SUMMARY
GB virus C/hepatitis G virus (GBV-C/HGV) is the most closely related human virus to hepatitis C virus (HCV). GBV-C is lymphotropic and not associated with any known disease, although it is associated with improved survival in HIV-infected individuals. In peripheral blood mononuclear cells, GBV-C induces the release of soluble ligands for HIV entry receptors (RANTES, MIP-1a, MIP-1b and SDF-1), suggesting that GBV-C may interact with lymphocytes to induce a chemokine and/or cytokine milieu that is inhibitory to HIV infection. Expression of GBV-C envelope glycoprotein E2 in CD4+ T cells or addition of recombinant E2 to CD4 cells recapitulates the HIV inhibition seen with GBV-C infection. Like HCV E2, GBV-C E2 is predicted to be post-translationally processed in the endoplasmic reticulum and is involved with cell binding. The C-termini of GBV-C E1 and E2 proteins contain predicted transmembrane domains sharing features with HCV TM domains. To date, cellular receptor(s) for GBV-C E2 have not been identified. GBV-C E2-mediated HIV inhibition is dose-dependent and HIV replication is blocked at the binding and/or entry step. In addition, a putative GBV-C E2 fusion peptide interferes with HIV gp41 peptide oligomerization required for HIV-1 fusion, further suggesting that GBV-C E2 may inhibit HIV entry. Additional work is needed to identify the GBV-C E2 cellular receptor, characterize GBV-C E2 domains responsible for HIV inhibition, and to examine GBV-C E2-mediated fusion in the context of the entire envelope protein or viral-particles. Understanding the mechanisms of action may identify novel approaches to HIV therapy.
Keywords: envelope glycoprotein, GBV-C, hepatitis G virus, HIV
HISTORY
Following the discovery of hepatitis C virus (HCV), it became clear that HCV was not detected in 10–20% of individuals with non-A, non-B hepatitis [1,2]. Virus discovery groups at Genelabs, Inc. and Abbott laboratories independently reported the identification of a virus in subjects with non-A, non-B, non-C hepatitis in 1995 and 1996 which shared several features with HCV [1,2]. Because a putative hepatitis F virus had been described [3], Genelabs named the virus they identified in a non-A, non-B, non-C hepatitis patient ‘Hepatitis G virus’ (HGV) [2], although there were limited epidemiological data to support an association with hepatitis. Abbott Laboratories had previously found two viruses in marmosets that had been inoculated with serum from a surgeon with non-A, non-B hepatitis whose initials were G.B. [4]. These viruses were also closely related to HCV and were called GB virus A and B (GBV-A, GBV-B) [4]. GBV-A and GBV-B were not found in humans, but using degenerate primers based on GBV-A and B, Abbott Laboratories subsequently identified a closely related virus in humans which they called GBV-C [1]. Sequence analysis revealed that HGV and GBV-C were two isolates of the same virus, and the proper taxonomic name is HGV/GBV-C (reviewed in [5]).
EPIDEMIOLOGY AND DISEASE ASSOCIATION
Numerous studies designed to determine if HGV/GBV-C represented an aetiological agent for acute or chronic hepatitis were reported between 1996 and 1998, and prospective and well-controlled retrospective studies did not observe an epidemiological association between this new virus and acute or chronic hepatitis (reviewed in [5–7]). Consequently, this virus is by definition not a ‘hepatitis’ virus and the term HGV is misleading. Similarly, no evidence exists to suggest that the surgeon G.B. was infected with GBV-C [5]. Thus, neither HGV nor GBV-C accurately describes this virus [1]. However, since the virus does not cause hepatitis, it will be referred to as GBV-C in this report. Since no convincing association between the virus and any disease entity has been identified (reviewed in [5,7]), it appears to be a nonpathogenic human virus.
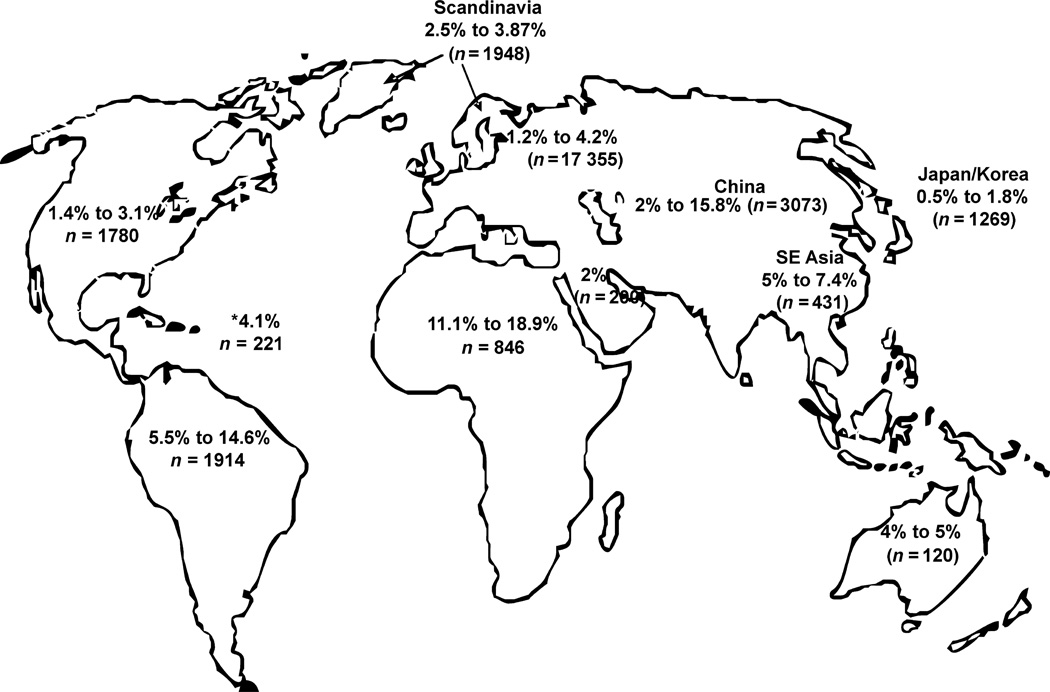
Like hepatitis C virus (HCV), GBV-C infection is found worldwide and is capable of persistent human infection [8,9]. GBV-C infection is common, and between 1 and 5% of healthy blood donors in developed countries are viraemic at the time of donation. The prevalence is higher in blood donors from developing countries (Fig. 1), and the prevalence of GBV-C is significantly higher in individuals with coexistent blood borne or sexually transmitted infections (reviewed in [5,7]). For example, the prevalence of GBV-C viraemia in HCV- and infected individuals is approximately 20 and 30%, respectively reviewed in [5–7]. Unlike HCV, antibodies to GBV-C proteins are not usually detected during viraemia, and antibody to the GBV-C envelope glycoprotein E2 develops following the clearance of viraemia [10]. E2 antibody appears to be somewhat protective against re-infection; thus, E2 antibody is thought to contain neutralizing activity [11]. Active viraemia is determined by the detection of viral RNA in sera or plasma using RT-PCR methods, while prior infection is inferred by the detection of GBV-C E2 antibodies (reviewed in [5,7,12]). Because E2 antibodies may disappear during longitudinal follow up, prevalence studies probably underestimate the rate of prior exposure [13].
Fig. 1.
GB virus C prevalence rates in blood donors from various regions of the world. GBV-C viraemia prevalence among 11 391 blood donors was summarized from 50 studies (references available upon request). Viraemia was detected by RT-PCR, and studies only included donors who passed screening procedures with the exception of one Scandinavian study that included donors with normal and high ALT (n = 393 high ALT, n = 184 with normal ALT; the GBV-C prevalence rate was similar in both groups). In developed countries, GBV-C RNA prevalence in blood donors ranged from 0.5 to 5%, compared to 5–18.9% in developing countries. *Caribbean (West Indies).
Although GBV-C viraemia may persist for decades, viraemia clears within 2 years following infection in the majority of individuals infected by blood transfusion (reviewed in [5,6]). By comparing the ratio of E2 antibody positive blood donors to donors with GBV-C viraemia, it appears that approximately 75% of GBV-C infections are spontaneously cleared by the infected host, at least in immune competent individuals [1,2,5–7]. Specifically, the prevalence of E2 antibody to GBV-C viraemia in blood donors is approximately 6:1, while the ratio of individuals with HCV antibody individuals without viraemia compared to those with viremia is approximately to 1:4, thus HCV infection is more likely to persist than GBV-C (reviewed in [5,6]). Among individuals with HIV infection, the ratio of E2 antibody prevalence to viraemia is generally less than 2:1, suggesting that GBV-C viral clearance is reduced in individuals with impaired cellular immunity [14].
