Summary
The formation and repair of DNA damage at specific locations in the genome is modulated by DNA sequence context, by DNA cytosine-5 methylation patterns, by the transcriptional status of the locus and by proteins associated with the DNA. The only method currently available to allow precise sequence mapping of DNA lesions in mammalian cells is the ligation-mediated polymerase chain reaction (LM-PCR) technique. We provide an update on technical details of LM-PCR. LM-PCR can be used, for example, for mapping of ultraviolet (UV) light-induced DNA photoproducts such as cyclobutane pyrimidine dimers.
Keywords: Ligation-mediated PCR, UV damage, cyclobutane pyrimidine dimers, (6-4) photoproducts
1. Introduction
The most abundant and well characterized lesions formed upon irradiation of DNA with ultraviolet (UV) light are the cyclobutane pyrimidine dimers (CPDs) and the pyrimidine (6-4) pyrimidone photoproducts [(6-4) photoproducts; (6-4)PPs] (1). CPDs are several-times more frequent than (6-4)PPs (2). Both photoproducts can cause mutations, but the CPD is the more mutagenic lesion in mammalian cells (3). CPDs persist much longer in mammalian DNA than (6-4)PPs owing to faster removal of (6-4)PPs (4,5). CPDs are subject to a specialized transcription-coupled repair pathway (6,7), which removes these lesions selectively from the template strand of genes transcribed by RNA polymerase II.
Nucleotide excision repair plays an important role in preventing UV-induced skin cancer. Cells from patients suffering from xeroderma pigmentosum (XP) are hypersensitive to UV light (8). The incidence of skin cancer in certain XP patients is increased by several thousand-fold relative to the normal population (9) and this probably is a consequence of a severe deficiency in repair of UV photolesions.
In previous work, we have developed a technique, based on the ligation-mediated PCR (LM-PCR) reaction, which can be used to analyze the formation and repair of UV photoproducts along specific human genes at the DNA sequence level (10–22). LM-PCR methods for the detection of ((6-4)PPs (12) and CPDs (13) are available. LM-PCR provides a sufficient level of sensitivity when physiologically relevant UV doses (equivalent to 10–20 J/m2 of UVC) are used for irradiation, and repair of CPDs can be measured reliably at these doses (10,11,14,17–21,23).
The ability of LM-PCR to detect DNA adducts depends on the specific conversion of adducts into strand breaks with a 5'-phosphate group. (6-4)PPs and their Dewar isomers can be converted by heating UV-irradiated DNA in piperidine (24). CPDs can be mapped at the DNA sequence level by cleavage with specific enzymes such as T4 endonuclease V (25,26). T4 endonuclease V cleaves the glycosidic bond of the 5' base in a pyrimidine dimer and also cleaves the sugar phosphate backbone between the two dimerized pyrimidines. The digestion products still contain a dimerized pyrimidine base at the cleavage site. We determined that these fragments could be amplified efficiently by LM-PCR after photoreversal of the cyclobutane ring with E. coli photolyase to result in a normal base on a 5' terminal sugar-phosphate (13).
This updated LM-PCR protocol describes the sequential steps employed for the detection of these DNA lesions, at the level of nucleotide resolution, in the genome of eukaryotic or prokaryotic cells (Figure 1). The updated protocol utilizes a computerized fluorescence-based labeling and detection method (LI-COR system; LI-COR; Lincoln, NE), which offers great advantage over the earlier versions of this protocol that included laborious and radioactivity-based steps, such as gel transfer, electroblot, 32P-containing probe synthesis, hybridization, and autoradiography. The sequential order of the steps involved in this updated protocol is as follows: (1) conversion of DNA lesions to ligatable single-stranded DNA breaks using chemical or enzymatic treatments; (2) primer extension towards the single stranded DNA breaks; (3) ligation of the extended fragments; (4) PCR amplification of the ligation products; (5) fluorescent-labeling of the PCR-amplified products; and (6) simultaneous gel electrophoresis and fluorescence detection of the labeled products. The updated protocol is a two-day procedure, although it may also be shortened to one day by replacing the overnight ligation step with a 2-hour ligation step. However, since the ligation efficiency depends upon the activity of ligase, we recommend the overnight ligation, which in our hands has shown high reproducibility. The protocol has been standardized and extensively validated in our laboratory for applications in DNA-lesion footprinting of various carcinogens in mammalian genomes. In the following sections, we describe the technical aspects of this updated protocol and specify its detailed steps. We highlight the applications of this protocol for footprinting of sunlight-induced DNA lesions with special focus on ultraviolet radiation-derived photodimers, including CPDs and (6-4)PPs (see Figure 2 for an example).
