Abstract
Particularly interesting new cysteine-histidine-rich protein (PINCH) is a LIM-domain-only adaptor protein involved in protein recruitment, subsequent assembly of multi-protein complexes, and subcellular localization of these complexes. PINCH is developmentally regulated and its expression is critical for proper cytoskeletal organization and extracellular matrix adhesion. Although PINCH has no catalytic abilities, the PIP (PINCH–ILK–parvin) complex serves as a link between integrins and components of growth factor receptor kinase and GTPase signaling pathways. Accordingly, PINCH-mediated signaling induces cell migration, spreading, and survival. Further research on the signaling cascades affected by PINCH is key to appreciating its biological significance in cell fate and systems maintenance, as the developmental functions of PINCH may extend to disease states and the cellular response to damage. PINCH is implicated in a diverse array of diseases including renal failure, cardiomyopathy, nervous system degeneration and demyelination, and tumorigenesis. This review presents evidence for PINCH's structural and functional importance in normal cellular processes and in pathogenesis. The current data for PINCH expression in nervous system disease is substantial, but due to the complex and ubiquitous nature of this protein, our understanding of its function in pathology remains unclear. In this review, an overview of studies identifying PINCH binding partners, their molecular interactions, and the potentially overlapping role(s) of PINCH in cancer and in nervous system diseases will be discussed. Many questions remain regarding PINCH's role in cells. What induces cell-specific PINCH expression? How does PINCH expression contribute to cell fate in the central nervous system? More broadly, is PINCH expression in disease a good thing? Clarifying the ambiguous functions of PINCH expression in the central nervous system and other systems is important to understand more clearly signaling events both in health and disease.
LIM-Domain Discovery
Over 20 years ago, separate discoveries of a common cysteine-rich amino acid motif led to the identification of a new class of proteins: the LIM-domain proteins. The LIM designation was derived from the names of genes whose expression led to the discovery of the cysteine-rich domain: mechanosensory-3 (MEC-3) (Way and Chalfie, 1988), cell lineage (LIN-11) (Freyd et al., 1990), and insulin gene enhancer binding protein (Isl-1) (Karlsson et al., 1990). For a more detailed description of LIM proteins, see the review by Kadrmas and Beckerle (2004).
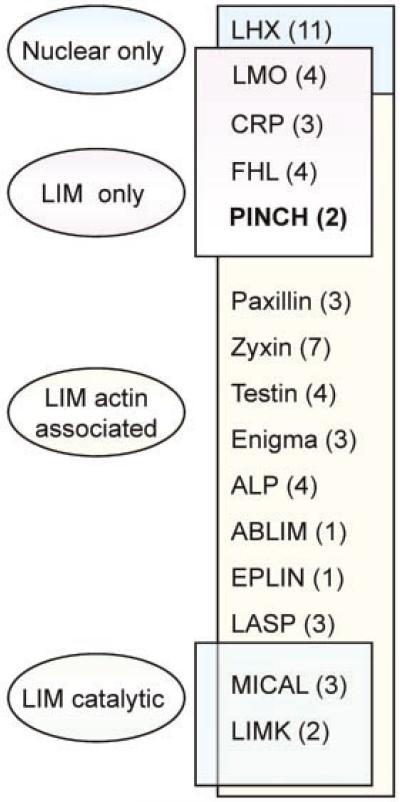
Human LIM proteins contain between one and five LIM domains. These domains may be located internally or near either termini. Four major founding groups of LIM domain-containing proteins have been described and consist of the (1) nuclear only, (2) LIM only, (3) LIM-actin-associated, and (4) LIM catalytic groups (Fig. 1). The focus of this review, particularly interesting new cysteine-histidine-rich protein (PINCH), is a LIM-only protein that lacks catalytic functions. PINCH is composed only of LIM domains, but other human LIM domain proteins contain LIM domains adjacent to kinase domains, homeodomains, cytoskeletal domains, and/or protein-binding domains (Fig. 1) (Kadrmas and Beckerle, 2004).
Fig. 1.
Human LIM proteins. Ovals represent the groups of LIM-containing proteins based on cellular location (nuclear only) or functional domain (LIM only, actin associated, or kinase catalytic). LIM families are listed in the box with the known number of members of each family in parentheses. LHX, LIM-homeodomain protein; LMO, LIM only; CRP, Cysteine-rich protein; FHL, four-and-a-half LIM; PINCH, particularly interesting new cysteine–histidine-rich protein; ALP, α-actinin-associated LIM protein; ABLIM, actin-binding LIM protein; EPLIN, epithelial protein lost in neoplasm; LASP, LIM and SH3 protein; MICAL, molecule interacting with CASL protein-1; LIMK, LIM kinase. Figure reproduced with permission from Kadrmas and Beckerle (2004).
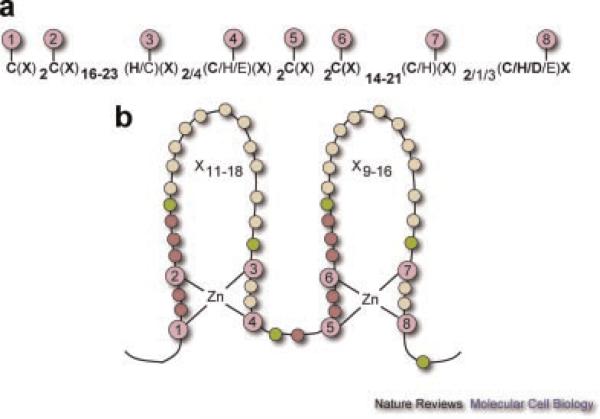
A LIM domain contains two zinc fingers, each of which binds one zinc ion (Michelsen et al., 1993) (Fig. 2). Each zinc ion is bound by four highly conserved cysteine/histidine amino acids in the fingers (Fig. 2, purple circles). A typical LIM domain consensus sequence contains approximately 55 amino acids with 8 highly conserved (cysteine/histidine) residues at defined intervals. From the human genome alone, at least 135 LIM sequences from at least 58 different genes have been identified (Kadrmas and Beckerle, 2004).
Fig. 2.
Typical LIM domain sequence and zinc fingers. a: Spacing and identity of the eight zinc-binding residues (1–8) based on an analysis of 135 human LIM sequences. Infrequently observed patterns, observed in about 10% of the cases, are denoted by un-bolded text. “X” denotes any amino acid. b: Topology of zinc coordination. Purple circles (1–8) indicate zinc-binding residues. Semi-conserved aliphatic/bulky residues are in green. Non-conserved residues with invariant spacing are in magenta and yellow circles indicate variable numbers of residues (X) that are possible in the sequence. Figure reproduced with permission from Kadrmas and Beckerle (2004).