Genome structure
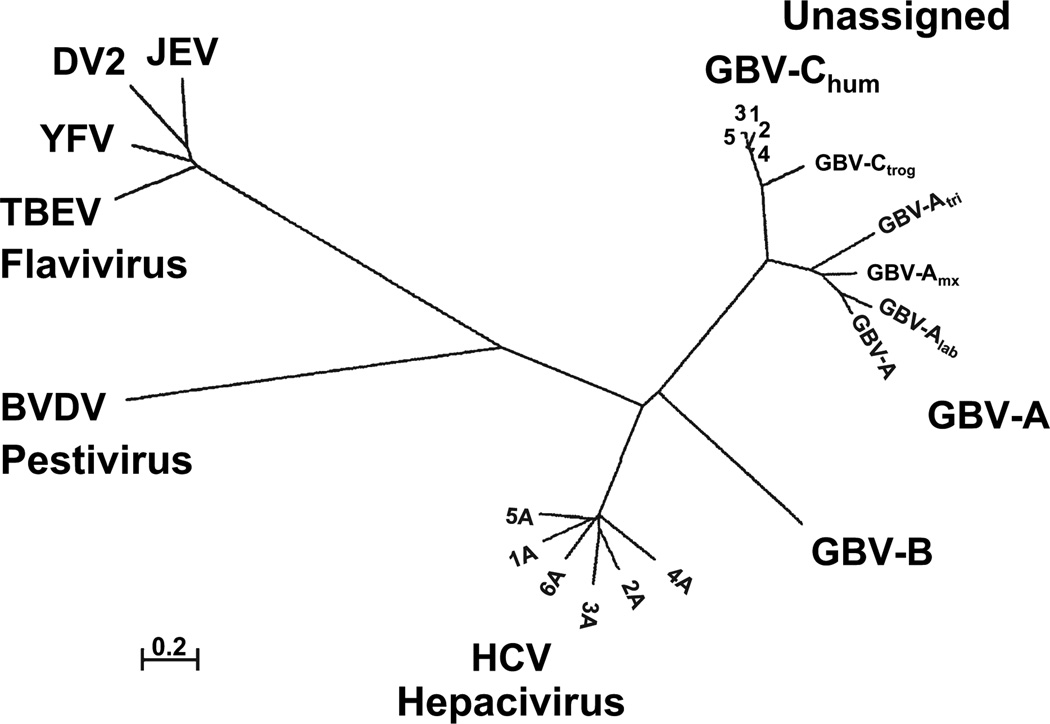
Based on nucleotide sequence and genome organization, GBV-C and HCV are classified as members of the family Flaviviridae, which has three known genera (flavi, pesti, and hepaci). The GB viruses are unassigned, although GBV-B has been proposed to be within the hepaciviruses. The phylogenetic relationships between GBV-A, GBV-B, GBV-C, HCV and representative members of the pesti and flavi genera are shown in Fig. 2. GBV-C shares considerable sequence identity with GBV-A (48%) and to a lesser extent with GBV-B and HCV (~30% for both) [15].
Fig. 2.
Phylogenetic relationships of the RNA-dependent, RNA polymerase sequences of several members of the family Flaviviridae. Three genera (flavivirus, pestivirus and hepacivirus) and the unassigned GB viruses are shown. Representative isolates from the six hepatitis C virus (HCV), five GBV-C, chimpanzee GBV-C (troglodyte) variant, and four GBV-A genotypes are depicted. The four GBV-A geno-types were identified in Sanguinus nigrocallis (GBV-A), Sanguinus labiatus (GBV-Alab), Aotus Trivirgatus (GBV-Alab), and a ‘callithrix hybrid’ (jacchus-penicillata cross) (GBV-Amyx). BVDV, bovine viral diarrhoea virus; BVDV; YFV, yellow fever virus (17D vaccine strain); TBEV, tick-borne encephalitis virus; DV, dengue virus (serotype 2) and JEV, Japanese encephalitis virus. 0.2, distance representing 0.2 amino acids substitutions per position.
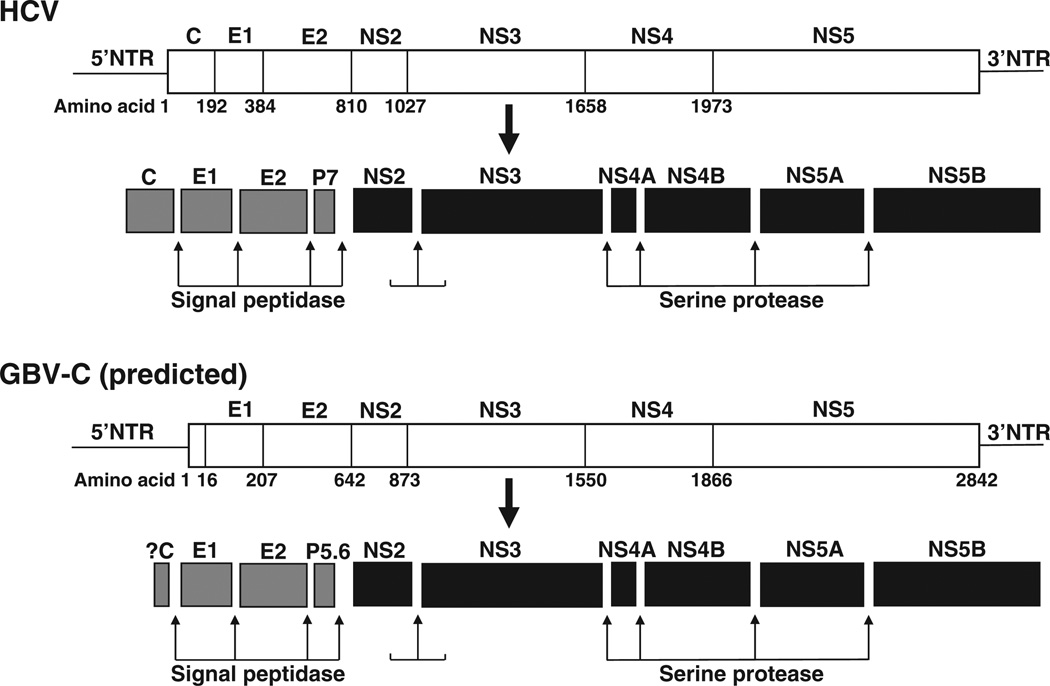
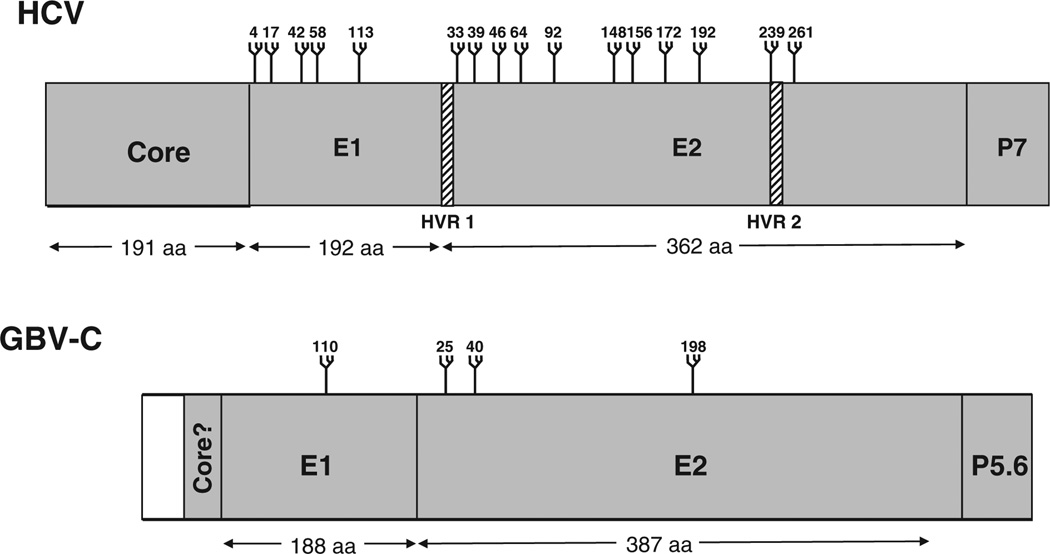
GBV-C contains a 9.4 kb single-stranded, positive sense RNA genome that is organized similarly to HCV (Fig. 3). HCV and GBV-C genomes contain 5′ and 3′ nontranslated regions (NTRs) and contain a long open reading frame (ORF) encoding approximately 3000 amino acids that is post-translationally cleaved into structural and nonstructural (NS) proteins [1,2,15]. The predicted 5′ntr of GBV-C (555 bp) is longer than the HCV 5′ntr (341 nt) and both contain an internal ribosome entry site (IRES) that directs translation of the mRNA, though the activity of the GBV-C IRES is considerably lower than that of HCV [16]. The GBV-C 3′ntr (~300 bp) is different from the HCV 3′ntr in that it does not contain poly-(A) or poly-(U) tracts, though it shares predicted structural elements [17]. HCV translation and protein processing have been experimentally demonstrated; however, GBV-C processing is largely based on predictions using sequence comparisons with HCV. The coding region for HCV and GBV-C structural proteins is found in the N-terminal one third of the ORF (reviewed in [18]). The envelope glycoproteins (E1 and E2) appear to form a heterodimer and are thought to be inserted into the viral envelope. The HCV pore-forming ion channel protein (p7) is predicted to be smaller (5.6 kDa) in GBV-C. Although the HCV ORF encodes a core protein upstream of E1 and GBV-C particles have similar biophysical characteristics with HCV, the coding region for a GBV-C core protein has not been definitively identified [19]. In addition, there is an alternative ORF within the HCV core coding region [20] which does not appear to exist in GBV-C.
