Abstract
Aldehyde dehydrogenase 1 A1 (ALDH1A1) has recently been suggested as a marker for cancer stem or stem-like cancer cells of some human malignancies. The purpose of this study was to investigate the stem cell-related function and clinical significance of the ALDH1A1 in bladder urothelial cell carcinoma. Aldefluor assay was used to isolate ALDH1A1+ cells from bladder cancer cells. Stem cell characteristics of the ALDH1A1+ cells were then investigated by in vitro and in vivo approaches. Immunohistochemistry (IHC) was performed for evaluating ALDH1A1 expression on 22 normal bladder tissues and 216 bladder tumor specimens of different stage and grade. The ALDH1A1+ cancer cells displayed higher in vitro tumorigenicity compared with isogenic ALDH1A1− cells. The ALDH1A1+ cancer cells could generate xenograft tumors that resembled histopathologic characteristics and heterogeneity of the parental cells. High ALDH1A1 expression was found in 26% (56 of 216) human bladder tumor specimens, and significantly related to advanced pathological stage, high histological grade, recurrence and progression, and metastasis of bladder urothelial cell carcinomas (all P < 0.05). Furthermore, ALDH1A1 expression was inversely associated with cancer-specific and overall survivals of the patients (P = 0.027 and P = 0.030, respectively). Therefore, ALDH1A1+ cell population could be enriched in tumor-initiating cells. ALDH1A1 may serve as a useful marker for monitoring the progression of bladder tumor and identifying bladder cancer patients with poor prognosis who might benefit from adjuvant and effective treatments.
Keywords: ALDH1A1, cancer stem cell, biomarker, bladder carcinoma, prognosis
Introduction
There are about 336,000 new cases of urothelial bladder carcinoma and 132,000 deaths annually all throughout the world (1). 60% of bladder carcinomas are low-grade (G1, G2) and noninvasive cancers (Ta and T1/pTis). 25% newly diagnosed bladder tumors are high-grade (G3)/muscle invasive lesions (greater than pT2) [1]. The patients with low grade and non-muscle-invasive carcinomas are routinely treated by endoscopic resection. After endoscopic resection, the majority of the patients develop cancer recurrences. The patients with high histological grade and muscle-invasive tumors receive more aggressive therapies including cystectomy and/or radiation/chemotherapy, at least half of the cases eventually progress with local and distant metastases. Periodic cystoscopy and urine cytology are currently used to monitor the patients for cancer recurrence or progression [2]. However, these clinical means are associated with high cost, substantial patient discomfort, and variable and poor accuracy. Therefore, novel and clinically applicable prognostic markers are urgently needed to identify patients at high risk for poor prognosis. To date, numerous potential markers have been identified by a variety of molecular biology and genetic studies [3–4]. However, the role of these molecular markers in clinical diagnosis and therapeutic decision making still remains uncertain. New diagnostic modalities have to be developed.
Accumulating evidence has proposed that tumor contains tumor-initiating cells or cancer stem cells (CSCs) that are responsible for its progression and relapse [5]. Although being present in a small population in tumor, CSCs can undergo self-renewal, recapitulate the phenotype of the cancer from which they were derived, proliferate, and drive continued expansion of malignant cells [5]. Bladder carcinoma growth and metastasis might also be promoted by CSCs that are responsible for its aggressiveness [6–10]. Therefore, analysis of molecular aberrations that are associated with CSCs would deep our understanding of the tumor biology of urothelial carcinoma. Most importantly, these molecular changes could be developed as a new diagnostic system for monitoring the progression of bladder tumor and offering the best opportunity to prevent its recurrence and probably cure the challenging malignancy [9–10].
Human cytosolic aldehyde dehydrogenase 1 (ALDH1) plays a role in the biosynthesis of retinoic acid (11). Altered metabolism of retinal to retinoic acid is likely to play a major role in stem cell biology [12–19]. ALDH1A1 is a major member of the ALDH1 family. Activation of ALDH1A1 has been found in stem cells (SCs) populations in multiple myeloma and acute myeloid leukemia [19–20]. Ginestier et al. [21] showed that ALDH1A1 was a marker of normal and malignant human mammary SCs and predictor of poor clinical outcome of breast cancer patients. We recently demonstrated that the ALDH1A1+ lung cancer cells could generate tumors that recapitulated the heterogeneity of lung carcinomas [22]. Furthermore, elevated ALDH1A1 expression was correlated with the stage and grade of lung tumors and associated to a poor prognosis for the patients [22]. However, no research has been reported concerning the role of ALDH1A1 in tumorigenesis of bladder urothelial cell carcinoma and its clinical importance in the challenge malignancy.
