Abstract
Background
Human herpesvirus type 8 (HHV-8) is hyperendemic in Amerindian populations, but its modes of transmission are unknown.
Methods
Antibodies against either HHV-8 lytic antigen or HHV-8 latency-associated nuclear antigen (LANA) were detected, by immunofluorescence assays, in 339 Amerindians and 181 non-Amerindians from the Brazilian Amazon. Serological markers of oro-fecal (hepatitis A), parenteral (hepatitis B and C), and sexual (herpes simplex virus type 2 and syphilis) transmission were measured by specific ELISAs. Salivary HHV-8 DNA was detected by use of a nested polymerase chain reaction assay and was sequenced.
Results
Antibodies against either lytic antigen or LANA were detected in 79.1% of Amerindians and in 6.1% of non-Amerindians (adjusted seroprevalence ratio [SR], 12.63 [95% confidence interval {CI}, 7.1–22.4]; P< .0001). HHV-8 seroprevalence increased with age among Amerindians (PTrend< .001) and already had high prevalence in childhood but was not sex specific in either population. The 2 populations did not differ in seroprevalence of oro-fecal or parenteral markers, but seroprevalence of markers of sexual transmission was lower among Amerindians. HHV-8 DNA in saliva was detected in 47 (23.7%) of 198 HHV-8 seropositive Amerindians. Detection of HHV-8 DNA decreased with age (PTrend< .04) and was more common in men (SR, 2.14 [95% CI, 1.3–3.5]; P= .003). A total of 36 (76.6%) of the 47 saliva HHV-8 DNA samples were sequenced, and all clustered as subtype E.
Conclusion
The data support the hypothesis of early acquisition and horizontal transmission, via saliva, of HHV-8 subtype E in Amerindian populations.
Human herpesvirus type 8 (HHV-8), also called “Kaposi sarcoma–associated herpesvirus” (KSHV), is the etiologic agent of all forms of Kaposi sarcoma (KS) [1, 2], primary effusion lymphoma [3], and the HIV-associated plasmablastic cell variant of multicentric Castleman disease [4, 5]. The modes of HHV-8 transmission have long remained unclear. Surveillance of KS cases at the beginning of the AIDS epidemic in the United States revealed that men who have sex with men (MSM) were at higher risk of KS than any other HIV-transmission group, suggesting a sexually transmissible pathogen [6]. Subsequent seroepidemiological studies have indicated that number of sex partners, HIV-seropositivity, and sexual practices involving exposure to saliva were associated with HHV-8 acquisition in MSM [7–9].
In the United States and most western European countries, areas where the occurrence of KS is rare out-side high-risk groups, HHV-8 serosurveys have shown that HHV-8 is acquired after puberty and that the seroprevalence remains low (2%–8%) in the general population [10, 11]. In contrast, in populations living around the Mediterranean basin [12–14] and in eastern and central African countries [15–18], areas where the incidence of KS is high, HHV-8 appears to be acquired early in childhood, and high seroprevalence has been reported. Recently, unusually high HHV-8 seroprevalences, ranging from 24% to 100%, have been described in Amerindian populations from Brazil, French Guiana, and Ecuador [19–22], for whom KS incidence data are not available, although the occurrence of classic KS has been documented in 1 HIV-negative Peruvian Amerindian [23]. In these endemic areas, the linear increase in HHV-8 seroprevalence before puberty [21, 24, 25], with modest increases in adulthood, and the association between HHV-8 seropositivity in children and having at least 1 seropositive first-degree relative [26] suggest nonsexual routes of transmission, probably via saliva, between siblings [24, 26].
Molecular studies have provided further insights into the epidemiology and transmission of this infection. Both the detection of HHV-8 DNA in the saliva of infected children [27] and the association between children’s infection status and their mother’s higher saliva HHV-8 loads [28] are consistent with the occurrence of HHV-8 transmission, via saliva, within families. A recent study has shown that 5 of 6 concordant polymerase chain reaction (PCR)–positive Ugandan mother-child pairs exhibited the same HHV-8 subtype, emphasizing the importance of intrafamilial transmission of HHV-8 in endemic areas [29]. A particular feature of the epidemiology of HHV-8 in Amerindians is that, to date, only 1 HHV-8 strain (subtype E) has been found in these populations [19, 20, 22], but data are lacking regarding its exact mode of transmission.
We conducted a study to investigate potential modes of transmission of HHV-8 in Amerindian populations of a remote region of the Brazilian Amazon. We estimated the age- and sex-specific seroprevalence of HHV-8 and other markers of orofecal, parenteral, or sexually transmissible pathogens in indigenous and nonindigenous groups living in the same area. Furthermore, we collected saliva samples from the Amerindian population, to investigate the age- and sex-specific occurrence of HHV-8 oral shedding and to conduct genotypic characterization of HHV-8 DNA in this population.
POPULATIONS, MATERIALS, AND METHODS
Study Setting and Populations
The study was conducted in the area of the Trombetas River, a tributary of the Amazon River, in Para State, Brazil. Amerindian populations originating from 4 different ethnic groups (Mawayana, WaiWai, Katwena, and Xerew) and comprising 1270 adults and children living in the Mapuera village were eligible for inclusion in the study. A group of ethnically mixed non-Amerindian populations, living in scattered houses on the banks of the Trombetas River, also were included in the study, as a comparison group to control for possible environmental factors of exposure.
The purpose and methods of the study were explained to community leaders and through community meetings, although informed-consent procedures were done individually, with the assistance of local indigenous health agents. In the Amerindian community, consent was provided mostly verbally and, for children under the age of 15 years, by parent or guardian, whereas the non-Amerindian participants provided it in written form.
