting that while GTP binding to Ffh and FtsY serves to prime interaction between the SRP GTPases, nucleotide binding alone is not accompanied by activating protein structural changes. Therefore, in the following, we can describe the structural changes between the GDP-bound monomeric protein and its GMPPCP-bound state in the heterodimeric complex as representative either of assembly or of disassembly of the complex. The structural changes of each loop are coupled across the protein interface—thus, for example, a ~2.0 Å shift between the DAGQ motif and the N-terminus of helix α3 that follows it accommodates rearrangement of the adjacent motif III loop (corresponding to the GTPase “switch 2”) as it adopts its “latched” configuration.30 The motif III shifts are large (Fig. 4)—switch 2 residue His197 moves by 8.0 Å across the protein surface, and motif III residue Leu196 by 2.3 Å, to contribute to a conserved and symmetric packing interaction at the heterodimer interface.30 In conjunction with this rearrangement a peptide flip between motif III residues Gly194 and Arg195 allows the arginine sidechain to interact with Glu205 along helix α2 and reorients the carbonyl oxygen of the conserved motif III glycine away from the active site.30 The latter releases a backbone hydrogen bond coupling motif III to motif I in the monomeric protein and allows motif I residue Asn111 to shift by 1.3 Å at the Cα, which opens up the motif I P-loop to engage the γ-phosphate oxygens of the GTP. Interestingly, it appears in this respect that FtsY mimics Ffh, as its structure in the monomeric state can serve to regulate the access of GTP to its canonical binding configuration in FtsY, allowing GTP binding to prime the GTPase, but not activate it.62
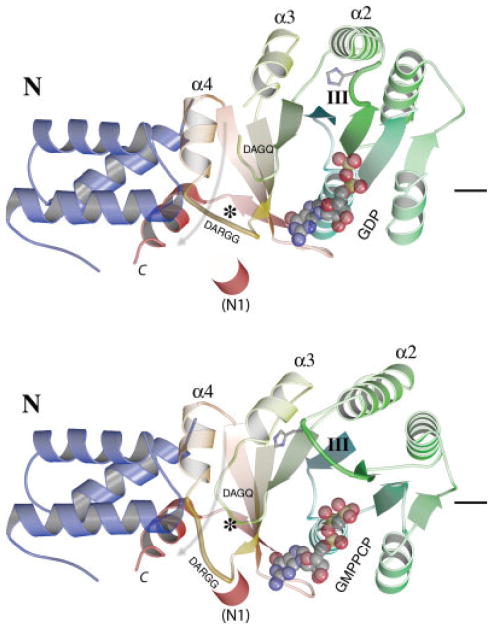
Fig. 4.
A rotation between the N and G domains of FtsY. The structures of the FtsY:GDP complex (top) and the FtsY:GMPPCP complex, the latter as observed in the assembled targeting heterodimer (1OKK, bottom), are compared following superposition over their N domains. The C-terminal helix and the α4 helix of FtsY remain fixed in the N domain frame, while the G domain undergoes a rotation relative to the N domain (ghosted arrow) that is accompanied by conformational changes of the DARGG loop (“*”), the motif III loop (labeled), and the DAGQ loop (between them), that generate the close-packed interactions of the “latched” configuration with Ffh at the heterodimer interface. The bound nucleotide is shown for each case, and the asterisk and the short line at right provide reference points. The shift of the DARGG loop necessarily displaces the N terminal helix extension (“N1”) if it were present in these structures.