To investigate the CSC-related function and clinical significance of ALDH1A1 in bladder urothelial cell carcinoma, we first isolated ALDH1A1+ cells from human bladder cancer cells by using Aldefluor assay and fluorescence-activated cell sorting. The ALDH1A1+ cells exhibited high in vitro tumorigenicity and initiated xenografts that gave rise to heterogeneous cell populations. Furthermore, immunohistochemistry analysis of clinical specimens showed that ALDH1A1 expression correlated significantly with poor patient prognosis. ALDH1A1 might therefore be a bladder CSC-associated marker and provides a potential prognostic factor for the malignant progression of bladder cancer.
Patients and Methods
Cell lines and cultures
Bladder cancer cell lines HTB-2, HTB-9, and HTB-4 were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cells were maintained in the culture medium recommended by ATCC and harvested by using treatment with 0.25% trypsin (Invitrogen, Carlsbad, CA) when they were in the logarithmic phase of growth for use in the following experiments.
Isolation of ALDH1A1+ cell population by Aldefluor assay and fluorescence-activated cell sorting (FACS)
An Aldefluor kit (StemCell Technologies, Durham, NC) optimized for interaction with human ALDH1A1 was used to identify ALDH1A1+ cells as described in our previous study [22]. Briefly, the brightly fluorescent ALDH1A1 expressing cells were detected by using an Aria cell sorter (BD Biosciences, San Jose, CA). Side-scatter and forward-scatter profiles were used to reduce cell doublets. Nonviable cells were eliminated using the viability dye 4',6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO). Specific ALDH1A1 activity was based on the difference between the presence/absence of the Aldefluor inhibitor diethylaminobenzaldehyde (DEAB) (Sigma).
FACS of CD44+ and CD44− cell population
Cells were stained for 15 min at 41°C in Tris-buffered saline (TBS) containing BSA and FITC-conjugated monoclonal anti-CD44 (BD PharMingen, San Jose, CA) and then used to sort out CD44+ and CD44− cells as previously described (23). For the positive population, only the top 10% most brightly stained cells were selected. For the negative population, only the bottom 5% most dimly stained cells were chosen. Data were analyzed by using Cell Quest software (B–D Biosciences). Each experiment was repeated three times.
Immunofluorescence analysis
ALDH1A1 and CD44 expressions in the bladder cancer cells were analyzed by using immunofluorescence assay with the primary antibodies against ALDH1A1 (Santa Cruz Biotechnology, Santa Cruz, CA) and CD44 (eBioscience, San Diego, CA) as previously described [22–3]. The cells were then stained with a fluorescently conjugated IgG (Abcam Inc. Cambridge, MA) and examined under a Leica microscope (Leica Microsystems, Inc. Buffalo, NY). Each batch of slides contained a positive control and a negative control.
Clonal analysis and clonogenic assays
To evaluate differences in cell survival and proliferation between ALDH1A1+ and ALDH1A1− cells. Cells were plated at density of 200 per well in a six-well tissue culture dish, as described in our previous report (22). Clones with >50 cells were scored at the end of week two. The percentage of cells that initiated a clone was presented as cloning efficiency. For clonogenic assays, cells were plated at 1,000 per well in six-well culture dishes coated with a thin layer of 1% solidified agar [22]. Spheres that arose within three weeks were considered as clonogenicity. For each cell type, triplicate samples were performed and the clones or spheres were counted by two individuals in a blind fashion.
Effects of ALDH1A1 knock-down on tumorigenicity/clonal growth
We made a siRNA plasmid construct to specifically knock down ALDH1A1 according to a protocol described by Moreb et al [24]. PSilencer 2.0-U6 siRNA plasmids (Ambion. Austin, TX) were used to produce ALDH1A1 LV siRNA vectors. The target site for ALDH1A1 (Genbank NM_000689) was 5'-gtagccttcacaggatcaa-3' (nt 777–795). Primers used to generate the siRNA construct were 5'ATA CGC GGA TCC CGT AGC CTT CAC AGG ATC AAT TCA AGA G AT TGA TC-3' and 5'CGC TAG ACT AGT TAA AAA AGT AGC CTT CAC AGG ATC AAT CTC TTG AA TTG ATC -3'. U6 promoterdriven LV-scrambled siRNA was also generated as previously described [25] and used as a control. The plasmids were transfected into the 293T cells to produce lentiviral vectors using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions. The lentiviral siRNA specific against ALDH1A1 was referred as lentiviral-ALDH1A1-siRNA. The sorted ALDH1A1+ cancer cells were transfected with lentiviral-ALDH1A1-siRNA and controls as previously described [24]. Effects of ALDH1A1 knock-down on tumorigenicity/clonal growth of these bladder cancer cells were investigated using the above assays.