The study was approved by the Amerindian traditional chiefs, the Indigenous National Agency of Brazil, and the ethics research committee of the Ministry of Health of Brazil.
Study Procedures
The fieldwork was conducted from March 2003 to April 2004. Participants’ characteristics were recorded by use of a simple interviewer-administered questionnaire. A 10-mL blood sample was collected for serological testing. Saliva samples were obtained from the Amerindians only, because of the expected low HHV-8 seropositivity among the non-Amerindians. Specimens were collected by rubbing a Dacron swab over the mucosa of both sides of the mouth. Each swab was expressed into a cryovial containing 3 mL of Hanks’ medium with 50 μg/mL vancomycin, 500 μg/mL imipenen, and 2 μg/mL amphotericin B. Samples were stored in liquid nitrogen, shipped to the São Paulo Instituto de Medicina Tropical, and stored at −80 °C until testing.
Laboratory Methods
Detection of antibodies against HHV-8
Antibodies against HHV-8 latency-associated nuclear antigen (LANA) and against HHV-8 lytic antigen were determined by immunofluorescence assay (IFA), at a 1:40 starting dilution, using a body cavity–based lymphoma (BCBL)–1 cell line. Punctuate nuclear staining in non-induced BCBL-1 cells was considered to indicate seropositivity for antibodies against LANA. The viral lytic cycle was induced by incubating BCBL-1 cells with 20 ng/mL 12-O-tetradecanoylphorbol-13–acetate (TPA) (Sigma) for 96 h. Entire-cell fluorescence in ~20% of TPA-treated cells was considered to indicate seropositivity for antibodies against lytic-phase antigen [30].
Surrogate markers of oro-fecal, blood-borne, and sexually transmitted infections
Serological surrogate markers of infections transmitted via different routes were identified to allow inferences to be made about HHV-8 transmission. The presence of antibodies against total anti–hepatitis A virus (HAV) was considered to be a surrogate marker of infectious agents transmitted via the oro-fecal route and was tested by use of Bioelisa HAV (Biokit); the presence of IgG antibodies against hepatitis C virus (HCV) was considered to be a surrogate marker of blood-borne agents and was tested by use of Ortho HCV 3.0 ELISA (Ortho Diagnostics); the presence of antibodies against Treponema pallidum and the presence of antibodies against herpes simplex virus type 2 (HSV-2) were considered to be surrogate markers for sexually transmitted infections and were tested, respectively, by Trepanostika ELISA (Biomérieux) and HerpeSelect ELISA (Focus Technologies) using a cutoff higher than that indicated by the manufacturer (i.e., OD >3.5 rather than OD >1.1), as suggested by others [31] and by our own performance evaluation [32]; and the presence of antibody against hepatitis B core antigen (HBcAg) was considered to be a marker of parenterally, sexually, or vertically transmissible agents and was tested by Ortho HBcAg ELISA (Ortho Diagnostics).
Detection of HHV-8 DNA
HHV-8 DNA from saliva samples was detected by nested PCR amplifying a 233-bp fragment from the HHV-8 minor capsid protein gene (open reading frame [ORF]26), as described elsewhere [30].
Molecular characterization of HHV-8
For molecular characterization of HHV-8 DNA isolates, 2 different fragments of the ORF-K1 variable–loop region, VR1 (380 bp) and VR2 (336 bp), were amplified by use of PCR primers described elsewhere [33]. Amplification was done in 50-μL reaction mixtures comprising 25 pmol of each primer, 200 μmol/L of each deoxynucleoside triphosphate, 10 mmol/L Tris-HCl (pH 9,0), 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 2.5 U of Taq platinum DNA polymerase (Invitrogen). Reactions were run in an Eppendorf Mastercycler gradient using a step-cycle program. After initial denaturation of DNA at 94 °C for 5 min, 35 cycles were run at 94 °C for 30 s, 61 °C for 45 s, and 72 °C for 1 min, with a subsequent extension, at 72 °C extension for 10 min, for VR1. For the second-round PCR, 2.5 μL of the first-round PCR product was used as template DNA, and the same conditions that had been used for producing a 380-bp DNA fragment were applied. For VR2, the outer primer pair produced a 373-bp DNA fragment, at an annealing temperature of 56 °C, and the inner primer pair produced a 336-bp DNA fragment, at an annealing temperature of 59 °C.
All laboratory procedures were performed under stringent conditions, to avoid contamination: 3 separate rooms were used to (1) prepare amplification mixes (buffer, MgCl2, deoxyribonucleoside triphosphate, expanded proofreading Taq DNA polymerase, primers, and distilled water), (2) apply DNA to the Eppendorf tubes, and (3) run the agarose gels. In addition, each PCR run contained several negative controls (i.e., water instead of DNA templates). PCR products were analyzed by gel electrophoresis in a 2% agarose gel and were visualized by exposure to UV light after being stained with ethidium bromide.
Amplicons were purified for direct sequencing, by use of Microcon100 Centrifugal Filter devices (Millipore). The sequencing mix was prepared by use of a Big Dye Terminator kit (Applied Biosystems), and the resulting labeled DNA was analyzed on an ABI PRISM 377 DNA sequencer (Applied Biosystems). Data were derived from both forward and reverse sequences of all PCR products.
The KSHV DNA sequences obtained were aligned by the ClustalW program in the BioEdit statistical package [34]. The translated amino acid sequences were classified by phylogenetic studies and by visual comparison with ORF-K1 prototype sequences described by Zong et al. [35]. Phylogenetic relationships between DNA sequences were analyzed by the neighbor-joining method [36] using the Kimura 2-parameter distance model [35] in the MEGA2 package [37]. The trees were determined in 1000 replicates, to conduct bootstrap analysis.