The comparison also reveals that the conformation of the guanine nucleotide twists between the GDP-bound monomeric and GMPPCP-bound complexed states of FtsY, as the ribose and guanine base of the bound nucleotides slide approximately 0.7–1.0 Å from one structure to the other (Fig. 5) while the α- and β-phosphate groups in common between the two structures remain fixed relative to the motif I P-loop. Because motif IV Asp258 tracks the shift of the N1 and 2-amino group nitrogens, the hydrogen bonding interactions with the base are likely maintained throughout. Such flexibility in nucleotide binding configuration, also observed in ultrahigh resolution structures of GDP and GMPPNP-bound Ffh (Ramirez et al., in preparation), belies the functional significance of deviation of active site elements from their canonical spatial relationship.13,31 It suggests instead that these binding interactions are maintained during assembly and disassembly of the GTPase heterodimer and accommodate large rearrangements, such as that of the DARGG motif loop adjacent to motif IV. There, a peptide flip between Lys262 and Gly263 allows the loop and the following α4 helix to maintain packing interactions during the acquisition or release of the “latched” configurations of the DAGQ motif, on one side, and the ALLEADV motif of the N domain, on the other (Figs. 1A and 4).
Fig. 5.
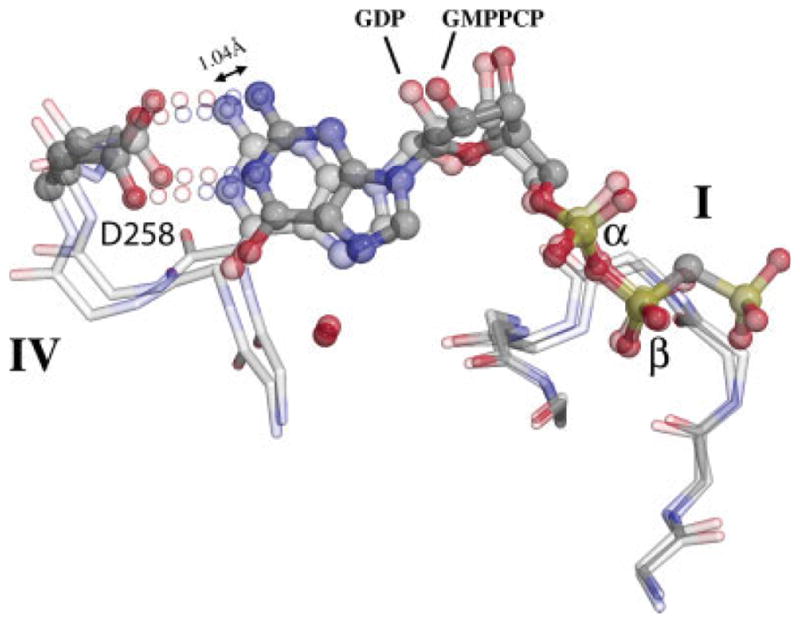
The nucleotide position slips between the GDP and GMPPCP states. Following superposition of the FtsY:GDP monomeric and FtsY:GMPPCP complexed states of FtsY over the motif I P-loop (at right), the α- and β-phosphate groups of GDP and GMPPCP (labeled) remain fixed relative to each other, but the ribose and the guanine base shift by ~1 Å between the two structures (1.04 Å at the 2-amino nitrogen, as indicated). This slip is tracked by movement of the motif IV loop and by the specificity determining carboxylate sidechain of Asp258.
