Abstract
Cyclophosphamide (CPA) is one of the most widely used chemotherapeutic prodrugs that undergoes hepatic bioactivation mediated predominantly by cytochrome P450 (CYP) 2B6. Given that the CYP2B6 gene is primarily regulated by the constitutive androstane receptor (CAR, NR1I3), we hypothesize that selective activation of CAR can enhance systemic exposure of the pharmacologically active 4-hydroxycyclophosamide (4-OH-CPA), with improved efficacy of CPA-based chemotherapy. In this study, we have developed a unique human primary hepatocyte (HPH)–leukemia cell coculture model; the chemotherapeutic effects of CPA on leukemia cells can be directly investigated in vitro in a cellular environment where hepatic metabolism was well maintained. Our results demonstrated that activation of CAR preferentially induces the expression of CYP2B6 over CYP3A4 in HPHs, although endogenous expression of these enzymes in leukemia cells remains negligible. Importantly, coadministration of CPA with a human CAR activator led to significantly enhanced cytotoxicity in leukemia cells by inducing the apoptosis pathways, without concomitant increase in the off-target hepatotoxicity. Associated with the enhanced antitumor activity, a time and concentration-dependent increase in 4-OH-CPA formation was observed in the coculture system. Together, our findings offer proof of concept that CAR as a novel molecular target can facilitate CPA-based chemotherapy by selectively promoting its bioactivation.
Key Points
The constitutive androstane receptor as a novel molecular target can facilitate cyclophosphamide-based chemotherapy.
Introduction
Hematopoietic malignancies are a group of heterogeneous disorders associated with considerably variable prognoses, depending in large part on the specific diagnosis and available treatment options. Cyclophosphamide (CPA), an alkylating prodrug, has been widely used in combination with other antineoplastics in the treatment of various cancers, including hematologic neoplasms, such as lymphoma and leukemia.1–4 Nevertheless, a significant number of patients succumb to their disease despite standard therapeutic regimens.5 The need for further optimization of current treatment paradigms is evident. Given that CPA is a prodrug requiring metabolic activation by hepatic drug-metabolizing enzymes, CPA-based chemotherapy could be improved by selectively enhancing the metabolic conversion of CPA to the pharmacologically active metabolite but not to the nontherapeutic byproducts.
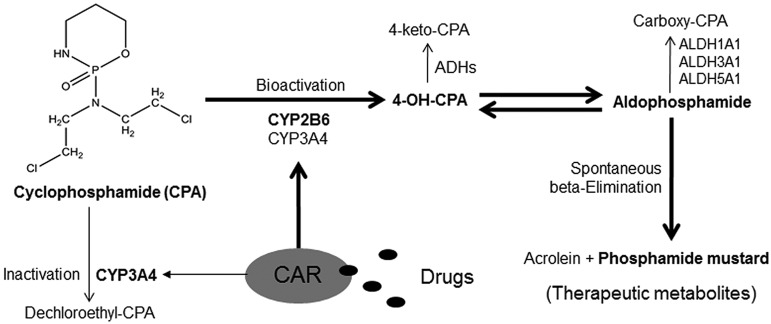
On administration, CPA undergoes hepatic oxidation to form the therapeutically active intermediate metabolite, 4-hydroxycyclophosphamide (4-OH-CPA), primarily mediated by cytochrome P450 (CYP) 2B6 and to a lesser extent by CYP3A4 and CYP2C9.6,7 4-OH-CPA is further tautomerized to aldophosphamide, followed by spontaneous β-elimination to release the phosphoramide mustard that exerts chemotherapeutic effects by attacking specific nucleophilic groups of DNA molecules in target cancer cells.8 Alternatively, CPA is subject to significant side-chain oxidation, primarily N-dechloroethylation to generate the inactive dechloroethyl-CPA and the neurotoxic chloroacetaldehyde predominantly by CYP3A4.9 In addition, 4-OH-CPA and aldophosphamide can be further deactivated by aldehyde dehydrogenases (ALDH) such as ALDH1A1, ALDH3A1, and ALDH5A1 (Figure 1).10 Notably, hydroxylation of CPA at the 4-carbon position represents the rate-limiting step of its bioactivation and the blood concentration of 4-OH-CPA has often been used as a biomarker monitoring the efficacy of CPA-based chemotherapy.11,12
Figure 1.
Schematic illustration of CPA metabolism and the proposed role of CAR in CPA bioactivation.
It is worth noting that although induction of CYP expression generally increases the elimination of drugs and leads to therapeutic failures, in the case of CPA, increasing CYP-mediated biotransformation can generate more cytotoxic intermediate metabolites with or without therapeutic potential, which may lead to comprehensive clinical ramifications.13,14 Currently, 2 orphan nuclear receptors, the constitutive androstane receptor (CAR, NR1I3) and the pregnane X receptor (PXR, NR1I2), are recognized as the primary regulators of drug-induced expression of CYP2B6 and CYP3A4, respectively, in the liver.15,16 Although activation of human (h) PXR induces both CYP3A4 and CYP2B6 with less discernible differences, recent evidence from our laboratory demonstrates that selective activation of hCAR leads to marked preferential induction of CYP2B6 over CYP3A4 in the liver.17 Based on the fact that hCAR asymmetrically cross-regulates the inductive expression of CYP2B6 and CYP3A4, we hypothesize that selective activation of hCAR can enhance the efficacy of CPA-based chemotherapy without simultaneously increasing off-target cytotoxicity.
Recognizing the specific role of CYP2B6 in the bioactivation of CPA, several studies have shown that local delivery of an adenovirus or retrovirus-encoding CYP2B expression cassette into tumor tissues can lead to enhanced intracellular CPA 4-hydroxylation and cytotoxicity.18,19 Although this strategy appears to be attractive in CPA-based treatment of localized solid tumors, it may not be applicable to hematologic malignancies, such as leukemia and lymphoma, in which systemic chemotherapy is necessary. Therefore, drugs affecting hepatic expression of CYP2B6 may profoundly influence the systemic exposure of CPA metabolites, as well as the efficacy and safety of CPA in the treatment of leukemia and lymphoma.
In this study, we present a novel human primary hepatocyte (HPH)–leukemia cell coculture model, which represents an excellent in vitro system mimicking the human in vivo situation, where hepatic drug metabolism and extrahepatic anticancer activity can be investigated concurrently. Human myeloid leukemia cells (HL-60) were used as a model target for CPA in the current study. Using gene expression assays, cytotoxicity experiments, cellular apoptosis analyses, and LC-MS/MS–based measurement of 4-OH-CPA formation, we provide experimental evidence demonstrating that selective activation of hCAR heightens the efficacy of CPA-mediated leukemia treatment without concomitant increase of off-target cytotoxicity.
Methods
Chemicals and biologic reagents
CPA, phenobarbital (PB), rifampicin (RIF), Hoechst 33342, semicarbazide hydrochloride (SCZ), and hexamethylphosphoramide (HMP) were purchased from Sigma-Aldrich. 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime (CITCO) was obtained from BIOMOL Research Laboratories. 4-hydroperoxycyclophosphamide was purchased from Enoresearch. Oligonucleotide primers were synthesized by Integrated DNA technologies. Matrigel, insulin, and ITS+ culture supplements were from BD Bioscience.
Culture and treatment of human primary hepatocytes
Human liver tissues were obtained after surgical resection by pathology staff after diagnostic criteria were met and with prior approval from the Institutional Review Board at the University of Maryland School of Medicine. The study was conducted in accordance with the Declaration of Helsinki. Hepatocytes were isolated from human liver specimens by a modification of the 2-step collagenase digestion method as previously described,20 or obtained from Life Technologies. Sandwich cultures of hepatocytes were maintained in 6-well or 12-well collagen-coated plates as described earlier.21 Thirty-six hours after seeding, hepatocytes were treated with vehicle control (0.1% DMSO), CITCO (1μM), or RIF (10μM) for another 24 hours or 72 hours before the detection of mRNA and protein, respectively. Cell culture medium was replaced on a daily basis.