PINCH
PINCH was identified from the screening of a human cDNA library with antibodies that recognized senescent red blood cells (Rearden, 1994). The new LIM protein (GenBank accession # U09284) consists of five LIM domains, each with unique sequences, and lacks a catalytic domain. In the first zinc finger of PINCH's LIM domains three and four, C2H2 is present rather than C2HC. Also, in the fifth LIM domain, a C4HC is substituted for C2HC, potentially altering the three-dimensional structure of the domain and therefore its binding specificity (Fig. 2). Following the discovery of PINCH (PINCH-1), a related protein PINCH-2, was characterized. The proteins share 82% amino acid sequence homology but are encoded by separate genes. Although PINCH-1 and -2 are co-expressed, they appear to be functionally distinct, with PINCH-2 potentially mediating the PINCH-1/integrin linked kinase (ILK) interaction (Zhang et al., 2002a).
Since its discovery in 1994 by Ann Rearden (Rearden, 1994), PINCH-1 has received much attention in both the basic science and clinical arenas. Studies identifying and characterizing molecular interactions of PINCH with its binding partners implicate PINCH in development of many tissues and organ systems (Fig. 3). PINCH appears to function in both a maintenance and a survival capacity (during health and disease, respectively). Moreover, aberrant PINCH-1 expression levels disrupted interactions with its binding partners, and changes in subcellular localization have been reported in a diverse array of diseases, including renal failure, cardiomyopathy, nervous system degeneration and white matter loss, and perhaps most notably, tumorigenesis.
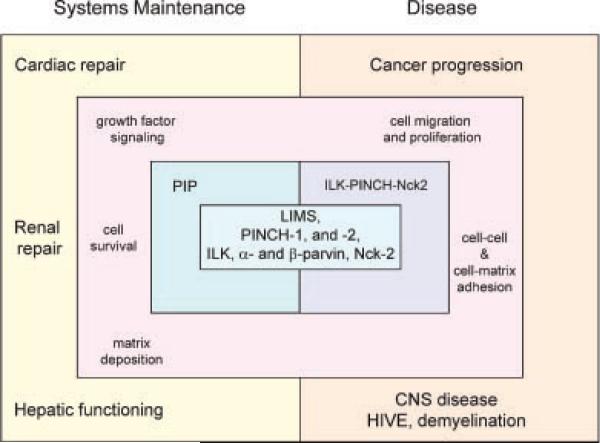
Fig. 3.
PINCH's proposed roles in the maintenance of multiple systems (cardiac, renal, and hepatic) and in pathological states including cancer and CNS diseases. With its binding partners ILK and Nck-2, PINCH is reported to function in growth factor signaling, cell survival, extracellular matrix deposition, migration, and attachment. PIP, PINCH–ILK–parvin; ILK, integrin-linked kinase; LIMS, cell lineage (LIN-11), insulin gene enhancer binding protein (Isl-1), mechanosensory-3 (MEC-3); CNS, central nervous system; HIV, human immunodeficiency virus.
PINCH Multi-Protein Complexes
PINCH participates in processes of cell–cell and cell–matrix adhesion, formation of multi-protein complexes, and facilitation of cell spreading, migration and survival. Although PINCH is not catalytic, it binds directly to other proteins (including ILK and Nck-2) (Fig. 4). Studies from Chuanyue Wu's group were the first to report that PINCH binds directly to Nck-2 and to ILK, thus connecting integrins with components of growth factor receptor kinase and GTPase signaling pathways (Tu et al., 1998, 1999). Proteins that bind to a PINCH-inclusive multi-protein complex (without binding PINCH directly) have also been identified. For instance, paxillin is a protein critical for the development of focal adhesions through its binding of ILK, and therefore through its indirect interactions with PINCH (Nikolopoulos and Turner, 2001). Research suggests that the PINCH–Nck-2 complex is an important component of focal adhesion assembly during integrin signaling (Velyvis et al., 2003). Since Nck-2 also recognizes components of growth factor receptor kinase pathways including PDGF-β1, EGF, and IRS-1, its connection with PINCH provides strong evidence linking PINCH to growth factor signaling systems. GST-pull down immunoprecipitation experiments indicated that PINCH provides a “bridge” for the ILK/PINCH/Nck-2 ternary complex, since without PINCH, ILK, and Nck-2 do not bind one another (Tu et al., 1999).
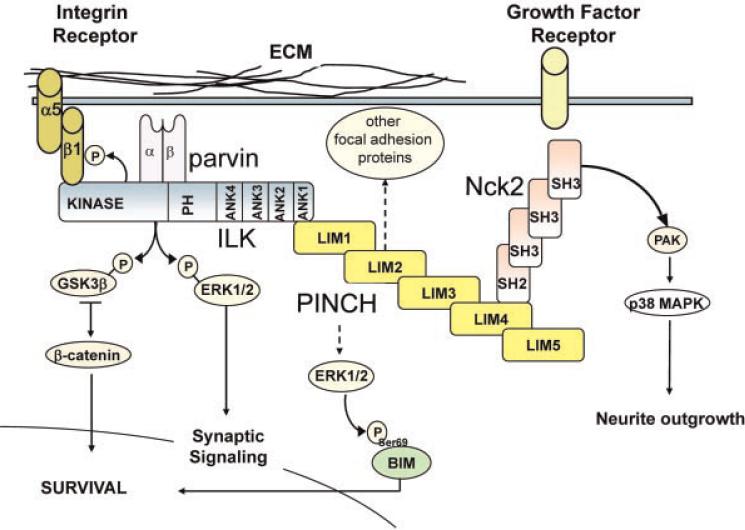
Fig. 4.
Hypothetical model of the PINCH signaling cascade. Composed of five LIM domains (yellow rectangles), PINCH binds to an SH domain of Nck-2 for growth factor receptor-mediated signaling, and to an ankyrin domain of ILK for focal adhesion and extracellular matrix crosstalk. The PINCH–ILK–parvin (PIP) complex is a downstream effector of PKC signaling. PINCH also influences the phosphorylation state of BIM leading to cell survival. ECM, extracellular matrix; PH, Pleckstrin homology domain; ANK, ankyrin; LIM, cell lineage (LIN-11), insulin gene enhancer binding protein (Isl-1), mechanosensory-3 (MEC-3); SH, Src homology; PAK, p21 activated kinase; BIM, pro-apoptotic protein.
ILK
Much of the attention that PINCH has received over the past two decades has centered on its interaction with ILK. ILK, a serine/threonine protein kinase, was first described as a β1 integrin-binding protein with the vital functions of mediating cell adhesion and cell migration (Hannigan et al., 1996). Subsequent research provided evidence that PINCH–ILK interactions were necessary for ILK to cluster and localize to sites of focal adhesion assembly, as well as for signaling between the extracellular matrix and cytoskeleton (Li et al., 1999; Zhang et al., 2002b; Fukuda et al., 2003).
Characterization of PINCH–ILK interactions revealed that the LIM1 domain at the N-terminal of PINCH interacts directly with ankyrin (ANK) repeats of ILK (Tu et al., 1999). ILK has three structurally distinct domains consisting of a C-terminal kinase domain, the binding site for β1 integrin cytoplasmic domain, and a pleckstrin homology (PH) domain (Hannigan et al., 1996). Adjacent to the C-terminal kinase domains of ILK, the (PH) domains bind phosphatidylinositol (3,4,5) triphosphate and likely aid in regulating kinase activity (Delcommenne et al., 1998). Four ANK repeats are located at the N-terminal domain of ILK, all of which are required for efficient PINCH binding (Tu et al., 1999).