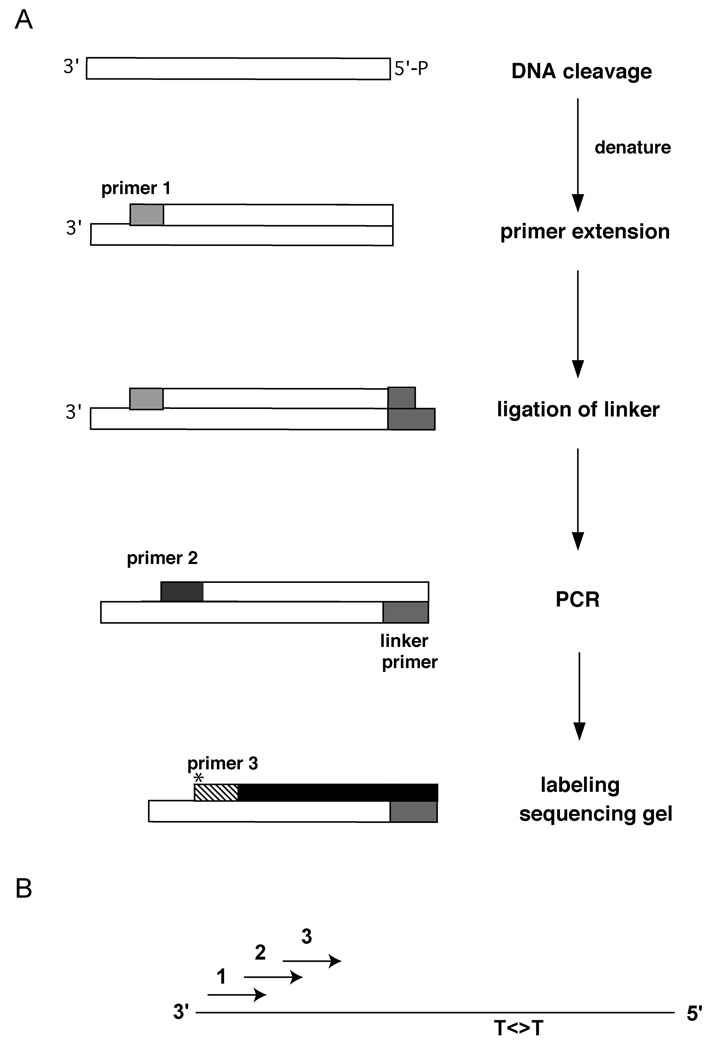
Figure 1. Outline of the LM-PCR procedure.
A. General scheme showing the LM-PCR approach for detection of strand breaks or DNA damage sites. DNA containing strand breaks introduced at the sites of UV damage is used in a primer extension reaction (with primer 1), followed by ligation of a linker, PCR (with primer 2) and a labeling step (using infrared-dye-labeled primer 3). B. Primer arrangement. The relative orientation of primers 1, 2, and 3 relative to the DNA template containing a pyrimidine dimer (T<>T) is shown.