Culture and treatment of leukemia and lymphoma cells
Leukemia and lymphoma cell lines including HL-60, SUDHL4, SUDHL6, K562, REH, OCI-LY-3, and Farage cells were grown in RPMI-1640 containing 10% fetal bovine serum and 1% penicillin/streptomycin. For primary cells, lymph node biopsy or bone marrow samples were obtained from cancer patients (2 T-cell lymphomas and 2 acute myeloid leukemias) on a protocol approved by the University of Maryland Medical School Institutional Review Board. Single-cell suspensions were created after cell density Ficoll-gradient separation. Cell number and viability were determined by trypan blue exclusion, with at least 70% to 80% viable cells before treatment exposure.
Human primary hepatocyte-HL-60 cell coculture model
HPHs were cultured in collagen-coated 6-well plates as described, with 3.0 μm polycarbonate membrane inserts from a 24-mm transwell plate (Corning) placed in the HPH-containing plates. Approximately 0.6 to 1.5 × 106 HL-60 cells were transferred into each insert-chamber, and a total of 4 mL of complete Williams E medium as aforementioned was shared by HPHs and HL-60 cells in each transwell. To evaluate the effects of hepatic metabolism on the anticancer activity of CPA in HL-60 cells, the cocultured cells were preincubated with vehicle control (0.1% DMSO) or CITCO (1μM) for 24 hours followed by coexposure of CPA at designated concentrations for up to 72 hours depending on experimental requirements.
Quantitative PCR analysis
Total RNA was isolated from control and treated hepatocytes or leukemia cells using the RNeasy Mini Kit (QIAGEN), and reverse transcribed using the High Capacity cDNA Archive kit (Applied Biosystems) following the manufacturers' instruction. Expression of CYP2B6, CYP3A4, ADH1B, ADH1C, ALDH1A1, ALDH3A1, ALDH5A1, and CAR mRNA was normalized against that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Quantitative real-time PCR assays were performed in 96-well optical plates on an ABI Prism 7000 sequence detection system with SYBR green PCR master mix (Applied Biosystems). Primer sequences for real-time PCR assays were acquired either from the PrimerBank (http://pga.mgh.harvard.edu/primerbank)22 or as previously reported,23 including: CYP2B6: 5′-AGACGCCTTCAATCCTGACC-3′ (forward), 5′-CCTTCACCAAGACAAATC-CGC-3′ (reverse); CYP3A4: 5′-GTGGGGCTTTTATGATGGTCA-3′ (forward), 5′-GCCTCAGATTTCTCACCAACACA-3′ (reverse); ADH1B: 5′-CCCGGAGAGCAACTACTGC-3′ (forward), 5′-AACCAGTCGAGAATCCACAGC-3′ (reverse); ADH1C: 5′-CTCGCCCCTGGAGAAAGTC-3′(forward), 5′-GGCCCCCAACTCTTTAGCC-3′ (reverse); ALDH1A1: 5′-CCGTGGCGTACTATGGATGC-3′ (forward), 5′-GCAGCAGACGATCTCTTTCGAT-3′ (reverse); ALDH3A1: 5′-CTCTGTGACCCCTCGATCCA-3′ (forward), 5′-TCTTCCCCGTAGAACTCTTTCA-3′ (reverse); ALDH5A1: 5′-AGTCATCACCCCGTGGAATTT-3′ (forward), 5′-GAGAAGGGCGTGTCTTCGG-3′ (reverse); hCAR: 5′-GAGCTGAGGAACTGTGTGGTA-3′ (forward), 5′-CTTTTGCTGACTGTTCTCCTGAA-3′ (reverse); and GAPDH: 5′-CCCATCACCATCTTCCAGGAG-3′ (forward), 5′-GTTGTCATGGATGACCTTGGC-3′ (reverse). Fold induction values were calculated according to the equation: fold over control = 2ΔΔCt, where ΔCt represents the differences in cycle threshold numbers between the target gene and GAPDH, and ΔΔCt represents the relative change in these differences between control and treatment groups.
Western blotting analyses
Homogenate proteins (20 μg) from treated HPHs or HL-60 cells were resolved on SDS-polyacrylamide gels, and electrophoretically transferred onto immobilon-P polyvinylidene difluoride membranes. Subsequently, membranes were incubated with specific antibodies against CYP2B6, CYP3A4 (Millipore-Chemicon) or cleaved caspase-3 (Cell Signaling Technology) diluted 1:4000, 1:5000, and 1:1000, respectively. β-actin was used for normalization of protein loading. After incubating with horseradish peroxidase goat anti–rabbit IgG antibody diluted 1:4000, membranes were developed using ECL Western blotting detection reagent (GE Healthcare).
Cell viability assay
After preincubation with vehicle control (0.1% DMSO) or CITCO (1μM) for 24 hours, HPH-HL-60 coculture was treated with 250, 500, or 1000μM of CPA in the presence or absence of CITCO (1μM), and 20 μL of HL-60 cells from each chamber were harvested at 12, 24, 36, and 48 hours. Prototypical trypan blue exclusion assays were carried out to determine cell viability using a Cellometer Auto T4 (Nexcelom Bioscience).
LC-MS/MS determination of 4-OH-CPA formation
Cell culture medium (200 μL) from each treatment group was mixed immediately with 20 μL of semicarbazide (SCZ; 2M) to derivatize 4-OH-CPA into 4-OH-CPA-SCZ. After vortexing for 10 minutes at room temperature, 2μM HMP was added as an internal standard and 200 μL acetonitrile was used for protein precipitation. After centrifugation, the supernatant was extracted twice with ethyl acetate. The organic phase was evaporated to dryness and reconstituted with mobile phase (water-acetonitrile, 50:50, vol/vol). The LC separation was performed on an Aquasil C18 column with water-acetonitrile (90:10, vol/vol) as the starting gradient, at a flow-rate of 0.4 mL/min with a total run time of 6.5 minutes. Mass transitions were monitored at m/z: 261→140 for CPA, m/z: 334→221 for 4-OH-CPA-SCZ, and m/z: 180→135 for HMP under the positive multiple reaction monitoring mode.
Hoechst 33342 staining
Treated HL-60 cells from the coculture system were fixed with 2% paraformaldehyde for 10 minutes at 4°C. After washing with PBS, HL-60 cells were stained with Hoechst 33342 and visualized under a fluorescence microscope (Nikon Eclipse TE-2000E). Cells that displayed intensely condensed and/or fragment nuclei staining were considered as apoptotic cells. The percentage of apoptosis was calculated from 200 cells randomly chosen from each treatment group.
Flow cytometry assay
HL-60 cells were harvested and processed with annexin V staining kit (eBioscience) according to the manufacturer's instruction. Briefly, HL-60 cells were washed once with PBS and the binding buffer, then suspended in 100 μL of binding buffer at a density of 5 × 106 cells/mL. Subsequently, cells were stained with fluorochrome-conjugated annexin V for 15 minutes at room temperature. After another round of washing, 200 μL of cells were further stained with propidium iodide and analyzed by MoFlo Legacy (Beckman Coulter).
DNA fragmentation
Genomic DNA from HL-60 cells was extracted and purified using the QIAamp DNA Blood Mini Kit (QIAGEN) following the manufacturer's instruction. From each treatment group, 4 μg of DNA were electrophoresed on a 2% agarose gel containing 0.375 μg/mL ethidium bromide in TAE buffer at 100 V for 1 hour. Images were acquired with ChemiDoc MP System (Bio-Rad) to visualize the “DNA ladder” formation.