Fig. 3.
Genome organization and proteolytic processing of hepatitis C virus (HCV) and GB virus C (GBV-C). HCV and GBV-C both contain 5′ nontranslated regions (NTR) containing internal ribosomal entry sites directing translation of polyproteins. The polyproteins are post-translationally processed into structural proteins [core (C), envelope glycoproteins (E1 and E2)] and an ion channel P7 (P5.6 for GBV-C) by cellular signal peptidases. Nonstructural proteins (NS) 2 and 3 are cleaved by NS2 protease, while the remaining cleavage sites are processed by the serine protease domain within NS3, in conjunction with the NS4A cofactor.
The C-terminal portion of the ORF encodes the NS proteins, NS2, NS3, NS4, NS5A and NS5B. The functions of the GBV-C NS proteins have not been experimentally characterized with one exception, but are proposed to be similar to HCV. GBV-C NS2 and NS3 proteins are predicted to function as a viral proteases, and the specific processing scheme for GBV-C is described in detail in the section on ‘Proteolytic Processing’ [15]. The C-terminal region of NS3 is predicted to have NTPase and helicase functions, and NS5B is an RNA-dependent RNA polymerase [15]. NS4B and 5A are less well characterized, but are predicted to function as a membrane alteration inducer and multifunctional phosphoprotein interfering with numerous cellular pathways respectively [18]. A summary of the HCV and predicted GBV-C genome organization and processing is shown in Fig. 3.
Genotypes
Based on phylogenetic analysis of more than 30 full-length GBV-C genome sequences, at least five, and possibly six genotypes of GBV-C have been identified [21]. The global distribution of different genotypes follows distinct patterns consistent with the migration patterns of humans out of Africa, suggesting that GBV-C has co-evolved with its human hosts [22]. Despite this presumed ancient history, GBV-C has a surprising lack of genetic diversity between GBV-C variants (<14%) compared with HCV (>30%) (Fig. 2) [22]. A variant of GBV-C was identified in chimpanzees (GBV-Ctrog) in 1998 and was significantly more diverse than human GBV-C isolates, further supporting species co-evolution [23,24]. GBV-Ctrog is predicted to have the same genome organization and protein function as human GBV-C and HCV, and the only full-length GBV-Ctrog sequence published (Accession number AF070476) demonstrates 83.6 and 32% identity with GBV-C and HCV respectively [23]. The phylo-genetic relationships between GBV-C genotypes and the human and troglodyte variants are shown in Fig. 2.
Tropism
Following translation and processing of the polyprotein, the GBV-C NS5B RNA dependent, RNA polymerase transcribes negative strand RNA from which positive strand RNA is subsequently transcribed. Thus, detection of negative strand RNA in cells is indicative of active viral RNA replication. Initial reports suggested that GBV-C negative strand RNA was present in liver tissue [25,26]; however, comparison of HCV and GBV-C RNA levels in liver and serum of co-infected individuals found that HCV RNA levels were consistently higher than the levels of GBV-C RNA in liver tissues, despite higher serum GBV-C RNA levels [27]. Furthermore, the median liver/serum ratio of GBV-C RNA was <1.0, consistent with serum contamination of liver tissue [27]. Laskus et al. were unable to demonstrate minus-strand GBV-C RNA in any of 10 liver samples tested [28] and in a clinical study, GBV-C serum RNA levels did not significantly decrease following liver transplantation, although this routinely occurs with HCV RNA levels [29]. These data support a non-hepatic source of GBV-C replication.
GBV-C RNA is found in, and is produced by, T and B lymphocytes removed from infected individuals [30,31]. Both CD4 + and CD8 + T cells contain GBV-C RNA, and the most widely-reported cell culture system for in vitro growth of GBV-C utilizes primary human peripheral blood mononuclear cells (PBMCs), suggesting that GBV-C is a lymphotropic virus (reviewed in 7, 30) and explaining the lack of association with hepatitis (reviewed in [5,7]). Nevertheless, questions remain about the primary site of GBV-C replication in humans. Negative strand GBV-C RNA is either very low in concentration or not detected in PBMCs of infected humans [31], and negative sense RNA was found in three of four bone marrow samples and two of three spleen samples in one study [32], suggesting that a lymphocyte progenitor cell may be the primary site of infection. Consistent with lymphotropism, GBV-C infection is transmitted by blood borne, vertical, and sexual routes (reviewed in [5,7,12]).
GBV-C–HIV INTERACTIONS
GBV-C research was initially performed by viral hepatitis research groups, and the realization that GBV-C did not cause hepatitis resulted in a marked reduction in research activity. However, in 1998, two groups reported that HIV-infected individuals who were co-infected with GBV-C survived longer than those without GBV-C, and these results were confirmed in several, though not all, subsequent studies (reviewed in [12,33]). It became clear that persistent viraemia with GBV-C was important for this association, as some individuals clear viraemia during follow up. These individuals have a worse prognosis compared to those who had never had GBV-C viraemia or in whom E2 antibodies were detected [14,34]. A meta-analysis of survival in studies of 1294 HIV-infected individuals in the era prior to effective combination anti-HIV (antiretroviral; ART) therapy found that persistent GBV-C viraemia is associated with a significant reduction in the risk of death (relative risk 0.41; 95% confidence interval 0.23–0.69) compared to those without GBV-C viraemia [33].
Most, though not all studies conducted after widespread use of combination ART have not identified a significant association between GBV-C and survival, presumably due to the reduction in mortality resulting from therapy (reviewed in [12]). This is true for other markers of delayed HIV disease progression as well, confirming that biological modifiers of HIV disease progression are generally modest by comparison to combination ART. Nevertheless, identifying variables that influence HIV disease progression are important for under-standing the natural history of HIV, and characterization of the mechanism(s) by which these variables influence HIV disease may identify new approaches for HIV therapy.
The finding that GBV-C replicates in CD4 + T cells in vitro 17] stimulated research on identifying potential interactions between GBV-C and HIV. GBV-C infection of PBMCs was shown to inhibit the replication of both CCR5-tropic and CXCR4-tropic HIV isolates in a co-infection model, and this effect was mediated at least in part by the induction of the chemokine ligands of the HIV entry receptors CCR5 and CXCR4 [35–38]. Consistent with this, HIV isolates representing all HIV Clades (A–H) and group O viruses were inhibited by GBV-C [38]. Inhibition of HIV did not depend on GBV-C replication, as transfection of either an infectious GBV-C RNA transcript or an RNA containing a deletion rendering the GBV-C replication incompetent inhibited HIV [17,38]. Furthermore, incubation of virus-free supernatants from GBV-C infected PBMCs inhibited HIV in vitro [38].
Cell culture studies found that GBV-C induces the release of soluble ligands for HIV entry receptors [RANTES, macro-phage inflammatory proteins (MIP)-1α and MIP-1β and SDF-1] [36,38,39]. In addition, CCR5 surface expression is decreased in GBV-C infected PBMCs, consistent with internalization of chemokine receptors upon increased ligand binding [36,38,39]. Consistent with this, a clinical study found that HIV-infected patients with GBV-C viraemia have significantly reduced CCR5 expression on their CD4 + T cells [39]. Epidemiological studies also found that either decreased CCR5 surface expression or increased serum levels of CCR5 and CXCR4 chemokine ligands is associated with prolonged survival in HIV-infected individuals [40]. A recent study found that the level of interferon activation in dendritic cells was significantly higher in GBV-C–HIV coinfected individuals compared to that in HIV-monoinfected people (as measured by endogenous levels of IFN-γ and PKR mRNA levels) [41]. In addition, interferon regulated gene expression correlated with IFN-γ expression, and dendritic cell activation (measured by CD80 expression) correlated with GBV-C viral load, supporting activation of innate immunity by GBV-C infection [41]. These multiple effects of GBV-C on immune cells provide a model in which GBV-C may inhibit HIV replication through the modulation of soluble ligands for the HIV coreceptor CCR5 and possibly CXCR4 and activation of innate immunity.
Given the in vitro interference between GBV-C and HIV replication, it is not surprising that GBV-C viraemia is associated with improved clinical response to antiretroviral therapy [42], and there is an inverse relationship between GBV-C and HIV plasma viral load [9,43]. Recent studies also found that GBV-C viraemia is associated with a block in CD4 + T cell proliferation following IL-2 therapy, suggesting that GBV-C also influences CD4 proliferation in response to IL-2 [44]. Taken together, these findings suggest that GBV-C modulates T cell homeostasis in ways that are beneficial for people infected with HIV.
Two viral proteins have been shown to inhibit HIV replication in vitro. Specifically, expression of NS5A downregulates CXCR4 and induces the expression of SDF-1 (the CXCR4 ligand) in a CD4 + T cell line, rendering the cells nearly completely resistant to HIV infection [45]. Expression of a 16 amino acid domain within NS5A is sufficient to inhibit HIV replication, and the addition of a synthetic peptide containing this domain resulted in dose–dependent HIV inhibition [46], suggesting therapeutic potential. During GBV-C infection, NS5A is only expressed in cells actively infected with GBV-C, and the protein is anchored in the endoplasmic reticulum, thus any effect on neighbouring cells would require the induction of soluble factors like chemokines to result in a widespread or potent effect [17].