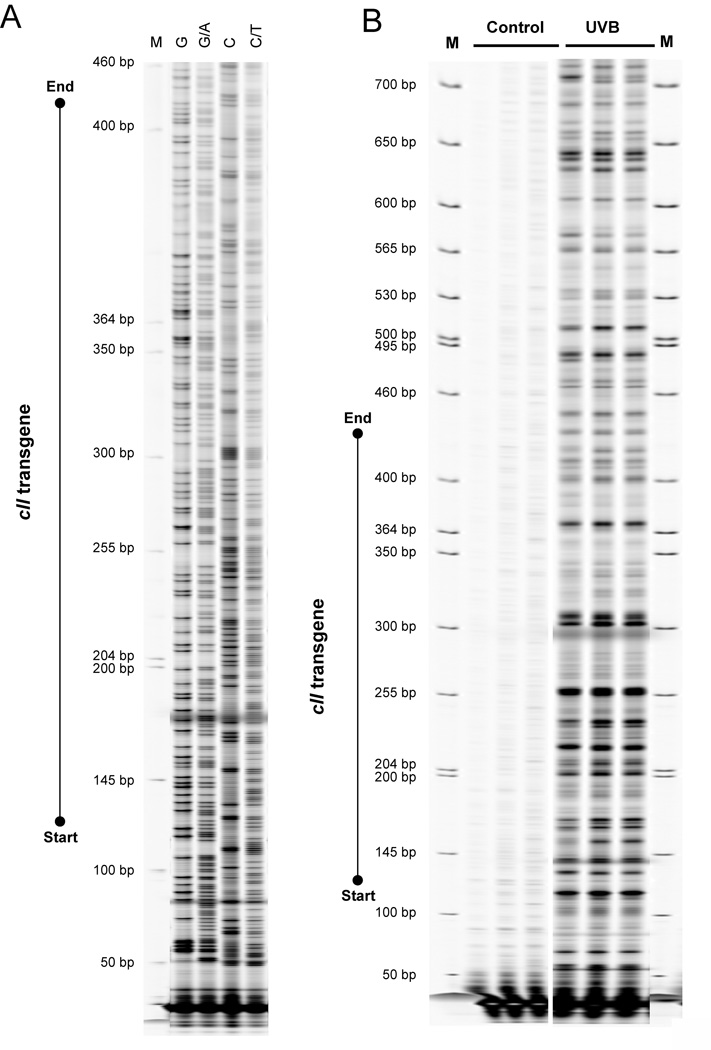
Figure 2. LM-PCR of the cII transgene gene in mouse cells.
A. Genomic DNA of transgenic Big Blue® mouse embryonic fibroblasts was subjected to standard Maxam and Gilbert chemical reactions, and subsequently DNA footprinting of the cII transgene was performed using our updated LM-PCR protocol, as described in the text. Individual Maxam/Gilbert sequencing ladders are: “G”, “G + A”, “C”, and “C + T”. B. Transgenic Big Blue® mouse embryonic fibroblasts were irradiated with ultraviolet light B (UVB) to produce CPDs. Control DNA was not irradiated. The cellular DNA was extracted and subsequently subjected to T4 endonuclease V cleavage and CPD photolyase reactivation, followed by LM-PCR to detect CPDs. M = Molecular size marker (IRDye® 700 Sizing Standard).
2. Materials
2.1. Cleavage of DNA at sites of UV photodamage
Piperidine (Fluka), 1 M, freshly prepared.
- 10×T4 endonuclease V buffer:
- 500 mM Tris-HCl, pH 7.6.
- 500 mM NaCl.
- 10 mM EDTA.
- 10 mM dithiothreitol (DTT).
- 1 mg/ml bovine serum albumin (BSA).
T4 endonuclease V. This enzyme is commercially available, for example, from Epicentre Technologies (Madison, WI), or from New England Biolabs (Ipswich, MA).
E. coli photolyase. This enzyme was kindly provided by Dr. A. Sancar (University of North Carolina at Chapel Hill) and can commercially be obtained from Trevigen (Gaithersburg, MD).
Two 360 nm black lights (Sylvania 15W F15T8).
TE buffer (10 mM Tris-HCl, pH 7.6, 1 mM EDTA).
2.2. Estimation of lesion frequency by alkaline agarose gels
Agarose.
50 mM NaCl, 4 mM EDTA.
Running buffer (30 mM NaOH, 2 mM EDTA).
Loading dye (50% glycerol, 1 M NaOH, 0.05% bromocresol green).
0.1 M Tris-Cl, pH 7.5.
Ethidium bromide (1 µg/ml).
2.3 Ligation-mediated PCR
2.3.1. Primer extension using a biotinylated primer 1
Recipe 1 (Primer extension mix)
| Component | 1× (µl) |
|---|---|
| H20 | 14.4 |
| 10 × Vent Buffer | 3.0 |
| 100 mM MgSO4 | 1.2 |
| 25 mM dNTP mix | 0.3 |
| 20 µM Primer 1 (Biotinylated) | 0.1 |
| Mix thoroughly by pipetting before adding the enzyme | --- |
| Vent(exo−) (2 Units/µl) | 1.0 |
| Total | 20.0 |
Vent(exo−), 10 × Vent Buffer, and MgSO4 (New England Biolabs (NEB); Ipswich, MA); Deoxynucleoside Triphosphate Set (Roche Diagnostics; Indianapolis, IN); add equal volumes of dATP, dCTP, dGTP, and dTTP (100 mM each) from this set to make the 25 mM dNTP.
Biotinylated primers can be synthesized in-house or alternatively purchased from various companies, such as Integrated DNA Technologies, Inc. (IDT; San Diego, CA).