Hepatocyte toxicity assays
A typical MTT assay was used to measure potential cytotoxicity in HPHs. In brief, HPHs were seeded in a collagen-coated 96-well plate at a density of 7.5 × 104 cells/well. After a 24 hours incubation, cells were treated with various concentrations of CPA in the presence and absence of CITCO (1μM) for 48 hours. An aliquot (20 μL) of MTT solution (5.0 mg/mL) was added to each well followed by 4 hours incubation, and the resulting crystals were dissolved in 150 μL of DMSO. Absorbance was analyzed at 580 nm using an iMark microplate reader (Bio-Rad Laboratories). In a separate experiment, the morphology of HPHs in the coculture plates was monitored under microscopy (Nikon Eclipse TS100). A toxic concentration of CPA (4000μM) was used as a positive control.
Statistical analysis
All data represent at least 3 independent experiments and are expressed as the mean ± SD. Statistical comparisons were made using 1-way analysis of variance (ANOVA) followed by a posthoc Dunnett test or Student t test where appropriate. Potential synergy between 2 drugs was analyzed using the nonlinear blending measurement as proposed by Peterson and Novick.24 The statistical significance was set at P < .05 or P < .01.
Results
Activation of CAR preferentially induces hepatic expression of CYP2B6 over CYP3A4, while exhibiting minor effects on the expression of ADHs and ALDHs
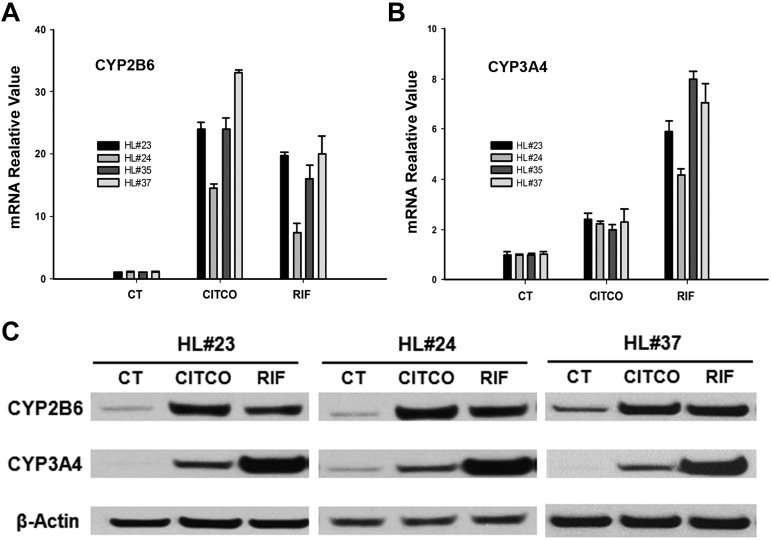
To investigate the effects of CAR on the expression of the major drug-metabolizing enzymes responsible for CPA biotransformation, HPHs were treated with the prototypical hCAR activator CITCO, or hPXR activator RIF as described in “Methods.” In 4 different preparations of human hepatocytes, CITCO (1μM) induced the expression of CYP2B6 mRNA by 14 to 34-fold, whereas CYP3A4 by 2 to 2.5-fold, respectively. On the other hand, RIF (10μM) induced CYP2B6 mRNA by 7- to 18-fold and CYP3A4 mRNA by 4- to 8-fold in these HPH cultures (Figure 2A-B). The robust and preferential induction of CYP2B6 by CITCO was further confirmed at the protein level in HPHs prepared from 3 different donors (Figure 2C). Clearly, activation of hCAR exhibits preferential induction of CYP2B6 over CYP3A4, whereas hPXR activation induced both CYP3A4 and CYP2B6 with less discernible differences.
Figure 2.
CAR-mediated induction of CYP2B6 and CYP3A4 in human primary hepatocytes. CYP2B6 and CYP3A4 mRNA and protein were measured in human primary hepatocytes prepared from 4 donors treated with CITCO (1μM), RIF (10μM), or vehicle control (0.1% DMSO) as outlined in “Methods.” (A-B) Induction of CYP2B6 and CYP3A4 mRNA analyzed with RT-PCR in HPHs from liver donors (HL#23, HL#24, HL#35, and HL#37). (C) Representative immunoblots of CYP2B6 and CYP3A4 proteins in HPHs from liver donors (HL#23, HL#24, and HL#37). RT-PCR data obtained from 3 independent experiments were expressed as mean ± SD normalized against vehicle control.
In addition to CYP2B6 and CYP3A4, several other drug-metabolizing enzymes such as ADH1B, ADH1C, ALDH1A1, ALDH3A1, and ALDH5A1 are also involved in the biotransformation and deactivation of CPA.25,26 Notably, activation of hCAR and hPXR by CITCO and RIF only exhibits modest influence on the expression of ADH1B, ADH1C, ALDH1A1, ALDH3A1, and ALDH5A1 in treated HPHs (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Together, these results indicate that selective activation of hCAR results in robust and selective induction of CYP2B6 that favor bioactivation of CPA, while exhibiting moderate effects on CYP3A4 and other enzymes associated with generation of nontherapeutic CPA metabolites.
Endogenous expression of drug-metabolizing enzymes associated with CPA metabolism in hematopoietic cancer cells
Given that in situ biotransformation of CPA in cancer cells could profoundly influence its therapeutic efficacy, we further analyzed the expression profile of genes that are associated with CPA metabolism in a diverse panel of leukemia and lymphoma cell lines, including HL-60, REH, K562, SUDHL4, SUDHL6, OCI-LY-3, and Farage, as well as in primary lymphoma and leukemia cells obtained from 4 patients. The relevant gene expression levels in these cancer cells were compared with that in HPHs by real-time PCR analysis. As shown in Figure 3A, endogenous expression of CYP2B6, CYP3A4, ADH1B, ADH1C, ALDH3A1, and CAR in all leukemia and lymphoma cells is negligible in comparison with that in HPHs, whereas expression of ALDH5A1 is relatively consistent across all tested cells. It is noteworthy that in contrast to other tested cancer cell lines, K562 cells express relatively high levels of ALDH1A1 that are comparable with that of HPHs, indicating that K562 may not be a CPA-sensitive target. Subsequently, we examined whether expression of the CYP2B6 and CYP3A4 genes was inducible in HL-60 and primary lymphoma/leukemia cells by CITCO (1μM) or RIF (10μM). As expected, expression of CYP2B6 and CYP3A4 mRNA remains negligible on these treatments in HL-60 and the primary cancer cells (Figure 3B-C). Collectively, these observations suggest that endogenous and inductive expressions of CPA-metabolizing enzymes are insignificant in most leukemia and lymphoma cells, and bioactivation of CPA for the treatment of hematopoietic malignancies relies predominantly on hepatic metabolism.
Figure 3.
Basal and induced expression of CYP2B6, CYP3A4, ADHs, ALDHs, and CAR in hematopoietic cancer cells. Total RNA was extracted from 7 hematopoietic cancer cell lines, namely, HL-60, REH, K562, SUDHL4, SUDHL6, OCI-LY-3, and Farage, and primary lymphoma and leukemia cells from 4 patients (T-cell lymphoma [TCL1 and TCL2], and acute myeloid leukemia [AML1 and AML2]). (A) The relevant gene abundance of CYP2B6, CYP3A4, ADH1B, ADH1C, ALDH1A1, ALDH3A1, ALDH5A1, and CAR in these cells was analyzed using RT-PCR in comparison with that in HPHs from donor HL#7 and HL#10, as detailed in “Methods.” (B-C) HL-60 cells, TCL1, AML1, and HPHs (HL#20) were treated with CITCO (1μM), RIF (10μM), or vehicle control (0.1% DMSO) for 24 hours. Subsequently, inductive expression of CYP2B6, CYP3A4, and CAR in these cells was measured by RT-PCR. RT-PCR data obtained from 3 independent experiments were expressed as mean ± SD normalized against vehicle control (**P < .01).