Nck-2
Nck-2, a Src homology (SH) adaptor protein was the first PINCH binding partner to be identified. Nck-2 contains three N-terminal SH3 domains and one C-terminal SH2 domain (Tu et al., 1998). PINCH–Nck-2 interactions are mediated through PINCH's LIM4 domain and the third SH3 domain of Nck-2. Point mutations of LIM4 residues in the SH3-binding interface disrupted LIM–SH3 interactions and impaired localization of PINCH to focal adhesions (Velyvis et al., 2003). Nuclear magnetic resonance structural analyses of the PINCH–Nck-2 interaction indicated a small, polar and weak interface between the LIM4 and the Nck-2 SH3 domains, which implies a transient Nck-2/PINCH association (Vaynberg et al., 2005). The dynamic nature of the PINCH–Nck2 interface was illustrated by genetic rescue experiments suggesting that this interaction is involved in mediating cell shape and migration during integrin signaling at focal adhesions (Xu et al., 2005).
PINCH–ILK–Parvin
One of the most well studied PINCH protein complexes is the PKC-dependent PINCH–ILK–Parvin (PIP) ternary complex (Fig. 4). When PIP is formed, ILK serves as a structural bridge, binding both α- (or β-) parvin through its C-terminal kinase-like domains and PINCH through its N-terminal ANK repeats (Fukuda et al., 2003). The PIP complex is a downstream effector of PKC, and its formation precedes integrin-mediated localization to cell adhesion sites, cell spreading, and focal adhesion assembly (Zhang et al., 2002b). For more information on the PIP complex, see the review by Wickstrom et al. (2009).
PINCH in Development
PINCH-1 is highly conserved and its homologue, uncoordinated-97 (Unc-97) was identified in the nematode, Caenorhabditis elegans. A search for mutants with altered body wall muscle structure in C. elegans, identified unc-97 as one of the genes involved in regulatory and contractile proteins in muscle (Zengel and Epstein, 1980). Further studies showed that Unc-97 was present in touch neurons, muscles, and co-localized with β-integrin to sites of attachment in muscles (Hobert et al., 1999). Disruption of Unc-97 function led to the disturbance of focal adhesion-like structures and to functional defects in the mechanosensory neurons (Hobert et al., 1999). Both the expression profile of Unc-97 during development and its embryonic lethal null phenotype indicate involvement of Unc-97 in embryonic development in the terminal differentiation of muscles, mechanosensory neurons and integrin-based focal adhesions (Norman et al., 2007). Other members of the Unc/LIM family of proteins have been identified in C. elegans and include Unc-89, -96, -98, LIM-8, and LIM-9 (Mercer et al., 2003; Miller et al., 2006; Qadota et al., 2007; Xiong et al., 2009).
In murine development, deletion of the PINCH-1 gene by homologous recombination showed that by day E5.5, embryos displayed decreased cell proliferation, excessive cell death, and a disorganized egg cylinder (Liang et al., 2005). Mice and embryonic stem cell-derived embryoid bodies lacking the PINCH-1 gene showed abnormal epiblast polarity, impaired cavitation, endodermal detachment, and cell death (Li et al., 2005). PINCH-1s involvement in normal neural crest-derived structures was shown to be regulated, at least in part, through transforming growth factor β (TGF-β) signaling (Liang et al., 2007).
PINCH in Tissue-Specific Development, Maturation, and Maintenance
Cardiac
In the cardiac system, the integrin-mediated organization of cardiac structures is required for optimal development and maintenance. The presence of the PIP complex and its formative protein PINCH are essential for cellular maturation, migration, and proper adhesion of the intercalated disks and costameres (Liang et al., 2009). The PIP complex is formed in response to signaling cues from well-known mediators of cardiac function: fibronectin, phenylephrine, and β1-integrin (Chen et al., 2005). Thymosin β-4 also forms a complex with PINCH and ILK to recruit G-actin during cardiomyocyte development and to promote cell survival via the AKT pathway (Bock-Marquette et al., 2004).
Some studies report that deleting PINCH-1 from myocytes has no phenotypic repercussions. These studies also report that PINCH-1 and -2 are expressed redundantly in myocardial tissue, and that PINCH and ILK are both necessary for cardiac development (Hannigan et al., 1996; Liang et al., 2009). Further evidence shows that redundancy exists for cardiac PIP since a binding mutation of ILK (ILK-C) does not prevent PINCH from localizing correctly to costameres (Chen et al., 2005). Fibronectin and phenylephrine respond to altered levels of ILK-C as though it were wild-type ILK (i.e., inducing hypertrophy in response to ILK-C deficiency) (Chen et al., 2005).
Mouse models presenting as cardiac-specific homozygous null for PINCH-1/2 showed structural inadequacy on the cellular level. Protein localization and expression were significantly disrupted in these knockout models, and the mice suffered heart failure by 4 weeks of age (Liang et al., 2009). In the cardiac nervous system, ILK-knockouts have underdeveloped sarcomeres and impaired hypertrophic signaling, while ILK-C-mutated models show hypertrophy, as well as disruption in cell adhesion and in cell survival signaling pathways (Chen et al., 2005; Hannigan et al., 2007).
Since the PIP complex facilitates both myocyte formation and repair, it is not surprising that both PINCH-1/2 and ILK cardiac-specific knockout models display structural and functional weaknesses, an inability to repair on a cellular level, and suffer cardiomyopathy and death (Chen et al., 2005; Hannigan et al., 2007; Liang et al., 2007). However, the unique binding abilities of PINCH and its influence upon anti-apoptotic signaling cascades allow for repair potential long after the recognized period of cardiac development is over. Both in vitro and in vivo experiments have shown that thymosin β-4 can upregulate ILK and increase AKT-mediated cell survival (Bock-Marquette et al., 2004), suggesting potential involvement of PINCH in this repair mechanism.
Renal
Glomerular cells, podocyte epithelial cells, and mesangial cells of the kidney depend on expression of PINCH-1/2, ILK, and α-parvin for proper development and renal health (Yang et al., 2005; Jung et al., 2007; Shi et al., 2008). Molecular mechanisms for cell adhesion, matrix deposition, cytoskeletal integrity, and cell-type-based differentiation involve an increase in levels of TGF-β-1 and PIP (Jung et al., 2007; Shi et al., 2008). An increase in PINCH-1 promotes adhesion, maturation, and podocyte differentiation (Yang et al., 2005), while an increase in PINCH-2 stabilizes the PIP complex, promoting anchorage and survival among mesangial cells and immature podocytes (Jung et al., 2007; Shi et al., 2008). However, this same increase in PINCH-2 initiates apoptosis of fully differentiated epithelial podocytes (Jung et al., 2007; Shi et al., 2008).