The ALLEADV motif at the N/G interface is shifted ~5.2 Å relative to the G domain between the two states as the G domain pivots relative to the N domain on as- sembly (Fig. 4). This shift, and the rearrangement of the DARGG motif loop that accompanies it, regulates the accessibility of a hydrophobic pocket at the N/G interface to the C-terminal helix. Four hydrophobic sidechains of the FtsY C-terminal helix (Phe297, Val298, Leu301, and Leu302) mediate its packing against the N domain. The hydrophobic character at each of these positions is conserved in the sequences of the NG GTPases.63 Interestingly, position 297 is phenylalanine or tyrosine in all sequences of FtsY,63 but is less conserved, and smaller, in sequences of Ffh. In FtsY, the sidechain of Phe297 is inserted between the α4 helix and the universally conserved Phe292 of the loop that precedes the C-terminal helix, and it maintains this packing relationship in both the monomeric and complexed states. This causes the C-terminal helix of T. aquaticus FtsY to maintain a rigid body relationship with the N domain between the two states (rmsd 0.59 Å), such that, in the N domain frame, the structural transition that occurs on assembly can be considered to be a rotation of the G domain against the N domain and C-terminal helix (Fig. 4). Further, the configuration adopted in the heterodimer complex with Ffh requires that the “N1” extension, when present (Fig. 3), must be displaced during assembly (Fig. 4). The heptad signature characteristic of the “N1” extension (Phe4, Leu7, and Leu11) occurs in the sequence of T. aquaticus FtsY,56 and a requirement for its displacement in the full length protein may explain the observation that cleavage of the N-terminal 20 residues of T. aquaticus FtsY must occur prior to formation of the stable heterodimeric T. aquaticus NG domain complex.9
Ffh and FtsY Adopt a Common Structure Through Different Paths
The heterodimer of the Ffh and FtsY NG domains exhibits pseudo-twofold symmetry, and seven loops contributed by each protein to the interface between them, as well as the C-helix of each protein, can be almost exactly superimposed on that of the other by a simple ~180° rotation.30 Five of these loops—motifs I, III, DARGG, IV, and ALLEADV—contribute to the “latch” interface. Although the DARGG loop, which also contributes directly to the N/G interface, is quite flexible in both Ffh and FtsY [Fig. 6(A)], it reorients on assembly to adopt a common structure that in both proteins directs the polar sidechains of the motif symmetrically across the heterodimer interface, and allows the respective C-terminal helix to pack against the glycine backbone atoms of the motif. In Ffh, as for FtsY, a spine of four hydrophobic residues along the C-terminal helix mediates packing against the hydrophobic face at the N/G interface. However, unlike in FtsY, the packing relationship with the N domain changes during the transition, as the sidechains of Leu287 and Ile291 shift away from the αN1 helix to pack against the glycine backbone of the DARGG motif and the C-terminal helix undergoes a rotation of ~25° relative to the N domain. Sequence conservation in Ffh is consistent with increased mobility of the C-terminal helix, as the residue corresponding to conserved Phe297 in FtsY (see above) is present as the more flexible methionine or leucine in most sequences of Ffh.63 Also, there is no “N1” extension in Ffh; instead, a phenylalanine residue is universally conserved near the N-terminus (Phe2 in T. aquaticus Ffh) that packs against the hydrophobic pocket at the N/G interface in the monomeric protein, and is displaced on assembly of the heterodimeric complex.30 It, like the “N1” helix of FtsY (see above), serves to protect the hydrophobic pocket of the monomeric protein prior to assembly of the heterodimer.
Fig. 6.
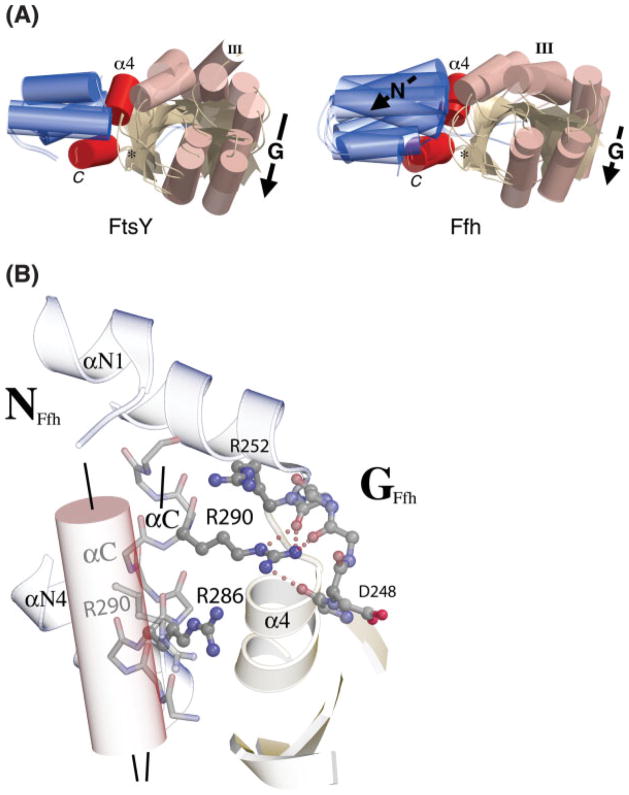