2.3.2. Preparation of magnetic beads
Streptavidin-coupled magnetic beads (Dynal Biotech ASA; Oslo, Norway).
2× magnetic bead wash buffer: 2 M NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA, pH 8.0.
2.3.3. Overnight ligation
Recipe 2 (Ligation mix)
| Component | 1× (µl) |
|---|---|
| H20 | 7.95 |
| 1 M Tris-HCl, pH 7.5 | 1.50 |
| 1 M MgCl2 | 0.30 |
| 1 M DTT | 0.30 |
| 100 mM ATP | 0.30 |
| 10 mg/ml BSA | 0.15 |
| 20 µM LM-PCR Linker | 3.00 |
| Mix thoroughly by pipetting before adding the enzyme | --- |
| T4 DNA Ligase (3 Units/µl) | 1.50 |
| Total | 15.00 |
ATP = Adenosine-5-triphosphate, lithium salt 100 mM, pH 7.0 (Roche Diagnostics).
BSA = Bovine serum albumin (NEB).
DTT = Dithiothreitol (Sigma-Aldrich Inc.; Saint Louis, MO).
T4 DNA Ligase (Promega; Madison, WI).
The LM-PCR linker is prepared in 250 mM Tris-HCl, pH 7.7, by annealing a 25-mer oligonucleotide (5’ –GCGGTGACCCGGGAGATCTGAATTC) to an 11-mer (5’ –GAATTCAGATC) (final concentration of both oligonucleotides: 20 pmol/µl). This mixture is heated to 95°C for 3 minutes, and subsequently cooled down to 4°C over a time period of at least 4 hours. The LM-PCR linker is aliquoted to working solution of 100 – 200 µl each, and stored in non-defrost −20°C freezers.
2.3.4. PCR amplification
Recipe 3 (PCR amplification mix)
| Component | 1× (µl) |
|---|---|
| H20 | 18.5 |
| Q Solution | 5.0 |
| 5× Taq Buffer | 10.0 |
| 25 mM MgCl2 | 4.0 |
| 25 mM dNTP | 0.5 |
| 20 µM Primer 2 | 0.5 |
| 20 µM LP25 | 0.5 |
| Mix thoroughly by pipetting before adding the enzyme | --- |
| AmpliTaq DNA polymerase (5 Units/µl) | 1.0 |
| Total | 40.0 |
5× Taq Buffer = 200 mM NaCl, 50 mM Tris-HCl, pH 8.9, and 0.05% (w/v) gelatin. AmpliTaq DNA polymerase and 25 mM MgCl2 (Applied Biosystems; Foster City, CA).
LP25 = This is a 25-mer universal linker primer with the following sequence: 5'-GCGGTGACCCGGGAGATCTGAATTC-3'.
Q Solution (Qiagen; Valencia, CA).
Primer 2 is the second gene specific primer, which is downstream of primer 1 but may overlap a few bases with primer 1 (see Figure 1B).
2.3.5. Labeling
Recipe 4 (Labeling mix)
| Component | 1× (µl) |
|---|---|
| H20 | 0.79 |
| Q Solution | 0.30 |
| 5× Taq Buffer | 0.60 |
| 25 mM MgCl2 | 0.18 |
| 25 mM dNTP | 0.03 |
| 1 µM Primer 3 (IR-Dye® 700/800) | 1.00 |
| Mix thoroughly by pipetting before adding the enzyme | --- |
| AmpliTaq DNA polymerase (5 Units/µl) | 0.10 |
| Total | 3.00 |
Fluorescence infrared dye-labeled primers (IR-Dye® 700/800 Primer 3) can be ordered from various companies, such as LI-COR Biosciences (LI-COR; Lincoln, NE) and Integrated DNA Technologies, Inc. (IDT; San Diego, CA). Primer 3 is 3’ (downstream) of primer 2 but can overlap a few bases with primer 2 (Figure 1B).
3. Methods
3.1. Cleavage of DNA at sites of UV photodamage
3.1.1. (6-4) Photoproducts
DNA from irradiated cells is isolated by standard methods, for example by phenol-chloroform extraction. To obtain DNA fragments with a 5' phosphate group at the positions of (6-4)PPs, DNA is heated in 1 M piperidine (12). This will destroy the photolesion and create strand breaks with 5' phosphate groups since the sugar residue at the 3'-base of the (6-4)PP is cleaved by beta-elimination.