Xenografting
ALDH1A1+, ALDH1A1−, ALDH1A1+/CD44+, ALDH1A1+/CD44−, ALDH1A1−/CD44+, ALDH1A1−/CD44−, CD44+, CD44−, and unsorted cancer cells were subcutaneously inoculated in 10 athymic Swiss nu/nu mice per each cell type at different dose (Table 1). The mice were observed to allow tumor growth and then euthanized under deep anesthesia with pentobarbital (Sigma) at the end of week four. The tumors were surgically removed, and their volume (mm3) was calculated as (W2 × L) / 2. A portion of each tumor tissue was fixed in 10% formaldehyde and embedded in paraffin for histopathologic examination. Additionally, a part of each engrafted tumor was dissociated with collagenase IV (Sigma) and incubated with mechanical disruption as previously described [22]. Serial transplantations of the bladder cancer xenografts were performed by regrafting the freshly disassociated cells into mice. The disaggregated cells of xenograft tumors from each passage were also reanalyzed by using Aldefluor assay for percentage of ALDH1A1+ cells as described above.
Table 1.
In vivo tumorigenicity of different numbers of bladder cancer cells a
| Cell type | No. of cells injected | Tumor incidence | Size of tumor (mm3) |
|---|---|---|---|
| ALDH1A1+ | 1 × 102 | 0/10 | |
| 1 × 103 | 8/10 | 19±1.2 | |
| 1 × 105 | 10/10 | 32±2.6 | |
| ALDH1A1− | 1 × 102 | 0/10 | |
| 1 × 103 | 0/10 | ||
| 1 × 105 | 1/10 | 5.5 | |
| ALDH1A1+/CD44+ | 1 × 102 | 0/10 | |
| 1 × 103 | 8/10 | 20±1.3 | |
| 1 × 105 | 10/10 | 33±2.3 | |
| ALDH1A1+/CD44− | 1 × 102 | 0/10 | |
| 1 × 103 | 8/10 | 19±1.1 | |
| 1 × 105 | 10/10 | 31±2.6 | |
| ALDH1A1−/CD44+ | 1 × 102 | 0/10 | |
| 1 × 103 | 0/10 | ||
| 1 × 105 | 4/10 | 13±0.9 | |
| CD44+ | 1 × 102 | 0/10 | |
| 1 × 103 | 3/10 | 11±0.5 | |
| 1 × 105 | 8/10 | 25±1.2 | |
| CD44− | 1 × 102 | 0/10 | |
| 1 × 103 | 0/10 | ||
| 1 × 105 | 2/10 | 5.4 | |
| ALDH1A1−/CD44− | 1 × 102 | 0/10 | |
| 1 × 103 | 0/10 | ||
| 1 × 105 | 1/10 | 5 | |
| Unsorted cells | 1 × 102 | 0/10 | |
| 1 × 103 | 1/10 | 7 | |
| 1 × 105 | 3/10 | 10±0.6 |
The table only shows results from HTB-2 cells.
Patients, clinical specimens, and immunohistochemistry (IHC) analysis
From 1992 to 2003, a total of 278 consecutive patients with bladder urothelial cell carcinoma underwent local transurethral resection (TUR) or cystectomy to remove the tumors at the Department of Urology, the School of Clinical Medicine of Southeast University in China. Under a protocol approved by the institutional review board for human subjects' research, we obtained complete medical records, follow-up data, and adequate formalin-fixed paraffin-embedded tissue blocks for 216 patients. The clinicopathological features of 216 bladder cancer cases are summarized in Table 2. No cancer patients received adjuvant chemotherapy or radiation therapy before surgery. All patients with noninvasive bladder carcinomas (Ta–T1) were treated with intravesical chemotherapy after TUR, while all patients with invasive disease (T2–T4) were treated with chemotherapy or radiotherapy after cystectomy. The median follow-up time was 98 (1–182) months. Ninety-two of 216 patients were dead in the follow-up period. Among the 92 patients who died, 65 died of bladder cancer and the remaining 27 died of other causes without evidence of tumor progression. During the follow-up period, tumor recurrences and progression were observed in 140 and 69 patients, respectively. Furthermore, 22 normal urothelium specimens were obtained by biopsy or from cystectomized bladders for other diseases than cancer, which were used as normal controls. Tissue sections (4 mm thick) were obtained from each block, stained with hematoxylin and eosin (H&E), and reviewed independently by two pathologists to confirm the diagnosis and the presence of tumor. The WHO 1973 criteria for grade and American Joint Cancer Committee 2002 TNM classification were used to evaluate patients’ histopathological features.