A degree of functional redundancy exists between PINCH-1 and -2. Over-expression of PINCH-2 or upregulation of TGF-β-1 indicates that PINCH-2 can replace PINCH-1 in the PIP complex (Shi et al., 2008). However, PINCH-1 and -2 have distinct functions with respect to podocyte survival, and variation in the normal molecular mechanisms (including knockdown of PIP, mutation of binding domains on ILK and inhibition of ILK) and can disrupt adhesion and lead to apoptosis (Yang et al., 2005; Jung et al., 2007; Shi et al., 2008). An increase in TGF-β-1 triggers a discriminatory signaling cascade for cell survival (Jung et al., 2007). The disruption of normal podocyte survival and adhesion contributes to proteinuria and is a fundamental step towards kidney failure (Yang et al., 2005; Jung et al., 2007).
Hepatic
In the liver, hepatocytes rely on PIP for proper matrix deposition, cytoskeletal structure, and differentiation (Gkretsi et al., 2007a,b). In vitro experiments show the extreme effects of ILK depletion and ILK over-expression in mouse hepatocytes. ILK depletion caused increased caspase-3 and apoptotic activity, and decreased PINCH and α-parvin levels, whereas, over-expression of ILK prevented apoptosis and returned PINCH and α-parvin levels to baseline (Gkretsi et al., 2007a,b). In these same studies, in vivo genetic alterations of ILK using Cre/Lox technology prompted hepatitis with fatty changes and necrosis. The specific role of PINCH was not addressed in the altered-ILK model.
PINCH in Disease
Along with the dissection of cellular and molecular mechanisms of PINCH's interactions with other proteins during development, evidence implicating PINCH in a diverse array of diseases has begun to emerge. Reports describing increased expression and changes in the subcellular localization of PINCH in cancer and in neurodegenerative disease have helped to link PINCH function with cell behavior.
Cancer
The first clues to PINCH's involvement in invasive tumorigenesis were the observations made by Tu et al. (1999) that PINCH localizes to peripheral ruffles during cell spreading. The first direct correlation of PINCH with cancer followed in 2002, when Wang-Rodriguez et al. reported the upregulation of PINCH at invasive edges in tumor-associated stromal cells. Specifically, increased PINCH expression was observed in breast, prostate, colon, lung, and skin cancers (Wang-Rodriguez et al., 2002). Later studies went on to define PINCH stromal labeling as an independent prognostic indicator in colorectal cancer (Gao et al., 2004). PINCH expression was also discovered in oral squamous cell carcinoma (Zhang et al., 2005; Zhu et al., 2008), gliomas (Wang et al., 2007; Zhu et al., 2008), and gastric cancer (Kim et al., 2006).
Several studies have contributed to understanding more clearly the molecular mechanism(s) by which PINCH contributes to cancer progression. As previously mentioned, a second LIM-only protein with approximately 92% similarity to PINCH-1 was identified (Zhang et al., 2002b; Braun et al., 2003). PINCH-1 mRNA was expressed at embryonic day 8.5 (E8.5) in the mouse embryo, whereas, PINCH-2 message was not detectable until E14.5 (Braun et al., 2003). Like PINCH-1, PINCH-2 was observed in the cytoplasm at cell-ECM contact sites and in the nucleus (Zhang et al., 2002a,b). Also, as observed for PINCH-1, deletion of the LIM1 domain of PINCH-2 abolished ILK binding. To further characterize the roles of PINCH-1 and -2, Fässler's group generated PINCH-2 null mice and found that unlike PINCH-1 deficient mice that die at the peri-implantation stage (Li et al., 2005), PINCH-2-null mice are viable, fertile and exhibit no overt phenotype (Stanchi et al., 2005). Over-expression of PINCH-2 significantly inhibited PINCH-1–ILK interactions, and reduced cell spreading and migration, suggesting that PINCH-2 regulates the interaction of PINCH-1 and ILK (Zhang et al., 2002a). More recent studies supported this finding by showing epigenetic silencing of PINCH-2 in 91% of gastric cancer cell lines and in 53% of primary gastric tumors (Kim et al., 2006).
One mechanism through which PINCH may contribute to invasion by cancerous cells involves the Ras suppressor, Rsu-1. In this context, detachment-mediated cell death via anoikis (Frisch and Francis, 1994) is disrupted in many tumor cells, thereby encouraging anchorage-independent growth and cellular metastatic potential (Dougherty et al., 2008). Recent studies showed that Rsu-1 binds the LIM5 domain of PINCH-1 (Kadrmas et al., 2004; Dougherty et al., 2005). Further studies examined interactions of Rsu-1 with the PIP complex during Ras activation (Dougherty et al., 2008). These studies showed that a truncated form of Rsu-1 (p29) is present in high-grade gliomas and that PINCH-1 does not bind to p29. On the other hand, association of full-length Rsu-1 with PIP correlates with reduced Ras transformation. Rsu-1–PIP complex formation appears to promote adhesion, thereby decreasing migration (Dougherty et al., 2008), pointing to the importance of PINCH in facilitating the protein–protein interactions, which ultimately mediate cell behavior and fate.
Ras-mediated transformation involves MAPK phosphorylation of BIM, a BH-3 protein, which leads to cell survival. Likewise, inhibition of MAPK activity prevents BIM phosphorylation results in induction of the apoptotic pathway. Studies by Chen et al. (2008) report an important link between PINCH-1 and BIM-mediated cell survival. In these studies, PINCH-1 depletion via siRNA knockdown in several cancer cell lines resulted in the induction of the apoptotic pathway. To investigate pathways involved, cells lacking the apoptotic proteins p53, PUMA, Smac, and Bax were transfected with PINCH-1 siRNA. PINCH-1 knockdown increased apoptosis in p53-, PUMA-, and Smac-knockout cells, but Bax knockout cells showed significantly less apoptosis in the absence of PINCH-1. This suggests that the Bax pathway is involved in PINCH-1-mediated cell survival, but it is not the direct target of PINCH-1. BIM is upstream of Bax in the apoptosis pathway and is regulated by Ras-ERK phosphorylation and by ubiquitination (Harada et al., 2004). Thus, BIM expression and phosphorylation levels were assessed in PINCH-1 knockdown cells. In the absence of PINCH-1, ERK-mediated phosphorylation of BIM Ser69 was significantly reduced and led to apoptosis (Chen et al., 2008). Over-expression of PINCH-1 in PINCH-1 depleted cells rescued the cells from apoptosis as BIM Ser69 phosphorylation levels increased. Further studies are needed to better characterize the role of PINCH in cell survival in cancer, and in other disease states.