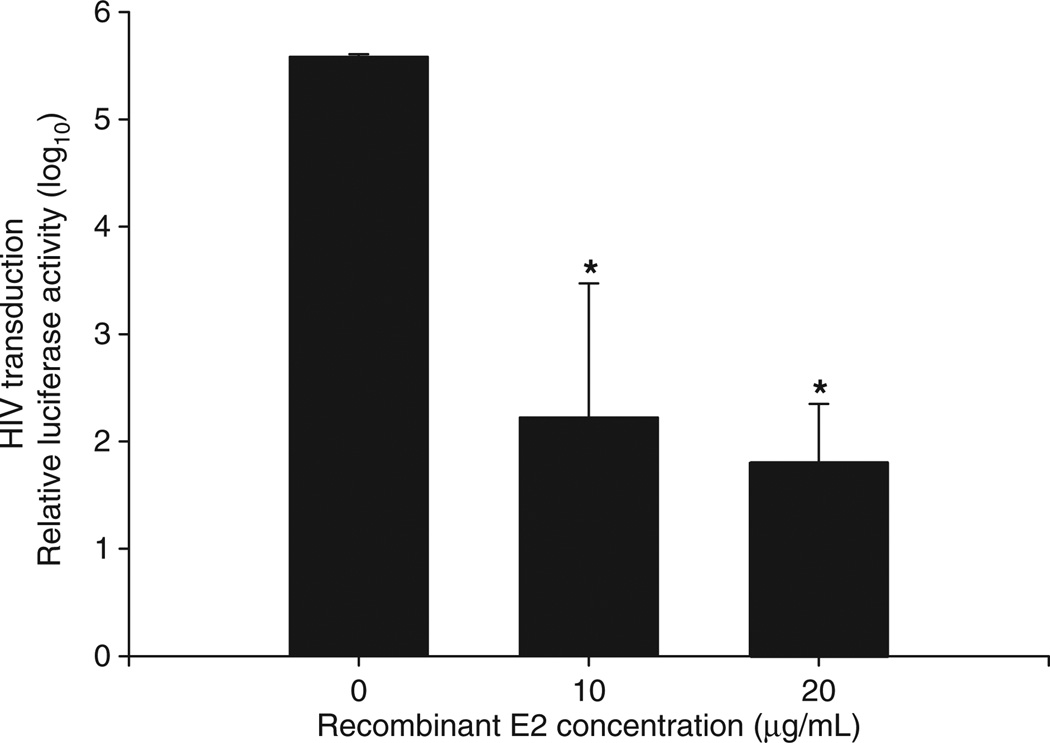
Recent data indicates that expression of the GBV-C E2 protein in CD4 + T cells or adding recombinant E2 protein to cells results in potent inhibition of HIV replication [39,47,48] and (Fig. 4). E2 is thought to mediate GBV-C binding to permissive cells, and thus it may be an important determinant of viral tropism [39,49,50]. Thus, it appears that engagement of the structural proteins with cellular receptors induces secretion of HIV inhibitory factors from PBMCs.
Fig. 4.
GB virus C envelope glycoprotein 2 (E2) inhibits early steps in the HIV life cycle. Ghost cells expressing CD4, CCR5 and CXCR4 were incubated with recombinant E2 at 4 °C for 4 h prior to transduction with HIV pseudotyped particles bearing a luciferase reporter. GBV-C E2 inhibited transduction of HIV pseudotyped particles in a dose–dependent fashion (*P < 0.01; t-test).
Since E2 is present on virions, and GBV-C titres are typically between 107 and 108 genome equivalents per millilitre plasma in infected individuals [9,43], this protein has ample access to HIV particles and infected cells. The remainder of this report will focus on current information related to GBV-C envelope glycoproteins in order to identify potential mechanisms by which the E2 protein is involved in HIV inhibition.
GB VIRUS C ENVELOPE GLYCOPROTEINS
Translation initiation
GBV-C contains a long 5′ntr that shares very little identity with the HCV 5′ntr [16]. Therefore, the location of the translation initiation site cannot be easily deduced by sequence comparison. The GBV-C 5′ntr has up to four AUG codons that are in-frame and could initiate translation, depending on the isolate [16,17]. Coupled in vitro transcription–translation studies with the GBV-C 5′ntr directing translation of a chloramphenicol acetyltransferase reporter gene demonstrated that the AUG codon at nt 555 (based on Genbank accession no. AF121950) is the site of translation initiation [16]. Nevertheless, the first in-frame AUG codon (nt 462) is highly conserved among GBV-C isolates [19]. There are no data to confirm if the translation initiator AUG codon identified in vitro is utilized in vivo.
Structural protein characteristics: proteolytic processing
A cellular signal peptidase is predicted to cleave E1-E2, E2-P7 and P7-NS2, based on consensus eukaryotic signal sequence cleavage sites [15]. The NS2 viral serine protease mediates cleavage at the NS2/NS3 [51] as in HCV (Fig. 3). The NS3 protease mediates NS3/NS4 and NS5A/NS5B cleavage and, together with NS4A, mediates cleavage of the NS4B/NS5A junction [15,51]. The protease responsible for mediating the cleavage of NS4A/NS4B was undetermined in these studies, but like HCV, the NS3 protease along with the NS4A cofactor is predicted to mediate this cleavage [51].
Because the processing sites of the structural proteins and the translation initiation AUG codon utilized in vivo have not been examined experimentally, the existence and genome region responsible for encoding a core protein are unclear. Biophysical studies demonstrate that HGV particles are similar to HCV particles, suggesting the existence of a nucleocapsid [19]. If the amino terminus of E1 is processed at a predicted signal peptide site (http://www.cbs.dtu.dk/services/SignalP/) using the predicted initiator AUG codon at nt 555, a 23 amino acid long, 2.2 kDa core protein with a pI of 5.4 would result. The size and pI are not consistent with other flavivirus core proteins. However, if the translation initiation site was located at nt 462, the core protein would be 5.4 kDa and the pI would be 11.0, similar to that observed in some other flaviviral core proteins [19]. The lack of a definitive AUG initiator codon makes numbering the proteins and their processing sites problematic, so in this review we number based on the predicted signal peptidase cleavage site. An alternative hypothesis has been proposed suggesting that the core protein is not part of the polyprotein. It has been hypothesized that the core protein is encoded on the negative strand of the GBV-C genome, or that GBV-C utilizes a cellular protein to form its nucleocapsid [52], although conclusive data are lacking. Thus, 14 years after discovery, the composition of the GBV-C nucleocapsid remains unknown.
Based on predicted structural similarities with HCV envelope glycoproteins, the GBV-C envelope glycoprotein ectodomains are proposed to be targeted to the ER lumen and the transmembrane (TM) domains inserted into the ER membrane to become type I TM proteins [18]. The GBV-C E1/E2 heterodimer is predicted to be retained in the ER because of the presence of N-linked oligosaccharides on E2 [53]. Based on predicted signal peptidase cleavage sites and sequence predictions, GBV-C E1 and E2 are predicted to be 20.7 and 41.8 kDa (without glycosylation). Treatment of secreted E2 lacking the C-terminal TM domain with N-glycanase reduces the molecular weight of protein, indicating that N-linked oligosaccharides are present [53]. Based on N-linked glycosylation predictions, HCV E1 and E2 contain considerably more sugars that GBV-C: HCV E2 has between 6 and 11 glycosylation sites (depending upon the isolate) [7,18] (Fig. 5). One of the targets of the host immune response for HCV infection is the first of two hypervariable regions which includes 27 amino acids at the N-terminus of E2 (HVR1). In contrast, GBV-C does not have hypervariable regions within E2 [12]. The combination of extensive glycosylation and hypervariable sequence domains on HCV E2 presumably contributes to immune evasion, and it is likely that these features explain, at least in part, the higher rate of HCV persistence compared to GBV-C. The C-terminus of E2 is followed by a putative signal peptidase site that would cleave a small, amphipathic protein (5.6 kDa) which appears to be a homolog of the HCV P7 protein. In HCV, this protein forms an ion channel [18].
Fig. 5.
Comparison of hepatitis C virus (HCV) and GB virus C (GBV-C) structural proteins and glycosylation sites. The relative size and predicted N-linked glycosylation sites on HCV and GBV-C core and envelope glycoproteins (E1 and E2) proteins are shown (http://www.cbs.dtu.dk/services/NetNGlyc/). The GBV-C core protein is depicted as residing at the N-terminus of the polyprotein. Grey represents the GBV-C Core if the AUG at position 462 serves as initiation codon, while blue represents initiation at the AUG at 555.