Dissolve 10 to 50 µg of UV-irradiated DNA in 100 µl of 1 M piperidine.
Heat the DNA at 90 °C for 30 min in a heat block (use lid locks to prevent tubes from popping). Cool samples briefly on ice after heating.
Add 10 µl of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethanol. Put on dry ice for 20 min.
Spin at 14,000 rpm in an Eppendorf centrifuge for 15 min.
Wash twice with 1 ml of 75% ethanol.
Remove traces of remaining piperidine by drying the sample overnight in a vacuum concentrator. Dissolve DNA in TE buffer to a concentration of approximately 0.5 to 1 µg/µl.
Determine the frequency of (6-4)PPs by separating 1 µg of the DNA on a 1.5% alkaline agarose gel along with appropriate size markers (see section 4.2).
3.1.2. Cyclobutane Pyrimidine Dimers
DNA is first incubated with T4 endonuclease V and then with E. coli photolyase to create fragments with 5'-phosphate groups and ligatable ends (13).
The UV-irradiated DNA (about 10 µg in 50 µl) is mixed with 10 µl of 10× T4 endonuclease V buffer and a saturating amount of T4 endonuclease V in a final volume of 100 µl. Saturating amounts of T4 endonuclease V activity can be determined by incubating UVC-irradiated (20 J/m2) genomic DNA with various enzyme dilutions and separating the cleavage products on alkaline agarose gels (see 4.2). Incubate at 37 °C for 1 h.
Add dithiotreitol to a final concentration of 10 mM. Add 5 µg of E. coli photolyase under yellow light.
Irradiate the samples in 1.5 ml tubes from two 360 nm UVA-emitting black lights (Sylvania 15W F15T8) filtered through 0.5 cm thick window glass for 1 h at room temperature at a distance of 3 cm.
Extract once with phenol-chloroform.
Precipitate the DNA by adding one tenth volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethanol. Leave on dry ice for 20 min. Centrifuge samples for 10 min at 14,000×g at 4 °C.
Wash pellets with 1 ml of 75% ethanol and air-dry.
Dissolve DNA in TE buffer to a concentration of about 0.5 to 1 µg/µl.
Determine the frequency of CPDs by running 1 µg of the samples on a 1.5% alkaline agarose gel.
3.2. Estimation of cleavage frequency by alkaline agarose gels
The approximate size of the fragments obtained after cleavage of UV-irradiated DNA is determined on an alkaline 1.5 % agarose gel.
Prepare a 1.5 % alkaline agarose gel by suspending agarose in 50 mM NaCl, 4 mM EDTA and microwaving. Pour the gel.
After the gel solidifies, soak it in running buffer for at least 2 h.
Dilute the DNA sample with one volume of loading dye. Incubate for 15 min at room temperature. Load the samples.
Run the gel at 40 V for 3–4 h.
Neutralize the gel by soaking for 60 min in 500 ml of 0.1 M Tris-Cl, pH 7.5.
Stain with ethidium bromide (1 µg/ml) for 30 min.
Destain in water for 30 min.
3.3. Ligation-mediated PCR
3.3.1. Primer extension using a biotinylated primer 1
Prepare the Primer extension mix (see section 2.3, Recipe 1) based on the number of samples to be analyzed (we recommend an additional 10% extra mix per preparation).
Add 20 µl of the primer extension mix to 10 µl of DNA digest (0.5 – 1.0 µg) in a siliconized 0.65-ml microcentrifuge tube.
Mix by pipetting (3–4 times).
- Place the samples in a pre-programmed thermocycler (paused at Step 1) and resume the run. The thermocycler is set up for the following program:
-
➢1st step: 95°C for 3 minutes;
-
➢2nd step: Tm of primer 1 (or up to 5°C higher) for 5 min;
-
➢3rd step: 72°C for 10 min; and
-
➢4th step: 4°C for cooling.
-
➢
3.3.2. Binding to streptavidin-coupled magnetic beads
Gently swirl the bottle containing the streptavidin-coupled magnetic beads (Dynal Biotech ASA; Oslo, Norway) to fully resuspend the beads.
Aliquot 20 µl of the beads per sample in a siliconized 0.65-ml microcentrifuge tube.
Place the microcentrifuge tubes containing the beads in a magnetic particle concentrator (MPC) (Dynal Biotech ASA), and allow for magnetic separation to occur (this may take approximately one minute).
Discard the supernatant, and wash the beads twice with 50 µl of 2× magnetic bead wash buffer (see Step 3 above).