Activation of CAR enhances CPA anticancer activity in the HPH-HL-60 coculture system through promoting CPA bioactivation
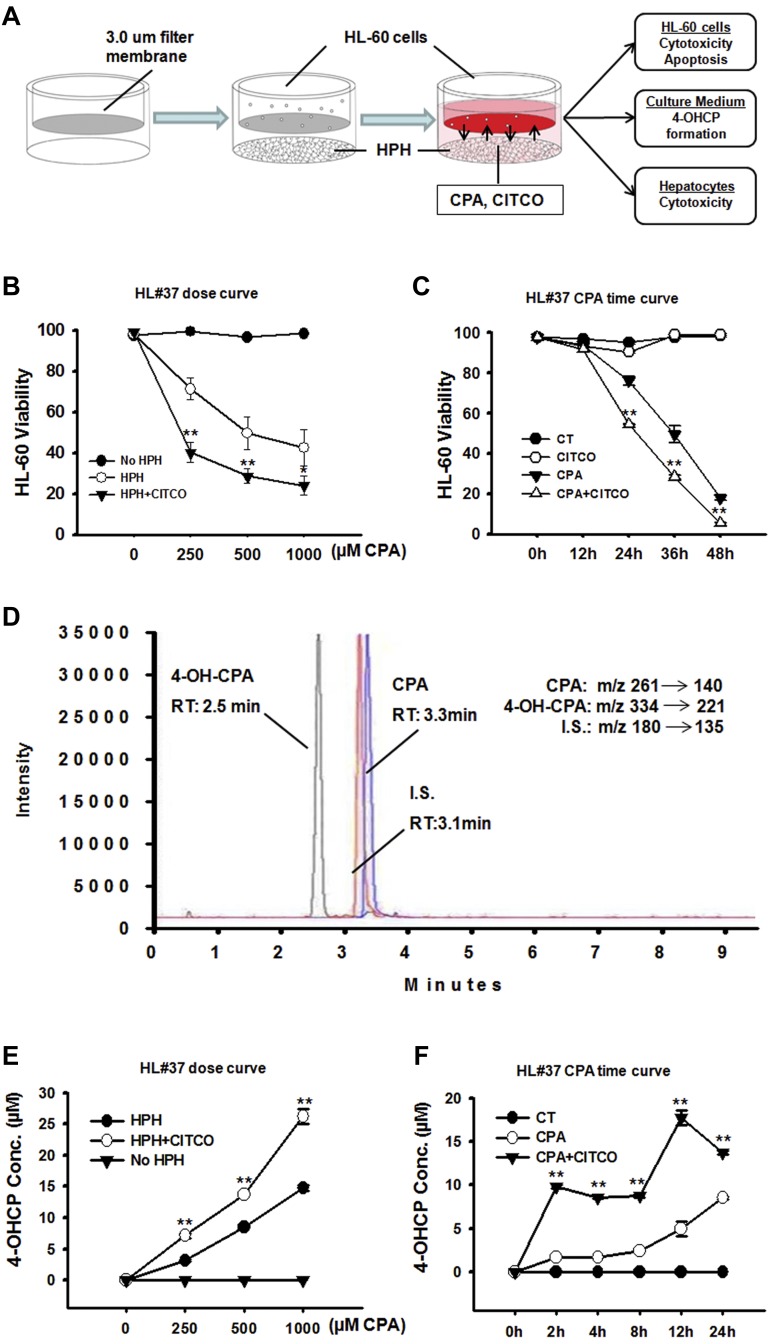
The HPH-HL-60 coculture model was established as depicted in Figure 4A. HPHs were preincubated with vehicle control (0.1% DMSO) or CITCO (1μM) for 24 hours to allow gene induction. Different concentrations of CPA were then applied to the cocultures with the presence of vehicle control or CITCO in the medium. CITCO at the concentration of 1μM produces optimal hCAR activation and CYP2B6 induction in HPHs.27,28 Therefore, unless otherwise indicated, CITCO was used at 1μM in the current studies. Results from the cocultures with HPHs prepared from a representative liver donor indicate that CPA at concentrations bracketing its pharmacologically relevant levels markedly decreased the viability of HL-60 cells in a concentration-dependent manner (Figure 4B). Notably, cotreatment with CITCO significantly enhanced CPA-mediated cytotoxicity in HL-60 cells, in that 250μM of CPA + CITCO could achieve equal to greater magnitudes of anticancer activity than CPA alone at the concentration of 1000μM. The temporal effects of CPA in the presence and absence of CITCO were also evaluated in parallel experiments. As shown in Figure 4C, CPA-induced cytotoxicity in HL-60 cells is time-dependent and the presence of CITCO can significantly enhance CPA-induced cytotoxicity as early as 24 hours after cotreatment. On the other hand, CITCO alone did not exhibit noticeable cytotoxicity in the cocultured HL-60 cells (supplemental Figure 2).
Figure 4.
Activation of CAR enhances CPA anticancer activity in the human primary hepatocyte-HL-60 cell coculture system. The anticancer activity of CPA was analyzed in a unique hepatocyte-cancer cell coculture model. (A) Illustration of the HPH-HL-60 coculture model and experimental scheme. (B) Effects of CAR activation on the concentration-dependent anticancer activity of CPA in HL-60 cells. As detailed in “Methods,” the HPH-HL-60 cocultures were treated with vehicle control (0.1% DMSO) or CPA (250, 500, and 1000μM) in the presence and absence of CITCO (1μM). Cell viability was analyzed 36 hours after the cotreatment. (C) Effects of CAR activation on the temporal changes of CPA-mediated anticancer activity in HL-60 cells. In the cocultures, cells were treated with vehicle control (0.1% DMSO) or CPA (500μM) with or without CITCO (1μM). Cell viability was measured at 0, 12, 24, 36, and 48 hours after the cotreatment. All viability data represent mean ± SD from 3 independent experiments and are expressed as percent viability of vehicle control (**P < .01). (D) The multiple-reaction monitoring chromatogram demonstrates separation and retention of 4-OH-CPA, CPA, and the internal standard from LC-MS/MS detection. (E) Effects of CAR activation on the concentration-dependent formation of 4-OH-CPA in the coculture under the treatment as outlined in panel B. (F) Effects of CAR activation on the temporal changes of 4-OH-CPA formation in cocultures under the treatment described in panel C. 4-OH-CPA concentrations represent mean ± SD of 3 LC-MS measurements (**P < .01).
To further evaluate the effects of CAR activation on the biotransformation of CPA in the HPH-HL-60 coculture system, culture medium from each treatment group as described was collected and subjected to LC-MS/MS quantification of 4-OH-CPA as outlined in “Methods.” The multiple-reaction monitoring chromatogram demonstrates clear separation and retention of 4-OH-CPA, CPA, and the internal standard (Figure 4D). As shown in Figure 4E, formation of 4-OH-CPA in the coculture system was increased in a concentration-dependent manner on CPA treatment; importantly, this trend was further enhanced in the presence of CITCO. Likewise, at a fixed concentration of CPA (500μM), 4-OH-CPA formation in the coculture medium increased through a time window from 2 to 24 hours, and CITCO markedly enhanced the 4-OH-CPA concentrations at all the time points measured (Figure 4F). These results indicate that the observed enhancement in the chemotherapeutic activity of CPA in HL-60 cells was attributed to CAR-mediated augmentation of 4-OH-CPA formation in the liver.