Hydrophobicity and C-terminal domain topology
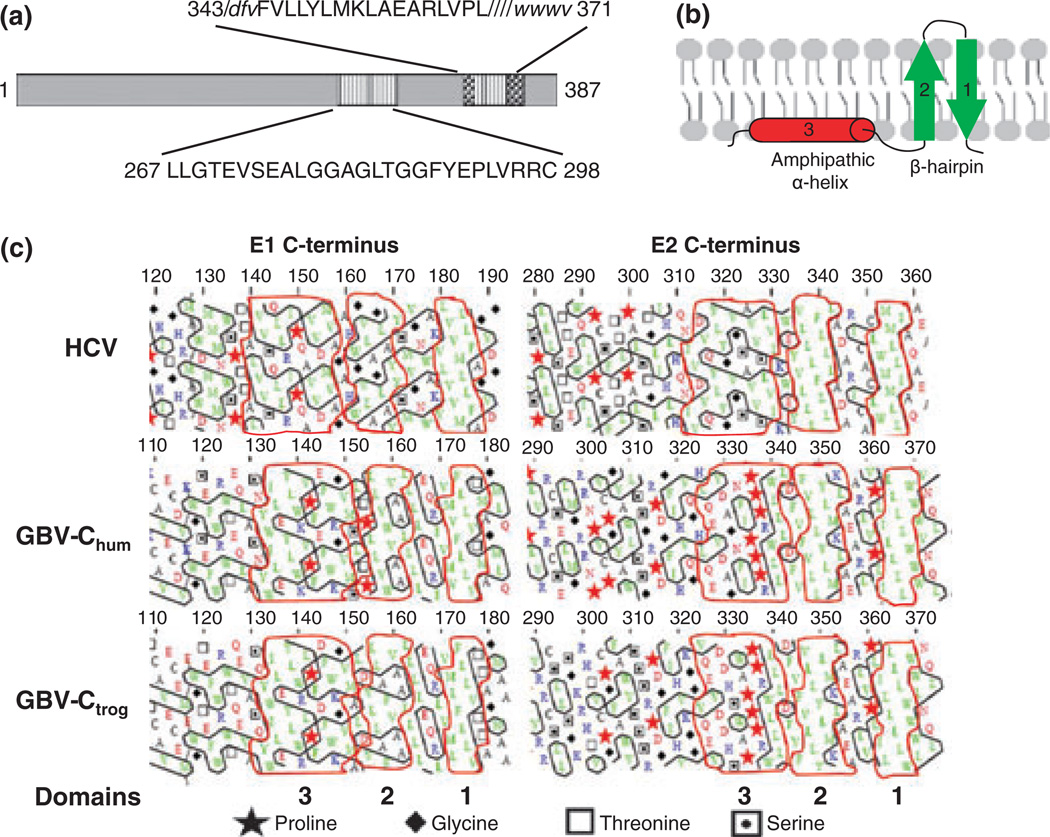
HCV envelope proteins E1 and E2 contain multifunctional TM regions in their C-terminal regions (Fig. 6). The TM domains function as membrane anchors and ER retention signals, and are involved in E1E2 heterodimerization [54]. The same features are predicted for GBV-C E1 and E2. While experimental studies examining the topology of the TM domains of human and troglodyte GBV-C have not been performed, computational analyses predict that the C-terminal domain of E1 may adopt an amphipathic α-helix followed by a β-hairpin topology and E2 may adopt a β-hairpin topology similar to that predicted for HCV [55] (Fig. 6b). Kyte and Doolittle hydrophobicity profiles of the E1 and E2 proteins of HCV, GBV-C and GBV-Ctrog identify hydrophobic regions at the C-termini suggestive of a TM domain with two potential membrane-spanning domains. Deletion of the GBV-C E2 C-terminal TM domain and adding an N-terminal secretory peptide results in secretion of the protein from CHO cells [49,56].
Fig. 6.
Functional mapping of the GB virus C envelope glycoprotein E2. The predicted GBV-C E2 transmembrane domain (TM) resides between amino acids 343 and 371 (a). Two putative peptide regions within E2 contain membrane interacting or fusion functions (347–364, within the TM) and 267–298. The M6 monoclonal antibody blocks E2 binding to cells, and recognizes a linear epitope including amino acids 276–292 within the putative fusion peptide (a). HCV hydrophobic TM regions on both E1 and E2 appear to be too short to serve as classical transmembrane α-helices, and are predicted to fold as two antiparallel β-strands in a hairpin structure (b). The amphiphilic α-helix upstream of the hairpin is predicted to stabilize the hairpin on the ER membrane (b). Hydrophobic cluster analysis of HCV, human and chimpanzee variants of GBV-C (GBVChum and GBV-Ctrog respectively) found that the E1 C-termini of these three viruses are similar (c) and share the putative structure shown in panel B. However, only the HCV E2 protein had this type of C terminus, while GBV-C contained helix breaking proline residues upstream from the TM region (c).
The biogenesis of the GBV-C TM domain topology is unknown, but based on sequence comparisons it appears to be formed in a similar manner to the HCV TM domain. Following translation of the HCV E1 and E2 proteins, the N-terminus of E1 is translocated in the ER lumen [54]. Because the E1 TM domain is also the signal sequence of E2, the E1 C-terminus is oriented towards the ER lumen, enabling E2 to be translocated into the ER lumen upon forming a putative hairpin structure [54]. Following signal sequence cleavage between E1 and E2, the E1 TM domain is reoriented to form a single membrane-spanning domain where the N-terminal ectodomain of E1 is in the ER lumen and the C-terminal domain is in the cytosol [54]. This conformational change of the TM domain from a hairpin structure to a single membrane-spanning domain following signal sequence cleavage also appears to occur with HCV E2 [54].
GBV-C E1, and possibly E2, are predicted to have a similar hairpin to single TM-spanning domain conformational change based on a hydrophobic cluster analysis (HCA) (Fig. 6). Specifically, the C-termini of HCV E1 and E2 suggest that the envelope glycoproteins share a similar pattern of a long amphiphilic region (18 or 19 residues) followed by two short hydrophobic stretches (7–10 residues) surrounded by hydrophilic residues [57]. Human and troglodyte GBV-C E1 and E2 C-termini show the same pattern of two short hydrophobic stretches, as suggested by the short vertical clusters on the HCA profile (http://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py?form=HCA), and GBV-C E1 shares the putative amphiphilic α-helix seen with HCV E1 and E2 (suggested by the long horizontal cluster on the HCA profile) [55]. The HCV hydrophobic regions are too short to be classical TM α-helices, and are predicted to fold as two antiparallel β-strands in a hairpin structure [57], and the amphiphilic α-helix upstream of the hairpin is predicted to stabilize the hairpin [57]. Because of the similarity of the cluster shapes and location in human and troglodyte GBV-C E1 to HCV E1, the human and troglodyte GBV-C E1 C-termini also appear to form a membrane-spanning hairpin structure (Fig. 6c). Although human and troglodyte GBV-C E2 and HCV E2 share a putative hairpin structure, the GBV-C E2 proteins do not contain a predicted amphiphilic α-helix domain, suggesting that there may be different membrane topologies compared to HCV E2 (Fig. 6c) [55].
This hairpin model of the HCV E1 and E2 membrane topology agrees with the topology of the TM domains in front of signal sequence cleavage sites suggested by others, except that the hairpins in other models are predicted to be located in the environment of protein complexes that transport proteins through the ER membrane rather than in the ER membrane [54,57]. Further experimental studies are necessary to determine the precise timing and location of the putative β-hairpin structures of these TM domains.
Fusion peptides
The envelope proteins of flaviviruses are class II fusion proteins which are predominantly non-helical, are not cleaved during biosynthesis and appear to have fusion peptides within internal loop structures far from the N-terminus [58]. Several studies have characterized putative fusion peptides within GBV-C E1 and E2 in recent years. For example, interactions of two overlapping peptides from GBV-C E2 with model membranes were studied: 267–284 (LLGTEVSEALGGAGLTGG) and 279–298 (AGLTGGFYEPL VRRCSELAG) [58,59]. Lipid mixing and red blood cell haemolysis studies identified the 279–298 peptide as more able to disrupt lipid bilayers, suggesting that it is more likely to be an internal fusion peptide than the 267–284 peptide [58,60]. Additional physiochemical analyses with Langmuir phospholipid monolayers demonstrate that the 279–298 peptide modifies the surface behaviour of phospholipid monolayers, further suggesting that this peptide may be involved in membrane fusion [61]. Of note, the 279–298 peptide is unstructured in aqueous solution, but forms an amphipathic helical structure in the presence of lipids and model membranes, further supporting a role in virus-cell fusion [62].
Recent studies examining peptide adsorption at air/water interfaces and their interaction with phospholipid mono-layers identified other potential fusion peptides within GBV-C E2: 267–284 LLGTEVSEVLGGAGLTGG [63] and 347–363 VLLYLMKLAEARLVPLI [64]. Two E1 fusion peptides were also proposed as internal fusion peptides based on predicted lipid-interacting structures: 53–66 AGLAVRPGKSAAQL [65] and 145–162 WKVPFDFWRGVISLTPLL [66]. Further studies of the candidate fusion peptides in the context of viral particles are needed to determine if these domains are truly involved in fusion events.