Distinct behavior of Ffh and FtsY on assembly. (A) The domain shifts of T. aquaticus Ffh and FtsY that occur during acquisition of the heterodimeric complex interface structure are different. Here, the structures of the GDP and GMPPCP (heterodimer) complexes of each protein are superimposed on a common C-terminal helix frame, which is approximately constant between the two pairs of structures. In FtsY, this frame is constant with the N domain, and the G domain shifts relative to it (Fig. 4). In Ffh, both the N- and G-domains shift relative to the helix; this is equivalent to movement of the C-terminal helix relative to the N domain on assembly into the heterodimer. In the structure of the Ffh:FtsY heterodimer the interface loops at the N/G interface (DARGG, ALLEADV) and within the G domain (I, II, III, and DAGQ) adopt symmetric configurations; their paths to acquisition of that common structure are distinct. (B) In Ffh, rearrangement of the C-terminal helix (αC) is stabilized by an interaction between Arg290 and the DARGG loop that links the position of the C-terminal helix and the sidechain of Ffh motif IV Asp248. The positions of αC in the apo (ghosted cylinder) and heterodimeric complex (backbone representation) configurations are indicated and their relative orientations highlighted by axial lines. Both Arg286 and Arg290 are conserved in the sequences of Ffh and they contribute to the formation of a basic “ladder” likely to underlay the M domain/4.5S components of the SRP in the heterodimeric state.
If the C-terminal helix is taken as the frame of reference, the cumulative effect of these differences between Ffh and FtsY can be distinguished as differences in the interdomain motion that occur during assembly and disassembly. In FtsY, the G domain rotates against the C-terminal helix frame [Figs. 4 and 6(A)], as rearrangements of the DARGG, DAGQ, and motif III loops generate the “latch” interactions that span the N/G subdomains at the heterodimer interface (Fig. 4). The relative shift of the G domain of FtsY is quite large, being ~6.2 Å for the motif I P-loop, for example (Fig. 4). A similar comparison of the structures of Ffh in its C-terminal helix frame reveals, as a consequence of the repacking of the C-terminal helix, quite different behavior [Fig. 6(A)]. Thus, the corresponding shift of the G domain is less than for FtsY (2.1 Å at P-loop), but the N domain helical bundle also shifts relative to this frame, by 5.9 Å at the ALLEADV motif [Fig. 6(A)]. Importantly, Ffh and FtsY adopt similar structural configurations at the interface of the heterodimer; therefore, the different behaviors of the N and G domain rearrangements in the two proteins provide two distinct paths between the structure of the monomeric protein and the same (or similar) structure adopted in the heterodimeric complex.
A Charged Surface is Organized at the Ffh N/G Domain Interface
There is an additional distinction in the behavior of the C-terminal helix between the two proteins. In Ffh, but not FtsY, two arginine residues, Arg286 and Arg290, located along the surface of the C-terminal helix are completely conserved in all prokaryotic sequences.63 In structures of the monomeric protein the sidechains are directed away from the protein surface and interact only with solvent. 6,47,58 The two arginine sidechains track the shift of the C-terminal helix on assembly of the heterodimer and adopt well-ordered configurations that, with the sidechain of conserved Arg252 of the DARGG loop, generate a positively charged surface across the heterodimer interface [Fig. 6(B)]. The resulting “ladder” of basic residues, precisely positioned by hydrophobic packing interactions of the C-terminal helix (e.g. Leu297), and by interactions of the Arg290 sidechain, is located where the signal sequence recognition subunit of Ffh, which comprises both the M domain and the 4.5S RNA, interacts with the surface of the assembled heterodimer.24 In the intact Ffh, the peptide that links the C-terminal helix of the NG domain to the M domain has substantial conformational freedom. 17,21,22 That the C-terminal helix of the NG GTPase moves independently of the N and G domains and exhibits highly conserved and well-defined charge interactions in the assembled state provides a mechanism by which the signal sequence recognition subunit of Ffh, the M domain/4.5S RNA complex, may modulate acquisition of a structural configuration competent for assembly with receptor.