Statistical Analyses
Statistical analyses were performed by use of Stata statistical software (version 8.2; StataCorp). The risk associated with HHV-8 infection and markers of oro-fecal, blood-borne, or sexual transmission were estimated by use of seroprevalence ratios (SRs) and their 95% confidence intervals (CIs), adjusted for age group (≤14 years, 15–24 years, 25–34 years, and ≥35 years) and for sex. Adjusted SRs were obtained by fitting a generalized linear model to estimate the risk, in Amerindians versus non-Amerindians, of infection with HHV-8 and other serological markers. χ2 Statistics, with Fisher’s exact test being used for small values, were computed for comparison of categorical variables. The χ2 test for linear trend was used to examine the variations in HHV-8 seroprevalence and HHV-8 oral shedding, adjusted for age group and sex.
RESULTS
A total of 339 Amerindians (195 [57.5%] of whom were female) from the Mapuera village (~27% of the total population), and 181 non-Amerindians living in neighboring riverside settlements (106 [58.6%] of whom were female) agreed to participate in the study. Median ages among the Amerindian and non-Amerindian populations were 22 years (interquartile range [IQR], 13–37 years) and 17 years (IQR, 9–35 years), respectively.
The age- and sex-adjusted seroprevalences of HHV-8 and other surrogate markers among Amerindian and non-Amerindian populations are presented in table 1. Overall, the seroprevalence of HHV-8 (according to either LANA or lyticantigen IFA) was 79.1% in Amerindians and 6.1% in non-Amerindians (SR, 12.63 [95% CI, 7.1–22.4]). Among Amerindians, antibodies against HHV-8 LANA were detected more often than antibodies against HHV-8 lytic antigen (76.7% vs. 62.2%; P < .0001); among non-Amerindians, the converse was observed (0.5% for antibodies against LANA vs. 6.1% for antibodies against lytic antigen; P < .001). These patterns were observed in all age groups.
Table 1.
Age- and sex-adjusted seroprevalences of markers for HHV–8 infection and surrogate markers for oro-fecal infection (HAV), parenteral infection (HBV and HCV), sexually transmissible infection (HSV-2; Treponema pallidum), in Mapuera Amerindians and non-Amerindian riverside residents of the Trombetas River, Amazon region of Brazil.
| Prevalence, no. (%) of subjects |
|||
|---|---|---|---|
| Serological marker | Amerindians (n = 339) |
Non-Amerindians (n = 181) |
SRa (95% CI) |
| HHV-8 | |||
| LANA | |||
| Negative | 79 (23.3) | 180 (99.4) | |
| Positive | 260 (76.7) | 1 (0.5) | 135.22 (19.1–955.5) |
| Lytic antigen | |||
| Negative | 128 (37.8) | 170 (93.9) | |
| Positive | 211 (62.2) | 11 (6.1) | 9.64 (3.9–23.8) |
| Overall (LANA or lytic antigen) | |||
| Negative | 71 (20.9) | 170 (93.9) | |
| Positive | 268 (79.1) | 11 (6.1) | 12.63 (7.1–22.4) |
| HAV | |||
| Negative | 6 (1.8) | 16 (8.8) | |
| Positive | 333 (98.2) | 165 (91.2) | 1.07 (1.0–1.1) |
| HBV | |||
| Negative | 316 (93.2) | 75 (83.3)b | |
| Positive | 23 (6.8) | 15 (16.7)b | 0.40 (0.2–0.7) |
| HCV | |||
| Negative | 339 (100) | 90 (100)b | |
| Positive | 0 | 0 | NC |
| HSV-2 | |||
| Negative | 314 (92.6) | 117 (64.6) | 0.17 (0.1–0.3) |
| Positive | 25 (7.4) | 64 (35.4) | NC |
| T. pallidum | |||
| Negative | 339 (100) | 171 (94.5) | |
| Positive | 0 | 10 (5.5) | NC |
CI, confidence interval; HAV, hepatitis A virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HHV-8, human herpesvirus type 8; HSV-2, herpes simplex virus type 2; LANA, latency-associated nuclear antigen; NC, not calculated; SR, seroprevalence ratio.
Adjusted for age group (≤14 years, 15–24 years, 25–34 years, and ≥35 years) and for sex.
Only 90 samples were tested.
In both Amerindians and non-Amerindians, there was a high seroprevalence of HAV, which was used as a surrogate marker for oro-fecal transmission. All but 1 of the Amerindians ≥15 years old (99%) had antibodies against HAV (data not shown). Conversely, in both populations there was a total absence of HCV, which was used as a surrogate marker for parenteral transmission. The seroprevalences of HSV-2 and T. pallidum, which were used as markers for sexual transmission, were significantly lower in the Amerindian population. The seroprevalence of antibodies against HBcAg, which was used as a surrogate marker of possible parenteral, or sexual transmission, also was significantly lower in the Amerindian population.
The seroprevalences of antibodies against either LANA or lytic antigen, adjusted for age and for sex, are presented in table 2. Among Amerindians, the seroprevalence of HHV-8 was age specific (PTrend< .001), being lower in children ≤14 years old than in individuals >15 years old (66.3% vs. 85.0%; P< .001), but there was no sex-specific difference (77.8% among males vs. 80.0% among females; P = .2). None of the 4 children who were ≤3 years old had antibodies against either LANA or lytic antigen. Among non-Amerindians, there were, in all age groups, only a few infections, with no statistical difference between the seroprevalence among younger individuals and that among older individuals (3.7% among children ≤14 years old vs. 7.9% among older individuals; P = .3) or between the seroprevalence in males and that in females (P = .3).