PINCH in the nervous system
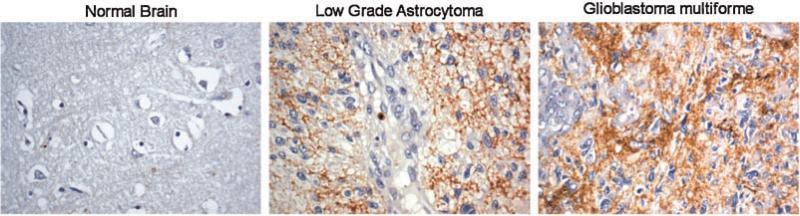
Studies in gliomas were the first to suggest a role for PINCH in CNS disease (Wang et al., 2007). Studies from the Sun group show a clear and significant correlation between PINCH expression and graded severity of glioma with greater PINCH expression in higher-grade tumors (Wang et al., 2007). These results suggest that PINCH may be involved in glioma development and differentiation. In support of these findings, recent data from our laboratory confirms that levels of PINCH expression correlate with stage severity of glioma (Fig. 5). PINCH immunoreactivity is undetectable in normal brain, but even low-grade astrocytoma has strong PINCH expression. Moreover, in glioblastoma multiforme, PINCH expression is significantly more intense than in the lower grade malignancy. The molecular basis for increased expression of PINCH in glioma is unclear. In other systems, PINCHs interaction with MAPK-mediated phosphorylation of BIM is reported to be a survival signal in transformed cells (Chen et al., 2008), however, the clinicopathological significance of this signaling cascade in glioma development is unknown.
Fig. 5.
Immunohistological analyses of human brain glioma tissues. Immunohistochemistry was performed using the avidin–biotin–peroxidase complex system. The sections of normal human brain tissue sections, low-grade astrocytoma and glioblastoma multiforme were immunolabeled with anti-PINCH-1 antibody and counterstained with hematoxylin. Microscopic evaluation was performed under the original magnification of 1,000×.
Most studies in the nervous system involving PINCH have focused on ILK and the potential contribution(s) of PINCH to maintenance of neurite extension (for review, see Ishii, 2005). Studies in a neuroblastoma cell line show that ILK activation is an early and critical event in integrin-mediated signaling. ILK activation via the p38 MAP kinase pathway was required for neurite extension (Ishii et al., 2001). Depletion of ILK by stable transfection of kinase-deficient ILK induced aberrant Tau phosphorylation (Ishii et al., 2003). ILK inactivation resulted in increased GSK3β activity and hyper-phosphorylation of Tau. Molecular and pharmacological studies by Mills et al. (2003) showed that in PC12 neuronal cells and in dorsal root ganglia neurons, inhibition of ILK resulted in reduced NGF-mediated neurite outgrowth. These studies also report Tau hyper-phosphorylation in the absence of functional ILK. However, the potential links among ILK, PINCH, GSK3β activity, and Tau phosphorylation were not addressed directly in these studies. It is unclear whether or not PINCH has a role in Tau phosphorylation. More recent studies addressing the mechanism of maladaptive plasticity in Alzheimer's disease show that LIMK-1, a member of the LIM kinase family of LIM proteins is involved in neuronal dysfunction (Heredia et al., 2006). In these studies, treatment of rat hippocampal neurons with fibrillar Aβ resulted in abnormal neurites, aberrant outgrowth, reduced axonal network, and actin filament reorganization. Further analysis revealed that LIMK-1 phosphorylation of the actin-depolymerizing factor, cofilin mediated Aβ-induced neuronal degeneration. Taken together, a clear role for ILK activation in neurite extension and GSK3β-mediated Tau-phosphorylation emerges. Potential roles for PINCH in these pathways are unclear since no direct assessments of PINCH expression were conducted.
PINCHs involvement in the peripheral nervous system was described by research showing robust expression of PINCH in the rat DRG after chronic constriction injury to the sciatic nerve (Campana et al., 2003). Prior to injury, immunolabeling detected low levels of PINCH at cell membranes. Five days after constriction injury, PINCH expression in DRG neurons, satellite cells and Schwann cells was dramatically increased and was localized in the cytoplasmic, nuclear, and peri-nuclear regions (see Fig. 2 in Campana et al., 2003). A putative leucine-rich nuclear export sequence and an overlapping basic nuclear localization sequence were identified as well, suggesting that PINCH shuttles between the nucleus and cytoplasm (Campana et al., 2003).
HIV
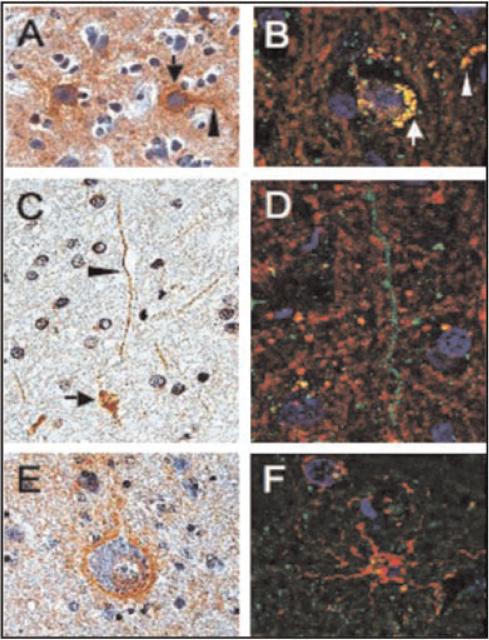
In 2008, the first report of PINCH expression in the CNS during HIV infection was described (Rearden et al., 2008). These studies reported the expression of PINCH in the neurons (Fig. 6A,B, arrows) and in the CSF of HIV-infected patients (Fig. 6). PINCH was nearly undetectable in the brains and CSF of uninfected control patients. Robust detection of PINCH was also observed in the white matter; however, PINCH was associated with neuronal processes in the white matter, but not with glial cells (Fig. 6C, arrowhead). Apart from neurons, no other cells in the brains of HIV-infected patients showed PINCH immunoreactivity.
Fig. 6.
Double immunofluorescence analyses of PINCH cellular localization in the frontal cortex of HIV patients. Panels A, C, and E are reacted with anti-PINCH antibody and counterstained with hematoxylin. Panels B, D, and F are double labeled with anti-PINCH antibody (green) and anti-neurofilament, -myelin basic protein, or -GFAP, (red, respectively). A: PINCH immunoreactivity (brown) is observed in neuronal cell bodies (arrow) and in neuronal processes (arrowhead) in an HIVE brain. B: Neuronal cells displayed both PINCH (green) and neurofilament (red) immunoreactivity with co-localization (yellow) observed in neuronal cell bodies (arrow) and in neuronal processes (arrowhead). C: PINCH (brown) immunoreactive neuronal cell bodies (arrow) and processes (arrowhead). D: Neuronal cell processes displayed PINCH immunoreactivity with a punctate, beadlike pattern (green), but did not co-localize with the surrounding myelin basic protein (red). E: Neuronal cell body and process displaying PINCH immunoreactivity (brown) surrounded by astroglial cell nuclei. F: Astroglial cell labeled with GFAP (red) showing no co-localization with PINCH (green). Scale bars = 10 μm. Reprinted with permission from Rearden et al. (2008).