DISCUSSION
The X-ray structure of the monomeric FtsY from T. aquaticus bound to GDP is similar to that of the apo FtsY (and Ffh) from other species, despite the deletion of 20 N-terminal amino acids in the crystallized protein [Fig. 3(A)]. A number of conformational changes suggested by the comparison of the crystal structures of FtsY with the structure in the assembled heterodimeric complex provide insight into the mechanisms available for the acquisition and regulation of the “latched” configuration observed in the complex of the SRP GTPases. We argued previously based on the published structures of the Ffh NG domain in its monomeric and complexed states that it was likely that the C-terminal helices of both Ffh and FtsY rearranged independently during assembly of the heterodimer interface.30 The observation of a rigid body rearrangement of the N domain/C-terminal helix in T. aquaticus FtsY contradicts this notion. Instead, interactions with the N/G domain interface of Ffh and FtsY are modulated by distinct structural mechanisms that exploit the different positions of the “NG” module within the context of each primary sequence (i.e. Ffh NG being N-terminal, and FtsY NG being C-terminal). Conservation of an N-terminal helix extension in the structures of the apo FtsY, and biochemical data that supports the notion that it plays a functional role in vivo both in targeting and as a substrate for cleavage, 12,26,27 suggests that the N-terminal extension may provide a mechanism for regulation of the heterodimeric “latched” configuration of FtsY, specifically by modulating the conformational change of the N domain relative to the G domain.
The surface of the N/G interface adjacent to the C-terminal helix has conserved hydrophobic character in both Ffh and FtsY,58 and it provides the packing context for the N-terminal extension of FtsY in the monomeric protein. 55,56 Positioning of the DARGG loop and elements of the G domain against the hydrophobic face of the C-terminal helix is coupled to conformational changes that extend across the loops of the “latch” interface.30,31 In Ffh, the positioning of the helix contributes to formation of a conserved positively charged surface that in the targeting heterodimer is oriented towards the M domain/4.5S SRP RNA complex.24 Interestingly, mutants that generate “half-of-sites” defects in the GTP hydrolysis activity of SRP:FtsY complexes locate to this region of the structure (the N/G interface),64 and mutation of the DARGG motifs of Ffh and FtsY cause specific, and distinct, defects along the targeting pathway in Ffh and FtsY.65,66 Further, the region, broadly defined, has been implicated in interaction with the signal peptide bound to SRP.67
That the T. aquaticus FtsY exhibits a canonical nucleotide binding configuration and minimal degeneracy of nucleotide hydrolysis (Fig. 2), apparently in contrast to the behavior of the E. coli FtsY,36 may arise from differences in the two protein constructs. In the E. coli work, the FtsY construct comprised an N-terminal GST-fusion to a truncated A-domain. The N-terminal peptide deleted in the T. aquaticus FtsY NGd20 construct directly interacts with the C-terminal helix and the motif IV recognition region in other structures of FtsY,56 and its deletion in the T. aquaticus protein allows rapid formation of a stable heterodimeric complex.9,37 Given the structural rearrangements we can infer for both proteins, this suggests a mechanism by which constraints on the packing of the C-terminal helix and the DARGG loop may inhibit adoption of a stable “latched” configuration at the interface. Such interactions might also modulate acquisition of a specific nucleotide binding conformation at motif IV, but the flexibility of the nucleotide binding conformation observed between the GDP and GMPPCP-bound complexed states (Fig. 5) suggests as well that it is the “protein-binding” specificity, the acquisition of the latched configuration of the heterodimer interface rather than the “nucleotide-binding” specificity of FtsY, that may be regulated by conformational change between the N and G subdomains of the SRP GTPases.