Resuspend the beads in 30 µl of 2× wash buffer and transfer the resuspended beads to the microcentrifuge tubes containing the primer extension products (prepared in Section 3.3.1.).
Immobilize the primer-extension products to the beads by rotating the mixture at room temperature for 15 to 60 minutes.
Pulse-spin the microcentrifuge tubes (at low speed for ~2 seconds), place them in a MCP, and allow for magnetic separation to occur.
Discard the supernatant, and wash the beads twice with 50 µl of 2× wash buffer.
Resuspend the beads in 15 µl of 0.1× TE buffer, pH 7.5 (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) (see Note 3).
3.3.3. Overnight ligation
Prepare the Ligation mix (see Recipe 2 in section 2.3.3.) based on the number of samples to be analyzed (we recommend an additional 10% extra mix per preparation) (see Note 4).
Add 15 µl of Ligation mix to each sample.
Mix by pipetting (3–4 times).
Overlay with 20 µl of mineral oil (see Note 1).
Place the samples in a thermocycler, and incubate overnight at 17°C.
3.3.4. PCR amplification
-
1.
Pulse-spin the microcentrifuge tubes containing the ligation products (samples prepared in section 3.3.3, and place them in a MCP.
-
2.
Discard the supernatant, and wash the beads three times with 100 µl of 1× TE buffer, pH 8.0 (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
-
3.
Resuspend the beads in 10 µl of 0.1× TE buffer, pH 8.0 (1 mM Tris-HCl, 0.1 mM EDTA, pH 8.0) (see Note 5).
-
4.
Prepare the PCR amplification mix (see section 2.3.4., Recipe 3) based on the number of samples to be analyzed (we recommend an additional 10% extra mix per preparation).
-
2.
Add 40 µl of the PCR amplification mix to each sample [ligation product] (see, Step 3 above).
-
3.
Mix by pipetting (3–4 times).
-
4.
Overlay with 20 µl of mineral oil (see Note 1).
-
5.Place the samples in a pre-programmed thermocycler (paused at cycle 1), and resume the run. The thermocycler is programmed for the following set up:
-
➢1st step: 95°C for 2 minutes;
-
➢2nd step: Tm of primer 2 (or 1°C lower) for 2 minutes;
-
➢3rd step: 72°C for 3 minutes;
-
➢4th step: twenty cycles of (95°C for 45 seconds, Tm of primer 2 (or 1°C lower) for 2 minutes, and 72°C for 3 minutes);
-
➢5th step: 95°C for 45 seconds;
-
➢6th step: Tm of primer 2 (or 1°C lower) for 2 minutes;
-
➢7th step: 72°C for 10 minutes; and
-
➢8th step: cool at 4°C.
-
➢
3.3.5. Labeling
Pulse-spin the microcentrifuge tubes (prepared in Section 3.3.4, and place them in a MCP, and allow for magnetic separation to occur.
Aspirate 10 µl of the supernatant (without touching the beads), and transfer it to a new microcentrifuge tube on ice (see Note 6).
Prepare the Labeling mix (see section 2.3.5, Recipe 4) based on the number of samples to be analyzed (we recommend an additional 10% extra mix per preparation).
Add 3 µl of the Labeling mix to each sample [PCR product] (see step 2 above).
Mix by pipetting (3–4 times).
Overlay with 20 µl of mineral oil (see Note 1).
- Place the samples in a pre-programmed thermocycler (paused at cycle 1), and resume the run. The thermocycler is programmed for the following set up:
-
➢1st step: 95°C for 2 minutes;
-
➢2nd step: Tm of primer 3 (or up 5°C higher) for 2 minutes;
-
➢3rd step: 72°C for 3 minutes;
-
➢4th step: three to six cycles of (95°C for 45 seconds, Tm of primer 3 (or up 5°C higher) for 2 minutes, and 72°C for 3 minutes). More cycles lead to lower signal to noise ratio, i.e., higher background and more nonspecific bands.
-
➢5th step: 95°C for 45 seconds;
-
➢6th step: Tm of primer 3 (or up 5°C higher) for 2 minutes;
-
➢7th step: 72°C for 10 minutes; and
-
➢8th step: cool at 4°C.