Table 2.
Seroprevalence of antibodies against human herpesvirus type 8 (HHV-8), in Mapuera Amerindians and non-Amerindian riverside residents of the Trombetas River, Amazon region of Brazil, according to age and sex, and detection of HHV-8 DNA in saliva from Amerindians.
| Non-Amerindians (n = 181): HHV-8, LANA or lytic-antigen IFA |
Amerindians (n = 339) |
||
|---|---|---|---|
| Characteristic | HHV-8, LANA or lytic-antigen IFA |
HHV-8 DNA, salivaa |
|
| Age group | |||
| ≤3 years | 0/12 (0) | 0/4 (0) | 0 |
| 4–9 years | 2/39 (5.1) | 28/39 (71.8) | 9/22 (40.9) |
| 10–14 years | 1/29 (3.4) | 39/58 (67.2) | 9/30 (30.0) |
| 15–19 years | 1/19 (5.3) | 36/54 (66.7) | 6/28 (21.4) |
| 20–24 years | 0/12 (0) | 31/37 (83.8) | 3/21 (14.3) |
| 25–29 years | 2/9 (22.2) | 22/24 (91.7) | 5/17 (29.4) |
| 30–34 years | 0/15 (0) | 22/24 (91.7) | 4/15 (26.7) |
| 35–39 years | 2/10 (20.0) | 23/26 (88.5) | 5/17 (29.4) |
| ≥40 years | 3/36 (8.3) | 67/73 (91.8) | 6/48 (12.5) |
| Total | 11/181 (6.1) | 268/339 (79.1) | 47/198 (23.7) |
| PTrend b | .2 | <.001 | .04 |
| Sex | |||
| Male | 3/75 (4.0) | 112/144 (77.8) | 28/82 (34.1) |
| Femalec | 8/106 (7.5) | 156/195 (80.0) | 19/116 (16.4) |
| Total | 11/181 (6.1) | 268/339 (79.1) | 47/198 (23.7) |
| SRd | 0.54 (0.1–1.9) | 0.94 (0.8–1.0) | 2.14 (1.3–3.5) |
| P | .3 | .2 | .003 |
NOTE. Data are no. of individuals testing positive/total no. of individuals tested (%), unless otherwise indicated. IFA, immunofluorescence assay; LANA, latency-associated nuclear antigen; SR, seroprevalence ratio.
Denominators include saliva samples from HHV-8–seropositive individuals only.
Adjusted for sex.
Reference group.
Adjusted for age, on the basis of the age groups listed.
Paired saliva-and-blood samples were obtained from 243 (71.7%) of the 339 Amerindians; 198 (81.5%) of these 243 were seropositive for antibodies against either LANA or lytic antigen. The seroprevalence of HHV-8 DNA in saliva was 23.7% (47/198) in this population (table 2), and HHV-8 DNA was not detected in the saliva of any of the 45 individuals who were seronegative for antibodies against LANA and against lytic antigen. It is noteworthy that all 47 individuals shedding HHV-8 were seropositive for antibodies against LANA but that only 37 (78.7%) of 47 were seropositive for antibody against lytic antigen (P = .001). Oral shedding decreased significantly with age (PTrend = .04) and was more frequent in HHV-8–seropositive males than in HHV-8–seropositive females (34.1% vs. 16.4%; P = .003), particularly among those ≤14 years old (14/23 [60.9%] in boys vs. 4/29 [13.8%] in girls; SR, 4.41 [95% CI, 1.7–11.6]; P = .003).
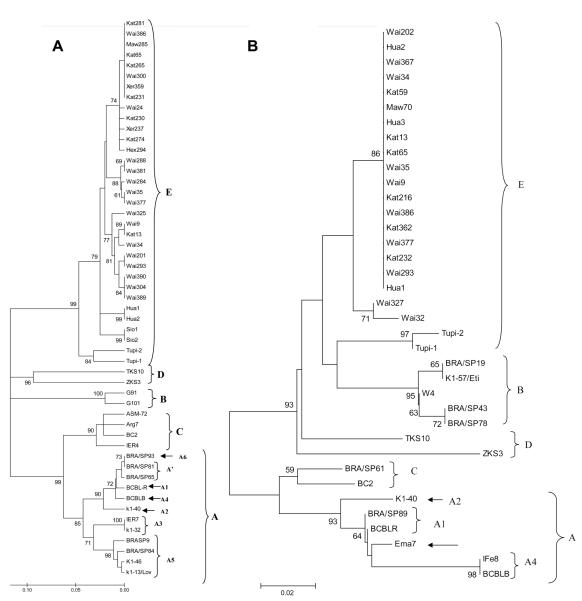
Genetic variability as determined by analysis of VR1 and VR2 fragments of the K1 gene was studied in 39 HHV-8 DNA sequences obtained from the 47 HHV-8 oral shedders. To allow for a more robust analysis, all samples were included in either VR1 or VR2 phylogenetic analysis; final trees with ≥70% bootstrap support are shown in figure 1. The simultaneous inclusion of DNA sequences in VR1 and VR2 phylogenetic trees was possible for 8 samples—Kat13, Kat65, Wai9, Wai34, Wai35, Wai293, Wai377, and Wai386. Of the 39 individual sequences, 36 could be included in phylogenetic analysis with a robust bootstrap and were submitted to GenBank (figure 1); all of these 36 samples clustered as subtype E. The majority of samples from Mapuera were closely related to isolates from the Huaorani Amerindians from Ecuador [20].