Activation of CAR promotes CPA-mediated apoptosis of HL-60 cells in the HPH-HL-60 coculture system
After CYP-mediated hydroxylation, CPA went through sequential metabolic processes and was converted to the ultimate active alkylating nitrogen mustard that attacks specific nucleophilic groups of DNA molecules in cancer cells, which often leads to cell apoptosis.29,30 To evaluate the effects of CAR activation on CPA-induced DNA damage in target cancer cells, a series of apoptosis experiments was carried out in the HPH-HL-60 coculture system.
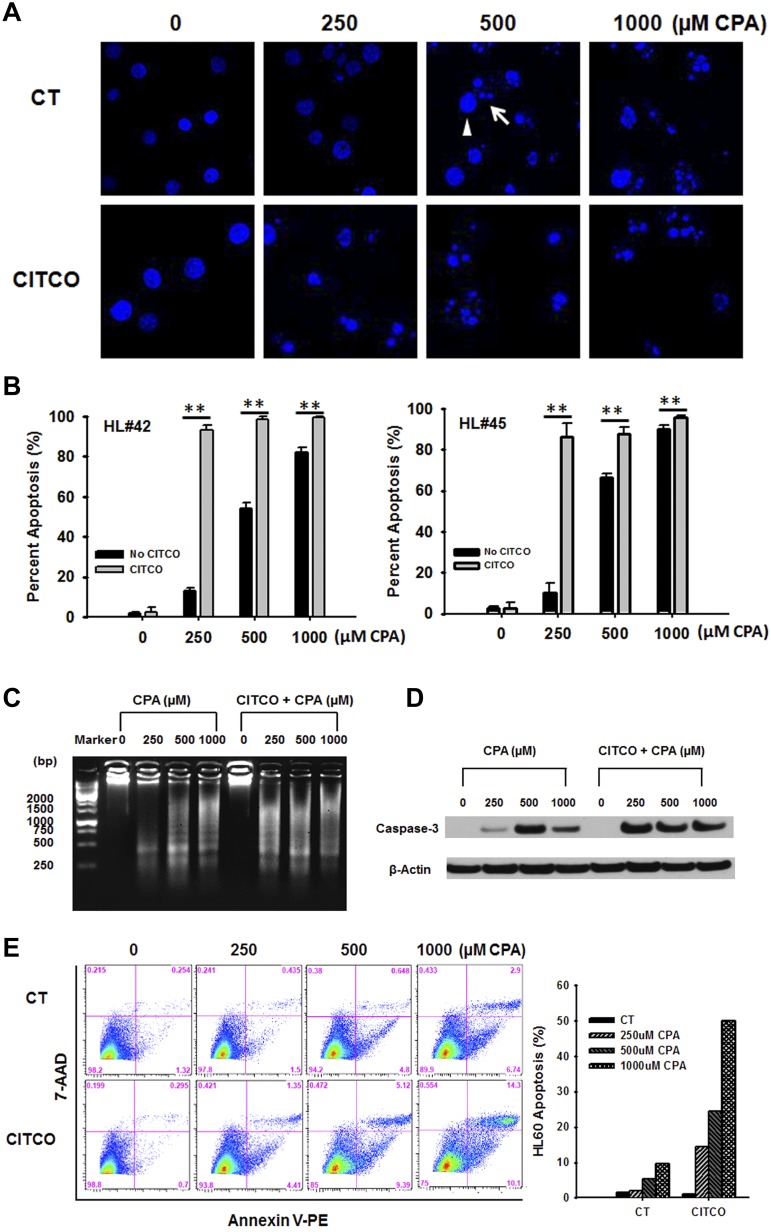
It is well known that formation of apoptotic bodies from chromatin condensation and nuclear fragmentation is one of the prototypical features of cell apoptosis.31 Examples of normal nuclei (arrow head) and apoptotic bodies (arrow) were demonstrated in Hoechst 33342-stained HL-60 cells under different treatments as indicated (Figure 5A). To quantify the accumulation of apoptotic bodies induced by CPA, 200 HL-60 cells from each treatment group were analyzed by fluorescence microscopy. As shown in Figure 5B, CPA at the concentration of 250, 500, and 1000μM resulted in 11% to 14%, 55% to 66%, and 82% to 90% of HL-60 apoptosis, respectively. When cotreated with CITCO, a significant increase of the apoptosis rate was observed at all 3 concentrations of CPA treatment. In particular, in the presence of CITCO, 250μM of CPA treatment resulted in approximately 90% of apoptotic HL-60 cells, a rate comparable with that induced by CPA at 1000μM alone. In parallel experiments, DNA ladder resulting from CPA-induced DNA fragmentation was also examined in HL-60 cells after treatment with CPA and CITCO as described. DNA fragmentation was clearly observed in HL-60 cells when treated with CPA at 500 and 1000μM in the coculture system (Figure 5C). Consistent with the previously mentioned chromatin condensation assays, CPA at 250μM alone only resulted in moderate DNA damage, whereas inclusion of CITCO has strongly potentiated CPA-mediated DNA fragmentation.
Figure 5.
Activation of CAR promotes CPA-mediated apoptosis of HL-60 cells in the HPH-HL-60 coculture model. In the coculture system, cells were treated with vehicle control (0.1% DMSO) or CPA (250, 500, and 1000μM) in the presence and absence of CITCO (1μM) as outlined in “Methods.” CPA-mediated apoptosis in HL-60 cells was analyzed with different assays. (A) Representative visualization of HL-60 cells stained with Hoechst 33342. Arrowhead and arrow depict normal nuclei and apoptotic bodies, respectively. (B) Quantitation of apoptotic bodies induced by CPA with/without CITCO. Two hundred HL-60 cells from each treatment group were calculated under fluorescent microscopy. Percent of apoptotic cells were expressed as mean ± SD obtained from 3 independent experiments (**P < .01). (C) DNA extracted from treated cells was loaded on an agarose gel to illustrate CPA-induced DNA fragmentation. (D) Caspase 3 activity was analyzed with Western blotting to detect the large fragment (17/19 kDa) of activated caspase 3 in HL-60 cells. β-actin was used to normalize protein loading. (E) Effects of CAR activation on CPA-mediated membrane translocation of phosphatidylserine during apoptosis were analyzed using flow cytometry as detailed in “Methods.”
Sequential activation of caspases plays a central role in cell apoptosis. Caspase 3 is considered to be one of the most important executive caspases in apoptosis pathways.32 To better understand the relationship between CAR activation and CPA-mediated apoptosis, we conducted Western blotting analysis to detect the large fragment (17/19 kDa) of activated caspase 3 in HL-60 cells treated with the same combination of CPA and CITCO as described in “Methods.” Induction of capsase 3 protein was observed in all CPA-treated groups, where 500μM of CPA resulted in the strongest induction and 250μM of CPA exhibited moderate enhancement of caspase 3 activation (Figure 5D). Similarly, when cotreated with CITCO, CPA (250μM)–induced caspase-3 activation was extensively augmented. Interestingly, activation of caspase 3 by CPA appears saturated at 500μM and further increase in CPA concentration did not result in additional expression of this cysteine protease.
It has been reported that apoptotic cells exhibit increased externalization of phosphatidylserine as a cell-surface modification.33 Next, we examined whether CAR activation affects CPA-mediated translocation of phosphatidylserine during apoptosis by flow cytometry analysis. As shown in Figure 5E, CPA induced HL-60 apoptosis in a concentration-dependent manner. With CITCO cotreatment, the same trend but significantly higher apoptosis rates were achieved at all 3 CPA concentrations. Taken together, results from these apoptosis analyses clearly establish that CITCO promotes CPA therapeutic effect in HL-60 cells by enhancing multiple apoptotic pathways.