-
➢
3.3.6. Detection
The IRDye® 700/800 fluorescence-labeled products (prepared in Section (3.3.5) are run on a polyacrylamide–urea gel electrophoresis system coupled to a computerized DNA sequencer (e.g., Long Read IR 4200 DNA Sequencing system (LI-COR)). The sequencer is equipped with a real time IRDye® 700/800-laser (dual) detector and data acquisition software, which enable simultaneous scanning of the sequencing gel during the electrophoresis run. A typical run consists of the following steps:
Prepare a 5–8% LI-COR sequencing gel, as instructed by the manufacturer (LI-COR).
Pre-run the gel for 20 minutes on a LI-COR DNA sequencer.
Preparation of samples for loading onto the gel: Add 4 µl of the IR2 Stop solution (LI-COR) to 13 µl of the IR-Dye® 700/800 labeled products (prepared in Section 3.3.5, and denature at 95°C for 2 minutes.
Cool the samples on ice for approximately 5 minutes.
Load 1.5–2.0 µl of samples onto each well of the pre-run gel.
Run the gel, as instructed by the manufacturer (LI-COR).
It is highly recommended that Maxam and Gilbert chemical reactions be prepared from the genomic sequence of interest, and run in parallel to the samples (see Figure 2A for an example of such sequencing lanes). Also, it is helpful to include the IRDye® 700/800 Sizing Standards (LI-COR), which contain labeled DNA fragments with different lengths, in all runs (Figure 2) (see Note 7). The inclusion of Maxam and Gilbert reactions and appropriate size markers in the sequencing run will help locate the exact position of each base in the sequence ladders from all samples. During the electrophoresis run, a solid-state laser diode excites the infrared dye present in the labeled DNA fragments as they migrate past the detector window. Simultaneously, a focusing fluorescence microscope containing a solid-state silicon avalanche photodiode scans back and forth across the width of the sequencing gel, and collects the data in real time. The raw data are processed and analyzed as the electrophoresis run progresses. The output image data are saved as TIFF files, and can be retrieved at any time during the electrophoresis run. Quantification of the image data can be achieved by specialized image analysis softwares, e.g., Gene ImagIR (Scanalytics Inc., Rockville, MD).
ACKNOWLEDGEMENT
This work was supported by a grant from the National Institute of Environmental Health Sciences (ES06070) to G.P.P.
Footnotes
Overlaying the samples with mineral oil is not needed if a thermocycler with hot-bonnet heated lid is used for the primer extension reaction. In such case, skip Step 4, pre-start the thermocycler run, and pause at 95°C (1st step) to allow the hot bonnet to equilibrate at this temperature.
Handling and processing of all samples in section 3.3.1. should be done on ice.
Handling and processing of all samples in section 3.3.2. can be done at room temperature.
Handling and processing of all samples in section 3.3.3. should be done on ice.
Handling and processing of all samples in Section 3.3.4 should be done on ice.
Preserve the leftover PCR product at 4°C (Do not freeze!). We have successfully used the leftover PCR products, which were stored at 4°C for several weeks, for subsequent labeling reactions.
Both the IR-Dye® 700/800 fluorescence primers and the sizing standards are light sensitive and should be handled under dimmed or yellow light.
REFERENCES
- 1.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 2.Yoon J-H, Lee C-S, O’Connor T, Yasui A, Pfeifer GP. The DNA damage spectrum produced by simulated sunlight. J Mol Biol. 2000;299:681–693. doi: 10.1006/jmbi.2000.3771. [DOI] [PubMed] [Google Scholar]
- 3.You YH, Lee DH, Yoon JH, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J Biol Chem. 2001;276:44688–44694. doi: 10.1074/jbc.M107696200. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell DL, Nairn RS. The biology of the (6-4) photoproduct. Photochem Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell DL, Fernandez AA. Different types of DNA damage play different roles in the etiology of sunlight-induced melanoma. Pigment Cell Melanoma Res. 2011;24:119–124. doi: 10.1111/j.1755-148X.2010.00789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 7.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 8.Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature (London) 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 9.Hanawalt PC, Sarasin A. Cancer-prone hereditary diseases with DNA processing abnormalities. Trends Genet. 1986;2:124–129. [Google Scholar]