Figure 1.
Unrooted phylogenetic trees constructed by the neighbor-joining method using the Kimura 2-parameter distance model with 1000 bootstrap replicates in the MEGA3.1 software package. The best tree was obtained from 368 bp of the VR1/K1 region of human herpesvirus type 8 (HHV-8) (A) and from 248 bp of the VR2/K1 region of HHV-8 (B). A, 27 VR1/K1 HHV-8 DNA sequences amplified from saliva samples from Brazilian Amerindians (identified as Kat, Maw, Wai, Xer, and Hex) and another 26 HHV-8 DNA sequences deposited in GenBank and representing 5 major HHV-8 groups: subtype E—Hua1, Hua2, Sio1, and Sio2 (Ecuador [20]) and Tupi1 and Tupi2 (Brazilian Amazon [19]); subtype D—ZKS3 (New Zealand [35]) and TKS10 (Taiwan [35]); subtype B—G91 and G101 (The Gambia [38]); subtype C—Arg7 (Argentine [39]), ASM-72 and BC2 (United States [35]), and IER4 (Italy [38]); subtype A—BRA/SP93 (variant A6) and BRA/SP81 and BRA/SP65 (variant A′) (South Brazil [40]); subtype A, variant A1—BCBL-R (United States [35]); subtype A, variant A4—BCBL-B (United States [35]); subtype A, variant A2—K1-40 (United States [41]); subtype A, variant A3—IER7 (Italy [38]) and K1-32/Bcb (United States [41]); and subtype A, variant A5—K1-13/Lov and K1-46 (French Guiana [42]) and BRA/SP9 and BRA/SP84 (South Brazil [40]). Open-reading-frame (ORF)K1/VR1 DNA sequences have been deposited in GenBank (accession numbers EF204999-EF205001, EF209410-EF209421, and EF375500-EF375513). B, 17 VR2/K1 HHV-8/ DNA sequences amplified from saliva samples from Brazilian Amerindians (identified as Kat, Maw, and Wai) and another 20 HHV-8 DNA sequences obtained elsewhere: subtype E—Hua1, Hua2, and Hua3 (Ecuador [20]) and Tupi1 and Tupi2 (Brazilian Amazon [19]); subtype D—ZKS3 (New Zealand [35]) and TKS10 (Taiwan [35] ); subtype B—BRA/SP19, BRA/SP43, and BRA/SP78 (South Brazil [40]), K1-57/Eti (French Guiana [41]), and W4 (Malawi [43]); subtype C—BRA/SP61 (South Brazil [40]) and BC2 (United States [35]); subtype A, variant A1— BRA/SP89 (South Brazil [40]) and BCBL-R (United States [35]); subtype A, variant A4—IFe8 (Italy [38]) and BCBL-B (United States [35]); and subtype A, variant A2—K1-40/Bc-1 (United States [41]) and Ema7 (Spain [38]). ORF K1/VR2 DNA sequences have been deposited in GenBank (accession numbers EF375514- EF375549).
DISCUSSION
The high seroprevalence of HHV-8 observed among Amerindians from the Para state of the Brazilian Amazon confirms previous reports showing HHV-8 to be hyperendemic among Amerindian populations from Brazil, French Guiana, and Ecuador [19–22]. The contrasting, 10-fold-lower seroprevalence of HHV-8 among non-Amerindian populations that are living in the same geographical area and under similar living conditions but are seldom in direct contact with each other suggests that HHV-8 infection in Amerindian populations is associated with specific risk factors or behaviors that have not yet been elucidated but that do not appear to be linked to the environment. Further anthropological and behavioral research is required to elucidate some of these factors. Another striking difference between the populations in the present study was the serological profile of HHV-8 infection. In the non-Amerindians, the seroprevalence of antibodies against HHV-8 lytic antigen was higher than that of antibodies against HHV-8 LANA.
A similar profile was found among children and adults from the general population and among high-risk groups in Brazil [44]. Conversely, the Amerindians exhibited a higher seroprevalence of antibodies against LANA than of antibodies against lytic antigen, a profile that has been reported in another Amerindian population, in Ecuador [20]. Further studies are needed to evaluate the significance of this finding and its possible connections to genetic characteristics of the virus, Amerindian populations, or both.
The high age- and sex-adjusted seroprevalences of HAV infection in both Amerindian and non-Amerindian populations reflect the rather poor sanitary conditions in the Trombetas River area. On the other hand, both the absence of serological markers of HCV and the lower age-adjusted seroprevalence of HBV infection among Amerindians suggest that cultural practices possibly associated with enhanced risk of parenteral transmission, such as the bloodletting, scarification, and tattooing practiced by Amerindian populations, do not play an important role in HHV-8 transmission. Likewise, both the absence of serological evidence of syphilis and the low age- and sex-adjusted seroprevalences of antibodies against HSV-2 and against HBV in Amerindians suggest that sexual activity is probably not a relevant route of HHV-8 transmission. The fact that two-thirds of children ≤14 years old were already infected with HHV-8 further supports the hypothesis that nonsexual transmission is an important mode of infection among Amerindians. HHV-8 seroprevalence rates of 35% and 41% in children ≤10 years old have been observed among other Amerindian populations from the Amazon region [19, 21], raising the possibility of both person-to-person and mother-to-child transmission of HHV-8 in this population [21]. Although none of the 4 3-year-old Amerindian children examined had antibodies against HHV-8, the design of the present study did not allow determination of the specific role played by vertical or perinatal transmission. Data from other studies indicate that mother-to-child HHV-8 transmission occurs in endemic populations [24–26, 28, 29] and in children born to HIV-infected mothers [45, 46], although it is not clear from these studies how such transmission actually occurs. However, the age-related increase in HHV-8 seroprevalence in some of these studies [24, 26–28, 46] highlights the importance of horizontal transmission of the virus, possibly including mother-to-child transmission. Strong correlations between HHV-8 seroprevalence in children and mother-child and sibling-sibling familial aggregation [24, 26] and high household density [27] have been observed, suggesting that transmission occurs mainly by close and intimate interpersonal contact, likely via saliva.