Antigenic structure
The antigenic structure of GBV-C E2 was investigated using monoclonal antibodies (MAbs) generated by DNA immunization [67]. These MAbs are comprised of four specificity groups that, based on competition studies, recognize overlapping epitopes [49,67]. The combination of all four groups of MAbs completely inhibits binding of human polyclonal anti-E2 antibodies to E2, indicating that this antigenic site is immunodominant during natural infection [49]. Two of the four groups of MAbs detect E2 bound to cells, suggesting that the epitopes for the other antibodies may be masked during E2 interactions with cells, or masked by conformation changes upon binding [49]. One group of MAbs (M6, M11) neutralized the binding of E2 to cells, indicating that this epitope may be involved in E2 cell binding and/or fusion. M6 is the only known MAb to recognize a linear epitope on E2 and all others described to date recognize conformation epitopes [49,67]. Human anti-E2 antibodies elicited during natural infection do not react with denatured E2, indicating that the M6 epitope is cryptic and not displayed during natural infection. The M6 epitope (GGAGLTGGFYEPLVRRC) resides between E2 amino acids 276 and 292 [49], which overlaps one of the proposed fusion peptides [58,63]. Sequential deletion analysis at the N- and C-terminus of the linear peptide recognized by M6 MAb determined that a core of six amino acids is required for binding. However, M6 only binds the peptide if at least four amino acids are included at either the C- or N-termini of these six amino acids, suggesting that there is both a size and a sequence requirement for optimal interaction [49].
Cell binding
Transient and inefficient replication has been documented in several cell lines including T cells, B cells and hepatocyte cell lines (reviewed in [7,30]). The most efficient system for growing GBV-C is in primary human PBMCs, although replication is inefficient, and GBV-C replication can only be detected in a small percentage of cells [17]. In contrast, E2 binds to a variety of cell types including Molt-4, HeLa, Jurkat, HEK 293 and murine 3T3 cells [49]. Thus, several cell types and more than human species appear to contain an E2 binding receptor [48,50]. Although E2 is able to attach to many cell types, viral entry may not be possible without additional receptor(s), which is similar to HCV [68]. CD81 is an HCV E2 attachment receptor that was proposed as a candidate receptor for GBV-C E2 [39]. However, GBV-C E2 did not bind to Daudi cells expressing high levels of CD81, and E2 binding was not blocked by competition with soluble human CD81 [50]. Thus, CD81 is not required for E2 binding. The GBV-C E2 cellular receptor has been implicated in CCR5 downregulation and HIV inhibition [38,39,47]; however, to date GBV-C receptor(s) have not been identified.
HIV inhibition and GBV-C envelope glycoproteins
As noted previously, incubation of human PBMCs with recombinant E2 protein or a recombinant E2-Fc fusion protein prior to HIV infection results in replication inhibition compared to untreated HIV-infected PBMCs and ghost cells (Fig. 4) [47,48]. In addition, transfection of T7-transcribed RNA encoding E1 and E2 in CD4+ T cells prior to HIV infection also results in HIV inhibition [47], while expression of the first part of E1 protein alone did not inhibit HIV, further supporting E2 as sufficient to inhibit HIV replication [47].
E2-mediated inhibition is dose–dependent and HIV replication is blocked at binding or entry by either truncated E2 or E2-Fc fusion proteins, as transduction of HIV pseudotyped particles are also inhibited [47,48]. The E2-Fc fusion protein inhibited both X4- and R5-enveloped HIV pseudoparticles (HIVpp) by 50% in PBMCs compared with Fc controls [47], and anti-E2 antibodies abrogate HIV inhibition indicating specificity [47]. In contrast, the E2-Fc fusion protein does not inhibit entry of VSV-G-pseudotyped particles, thus E2 inhibition of HIV replication appears to target early steps in the HIV replication cycle involving HIV gp120 or gp41 [47]. Recent work localizes the step of HIV replication inhibition even further than an early entry step. Single cycle infections of T cell lines expressing E2 and cell-virus fusion experiments on HEK 293T cells found that GBV-C E2 inhibited HIV replication soon after the gp120/CD4 dependent entry and prior to reverse transcription [69].
E2 fusion peptides inhibit HIV gp41 fusion peptides
Studies of putative GBV-C fusion peptides and HIV-1 gp41 suggest that a candidate E2 fusion peptide (269–286 GTEVSEALGGAGLTGGFY) inhibits HIV fusion [70]. Vesicle leakage assays conducted with synthetic overlapping peptides found that the GBV-C E2 269–286 peptide inhibited membrane leakage induced by the HIV-1 gp41 fusion peptide by at least 55% compared to peptides synthesized from adjacent E2 sequences [70]. The inhibition was specific for the HIV-1 fusion peptide, as the E2 peptide did not inhibit membrane lytic activity of a control fusion protein (melittin). The E2 peptide also inhibits HIV-1 peptide-induced lipid mixing and binds to the HIV-1 gp41 fusion peptide in a 1:1 ratio in an energetically favourable manner. When both peptides are mixed together in a membrane-mimicking environment (TFE), their conformation changes from α-helices to β-turns and random structures indicating that their interaction leads to conformational changes of both peptides. NMR spectra of the mixed peptides indicate that they interact and form high-molecular-weight aggregates. The authors suggest that the binding of the E2 peptide may prevent oligomerization of the HIV-1 fusion peptide upon membrane interaction. This would inhibit the membrane destabilization effect necessary for HIV-1 membrane fusion. The inhibition of HIV-1 fusion by a GBV-C E2 peptide raises more questions about HIV inhibition in larger systems: does recombinant E2 interact with HIV gp41 and prevent HIV fusion even though its putative fusion peptide is not exposed prior to low-pH conditions? If so, what are the kinetics of this interaction? Does E2 interaction with cellular receptors lead to a conformational change that leads to exposure of this peptide domain to the cell membrane? Do GBV-C particles interact with HIV particles in vivo before either interacts with a lipid membrane?
One could argue that these in vitro studies with isolated fusion peptides or cell lines expressing E2 may not completely reflect the main method of HIV replication inhibition in vivo because fewer than 1 in 5000 peripheral-blood lymphocytes contain GBV-C RNA and are expressing E2. However, the average titre of GBV-C particles in plasma is roughly 1000-fold higher than that of HIV, and GBV-C particles are predicted to contain multiple copies of E1–E2 heterodimers on their envelope. Thus, GBV-C E2 interactions with lympyhocytes are constant. With the high titre of E2 in infected humans, GBV-C may well interact with lymphocytes to induce a chemokine and/or cytokine milieu that is inhibitory to HIV infection in both infected and uninfected-bystander cells (without entering). It will be important to examine E2 peptide-HIV interactions in larger model systems in which HIV and GBV-C particles are mixed, and then assess their effects on PBMCs and HIV fusion.
CONCLUSION
GBV-C is an interesting virus that is closely related to HCV and yet is not associated with any disease. GBV-C grows in PBMCs rather than hepatocytes, and the titre of virus in plasma is very high, averaging more than 107 genome equivalents/ml in several cohort studies. Epidemiological studies found an association of decreased morbidity and mortality in HIV-infected individuals who are co-infected with GBV-C, and in vitro co-infection of PBMCs demonstrates that GBV-C inhibits HIV replication. Two GBV-C proteins have been identified to date that specifically inhibit HIV replication: NS5A and E2. In this review we focused on the characterization of E2 protein and its interactions with CD4 + T cells and with HIV particles that may result in HIV inhibition. E2 appears to inhibit an early step in HIV infection and may include an internal fusion peptide that inhibits HIV gp41 fusion. Further work is needed to identify the E2 cellular receptor, characterize the domains of E2 responsible for HIV inhibition, and examine E2-mediated fusion in the context of the whole protein or pseudoparticles. These studies to elucidate E2-cell interactions may identify novel approaches to HIV therapy.
ACKNOWLEDGEMENTS
This work was supported in part by a Merit Review grant from the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development (JTS), a grant from the National Institutes of Health (RO1 AI-58740, JTS), and Emma Mohr received support from the University of Iowa MSTP program and Virology NIH training Grant (T-32).