More recent studies to address potential triggers for PINCH induction in HIV patients’ brains used an in vitro system mimicking some aspects of HIV infection of the CNS (Jatiani et al., 2010). Neuronal PINCH expression, subcellular distribution and biological consequences of PINCH sequestration upon challenge with Tat, gp120, and TNF-α were assessed. TNF-α or Tat treatment of neurons increased PINCH expression and changed its subcellular localization. The elevated PINCH expression that was observed following Tat treatment may be an indirect effect resulting from Tat's induction of TNF-α. Similar increases in ILK levels following exposure to Tat or TNF-α were also observed, indicative of an interaction between ILK and PINCH. On the other hand, gp120 had no effect on PINCH levels in neurons. Interestingly, sequestering PINCH using antibody-mediated mobility-disruption resulted in fewer and shorter neurite processes following treatment with TNF-α, indicating that not only is PINCH increased upon stimulation with TNF-α, but it must also be free to migrate throughout the cell to maintain neurite extensions (Jatiani et al., 2010).
White matter
As the CNS ECM is unique from the ECM of tissues and organs in the rest of the body, so are the interactions among cells of the CNS with their ECM. Tightly linked crosstalk between neurons and oligodendrocytes is a unique aspect of the CNS. For example, CNS laminin-2 (LN-2; merosin; α2β1γ1) is expressed on the axonal surface and stimulates oligodendrocytes to extend lamellae to spiral around axons (Buttery and ffrench-Constant, 1999). LN-2 deficient mice have less myelin and fewer mature oligodendrocytes, implying a distinct role for LN-2 within the CNS (Chun et al., 2003). Functional interactions of oligodendrocytes with LN-2 are required to stimulate ILK activity. It is in this context that studies are underway to address the role of PINCH in white matter diseases.
Conclusion
The functions of PINCH-mediated signaling (both in cells functioning normally and in cells responding to damage) encompass cell migration, cell spreading, and cell survival. PINCH plays a pivotal role in the adult human CNS, and may be especially important in diseases that affect neurons. The molecular events that coincide with (and that may be dependent upon) PINCH expression are beginning to be clarified. Likewise, the role of PINCH-2 in CNS diseases has yet to be addressed. Considering the importance of both PINCH-1 and -2 in development, cell migration and survival, understanding the potential contribution(s) of PINCH-2 to cellular response during CNS disease may shed light onto host-mediated attempts at cell recovery. The unique binding capacities of PINCH and its regulation of repair after cellular differentiation have implications for its contributions to CNS cell recovery. Understanding both the upstream and downstream signaling cascades involving PINCH is a first step towards defining the biological significance of PINCH in cellular response to challenge. Many questions remain regarding PINCHs role in cells of the CNS. What induces cell-specific PINCH expression? How does PINCH expression contribute to cell fate and behavior in the CNS? And more broadly, is PINCH expression a good thing? These questions continue to prompt in-depth investigations into signaling events facilitated by PINCH. Research that focuses on the molecular mechanisms of PINCH expression in the CNS will contribute to understanding of cellular processes in both health and in disease.
Materials and Methods
Immunohistochemistry
Formalin fixed, paraffin embedded human brain samples from normal, low-grade astrocytoma and glioblastoma multiforme (US BIOmax, Inc., Rockville, MD) were histologically classified according to the World Health Organization Classification of Tumors of the Nervous System. Immunohistochemistry was performed using the avidin–biotin–peroxidase complex system, according to the manufacturer's instructions (Vectastain Elite ABC Peroxidase Kit; Vector Laboratories, Burlingame, CA). Briefly, sections were deparaffinized in xylene and re-hydrated through descending alcohols to water. For non-enzymatic antigen retrieval, sections were heated in 0.01 M sodium citrate buffer (pH 6.0) to 95°C for 40 min and allowed to cool for 20 min at room temperature. To quench endogenous peroxidase, slides were then rinsed with PBS and incubated in MeOH/3% H2O2 for 20 min. Sections were then washed with PBS and blocked in PBS containing 0.1% BSA, 5% normal horse or goat serum for 2 h at room temperature, and incubated overnight at room temperature with primary antibodies. Sections were labeled with anti-PINCH rabbit polyclonal antibody (1:500) as previously described (Rearden et al., 2008). Secondary antibody was biotin-conjugated goat anti-rabbit IgGs (Vector Laboratories). Sections were developed with a diaminobenzidine substrate (Sigma, St. Louis, MO), counterstained with hematoxylin and mounted with Permount.
Double immunofluorescence labeling
Double immunofluorescence labeling was conducted as previously described (Rearden et al., 2008). Antibodies included anti-glial fibrillary acidic protein (GFAP, clone 6F2, 1:100 dilution, DAKO, Carpinteria, CA) for astrocytes, pan-neuronal cocktail for phosphorylated neurofilaments (clone SMI-311, 1:2000 dilution, Sternberger Monoclonals, Berkeley, CA) for neurons, and myelin basic protein (MBP, clone SMI-99, 1:1000 dilution, Sternberger Monoclonals, Baltimore, MD) for myelin. Sections were then incubated with either fluorescein isothiocynate (FITC)-or rhodamine-tagged secondary antibodies and cover slipped with an aqueous mounting media containing DAPI for nuclear labeling (Vectashield; Vector Laboratories). Images were visualized with a Nikon ultraviolet inverted microscope and processed with deconvolution software (Slidebook 4.0, Intelligent Imaging Innovations, Denver, Colorado). Deconvolution of selected micrographs is performed using the imaging software SlideBook 4.0 (Intelligent Imaging Innovations).
Acknowledgments
This work was supported by NIMH085602 to TDL. We would like to thank Dr. Luis Del Valle for performing the immunolabeling of tissue sections and the descriptive neuropahtological analyses included in this review.