HHV-8 DNA is commonly detected in the saliva of HIV-infected men [47–49] and women [50]. Epidemiological evidence has pointed to particular sexual behaviors involving saliva, such as deep kissing and oro-anal contact, as being significant risk factors for HHV-8 infection [48, 49]. In a hyperendemic population in South Africa, higher HHV-8 seroprevalence among children was correlated with higher HHV-8 loads in maternal saliva [28]. The fact that nearly one-fourth of HHV-8–seropositive Amerindians were shedding HHV-8 in saliva suggests that person-to-person contact might be a major mechanism of HHV-8 transmission in these populations. The significant age-related decrease in HHV-8 DNA in saliva suggests that virus transmission via saliva is more frequent among children, particularly in those ≤14 years old. The reasons for higher virus shedding in young males will require further exploration; however, it can be hypothesized that the apparently poorer host control of HHV-8 replication in males, indicated by increased oral shedding, may be linked to a higher incidence of KS later in life, as has been noted in other populations [2], although these data are lacking for Amerindian populations and should be further investigated
Because saliva samples were collected only once, and because of the intermittent nature of oral viral shedding [48, 49], it is quite likely that the observed rates of shedding are an underestimation. Moreover, we have not collected samples from other possible shedding sources (e.g., genital secretions, urine, or stool), although it is known that such sources may play a minor role compared with that played by oral shedding [48, 50]. Thus, the overall frequency of HHV-8 shedding in this population, although not known, is likely to be very high.
The finding that 100% of Amerindians shedding HHV-8 were seropositive for antibody against LANA but that only 77% of such individuals were seropositive for antibody against lytic antigen indicates that, in this population, the presence of antibody against LANA may be more closely related to HHV-8 replication than is the lytic antigen–antibody response. This result contrasts with findings from studies in Africa, in which the presence of HHV-8 DNA in saliva was found to be associated with either seropositivity for [27] or higher titers of [28] antibodies against lytic antigen. Such differences could be related to the main HHV-8 strain (i.e., subtype E) circulating in this population, host genetic factors characteristic of the Amerindian populations, or particular virus-host interactions in these populations.
In conclusion, we have observed very high levels of infection with, and oral shedding of, HHV-8 in Amerindians but not in non-Amerindians living in the same area of the Brazilian Amazon. Among Amerindians, HHV-8 is acquired at a young age, with no sex-specific difference in frequency, supporting the hypothesis of horizontal transmission of the virus. Oral excretion of the virus—exclusively subtype E—is more frequent in young males. Factors that govern acquisition and transmission of HHV-8 in Amerindian populations should be further studied.
Acknowledgments
We thank Helen Weiss (London School of Hygiene & Tropical Medicine) and Martin Plummer (International Agency for Research on Cancer) for statistical advice.
Financial support: Wellcome Trust (contract 075454/B/04/2); Conselho Nacional de Desenvolvimento Científico e Technológico (contract 304879/2003-7); Fundação Faculdade de Medicina, University of São Paulo, Brazil.
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–9. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Boshoff C, Weiss RA. Epidemiology and pathogenesis of Kaposi’s sarcoma-associated herpesvirus. Philos Trans R Soc Lond B Biol Sci. 2001;356:517–34. doi: 10.1098/rstb.2000.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–91. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4.Soulier J, Grollet L, Oksenhendler E, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–80. [PubMed] [Google Scholar]
- 5.Dupin N, Diss TL, Kellam P, et al. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood. 2000;95:1406–12. [PubMed] [Google Scholar]
- 6.Beral V, Peterman TA, Berkelman R, Jaffe HW. Kaposi’s sarcoma among patients with AIDS: a sexually transmitted infection? Lancet. 1990;335:123–8. doi: 10.1016/0140-6736(90)90001-l. [DOI] [PubMed] [Google Scholar]
- 7.Martin JN, Ganem DE, Osmond DH, Page-Shafer KA, Macrae D, Kedes DH. Sexual transmission and the natural history of human herpesvirus 8 infection. N Engl J Med. 1998;338:948–54. doi: 10.1056/NEJM199804023381403. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien TR, Kedes D, Ganem D, et al. Evidence for concurrent epidemics of human herpesvirus 8 and human immunodeficiency virus type 1 in US homosexual men: rates, risk factors, and relationship to Kaposi’s sarcoma. J Infect Dis. 1999;180:1010–7. doi: 10.1086/315039. [DOI] [PubMed] [Google Scholar]