Abbreviations
- IRES
internal ribosome entry site
- MIP
macrophage inflammatory proteins
- NTRs
nontranslated regions
- ORF
open reading frame
- PBMCs
peripheral blood mononuclear cells
- TM
transmembrane
REFERENCES
- 1.Simons JN, Leary TP, Dawson GJ, et al. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 2.Linnen J, Wages J, Zhang-Keck Z-Y, et al. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 3.Deka N, Sharma MD, Mukerjee R. Isolation of the novel agent from human stool samples that is associated with sporadic non-A, non-B hepatitis. J Virol. 1994;68:7810–7815. doi: 10.1128/jvi.68.12.7810-7815.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaluder GG, Dawson GJ, Simons JN, et al. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J Med Virol. 1995;46:81–90. doi: 10.1002/jmv.1890460117. [DOI] [PubMed] [Google Scholar]
- 5.Alter HJ. G-pers creepers, where’d you get those papers? A reassessment of the literature on the hepatitis G virus. Transfusion. 1997;37:569–572. doi: 10.1046/j.1537-2995.1997.37697335149.x. [DOI] [PubMed] [Google Scholar]
- 6.Dickens T, Lemon SM. GB Virus C, Hepatitis G Virus, or Human Orphan Flavivirus? Hepatology. 1997;25:1285–1286. doi: 10.1002/hep.510250541. [DOI] [PubMed] [Google Scholar]
- 7.Stapleton JT. GB virus type C/Hepatitis G virus. Semin Liver Dis. 2003;23:137–148. doi: 10.1055/s-2003-39943. [DOI] [PubMed] [Google Scholar]
- 8.Lefrère J-J, Loiseau P, Maury J, et al. Natural history of GBV-C/hepatitis G virus infection through the follow-up of GBV-C/hepatitis G virus-infected blood donors and recipients studied by RNA polymerase chain reaction and anti-E2 serology. Blood. 1997;90:3776–3780. [PubMed] [Google Scholar]
- 9.Barnes A, Allen JB, Klinzman D, Wang Z, Chaloner K, Stapleton JT. GBV-C persistence does not require CD4 + T cell preservation, and GBV-C viral load (VL) is weakly inversely related to HIV VL. 4th IAS Conference on HIV Pathogenesis, Treatment and Prevention; Sydney. 2007. [Google Scholar]
- 10.Tacke M, Schmolke S, Schlueter V, et al. Humoral immune response to the E2 protein of hepatitis G virus is associated with long-term recovery from infection and reveals a high frequency of hepatitis G virus exposure among healthy blood donors. Hepatology. 1997;26:1626–1633. doi: 10.1002/hep.510260635. [DOI] [PubMed] [Google Scholar]
- 11.Tillmann HL, Heringlake S, Trauwein C, et al. Antibodies against the GB virus C envelope 2 protein before liver transplantation protect against GB virus C de novo infection. Hepatology. 1998;28:379–384. doi: 10.1002/hep.510280213. [DOI] [PubMed] [Google Scholar]
- 12.Stapleton JT, Williams CF, Xiang J. GB Virus C: A beneficial infection? J Clin Microbiol. 2004;42:3915–3919. doi: 10.1128/JCM.42.9.3915-3919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark K, Bienzle U, Hess G, Engel AM, Hegenscheid B, Schlueter V. Detection of the hepatitis G virus genome among injecting drug users, homosexual and bisexual men, and blood donors. JID. 1996;174:1320–1323. doi: 10.1093/infdis/174.6.1320. [DOI] [PubMed] [Google Scholar]
- 14.Williams CF, Klinzman D, Yamashita TE, et al. Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med. 2004;350:981–990. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- 15.Leary TP, Muerhoff AS, Simons JN, et al. Sequence and genomic organization of GBV-C: A novel member of the Flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 16.Simons JN, Desai SM, Schultz DE, Lemon SM, Mushahwar IK. Translation initiation in GB viruses A and C: evidence for internal ribosome entry and implications for genomic organization. J Virol. 1996;70:6126–6135. doi: 10.1128/jvi.70.9.6126-6135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang J, Wunschmann S, Schmidt WN, SHao J, Stapleton JT. Full-length GB Virus C (Hepatitis G Virus) RNA transcripts are infectious in primary CD4-Positive T cells. J Virol. 2000;74:9125–9133. doi: 10.1128/jvi.74.19.9125-9133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 19.Xiang J, Klinzman D, McLinden J, et al. Characterization of hepatitis G virus (GB-C Virus) particles: evidence for a nucleocapsid and expression of sequences upstream of the E1 protein. J Virol. 1998;72:2738–2744. doi: 10.1128/jvi.72.4.2738-2744.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walewski JL, Keller TR, Stump DD, Branch AD. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA-A Publ RNA Soc. 2001;7:710–721. doi: 10.1017/s1355838201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muerhoff AS, Dawson GJ, Desai SM. A previously unrecognized sixth genotype of GB virus C revealed by analysis of 5’untranslated region sequences. J Med Virol. 2006;78:1729–1735. doi: 10.1002/jmv.20510. [DOI] [PubMed] [Google Scholar]
- 22.Pavesi A. Origin and evolution of GBV-C/Hepatits G Virus and relationships with ancient human migrations. J Mol Evol. 2001;53:104–113. doi: 10.1007/s002390010198. [DOI] [PubMed] [Google Scholar]
- 23.Birkenmeyer LG, Desai SM, Muerhoff AS, et al. Isolation of a GB virus-related genome from a chimpanzee. J Med Virol. 1998;56:44–51. doi: 10.1002/(sici)1096-9071(199809)56:1<44::aid-jmv8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 24.Adams NJ, Prescott LE, Jarvis LM, et al. Detection in chimpanzees of a novel flavivirus related to GB virus-C/hepatitis G virus. J Gen Virol. 1998;79:1871–1877. doi: 10.1099/0022-1317-79-8-1871. [DOI] [PubMed] [Google Scholar]
- 25.Madejon A, Fogeda M, Bartolome J, et al. GB Virus C RNA in serum, liver and peripheral blood mononuclear cells from patients with chronic hepatitis B, C and D. Gastroenterology. 1997;113:573–578. doi: 10.1053/gast.1997.v113.pm9247478. [DOI] [PubMed] [Google Scholar]
- 26.Saito S, Tanaka K, Kondo M, et al. Plus- and minus-stranded hepatitis G virus RNA in liver tissue and in peripheral blood mononuclear cells. Biochem Biophys Res Commun. 1997;237:288–291. doi: 10.1006/bbrc.1997.7103. [DOI] [PubMed] [Google Scholar]
- 27.Pessoa MG, Terrault NA, Detmer J, et al. Quantitation of hepatitis G and C viruses in the liver: evidence that hepatitis G virus is not hepatotropic. Hepatology. 1998;27:877–880. doi: 10.1002/hep.510270335. [DOI] [PubMed] [Google Scholar]
- 28.Laskus T, Radkowski M, Wang L-F, Vargas H, Rakela J. Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C virus and G virus. J Virol. 1997;71:7804–7806. doi: 10.1128/jvi.71.10.7804-7806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bert T, Muller AR, Platz KP, et al. Dynamics of GB virus C viremia early after orthotopic liver transplantation indicates extrahepatic tissues as the predominant site of GB virus C replication. Hepatology. 1999;29:245–249. doi: 10.1002/hep.510290121. [DOI] [PubMed] [Google Scholar]
- 30.George SL, Varmaz D, Stapleton JT. GB virus C replicates in primary T and B lymphocytes. J Infect Dis. 2006;193:451–454. doi: 10.1086/499435. [DOI] [PubMed] [Google Scholar]
- 31.Mellor J, Haydon G, Blair C, Livingstone W, Simmonds P. Low level or absent in vitro replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J Gen Virol. 1998;79:705–714. doi: 10.1099/0022-1317-79-4-705. [DOI] [PubMed] [Google Scholar]
- 32.Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Detection of hepatitis G virus replication sites by using highly strand-specific Tth-based reverse transcriptase PCR. J Virol. 1998;72:3072–3075. doi: 10.1128/jvi.72.4.3072-3075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Chaloner K, Tillmann HL, Williams CF, Stapleton JT. Effect of early and late GBV-C viremia on survival of HIV infected individuals: A meta-analysis. HIV Med. 2006;7:173–180. doi: 10.1111/j.1468-1293.2006.00366.x. [DOI] [PubMed] [Google Scholar]
- 34.Van der Bij AK, Kloosterboer N, Prins M, et al. GB Virus C coinfection and HIV-1 disease progression: the Amsterdam cohort study. J Infect Dis. 2005;191:678–685. doi: 10.1086/427559. [DOI] [PubMed] [Google Scholar]
- 35.Xiang J, Wunschmann S, Diekema DJ, et al. Effect of coinfection with GB Virus C (Hepatitis G virus) on survival among patients with HIV infection. N Engl J Med. 2001;345:707–714. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- 36.Xiang J, George SL, Wunschmann S, Chang Q, Klinzman D, Stapleton JT. Inhibition of HIV-1 replication by GB virus C infection through increases in RANTES, MIP-1α, MIP-1β, and SDF-1. Lancet. 2004;363:2040–2046. doi: 10.1016/S0140-6736(04)16453-2. [DOI] [PubMed] [Google Scholar]
- 37.Xiang J, Sathar MA, McLinden JH, Klinzman D, Chang Q, Stapleton JT. South African GB virus C isolates: interactions between genotypes 1 and 5 GBV-C isolates and the human immunodeficiency virus. J Infect Dis. 2005;192:2147–2151. doi: 10.1086/498170. [DOI] [PubMed] [Google Scholar]