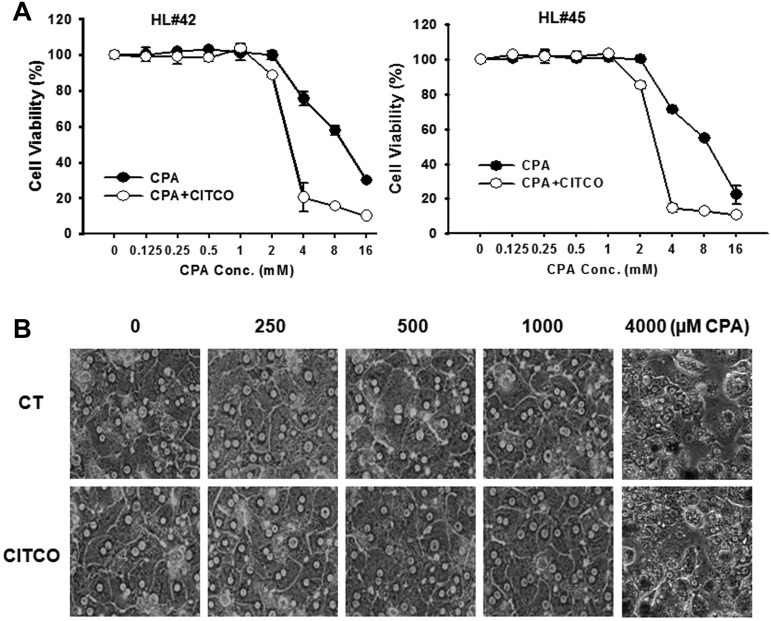
Cytotoxicity in human primary hepatocytes
MTT assays were performed in HPHs after 48 hours incubation of CPA at a wide range of concentrations, in the presence or absence of CITCO. As shown in Figure 6A, the viability of HPHs remained more than 90% at CPA concentrations up to 1mM, which represents the highest concentration used in all aforementioned studies and, cotreatment with CITCO did not increase hepatotoxicity within this concentration (0.125-1mM) range. However, when CPA concentration reaches 2mM and beyond, significant hepatotoxicity was observed in HPHs and this toxicity was further enhanced by CITCO (Figure 6A). In addition, we have monitored the morphologic changes of HPHs in the coculture model. Consistent with the MTT assay results, CPA exhibits no hepatotoxicity at the concentration of 250, 500, and 1000μM regardless of the presence or absence of CITCO (Figure 6B). CPA at 4000μM was applied as a positive control. These results indicate that CPA at concentrations applied in the current studies (250-1000μM) is nontoxic to the nontarget hepatocytes with or without the presence of CITCO.
Figure 6.
Cytotoxicity of CPA and CITCO in human primary hepatocytes. (A) HPHs from donors (HL#42 and HL#45) in 96-well collagen coated plate were treated with CPA at the wide range of concentrations in the presence and absence of CITCO (1μM). Prototypical MTT assays were performed 48 hours after the treatments as described in “Methods.” Cell viability data represent mean ± SD from 3 independent measurements and are expressed as percent of vehicle control. (B) Morphologic changes of HPHs in the coculture model were monitored under microscopy after various treatments. CPA at 4000μM was used as a positive control demonstrating toxic morphology damages to the HPHs.
Discussion
CPA belongs to the oxazaphosphorine-type of alkylating agents that have a broad spectrum of antineoplastic activity against various types of cancers.1,3 Designed as a prodrug, the therapeutic efficacy of CPA largely relies on drug-metabolizing enzyme-mediated bioactivation to generate the DNA-crosslinking metabolite. Among others, CYP2B6 enzyme is predominantly responsible for the hydroxylation of CPA at the 4-carbon position as the initial and rate-limiting step of CPA bioactivation.34 Therefore, selective increase of the expression and activity of CYP2B6 in cancer patients may benefit CPA-based chemotherapy. In this study, we showed that CAR is a novel therapeutic target that can facilitate CPA-based anticancer activity by preferentially increasing hepatic expression of CYP2B6 and the formation of 4-OH-CPA.
As one of the key mechanisms of action, biotransformation of CPA has been extensively studied and many drug-metabolizing enzymes that differentially contribute to the bioactivation and deactivation of CPA have been identified.5,13 Specifically, the CYP2B6 enzyme contributes primarily to the bioactivation of CPA, whereas CYP3A4 was responsible for approximately 95% of CPA N-dechloroethylation, generating the inactive dechloroethyl-CPA and the neurotoxic chloroacetaldehyde.6,34 In addition, several ALDHs convert the aldophosphamide to the carboxyphosphamide which represents a major stable nontherapeutic metabolite of CPA.26,35 Notably, although liver is often not the target for CPA-based chemotherapy, biotransformation of CPA to its active forms occurs predominantly in this metabolic organ before reaching the targeted cancer cells.36,37 In accord with this concept, our results revealed that endogenous expression of the major drug-metabolizing enzymes responsible for CPA metabolism is negligible in several leukemia and lymphoma cells, suggesting that in situ bioactivation of CPA in these cancer cells is generally insignificant. However, it is noteworthy that expression of ALDH1A1 in K562 cells is comparable with that in HPHs, and is markedly greater than the expression in other cancer cells tested. Given the importance of ALDH1A1 in the deactivation of CPA, this result indicates that K562 cells might be less sensitive to CPA than other leukemia cells. Indeed, previous studies showed that cancer stem cells expressing higher levels of ALDH1A1 are resistant to CPA treatment38; and selective knockdown of ALDH1A1 expression in K562 cells sensitized their response to CPA.39
To date, it has been widely accepted that inductive expression of CYP2B6, the rate-limiting enzyme for CPA bioactivation, is primarily regulated at the transcriptional level by the nuclear receptors CAR and PXR in a tissue-specific manner.15,40,41 However, many drugs induce both CYP2B6 and CYP3A4 expression with less discernible differences in HPHs through activation of PXR only (eg, RIF, hyperforin, and SR12813), activation of both PXR and CAR (eg, PB, and artemisinin), or activation of PXR but deactivation of CAR (eg, clotrimazole, ethynyl estradiol, and PK11195).28,42–44 On the other hand, in line with our previous findings,17 we showed that selective activation of human CAR by CITCO leads to preferential induction of CYP2B6 over CYP3A4 in HPHs but not in leukemia and lymphoma cells. Given the favorable effects of CYP2B6 in CPA bioactivation, these observations support our hypothesis that selective activation of CAR may benefit CPA-based chemotherapy of hematopoietic malignancies, in which CPA continues to be an important component of front-line regimens.
Tumor-specific expression of drug-metabolizing enzymes, particularly CYP2B6, would significantly contribute to the “selective cytotoxicity” of CPA and its therapeutic efficacy. In this effort, several studies have shown that using retroviral, replicating herpes viral, and adenoviral vectors, regional delivery of CYP2B expression cassette into tumor tissues significantly increased cancer cell cytotoxicity and intracellular 4-OH-CPA formation.18,19,45,46 Whereas this strategy may benefit patients with localized solid tumors, in the case of hematologic malignancies, systemic treatment is required and bioactivation of CPA primarily relies on hepatic drug-metabolizing enzymes. Our HPH-leukemia cell coculture model represents an excellent in vitro system mimicking human in vivo condition that allows simultaneous investigation of hepatic metabolism and extrahepatic anti-cancer activity under a shared cellular environment. We found that coculture of HPHs with HL-60 cells markedly increased the cytotoxicity of CPA in HL-60 cells, as well as the formation of 4-OH-CPA in the shared culture medium, in concentration and time-dependent manners. Of importance, activation of CAR by CITCO synergistically enhanced the anti-cancer activity of CPA in HL-60 cells, particularly, 250μM of CPA in the presence of CITCO (1μM) led to an increased anti-cancer activity that could challenge what was achieved by 1000μM of CPA alone. On the other hand, inclusion of CITCO did not alter the cytotoxicity in cocultured HPHs on treatment with CPA up to 1000μM. This selective toxicity of 4-OH-CPA in HL-60 cells over HPHs may be attributable to the quiescent nature of HPHs, as well as the fact that HPHs express abundant enzymes associated with both activation and deactivation of CPA. Given that chemotherapeutics are generally more toxic than other drugs, including CAR activators, it is reasonable to speculate that concurrent administration of a selective hCAR activator with a lower dose of CPA would benefit cancer patients receiving CPA-based chemotherapy.