- 9.Blackbourn DJ, Osmond D, Levy JA, Lenette ET. Increased human herpesvirus 8 seroprevalence in young homosexual men who have multiple sex contacts with different partners. J Infect Dis. 1999;179:237–9. doi: 10.1086/314570. [DOI] [PubMed] [Google Scholar]
- 10.Gao SJ, Kingsley L, Li M, et al. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2:925–8. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 11.Simpson GR, Schulz TF, Whitby D, et al. Prevalence of Kaposi’s sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–8. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 12.Calabro ML, Sheldon J, Favero A, et al. Seroprevalence of Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 in several regions of Italy. J Hum Virol. 1998;1:207–13. [PubMed] [Google Scholar]
- 13.Whitby D, Luppi M, Barozzi P, Boshoff C, Weiss RA, Torelli G. Human herpesvirus 8 seroprevalence in blood donors and lymphoma patients from different regions of Italy. J Natl Cancer Inst. 1998;90:395–7. doi: 10.1093/jnci/90.5.395. [DOI] [PubMed] [Google Scholar]
- 14.Whitby D, Luppi M, Sabin C, et al. Detection of antibodies to human herpesvirus 8 in Italian children: evidence for horizontal transmission. Br J Cancer. 2000;82:702–4. doi: 10.1054/bjoc.1999.0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayama S, Cuevas LE, Sheldon J, et al. Prevalence and transmission of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in Ugandan children and adolescents. Int J Cancer. 1998;77:817–20. doi: 10.1002/(sici)1097-0215(19980911)77:6<817::aid-ijc2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 16.de-Thé G, Bestetti G, van Beveren M, Gessain A. Prevalence of human herpesvirus 8 infection before the acquired immuno-deficiency disease syndrome-related epidemic of Kaposi’s sarcoma in East Africa. J Natl Cancer Inst. 1999;91:1888–9. doi: 10.1093/jnci/91.21.1888. [DOI] [PubMed] [Google Scholar]
- 17.Rezza G, Tchangmena OB, Andreoni M, et al. Prevalence and risk factors for human herpesvirus 8 infection in northern Cameroon. Sex Transm Dis. 2000;27:159–64. doi: 10.1097/00007435-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Newton R, Ziegler J, Bourboulia D, et al. The sero-epidemiology of Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in adults with cancer in Uganda. Int J Cancer. 2003;103:226–32. doi: 10.1002/ijc.10817. [DOI] [PubMed] [Google Scholar]
- 19.Biggar RJ, Whitby D, Marshall V, Linhares AC, Black F. Human herpesvirus 8 in Brazilian Amerindians: a hyperendemic population with a new subtype. J Infect Dis. 2000;181:1562–8. doi: 10.1086/315456. [DOI] [PubMed] [Google Scholar]
- 20.Whitby D, Marshall VA, Bagni RK, et al. Genotypic characterization of Kaposi’s sarcoma-associated herpesvirus in asymptomatic infected subjects from isolated populations. J Gen Virol. 2004;85:155–63. doi: 10.1099/vir.0.19465-0. [DOI] [PubMed] [Google Scholar]
- 21.Cunha AMG, Caterino-de-Araujo A, Costa SCB, et al. Increasing seroprevalence of human herpesvirus 8 (HHV-8) with age confirms HHV-8 endemicity in Amazon Amerindians from Brazil. J Gen Virol. 2005;86:2433–7. doi: 10.1099/vir.0.81087-0. [DOI] [PubMed] [Google Scholar]
- 22.Kazanji M, Dussart P, Duprez R, et al. Serological and molecular evidence that human herpesvirus 8 is endemic among Amerindians in French Guiana. J Infect Dis. 2005;192:1525–9. doi: 10.1086/491744. [DOI] [PubMed] [Google Scholar]
- 23.Mohanna S, Maco V, Gown A, Morales D, Bravo F, Gotuzzo E. Is classic Kaposi’s sarcoma endemic in Peru? Report of a case in an indigenous patient. Am J Trop Med Hyg. 2006;75:324–6. [PubMed] [Google Scholar]
- 24.Plancoulaine S, Abel L, van Beveren M, et al. Human herpesvirus 8 transmission from mother to child and between siblings in an endemic population. Lancet. 2000;356:1062–5. doi: 10.1016/S0140-6736(00)02729-X. [DOI] [PubMed] [Google Scholar]
- 25.Andreoni M, el-Sawaf G, Rezza G, et al. High seroprevalence of antibodies to human herpesvirus-8 in Egyptian children: evidence of nonsexual transmission. J Natl Cancer Inst. 1999;91:465–9. doi: 10.1093/jnci/91.5.465. [DOI] [PubMed] [Google Scholar]
- 26.Mbulaiteye SM, Pfeiffer RM, Whitby D, Brubaker GR, Shao J, Biggar RJ. Human herpesvirus 8 infection within families in rural Tanzania. J Infect Dis. 2003;187:1780–5. doi: 10.1086/374973. [DOI] [PubMed] [Google Scholar]
- 27.Mbulaiteye SM, Pfeiffer RM, Engels EA, et al. Detection of Kaposi’s sarcoma-associated herpesvirus DNA in saliva and buffy-coat samples from children with sickle cell disease in Uganda. J Infect Dis. 2004;190:1382–6. doi: 10.1086/424489. [DOI] [PubMed] [Google Scholar]
- 28.Dedicoat M, Newton R, Alkharsah KR, et al. Mother-to-child transmission of human herpesvirus-8 in South Africa. J Infect Dis. 2004;190:1068–75. doi: 10.1086/423326. [DOI] [PubMed] [Google Scholar]
- 29.Mbulaiteye S, Marshall V, Bagni RK, et al. Molecular evidence for mother-to-child transmission of Kaposi sarcoma-associated herpesvirus in Uganda and K1 gene evolution within the host. J Infect Dis. 2006;193:1250–7. doi: 10.1086/503052. [DOI] [PubMed] [Google Scholar]