- 10.Dammann R, Pfeifer GP. Lack of gene- and strand-specific DNA repair in RNA polymerase III transcribed human tRNA genes. Mol Cell Biol. 1997;17:219–229. doi: 10.1128/mcb.17.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao S, Drouin R, Holmquist GP. DNA repair rates mapped along the human PGK-1 gene at nucleotide resolution. Science. 1994;263:1438–1440. doi: 10.1126/science.8128226. [DOI] [PubMed] [Google Scholar]
- 12.Pfeifer GP, Drouin R, Riggs AD, Holmquist GP. In vivo mapping of a DNA adduct at nucleotide resolution: detection of pyrimidine (6-4) pyrimidone photoproducts by ligation-mediated polymerase chain reaction. Proc Natl Acad Sci USA. 1991;88:1374–1378. doi: 10.1073/pnas.88.4.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeifer GP, Drouin R, Riggs AD, Holmquist GP. Binding of transcription factors creates hot spots for UV photoproducts in vivo. Mol Cell Biol. 1992;12:1798–1804. doi: 10.1128/mcb.12.4.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tornaletti S, Pfeifer GP. Slow repair of pyrimidine dimers at p53 mutation hotspots in skin cancer. Science. 1994;263:1436–1438. doi: 10.1126/science.8128225. [DOI] [PubMed] [Google Scholar]
- 15.Tornaletti S, Pfeifer GP. Ligation-mediated PCR for analysis of UV damage. In: Pfeifer GP, editor. Technologies for Detection of DNA Damage and Mutations. New York: Plenum Press; 1996. pp. 199–209. [Google Scholar]
- 16.Törmänen VT, Pfeifer GP. Mapping of UV photoproducts within ras protooncogenes in UV-irradiated cells: correlation with mutations in human skin cancer. Oncogene. 1992;7:1729–1736. [PubMed] [Google Scholar]
- 17.Tu Y, Tornaletti S, Pfeifer GP. DNA repair domains within a human gene: selective repair of sequences near the transcription initiation site. EMBO J. 1996;15:675–683. [PMC free article] [PubMed] [Google Scholar]
- 18.Tommasi S, Oxyzoglou AB, Pfeifer GP. Cell cycle-independent removal of UV-induced pyrimidine dimers from the promoter and the transcription initiation domain of the human CDC2 gene. Nucleic Acids Res. 2000;28:3991–3998. doi: 10.1093/nar/28.20.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu W, Feng Z, Chasin LA, Tang MS. Transcription-coupled and transcription-independent repair of cyclobutane pyrimidine dimers in the dihydrofolate reductase gene. J Biol Chem. 2002;277:38305–38310. doi: 10.1074/jbc.M206375200. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Q, Wani MA, El-Mahdy M, Wani AA. Decreased DNA repair efficiency by loss or disruption of p53 function preferentially affects removal of cyclobutane pyrimidine dimers from non-transcribed strand and slow repair sites in transcribed strand. J Biol Chem. 2000;275:11492–11497. doi: 10.1074/jbc.275.15.11492. [DOI] [PubMed] [Google Scholar]
- 21.Tu Y, Bates S, Pfeifer GP. Sequence-specific and domain-specific DNA repair in xeroderma pigmentosum and Cockayne syndrome cells. J Biol Chem. 1997;272:20747–20755. doi: 10.1074/jbc.272.33.20747. [DOI] [PubMed] [Google Scholar]
- 22.Hendriks G, Calleja F, Besaratinia A, Vrieling H, Pfeifer GP, Mullenders LH, Jansen JG, de Wind N. Transcription-dependent cytosine deamination is a novel mechanism in ultraviolet light-induced mutagenesis. Curr Biol. 2010;20:170–175. doi: 10.1016/j.cub.2009.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besaratinia A, Kim SI, Pfeifer GP. Rapid repair of UVA-induced oxidized purines and persistence of UVB-induced dipyrimidine lesions determine the mutagenicity of sunlight in mouse cells. FASEB J. 2008;22:2379–2392. doi: 10.1096/fj.07-105437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippke JA, Gordon LK, Brash DE, Haseltine WA. Distribution of UV light-induced damage in a defined sequence of human DNA: detection of alkaline-sensitive lesions at pyrimidine nucleoside-cytidine sequences. Proc Natl Acad Sci USA. 1981;78:3388–3392. doi: 10.1073/pnas.78.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon LK, Haseltine WA. Comparison of the cleavage of pyrimidine dimers by the bacteriophage T4 and Micrococcus luteus UV-specific endonucleases. J Biol Chem. 1980;255:12047–12050. [PubMed] [Google Scholar]
- 26.Radany EH, Friedberg EC. A pyrimidine dimer-DNA glycosylase activity associated with the v gene product of bacterophage T4. Nature. 1980;286:182–185. doi: 10.1038/286182a0. [DOI] [PubMed] [Google Scholar]