The SRP GTPases undergo a coordinate rearrangement during the transition from the quiescent monomeric state of each protein to the assembled heterodimeric SRP targeting complex. GTP binding primes the assembly of the complex, enabling it to interact with and direct the signal peptide to the membrane translocon, and GTP hydrolysis, activated by interactions subsequent to release of the signal peptide, then disengages SRP from its receptor. Recent studies have mapped the 4.5S RNA in the assembled complex to extend across a face of the heterodimer24 that is defined by highly conserved surface-exposed sequence elements contributed by both Ffh and FtsY.37 The distinct conformational changes between the monomeric and complexed states of T. aquaticus Ffh and FtsY suggest that the interactions of the C-terminal helix can provide two different means by which assembly of the GTPase targeting complex can be regulated. In the case of FtsY, rotation of the G domain of FtsY relative to the C-terminal helix and N domain framework must be coupled to displacement of the “N1”-linked N-terminal extension, the likely site of membrane interaction26; this is equivalent to the N-terminal extension inhibiting rotation of the G domain, and, thus, assembly of the targeting heterodimer. In the case of Ffh, the arginine sidechain interactions contributed by the C-terminal helix on assembly, some with the DARGG loop, provide a “handle” by which the M domain or the anionic 4.5S RNA could gain purchase to regulate the structure and assembly of the Ffh heterodimer interface. Thus, the symmetric interaction between the two SRP GTPases can be regulated asymmetrically by the other components of the SRP targeting complex that provide distinct contexts, or environments, for each.
Acknowledgments
Grant sponsor: NIH; Grant number: GM058500; Grant sponsor: R.H. Lurie Cancer Center; Grant sponsor: National Institutes of Health, National Center for Research Resources; Grant number: RR07707.
D.M.F. thanks Dr. Elizabeth A. Wiatr for insightful discussion. J.G.S. was responsible for mutagenesis, expression, characterization and crystallization studies; J.S.C. V carried out the GTP hydrolysis assays; P.J.F. collected the diffraction data and D.M.F. was responsible for the subsequent crystallographic work; D.M.F. and P.J.F. wrote the manuscript.
References
- 1.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Batey RT. Structural insights into the signal recognition particle. Annu Rev Biochem. 2004;73:539–557. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- 3.Koch HG, Moser M, Muller M. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev Physiol Biochem Pharmacol. 2003;146:55–94. doi: 10.1007/s10254-002-0002-9. [DOI] [PubMed] [Google Scholar]
- 4.Wild K, Rosendal KR, Sinning I. A structural step into the SRP cycle. Mol Microbiol. 2004;53:357–363. doi: 10.1111/j.1365-2958.2004.04139.x. [DOI] [PubMed] [Google Scholar]
- 5.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 6.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 7.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 8.Luirink J, ten Hagen-Jongman CM, van der Weijden CC, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Eschericia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shepotinovskaya IV, Freymann DM. Conformational change of the N-domain on formation of the complex between the GTPase domains of Thermus aquaticus Ffh and FtsY. Biochim Biophys Acta. 2002;1597:107–114. doi: 10.1016/s0167-4838(02)00287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haddad A, Rose RW, Pohlschroder M. The Haloferax volcanii FtsY homolog is critical for haloarchaeal growth but does not require the A domain. J Bacteriol. 2005;187:4015–4022. doi: 10.1128/JB.187.12.4015-4022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelazny A, Seluanov A, Cooper A, Bibi E. The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc Natl Acad Sci USA. 1997;94:6025–6029. doi: 10.1073/pnas.94.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herskovits AA, Seluanov A, Rajsbaum R, ten Hagen-Jongman CM, Henrichs T, Bochkareva ES, Phillips GJ, Probst FJ, Nakae T, Ehrmann M, Luirink J, Bibi E. Evidence for coupling of membrane targeting and function of the signal recognition particle (SRP) receptor FtsY. EMBO Rep. 2001;2:1040–1046. doi: 10.1093/embo-reports/kve226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montoya G, Svensson C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–369. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 14.Freymann DM, Keenan RJ, Stroud RM, Walter P. Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP. Nat Struct Biol. 1999;6:793–801. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- 15.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 16.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 17.Zopf D, Bernstein HD, Johnson AE, Walter P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be cross-linked to a signal sequence. EMBO J. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batey RT, Sagar MB, Doudna JA. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J Mol Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- 19.Rosendal KR, Wild K, Montoya G, Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc Natl Acad Sci USA. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buskiewicz I, Peske F, Wieden HJ, Gryczynski I, Rodnina MV, Wintermeyer W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J Mol Biol. 2005;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 22.Lütcke H, High S, Römisch K, Ashford AJ, Dobberstein B. The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J. 1992;11:1543–1551. doi: 10.1002/j.1460-2075.1992.tb05199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buskiewicz I, Kubarenko A, Peske F, Rodnina MV, Wintermeyer W. Domain rearrangement of SRP protein Ffh upon binding 4. 5S RNA and the SRP receptor FtsY. RNA. 2005;11:947–957. doi: 10.1261/rna.7242305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spanggord RJ, Siu F, Ke A, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat Struct Mol Biol. 2005;12:1116– 1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 25.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Leeuw E, Poland D, Mol O, Sinning I, ten Hagen-Jongman CM, Oudega B, Luirink J. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 1997;416:225–229. doi: 10.1016/s0014-5793(97)01238-6. [DOI] [PubMed] [Google Scholar]
- 27.de Leeuw E, te Kaat K, Moser C, Menestrina G, Demel R, de Kruijff B, Oudega B, Luirink J, Sinning I. Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 2000;19:531–541. doi: 10.1093/emboj/19.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz T, Blobel G. Structural basis for the function of the β subunit of the eukaryotic signal recognition particle receptor. Cell. 2003;112:793–803. doi: 10.1016/s0092-8674(03)00161-2. [DOI] [PubMed] [Google Scholar]
- 29.Schlenker O, Hendricks A, Sinning I, Wild K. The structure of the mammalian SRP receptor as prototype for the interaction of small GTPases with longin domains. J Biol Chem. 2006;281:8898–8906. doi: 10.1074/jbc.M512415200. [DOI] [PubMed] [Google Scholar]
- 30.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Z, Jiang Y, Mandon EC, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 34.Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;472:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 35.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, III, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318– 324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shan SO, Walter P. Induced nucleotide specificity in a GTPase. Proc Natl Acad Sci USA. 2003;100:4480–4485. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Focia PJ, Gawronski-Salerno J, Coon VJ, Freymann DM. Structure of a GDP:AlF(4) complex of the SRP GTPases Ffh and FtsY, and identification of a peripheral nucleotide interaction site. J Mol Biol. 2006;360:631–643. doi: 10.1016/j.jmb.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepotinovskaya IV, Focia PJ, Freymann DM. Crystallization of the GMPPCP complex of the NG domains of T. aquaticus Ffh and FtsY. Acta Crystallogr D Biol Crystallogr. 2003;59:1834–1837. doi: 10.1107/s0907444903016573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng T-Y. Mounting of crystals for macromolecular crystallography in a free-standing thin film. J Appl Crystallogr. 1990;23:387– 391. [Google Scholar]
- 40.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;(26) [Google Scholar]
- 41.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 43.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Cystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247– 255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 45.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 46.Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