Literature Cited
- Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432:466–472. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- Braun A, Bordoy R, Stanchi F, Moser M, Kostka GG, Ehler E, Brandau O, Fassler R. PINCH2 is a new five LIM domain protein, homologous to PINCH and localized to focal adhesions. Exp Cell Res. 2003;284:239–250. doi: 10.1016/s0014-4827(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Buttery PC, ffrench-Constant C. Laminin-2/integrin interactions enhance myelin membrane formation by oligodendrocytes. Mol Cell Neurosci. 1999;14:199–212. doi: 10.1006/mcne.1999.0781. [DOI] [PubMed] [Google Scholar]
- Campana WM, Myers RR, Rearden A. Identification of PINCH in Schwann cells and DRG neurons: Shuttling and signaling after nerve injury. Glia. 2003;41:213–223. doi: 10.1002/glia.10138. [DOI] [PubMed] [Google Scholar]
- Chen H, Huang XN, Yan W, Chen K, Guo L, Tummalapali L, Dedhar S, St-Arnaud R, Wu C, Sepulveda JL. Role of the integrin-linked kinase/PINCH1/alpha-parvin complex in cardiac myocyte hypertrophy. Lab Invest. 2005;85:1342–1356. doi: 10.1038/labinvest.3700345. [DOI] [PubMed] [Google Scholar]
- Chen K, Tu Y, Zhang Y, Blair HC, Zhang L, Wu C. PINCH-1 regulates the ERK-Bim pathway and contributes to apoptosis resistance in cancer cells. J Biol Chem. 2008;283:2508–2517. doi: 10.1074/jbc.M707307200. [DOI] [PubMed] [Google Scholar]
- Chun SJ, Rasband MN, Sidman RL, Habib AA, Vartanian T. Integrin-linked kinase is required for laminin-2-induced oligodendrocyte cell spreading and CNS myelination. J Cell Biol. 2003;163:397–408. doi: 10.1083/jcb.200304154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcommenne M, Tan C, Gray V, Rue L, Woodgett J, Dedhar S. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc Natl Acad Sci USA. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty GW, Chopp T, Qi SM, Cutler ML. The Ras suppressor Rsu-1 binds to the LIM 5 domain of the adaptor protein PINCH1 and participates in adhesion-related functions. Exp Cell Res. 2005;306:168–179. doi: 10.1016/j.yexcr.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Dougherty GW, Jose C, Gimona M, Cutler ML. The Rsu-1-PINCH1-ILK complex is regulated by Ras activation in tumor cells. Eur J Cell Biol. 2008;87:721–734. doi: 10.1016/j.ejcb.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyd G, Kim SK, Horvitz HR. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Chen K, Shi X, Wu C. PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J Biol Chem. 2003;278:51324–51333. doi: 10.1074/jbc.M309122200. [DOI] [PubMed] [Google Scholar]
- Gao J, Arbman G, Rearden A, Sun XF. Stromal staining for PINCH is an independent prognostic indicator in colorectal cancer. Neoplasia. 2004;6:796–801. doi: 10.1593/neo.04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkretsi V, Bowen WC, Yang Y, Wu C, Michalopoulos GK. Integrin-linked kinase is involved in matrix-induced hepatocyte differentiation. Biochem Biophys Res Commun. 2007a;353:638–643. doi: 10.1016/j.bbrc.2006.12.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkretsi V, Mars WM, Bowen WC, Barua L, Yang Y, Guo L, St-Arnaud R, Dedhar S, Wu C, Michalopoulos GK. Loss of integrin linked kinase from mouse hepatocytes in vitro and in vivo results in apoptosis and hepatitis. Hepatology. 2007b;45:1025–1034. doi: 10.1002/hep.21540. [DOI] [PubMed] [Google Scholar]
- Hannigan GE, Leung-Hagesteijn C, Fitz-Gibbon L, Coppolino MG, Radeva G, Filmus J, Bell JC, Dedhar S. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- Hannigan GE, Coles JG, Dedhar S. Integrin-linked kinase at the heart of cardiac contractility, repair, and disease. Circ Res. 2007;100:1408–1414. doi: 10.1161/01.RES.0000265233.40455.62. [DOI] [PubMed] [Google Scholar]
- Harada H, Quearry B, Ruiz-Vela A, Korsmeyer SJ. Survival factor-induced extracellular signal-regulated kinase phosphorylates BIM, inhibiting its association with BAX and proapoptotic activity. Proc Natl Acad Sci USA. 2004;101:15313–15317. doi: 10.1073/pnas.0406837101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia L, Helguera P, de Olmos S, Kedikian G, Sola Vigo F, LaFerla F, Staufenbiel M, de Olmos J, Busciglio J, Caceres A, Lorenzo A. Phosphorylation of actindepolymerizing factor/cofilin by LIM-kinase mediates amyloid beta-induced degeneration: A potential mechanism of neuronal dystrophy in Alzheimer's disease. J Neurosci. 2006;26:6533–6542. doi: 10.1523/JNEUROSCI.5567-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Moerman DG, Clark KA, Beckerle MC, Ruvkun G. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T. Role of integrin-linked kinase in neuronal cells. Current Enzyme Inhibition. 2005;1:3–10. [Google Scholar]
- Ishii T, Satoh E, Nishimura M. Integrin-linked kinase controls neurite outgrowth in N1E-115 neuroblastoma cells. J Biol Chem. 2001;276:42994–43003. doi: 10.1074/jbc.M105198200. [DOI] [PubMed] [Google Scholar]
- Ishii T, Furuoka H, Muroi Y, Nishimura M. Inactivation of integrin-linked kinase induces aberrant tau phosphorylation via sustained activation of glycogen synthase kinase 3beta in N1E-115 neuroblastoma cells. J Biol Chem. 2003;278:26970–26975. doi: 10.1074/jbc.M304113200. [DOI] [PubMed] [Google Scholar]
- Jatiani A, Pannizzo P, Gualco E, Del Valle L, Langford D. Neuronal PINCH is regulated by TNF-a and is required for neurite extension. J Neuroimmune Pharmacol. 2010 doi: 10.1007/s11481-010-9236-5. DOI: 10.1007/s11481-010-9236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KY, Chen K, Kretzler M, Wu C. TGF-beta1 regulates the PINCH-1-integrin-linked kinase-alpha-parvin complex in glomerular cells. J Am Soc Nephrol. 2007;18:66–73. doi: 10.1681/ASN.2006050421. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: From the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Smith MA, Clark KA, Pronovost SM, Muster N, Yates JR, III, Beckerle MC. The integrin effector PINCH regulates JNK activity and epithelial migration in concert with Ras suppressor 1. J Cell Biol. 2004;167:1019–1024. doi: 10.1083/jcb.200408090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990;344:879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- Kim SK, Jang HR, Kim JH, Noh SM, Song KS, Kim MR, Kim SY, Yeom YI, Kim NS, Yoo HS, Kim YS. The epigenetic silencing of LIMS2 in gastric cancer and its inhibitory effect on cell migration. Biochem Biophys Res Commun. 2006;349:1032–1040. doi: 10.1016/j.bbrc.2006.08.128. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang Y, Wu C. Integrin-linked kinase is localized to cell-matrix focal adhesions but not cell-cell adhesion sites and the focal adhesion localization of integrin-linked kinase is regulated by the PINCH-binding ANK repeats. J Cell Sci. 1999;112:4589–4599. doi: 10.1242/jcs.112.24.4589. [DOI] [PubMed] [Google Scholar]