Many chemotherapeutics trigger necrosis or apoptosis by attacking cellular macromolecules in actively proliferating cancer cells. In the case of CPA, formation of the active DNA-crosslinking metabolite, the phosphoramide mustard, primarily provokes cancer cell apoptosis.29 We found that hepatic activation of CPA resulted in multifaceted apoptotic reactions in the targeted HL-60 cells, including the formation of apoptotic bodies, DNA fragmentation, cleavage of caspase 3, and plasma membrane blebbing. In line with the aforementioned cytotoxicity results in HL-60 cells, activation of hCAR markedly enhanced CPA-mediated apoptosis in these cells. In particular, a synergistic response was observed between CITCO and the lower concentration of CPA, further signifying the potentially plausible effects of CAR activation in CPA treatment.
In summary, our results demonstrate that selective activation of hCAR can facilitate CPA-based antineoplastic activity in leukemia cells by preferentially promoting CYP2B6–mediated CPA bioactivation, without increasing cytotoxicity in cocultured HPHs. Moreover, we have established an innovative HPH-leukemia cell coculture system and demonstrated it to be a useful in vitro model for studying the biotransformation and therapeutic effects of prodrugs in an environment that closely mimics human in vivo conditions. Our findings thus far suggest a provocative notion that inclusion of a selective hCAR activator in CPA-based regimen may benefit cancer patients with hematologic malignancies. Conversely, we are aware that the current discoveries are still at the preclinical stage and selective hCAR activators identified thus far from clinically used drugs are extremely limited. It is interesting to note that previous studies have shown that administration of PB (activator of both hCAR and hPXR) was associated with acceleration of CPA biotransformation, but with little impact on drug efficacy.47,48 Notably, although PB is a potent tumor promoter by virtue of stimulating cell proliferation, such effect was not associated with CITCO (data not shown), emphasizing a need to identify novel and selective hCAR modulators with eliminated undesirable effects from a therapeutic perspective. Given that improving the therapeutic index of CPA continues to have high clinical relevance, future in vivo and clinical studies are warranted.
Supplementary Material
Acknowledgments
The authors thank Dr Maureen Kane (Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, MD) for advising on the development of LC-MS/MS method; Dr Hongbin Fang (Division of Biostatistics, University of Maryland Greenebaum Cancer Center) for advising on the statistical analysis; and Ms Carol Arca (University of Maryland Medical Center, Baltimore, MD) for providing the human liver samples used in this study. The authors are also grateful to members of the Wang laboratory for discussions and comments on the paper.
This work was supported in part by the National Institutes of Health (grant R01 DK061652) and a Seed Grant from University of Maryland Greenebaum Cancer Center.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.W. designed and performed the experiments with the assistance of L.L. and H.Y., analyzed the data, and wrote the paper; S.S.F. provided new reagents; R.B.G. and M.R.B. provided new reagents and advice on the experiments; H.W. designed and supervised all of the research and revised the paper; and all authors read and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hongbing Wang, Dept of Pharmaceutical Sciences, University of Maryland School of Pharmacy, 20 Penn St, Baltimore, MD 21201; e-mail: hwang@rx.umaryland.edu.
References
- 1.Bonadonna G, Valagussa P, Moliterni A, Zambetti M, Brambilla C. Adjuvant cyclophosphamide, methotrexate, and fluorouracil in node-positive breast cancer: the results of 20 years of follow-up. N Engl J Med. 1995;332(14):901–906. doi: 10.1056/NEJM199504063321401. [DOI] [PubMed] [Google Scholar]
- 2.Coiffier B. Treatment of diffuse large B-cell lymphoma. Curr Hematol Rep. 2005;4(1):7–14. [PubMed] [Google Scholar]
- 3.Mounier N, Briere J, Gisselbrecht C, et al. Rituximab plus CHOP (R-CHOP) overcomes bcl-2–associated resistance to chemotherapy in elderly patients with diffuse large B-cell lymphoma (DLBCL). Blood. 2003;101(11):4279–4284. doi: 10.1182/blood-2002-11-3442. [DOI] [PubMed] [Google Scholar]
- 4.Casak SJ, Lemery SJ, Shen YL, et al. U.S. Food and drug administration approval: rituximab in combination with fludarabine and cyclophosphamide for the treatment of patients with chronic lymphocytic leukemia. Oncologist. 2011;16(1):97–104. doi: 10.1634/theoncologist.2010-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraud B, Hebert G, Deroussent A, Veal GJ, Vassal G, Paci A. Oxazaphosphorines: new therapeutic strategies for an old class of drugs. Expert Opin Drug Metab Toxicol. 2010;6(8):919–938. doi: 10.1517/17425255.2010.487861. [DOI] [PubMed] [Google Scholar]
- 6.Huang Z, Roy P, Waxman DJ. Role of human liver microsomal CYP3A4 and CYP2B6 in catalyzing N-dechloroethylation of cyclophosphamide and ifosfamide. Biochem Pharmacol. 2000;59(8):961–972. doi: 10.1016/s0006-2952(99)00410-4. [DOI] [PubMed] [Google Scholar]
- 7.Roy P, Yu LJ, Crespi CL, Waxman DJ. Development of a substrate-activity based approach to identify the major human liver P-450 catalysts of cyclophosphamide and ifosfamide activation based on cDNA-expressed activities and liver microsomal P-450 profiles. Drug Metab Dispos. 1999;27(6):655–666. [PubMed] [Google Scholar]
- 8.Fenselau C, Kan MN, Rao SS, Myles A, Friedman OM, Colvin M. Identification of aldophosphamide as a metabolite of cyclophosphamide in vitro and in vivo in humans. Cancer Res. 1977;37(8 Pt 1):2538–2543. [PubMed] [Google Scholar]
- 9.Yu L, Waxman DJ. Role of cytochrome P450 in oxazaphosphorine metabolism. Deactivation via N-dechloroethylation and activation via 4-hydroxylation catalyzed by distinct subsets of rat liver cytochromes P450. Drug Metab Dispos. 1996;24(11):1254–1262. [PubMed] [Google Scholar]
- 10.Dockham PA, Sreerama L, Sladek NE. Relative contribution of human erythrocyte aldehyde dehydrogenase to the systemic detoxification of the oxazaphosphorines. Drug Metab Dispos. 1997;25(12):1436–1441. [PubMed] [Google Scholar]
- 11.Ekhart C, Gebretensae A, Rosing H, Rodenhuis S, Beijnen JH, Huitema AD. Simultaneous quantification of cyclophosphamide and its active metabolite 4-hydroxycyclophosphamide in human plasma by high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry (LC-MS/MS). J Chromatogr B Analyt Technol Biomed Life Sci. 2007;854(1-2):345–349. doi: 10.1016/j.jchromb.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 12.Kaijser GP, Beijnen JH, Jeunink EL, et al. Determination of chloroacetaldehyde, a metabolite of oxazaphosphorine cytostatic drugs, in plasma. J Chromatogr. 1993;614(2):253–259. doi: 10.1016/0378-4347(93)80316-v. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Tian Q, Yung Chan S, et al. Metabolism and transport of oxazaphosphorines and the clinical implications. Drug Metab Rev. 2005;37(4):611–703. doi: 10.1080/03602530500364023. [DOI] [PubMed] [Google Scholar]