- 47.Focia PJ, Alam H, Lu T, Ramirez UD, Freymann DM. Novel protein and Mg2+ configurations in the Mg2+GDP complex of the SRP GTPase ffh. Proteins. 2004;54:222–230. doi: 10.1002/prot.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher DK, Higgins TJ. A sensitive, high-volume, colorimetric assay for protein phosphatases. Pharm Res. 1994;11:759–763. doi: 10.1023/a:1018996817529. [DOI] [PubMed] [Google Scholar]
- 49.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 50.Ng DH, Harder KW, Clark-Lewis I, Jirik F, Johnson P. Non-radioactive method to measure CD45 protein tyrosine phosphatase activity isolated directly from cells. J Immunol Methods. 1995;179:177–185. doi: 10.1016/0022-1759(94)00281-z. [DOI] [PubMed] [Google Scholar]
- 51.Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 52.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 53.Moll R, Schmidtke S, Petersen A, Schäfer G. The signal recognition particle receptor a subunit of the hyperthermophilic archaeon Acidianus ambivalens exhibits an intrinsic GTP-hydrolyzing activity. Biochem Biophys Acta. 1997;1335:218–230. doi: 10.1016/s0304-4165(96)00141-9. [DOI] [PubMed] [Google Scholar]
- 54.Moser C, Mol O, Goody RS, Sinning I. The signal recognition particle receptor of Escherichia coli (FtsY) has a nucleotide exchange factor built into the GTPase domain. Proc Natl Acad Sci USA. 1997;94:11339–11344. doi: 10.1073/pnas.94.21.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joint Center for Structural Genomics. Crystal structure of cell division protein ftsY (TM0570) from Thermotoga maritima at 1.60 Å resolution. PDB No. 1vma.pdb. [Google Scholar]
- 56.Gariani T, Samuelsson T, Sauer-Eriksson AE. Conformational variability of the GTPase domain of the signal recognition particle receptor FtsY. J Struct Biol. 2006;153:85–96. doi: 10.1016/j.jsb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Ramirez UD, Minasov G, Focia PJ, Stroud RM, Walter P, Kuhn P, Freymann DM. Structural basis for mobility in the 1.1 Å crystal structure of the NG domain of Thermus aquaticus Ffh. J Mol Biol. 2002;320:783–799. doi: 10.1016/s0022-2836(02)00476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montoya G, Kaat KT, Moll R, Schäfer G, Sinning I. The crystal structure of the conserved GTPase of SRP54 from the archeon Acidianus ambivalens and its comparison with related structures suggests a model for the SRP-SRP receptor complex. Structure. 2000;8:515–525. doi: 10.1016/s0969-2126(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 59.Millman JS, Andrews DW. A site-specific, membrane-dependent cleavage event defines the membrane binding domain of FtsY. J Biol Chem. 1999;274:33227–33234. doi: 10.1074/jbc.274.47.33227. [DOI] [PubMed] [Google Scholar]
- 60.Rapiejko PJ, Gilmore R. Empty site forms of the SRP54 and SR α GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell. 1997;89:703– 713. doi: 10.1016/s0092-8674(00)80253-6. [DOI] [PubMed] [Google Scholar]
- 61.Song W, Raden D, Mandon EC, Gilmore R. Role of Sec61α in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- 62.Padmanabhan S, Freymann DM. The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase. Structure. 2001;9:859–867. doi: 10.1016/s0969-2126(01)00641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zwieb C, Samuelsson T. SRPDB (signal recognition particle database) Nucleic Acids Res. 2000;28:171–172. doi: 10.1093/nar/28.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shan SO, Stroud RM, Walter P. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2004;2:E320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Newitt JA, Bernstein HD. The N-domain of the signal recognition particle 54-kDa subunit promotes efficient signal sequence binding. Eur J Biochem. 1997;245:720–729. doi: 10.1111/j.1432-1033.1997.00720.x. [DOI] [PubMed] [Google Scholar]
- 66.Lu Y, Qi HY, Hyndman JB, Ulbrandt ND, Teplyakov A, Tomasevic N, Bernstein HD. Evidence for a novel GTPase priming step in the SRP protein targeting pathway. EMBO J. 2001;20:6724–6734. doi: 10.1093/emboj/20.23.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cleverley RM, Gierasch LM. Mapping the signal sequence-binding site on SRP reveals a significant role for the NG domain. J Biol Chem. 2002;277:46763–46768. doi: 10.1074/jbc.M207427200. [DOI] [PubMed] [Google Scholar]