- 38.Jung S, Knauer O, Donhauser N, et al. Inhibition of HIV strains by GB Virus C in cell culture can be mediated by CD4 and CD8 T-lymphocyte derived soluble factors. AIDS. 2005;19:12567–1272. doi: 10.1097/01.aids.0000180097.50393.df. [DOI] [PubMed] [Google Scholar]
- 39.Nattermann J, Nischalke HD, Kupfer B, et al. Regulation of CC chemokine receptor 5 in Hepatitis G virus infection. AIDS. 2003;17:1457–1462. doi: 10.1097/00002030-200307040-00006. [DOI] [PubMed] [Google Scholar]
- 40.Rowland-Jones S. The role of chemokine receptors in HIV infection. Sexually Transmitted Infect. 1999;75:148–511. doi: 10.1136/sti.75.3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lalle E, Sacchi A, Abbate I, et al. Activation of interferon response genes and of plasmacytoid dendritic cells in HIV-1 positive subjects with GB virus C co-infection. Int J Immunopathol Pharm. 2008;21:161–171. doi: 10.1177/039463200802100118. [DOI] [PubMed] [Google Scholar]
- 42.Souza IE, Zhang W, Diaz RS, Chaloner K, Klinzman D, Stapleton JT. Effect of GB Virus C on response to antiretroviral therapy in HIV infected Brazilians. HIV Med. 2006;7:25–31. doi: 10.1111/j.1468-1293.2005.00339.x. [DOI] [PubMed] [Google Scholar]
- 43.Tillmann HL, Heiken H, Knapik-Botor A, et al. Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med. 2001;345:715–724. doi: 10.1056/NEJMoa010398. [DOI] [PubMed] [Google Scholar]
- 44.Stapleton JT, Chaloner K, Zhang J, et al. GB virus C viremia is associated with reduced CD4 expansion following interleukin 2 therapy in HIV-infected people receiving HAART. AIDS. 2008;23:605–610. doi: 10.1097/QAD.0b013e32831f1b00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang J, McLinden JH, Chang Q, Kaufman TM, Stapleton JT. An 85 amino acid segment of the GB Virus type C NS5A phosphoprotein inhibits HIV-1 replication in CD4+ Jurkat T-cells. Proc Natl Acad Sci USA. 2006;103:15570–15575. doi: 10.1073/pnas.0604728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang J, McLinden JH, Chang Q, Jordan EL, Stapleton JT. Characterization of a peptide domain within the GB virus C NS5A phosphoprotein that inhibits HIV replication. PLoS ONE. 2008;3:e2580, 1–10. doi: 10.1371/journal.pone.0002580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung S, Eichenmueller M, Donhauser N, et al. HIV entry inhibition by the envelope 2 glycoprotein of GB virus C. AIDS. 2007;21:645–647. doi: 10.1097/QAD.0b013e32803277c7. [DOI] [PubMed] [Google Scholar]
- 48.Mohr EL, McLinden JH, Xiang J, Stapleton JT. GBV-C glycoproteins mediate cell entry of pseudotyped retroviral particles. 15th International Symposium on Hepatitis C and Related Viruses; San Antonio, TX. 2008. [Google Scholar]
- 49.McLinden JH, Kaufman TM, Xiang J, et al. Characterization of an immunodominant antigenic site on GB virus C glycoprotein E2 that is involved in cell binding. J Virol. 2006;80:12131–12140. doi: 10.1128/JVI.01206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaufman TM, McLinden JH, Xiang J, Engel AM, Stapleton JT. The GBV-C envelope glycoprotein E2 does not interact specifically with CD81. AIDS. 2007;21:1045–1048. doi: 10.1097/QAD.0b013e3280f77412. [DOI] [PubMed] [Google Scholar]
- 51.Belyaev AS, Chong S, Novikov a, et al. Hepatitis G virus encodes protease activities which can effect processing of the virus putative nonstructural proteins. J.Virol. 1998;72:868–872. doi: 10.1128/jvi.72.1.868-872.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theodore D, Lemon SM. GB virus C, Hepatitis G virus, or human orphan flavivirus? Hepatology. 1997:1285–1286. doi: 10.1002/hep.510250541. [DOI] [PubMed] [Google Scholar]
- 53.Pilot-Matias TJ, Carrick RJ, Coleman PF, et al. Expression of the GB virus C E2 glycoprotein using the Semliki Forest virus vector system and its utility as a serologic marker. Virol. 1996;225:282–292. doi: 10.1006/viro.1996.0602. [DOI] [PubMed] [Google Scholar]
- 54.Cocquerel L, Op dB, Lambot M, et al. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 2002;21:2893–2902. doi: 10.1093/emboj/cdf295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jordan EL, McLinden J, Murthy KK, Stapleton JT. Comparison and predicted membrane topology of human and troglodyte GBV-C E2 sequences. 14th International Symposium on Hepatitis C Virus & Related Viruses; Glascow. 9 September 2007. [Google Scholar]
- 56.Surowy TK, Leary TP, Carrick RJ, et al. GB virus C E2 glycoprotein: expression in the CHO cells, purification and characterization. J Gen Virol. 1997;78:1851–1859. doi: 10.1099/0022-1317-78-8-1851. [DOI] [PubMed] [Google Scholar]
- 57.Charloteaux B, Lins L, Moereels H, Brasseur R. Analysis of the C-terminal membrane anchor domains of hepatitis C virus glycoproteins E1 and E2: toward a topological model. J Virol. 2002;76:1944–1958. doi: 10.1128/JVI.76.4.1944-1958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larios C, Casas J, Alsina MA, Mestres C, Gomara MJ, Haro I. Characterization of a putatitve fusogenic sequence in the E2 hepatitis G protein. Arch Biochem Biophys. 2005;442:149–159. doi: 10.1016/j.abb.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 59.Larios C, Casas J, Mestres C, Haro I, Alsina MA. Perturbations induced by synthetic peptides from hepatitis G virus structural proteins in lipid model membranes: a fluorescent approach. Luminescence. 2005;20:279–281. doi: 10.1002/bio.850. [DOI] [PubMed] [Google Scholar]
- 60.Larios C, Christiaens B, Gomara MJ, Alsina MA, Hara I. Interaction of synthetic peptides corresponding to hepatitis G virus (HGV/GBV-C) E2 structural protein with phospholipid vesicles. FEBS J. 2005;272:2456–2466. doi: 10.1111/j.1742-4658.2005.04666.x. [DOI] [PubMed] [Google Scholar]
- 61.Larios C, Minones J, Jr, Haro I, Alsina MA, Busquets MA, Trillo JM. Study of adsorption and penetration of E2(279-298) peptide into Langmuir phospholipid monolayers. J Phys Chem B. 2006;110:23292–23299. doi: 10.1021/jp0628582. [DOI] [PubMed] [Google Scholar]
- 62.Mazzini S, Fernandez-Vidal M, Galbusera V, et al. 3D-Structure of the interior fusion peptide of HGV/GBV-C by 1H NMR, CD and molecular dynamics studies. Arch Biochem Biophys. 2007;465:187–196. doi: 10.1016/j.abb.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 63.Casas J, Espina M, Haro M, et al. Interfacial properties of a synthetic peptide derived from hepatitis G virus E2 protein: interaction with lipid monolayers. Langmuir. 2006;22:246–254. doi: 10.1021/la051812h. [DOI] [PubMed] [Google Scholar]
- 64.Perez-Lopez S, Nieto-Suarez M, Mestres C, Alsina MA, Haro I, Vila-Romeu N. Behaviour of a peptide sequence from the GB virus C/hepatitis G virus E2 protein in Langmuir monolayers: its interaction with phospholipid membrane models. Biophys Chem. 2009;141:153–161. doi: 10.1016/j.bpc.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 65.Perez-Lopez S, Vila-Romeu N, Alsina Esteller MA, Espina M, Haro I, Mestres C. Interaction of GB virus C/hepatitis G virus synthetic peptides with lipid langmuir monolayers and large unilamellar vesicles. J Phys Chem B. 2008;113:319–327. doi: 10.1021/jp806938y. [DOI] [PubMed] [Google Scholar]
- 66.Sanchez-Martin MJ, Amigo JM, Pujol M, Haro I, Alsina MA, Busquets MA. Fluorescence study of the dynamic interaction between E1(145-162) sequence of hepatitis GB virus C and liposomes. Anal Bioanal Chem. 2009;394:1003–1010. doi: 10.1007/s00216-008-2593-8. [DOI] [PubMed] [Google Scholar]
- 67.Schmolke S, Tacke M, Schmitt U, Engel AM, Ofenloch- Haehnle B. Identification of Hepatitis G virus particles in human serum by E2-specific monoclonal antibodies generated by DNA immunization. J Virol. 1998;72:4541–4545. doi: 10.1128/jvi.72.5.4541-4545.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dubuisson J, Helle F, Cocquerel L. Early steps of the hepatitis C virus life cycle. Cell Microbiol. 2008;10:821–827. doi: 10.1111/j.1462-5822.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- 69.Hanel K, Jung S, Wend H, Fleckenstein B. Reil H Localization of HIV replication steps influenced by different GB virus C proteins. 16th Conference on Retroviruses and Opportunistic Infections; Sydney. 2 August 2009. [Google Scholar]
- 70.Herrera E, Gomara MJ, Mazzini S, Ragg E, Haro I. Synthetic peptides of hepatitis G virus (GBV-C/HGV) in the selection of putative peptide inhibitors of the HIV-1 fusion peptide. J Phys Chem B. 2009;113:7383–7391. doi: 10.1021/jp900707t. [DOI] [PubMed] [Google Scholar]