- 30.Camera Pierrotti L, Sumita LM, Santos Freire W, Hehl Caiaffa Filho H, de Souza VA. Detection of human herpesvirus 8 DNA and antibodies to latent nuclear and lytic-phase antigens in serial samples from AIDS patients with Kaposi’s sarcoma. J Clin Virol. 2000;16:247–51. doi: 10.1016/s1386-6532(99)00078-5. [DOI] [PubMed] [Google Scholar]
- 31.Ashley-Morrow R, Nollkamper J, Robinson NJ, Bishop N, Smith J. Performance of FOCUS ELISA tests for herpes simplex virus type 1 (HSV-1) and HSV-2 antibodies among women in ten diverse geographical locations. Clin Microbiol Infect. 2004;10:530–6. doi: 10.1111/j.1469-0691.2004.00836.x. [DOI] [PubMed] [Google Scholar]
- 32.Nascimento MC, Ferreira S, Sabino E, et al. Performance of the HerpeSelect (Focus) and Kalon enzyme-linked immunosorbent assays for detection of antibodies against herpes simplex virus type 2 by use of monoclonal antibody-blocking enzyme immunoassay and clinicovirological reference standards in Brazil. J Clin Microbiol. 2007;45:2309–11. doi: 10.1128/JCM.00144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stebbing J, Bourboulia D, Johnson M, et al. Kaposi’s sarcoma-associated herpesvirus cytotoxic T lymphocytes recognize and target Darwinian positively selected autologous K1 epitopes. J Virol. 2003;77:4306–14. doi: 10.1128/JVI.77.7.4306-4314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 35.Zong JC, Ciufo DM, Alcendor DJ, et al. High-level variability in the ORF K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–70. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–5. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 38.Cook PM, Whitby D, Calabro ML, et al. Variability and evolution of Kaposi’s sarcoma-associated herpesvirus in Europe and Africa. AIDS. 1999;13:1165–76. doi: 10.1097/00002030-199907090-00004. [DOI] [PubMed] [Google Scholar]
- 39.Meng YX, Sata T, Stamey FR, et al. Molecular characterization of strains of Human herpesvirus 8 from Japan, Argentina, and Kuwait. J Gen Virol. 2001;82:499–506. doi: 10.1099/0022-1317-82-3-499. [DOI] [PubMed] [Google Scholar]
- 40.Nascimento MC, Wilder N, Pannuti CS, Weiss HA, Mayaud P. Molecular characterization of Kaposi’s sarcoma associated herpesvirus (KSHV) from patients with AIDS-associated Kaposi’s sarcoma in São Paulo, Brazil. J Clin Virol. 2005;33:52–9. doi: 10.1016/j.jcv.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 41.Lacoste V, Judde JG, Brière J, et al. Molecular epidemiology of human herpesvirus 8 in Africa: both B and A5 K1 genotypes, as well as the M and P genotypes of K14.1/K15 loci, are frequent and widespread. Virology. 2000;278:60–74. doi: 10.1006/viro.2000.0629. [DOI] [PubMed] [Google Scholar]
- 42.Fouchard N, Lacoste V, Couppie P, et al. Detection and genetic polymorphism of human herpes virus type 8 in endemic or epidemic Kaposi’s sarcoma from West and Central Africa, and South America. Int J Cancer. 2000;85:166–70. [PubMed] [Google Scholar]
- 43.Cook RD, Hodgson TA, Waugh ACW, et al. Mixed patterns of transmission of human herpesvirus-8 (Kaposi’s sarcoma-associated herpesvirus) in Malawian families. J Gen Virol. 2002;83:1613–9. doi: 10.1099/0022-1317-83-7-1613. [DOI] [PubMed] [Google Scholar]
- 44.Souza VA, Sumita LM, Freire W, et al. Prevalence of antibodies to human herpesvirus-8 in populations with and without risk for infection in São Paulo State. Braz J Med Biol Res. 2004;37:123–7. doi: 10.1590/s0100-879x2004000100017. [DOI] [PubMed] [Google Scholar]
- 45.Bourboulia D, Whitby D, Boshoff C, et al. Serologic evidence for mother-to-child transmission of Kaposi sarcoma-associated herpesvirus infection. JAMA. 1998;280:31–2. doi: 10.1001/jama.280.1.31-a. [DOI] [PubMed] [Google Scholar]
- 46.Lyall EG, Patton GS, Sheldon J, et al. Evidence for horizontal and not vertical transmission of human herpesvirus 8 in children born to human immunodeficiency virus-infected mothers. Pediatr Infect Dis J. 1999;18:795–9. doi: 10.1097/00006454-199909000-00010. [DOI] [PubMed] [Google Scholar]
- 47.Koelle DM, Huang ML, Chandran B, Vieira J, Piepkorn M, Corey L. Frequent detection of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in saliva of human immunodeficiency virus-infected men: clinical and immunologic correlates. J Infect Dis. 1997;176:94–102. doi: 10.1086/514045. [DOI] [PubMed] [Google Scholar]
- 48.Pauk J, Huang ML, Brodie SJ, et al. Mucosal shedding of human herpesvirus 8 in men. N Engl J Med. 2000;343:1369–77. doi: 10.1056/NEJM200011093431904. [DOI] [PubMed] [Google Scholar]
- 49.Corey L, Brodie S, Huang ML, Koelle DM, Wald A. HHV-8 infection: a model for reactivation and transmission. Rev Med Virol. 2002;12:47–63. doi: 10.1002/rmv.341. [DOI] [PubMed] [Google Scholar]
- 50.Taylor MM, Chohan B, Lavreys L, et al. Shedding of human herpesvirus 8 in oral and genital secretions from HIV-1-seropositive and –sero-negative Kenyan women. J Infect Dis. 2004;190:484–8. doi: 10.1086/421466. [DOI] [PMC free article] [PubMed] [Google Scholar]