- 14.Wang DW, H Oxazaphosphorine bioactivation and detoxification: the role of xenobiotic receptors. Acta Pharmaceutica Sinica B. 2012;2(2):105–115. doi: 10.1016/j.apsb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie W, Barwick JL, Simon CM, et al. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 2000;14(23):3014–3023. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, LeCluyse EL. Role of orphan nuclear receptors in the regulation of drug-metabolising enzymes. Clin Pharmacokinet. 2003;42(15):1331–1357. doi: 10.2165/00003088-200342150-00003. [DOI] [PubMed] [Google Scholar]
- 17.Faucette SR, Sueyoshi T, Smith CM, Negishi M, Lecluyse EL, Wang H. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther. 2006;317(3):1200–1209. doi: 10.1124/jpet.105.098160. [DOI] [PubMed] [Google Scholar]
- 18.Jounaidi Y, Hecht JE, Waxman DJ. Retroviral transfer of human cytochrome P450 genes for oxazaphosphorine-based cancer gene therapy. Cancer Res. 1998;58(19):4391–4401. [PubMed] [Google Scholar]
- 19.Schwartz PS, Chen CS, Waxman DJ. Sustained P450 expression and prodrug activation in bolus cyclophosphamide-treated cultured tumor cells. Impact of prodrug schedule on P450 gene-directed enzyme prodrug therapy. Cancer Gene Ther. 2003;10(8):571–582. doi: 10.1038/sj.cgt.7700601. [DOI] [PubMed] [Google Scholar]
- 20.LeCluyse EL, Alexandre E, Hamilton GA, et al. Isolation and culture of primary human hepatocytes. Methods Mol Biol. 2005;290:207–229. doi: 10.1385/1-59259-838-2:207. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Chen T, Cottrell J, Wang H. Nuclear translocation of adenoviral-enhanced yellow fluorescent protein-tagged-human constitutive androstane receptor (hCAR): a novel tool for screening hCAR activators in human primary hepatocytes. Drug Metab Dispos. 2009;37(5):1098–1106. doi: 10.1124/dmd.108.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–799. doi: 10.1093/nar/gkp1005. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Li L, Fuhrman J, Ferguson S, Wang H. The role of constitutive androstane receptor in oxazaphosphorine-mediated induction of drug-metabolizing enzymes in human hepatocytes. Pharm Res. 2011;28(8):2034–2044. doi: 10.1007/s11095-011-0429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson JJ, Novick SJ. Nonlinear blending: a useful general concept for the assessment of combination drug synergy. J Recept Signal Transduct Res. 2007;27(2-3):125–146. doi: 10.1080/10799890701417576. [DOI] [PubMed] [Google Scholar]
- 25.von Eitzen U, Meier-Tackmann D, Agarwal DP, Goedde HW. Detoxification of cyclophosphamide by human aldehyde dehydrogenase isozymes. Cancer Lett. 1994;76(1):45–49. doi: 10.1016/0304-3835(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 26.Sladek NE. Aldehyde dehydrogenase-mediated cellular relative insensitivity to the oxazaphosphorines. Curr Pharm Des. 1999;5(8):607–625. [PubMed] [Google Scholar]
- 27.Maglich JM, Parks DJ, Moore LB, et al. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278(19):17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 28.Faucette SR, Wang H, Hamilton GA, et al. Regulation of CYP2B6 in primary human hepatocytes by prototypical inducers. Drug Metab Dispos. 2004;32(3):348–358. doi: 10.1124/dmd.32.3.348. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz PS, Waxman DJ. Cyclophosphamide induces caspase 9-dependent apoptosis in 9L tumor cells. Mol Pharmacol. 2001;60(6):1268–1279. doi: 10.1124/mol.60.6.1268. [DOI] [PubMed] [Google Scholar]
- 30.Hickman JA. Apoptosis induced by anticancer drugs. Cancer Metastasis Rev. 1992;11(2):121–139. doi: 10.1007/BF00048059. [DOI] [PubMed] [Google Scholar]
- 31.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunez G, Benedict MA, Hu Y, Inohara N. Caspases: the proteases of the apoptotic pathway. Oncogene. 1998;17(25):3237–3245. doi: 10.1038/sj.onc.1202581. [DOI] [PubMed] [Google Scholar]
- 33.Kenis H, Reutelingsperger C. Targeting phosphatidylserine in anti-cancer therapy. Curr Pharm Des. 2009;15(23):2719–2723. doi: 10.2174/138161209788923903. [DOI] [PubMed] [Google Scholar]
- 34.May-Manke A, Kroemer H, Hempel G, et al. Investigation of the major human hepatic cytochrome P450 involved in 4-hydroxylation and N-dechloroethylation of trofosfamide. Cancer Chemother Pharmacol. 1999;44(4):327–334. doi: 10.1007/s002800050985. [DOI] [PubMed] [Google Scholar]
- 35.Giorgianni F, Bridson PK, Sorrentino BP, Pohl J, Blakley RL. Inactivation of aldophosphamide by human aldehyde dehydrogenase isozyme 3. Biochem Pharmacol. 2000;60(3):325–338. doi: 10.1016/s0006-2952(00)00344-0. [DOI] [PubMed] [Google Scholar]
- 36.Brock N, Hilgard P, Peukert M, Pohl J, Sindermann H. Basis and new developments in the field of oxazaphosphorines. Cancer Invest. 1988;6(5):513–532. doi: 10.3109/07357908809082119. [DOI] [PubMed] [Google Scholar]
- 37.Colvin OM. An overview of cyclophosphamide development and clinical applications. Curr Pharm Des. 1999;5(8):555–560. [PubMed] [Google Scholar]
- 38.Moreb JS, Ucar D, Han S, et al. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem Biol Interact. 2012;195(1):52–60. doi: 10.1016/j.cbi.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreb JS, Maccow C, Schweder M, Hecomovich J. Expression of antisense RNA to aldehyde dehydrogenase class-1 sensitizes tumor cells to 4-hydroperoxycyclophosphamide in vitro. J Pharmacol Exp Ther. 2000;293(2):390–396. [PubMed] [Google Scholar]
- 40.Sueyoshi T, Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol. 2001;41:123–143. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- 41.Qatanani M, Moore DD. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr Drug Metab. 2005;6(4):329–339. doi: 10.2174/1389200054633899. [DOI] [PubMed] [Google Scholar]
- 42.Goodwin B, Moore LB, Stoltz CM, McKee DD, Kliewer SA. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60(3):427–431. [PubMed] [Google Scholar]
- 43.Makinen J, Frank C, Jyrkkarinne J, Gynther J, Carlberg C, Honkakoski P. Modulation of mouse and human phenobarbital-responsive enhancer module by nuclear receptors. Mol Pharmacol. 2002;62(2):366–378. doi: 10.1124/mol.62.2.366. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol Pharmacol. 2008;74(2):443–453. doi: 10.1124/mol.108.046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CS, Jounaidi Y, Su T, Waxman DJ. Enhancement of intratumoral cyclophosphamide pharmacokinetics and antitumor activity in a P450 2B11-based cancer gene therapy model. Cancer Gene Ther. 2007;14(12):935–944. doi: 10.1038/sj.cgt.7701092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kan O, Griffiths L, Baban D, et al. Direct retroviral delivery of human cytochrome P450 2B6 for gene-directed enzyme prodrug therapy of cancer. Cancer Gene Ther. 2001;8(7):473–482. doi: 10.1038/sj.cgt.7700329. [DOI] [PubMed] [Google Scholar]
- 47.Chen L, Waxman DJ, Chen D, Kufe DW. Sensitization of human breast cancer cells to cyclophosphamide and ifosfamide by transfer of a liver cytochrome P450 gene. Cancer Res. 1996;56(6):1331–1340. [PubMed] [Google Scholar]
- 48.Moore MJ. Clinical pharmacokinetics of cyclophosphamide. Clin Pharmacokinet. 1991;20(3):194–208. doi: 10.2165/00003088-199120030-00002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.