- Li S, Bordoy R, Stanchi F, Moser M, Braun A, Kudlacek O, Wewer UM, Yurchenco PD, Fassler R. PINCH1 regulates cell-matrix and cell-cell adhesions, cell polarity and cell survival during the peri-implantation stage. J Cell Sci. 2005;118:2913–2921. doi: 10.1242/jcs.02422. [DOI] [PubMed] [Google Scholar]
- Liang X, Zhou Q, Li X, Sun Y, Lu M, Dalton N, Ross J, Jr, Chen J. PINCH1 plays an essential role in early murine embryonic development but is dispensable in ventricular cardiomyocytes. Mol Cell Biol. 2005;25:3056–3062. doi: 10.1128/MCB.25.8.3056-3062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Sun Y, Schneider J, Ding JH, Cheng H, Ye M, Bhattacharya S, Rearden A, Evans S, Chen J. Pinch1 is required for normal development of cranial and cardiac neural crest-derived structures. Circ Res. 2007;100:527–535. doi: 10.1161/01.RES.0000259041.37059.8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Sun Y, Ye M, Scimia MC, Cheng H, Martin J, Wang G, Rearden A, Wu C, Peterson KL, Powell HC, Evans SM, Chen J. Targeted ablation of PINCH1 and PINCH2 from murine myocardium results in dilated cardiomyopathy and early postnatal lethality. Circulation. 2009;120:568–576. doi: 10.1161/CIRCULATIONAHA.109.864686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol Biol Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen JW, Schmeichel KL, Beckerle MC, Winge DR. The LIM motif defines a specific zinc-binding protein domain. Proc Natl Acad Sci USA. 1993;90:4404–4408. doi: 10.1073/pnas.90.10.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Qadota H, Landsverk ML, Mercer KB, Epstein HF, Benian GM. UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills J, Digicaylioglu M, Legg AT, Young CE, Young SS, Barr AM, Fletcher L, O'Connor TP, Dedhar S. Role of integrin-linked kinase in nerve growth factor-stimulated neurite outgrowth. J Neurosci. 2003;23:1638–1648. doi: 10.1523/JNEUROSCI.23-05-01638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulos SN, Turner CE. Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J Biol Chem. 2001;276:23499–23505. doi: 10.1074/jbc.M102163200. [DOI] [PubMed] [Google Scholar]
- Norman KR, Cordes S, Qadota H, Rahmani P, Moerman DG. UNC-97/PINCH is involved in the assembly of integrin cell adhesion complexes in Caenorhabditis elegans body wall muscle. Dev Biol. 2007;309:45–55. doi: 10.1016/j.ydbio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rearden A. A new LIM protein containing an autoepitope homologous to “senescent cell antigen”. Biochem Biophys Res Commun. 1994;201:1124–1131. doi: 10.1006/bbrc.1994.1822. [DOI] [PubMed] [Google Scholar]
- Rearden A, Hurford R, Luu N, Kieu E, Sandoval M, Perez-Liz G, Del Valle L, Powell H, Langford TD. Novel expression of PINCH in the central nervous system and its potential as a biomarker for human immunodeficiency virus-associated neurodegeneration. J Neurosci Res. 2008;86:2535–2542. doi: 10.1002/jnr.21701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Qu H, Kretzler M, Wu C. Roles of PINCH-2 in regulation of glomerular cell shape change and fibronectin matrix deposition. Am J Physiol Renal Physiol. 2008;295:F253–F263. doi: 10.1152/ajprenal.00070.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanchi F, Bordoy R, Kudlacek O, Braun A, Pfeifer A, Moser M, Fassler R. Consequences of loss of PINCH2 expression in mice. J Cell Sci. 2005;118:5899–5910. doi: 10.1242/jcs.02686. [DOI] [PubMed] [Google Scholar]
- Tu Y, Li F, Wu C. Nck-2, a novel Src homology2/3-containing adaptor protein that interacts with the LIM-only protein PINCH and components of growth factor receptor kinase-signaling pathways. Mol Biol Cell. 1998;9:3367–3382. doi: 10.1091/mbc.9.12.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Li F, Goicoechea S, Wu C. The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol Cell Biol. 1999;19:2425–2434. doi: 10.1128/mcb.19.3.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaynberg J, Fukuda T, Chen K, Vinogradova O, Velyvis A, Tu Y, Ng L, Wu C, Qin J. Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell. 2005;17:513–523. doi: 10.1016/j.molcel.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Velyvis A, Vaynberg J, Yang Y, Vinogradova O, Zhang Y, Wu C, Qin J. Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling. Nat Struct Biol. 2003;10:558–564. doi: 10.1038/nsb938. [DOI] [PubMed] [Google Scholar]
- Wang MW, Gu P, Zhang ZY, Zhu ZL, Li YM, Zhao HX, Sun XF. Expression of PINCH protein in gliomas and its clinicopathological significance. Oncology. 2007;72:343–346. doi: 10.1159/000113064. [DOI] [PubMed] [Google Scholar]
- Wang-Rodriguez J, Dreilinger AD, Alsharabi GM, Rearden A. The signaling adapter protein PINCH is up-regulated in the stroma of common cancers, notably at invasive edges. Cancer. 2002;95:1387–1395. doi: 10.1002/cncr.10878. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- Wickstrom SA, Lange A, Montanez E, Fassler R. The ILK/PINCH/parvin complex: The kinase is dead, long live the pseudokinase. EMBO J. 2009;29:281–291. doi: 10.1038/emboj.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, Qadota H, Mercer KB, McGaha LA, Oberhauser AF, Benian GM. A LIM-9 (FHL)/SCPL-1 (SCP) complex interacts with the C-terminal protein kinase regions of UNC-89 (obscurin) in Caenorhabditis elegans muscle. J Mol Biol. 2009;386:976–988. doi: 10.1016/j.jmb.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Fukuda T, Li Y, Quin J, Wu C. Molecular dissection of PINCH-1 reveals a mechanism of coupling and uncoupling of cell shape modulation and survival. J Biol Chem. 2005;280:27361–27637. doi: 10.1074/jbc.M504189200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Guo L, Blattner SM, Mundel P, Kretzler M, Wu C. Formation and phosphorylation of the PINCH-1-integrin linked kinase-alpha-parvin complex are important for regulation of renal glomerular podocyte adhesion, architecture, and survival. J Am Soc Nephrol. 2005;16:1966–1976. doi: 10.1681/ASN.2004121112. [DOI] [PubMed] [Google Scholar]
- Zengel JM, Epstein HF. Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Guo L, Wu C. Characterization of PINCH-2, a new focal adhesion protein that regulates the PINCH-1-ILK interaction, cell spreading, and migration. J Biol Chem. 2002a;277:38328–38338. doi: 10.1074/jbc.M205576200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Tu Y, Velyvis A, Yang Y, Qin J, Wu C. Assembly of the PINCH-ILK-CH-ILKBP complex precedes and is essential for localization of each component to cell-matrix adhesion sites. J Cell Sci. 2002b;115:4777–4786. doi: 10.1242/jcs.00166. [DOI] [PubMed] [Google Scholar]
- Zhang JT, Li QX, Wang D, Zhu ZL, Yang YH, Cui DS, Wang MW, Sun XF. Up-regulation of PINCH in the stroma of oral squamous cell carcinoma predicts nodal metastasis. Oncol Rep. 2005;14:1519–1522. [PubMed] [Google Scholar]
- Zhu Z, Yang Y, Zhang Y, Wang Z, Cui D, Zhang J, Wang M, Sun XF. PINCH expression and its significance in esophageal squamous cell carcinoma. Dis Markers. 2008;25:75–80. doi: 10.1155/2008/473860. [DOI] [PMC free article] [PubMed] [Google Scholar]