Table 2.
Association ALDH1A1 expression with clinicopathological characteristics of the bladder cancer patients
| Characteristics | No. of cases (%) | High ALDH1A expression (%) |
p-value |
|---|---|---|---|
| All cases | 216 | ||
| Age | 0.392 | ||
| <60 | 82 (38%) | 19 (23%) | |
| ≥60 | 134 (62%) | 37 (28%) | |
| Gender | 0.413 | ||
| Male | 164 (76%) | 41 (25%) | |
| Female | 52 (24%) | 15 (29%) | |
| Tumor grade | 0.001 | ||
| 1 | 84 (39%) | 12 (14%) | |
| 2 | 82 (38%) | 19 (23%) | |
| 3 | 50 (23%) | 25 (50%) | |
| Stage | 0.002 | ||
| Ta | 86 (40%) | 10 (12%) | |
| T1/pTis | 74 (34%) | 23 (31%) | |
| T2–4 | 56 (26%) | 23 (41%) | |
| Lymph node status | <0.001 | ||
| pN0 | 197 (91%) | 43 (22%) | |
| pN1–2 | 19 (8%) | 13 (68%) | |
| Tumor size | 0.436 | ||
| ≤3 cm | 112 (52%) | 30 (27%) | |
| >3cm | 104 (48%) | 26 (25%) | |
| Tumor multiplicity | 0.489 | ||
| Unifocal | 145 (67%) | 39 (27%) | |
| Multifocal | 71 (33%) | 17 (24%) | |
| Treatment | 0.512 | ||
| BCG | 132 (61%) | 34 (26%) | |
| Adjuvant radiation therapy | 39 (18%) | 10 (26%) | |
| Adjuvant chemotherapy | 45 (21%) | 12 (27%) | |
| Recurrence | 0.007 | ||
| Yes | 140 (65%) | 45 (32%) | |
| No | 76 (35%) | 11 (15%) | |
| Progression | 0.001 | ||
| Yes | 69 (32%) | 38 (55%) | |
| No | 147 (68%) | 18 (12%) |
IHC studies were performed using a protocol as previously described [22–3, 26–7]. Briefly, after being deparaffinized in xylene and retrieved by boiling in a steamer with sodium citrate buffer, tissue sections were incubated with 10% normal horse serum/TBS for 30 min at room temperature. Antibodies against ALDH1A1 (Santa Cruz Biotechnology) and CD44 (eBioscience) were mounted for 1 h at room temperature. Respective secondary antibodies from the EnVision system (Dako Carpinteria, CA, USA) were applied on the slides for 30 min. The slides were then rinsed in the 1 X TBS and visualized by a 5-min incubation with liquid 3.3’-diaminobenzidine in buffered substrate (Dako) and counterstained with H&E (Dako). Immunoreactive staining intensity for each antibody was rated according to the following scale: no visible staining = 0, faint staining = 1, moderate staining = 2, and strong staining = 3. The total number of cells with positive staining for the antibodies was quantized in 20 fields on each tissue section. Percentage of cells with positive staining was graded as 0%, <10%, 10—25%, 25—50%, 50—75% or higher. An overall score was assigned by multiplying the intensity score by the mean percentage of cells staining.
Statistical analyses
The clinical parameters of the patients, including tumor recurrence and progression, cancer-specific survival, and overall survival were determined as previously described [27]. Data on various treatments including TUR, cystectomy, intravesical Bacillus Calmette-Guérin (BCG) treatment, radiation, and chemotherapy were also collected during the time of each follow-up category. The patients’ age, gender, grade, stage, tumor size, multiplicity, recurrence, progression, and various treatment modalities were collected as baseline variables. The X2 test was applied to evaluate association between ALDH1A1 expression and the clinicopathologic parameters. The X2 test was also used for comparison of ordinal variables. Cancer-specific and overall survival rates were estimated by Kaplan-Meier method and evaluated with the use of log-rank test for univariate analysis. Univariate and multivariate survival analyses were performed using the Cox proportional hazards regression model. Statistical significance was set as P ≤ .05. All reported P values were 2-sided.
Results
Identification of a small ALDH1A1+ subpopulation in bladder cancer cell lines with increased properties of clonogenicity
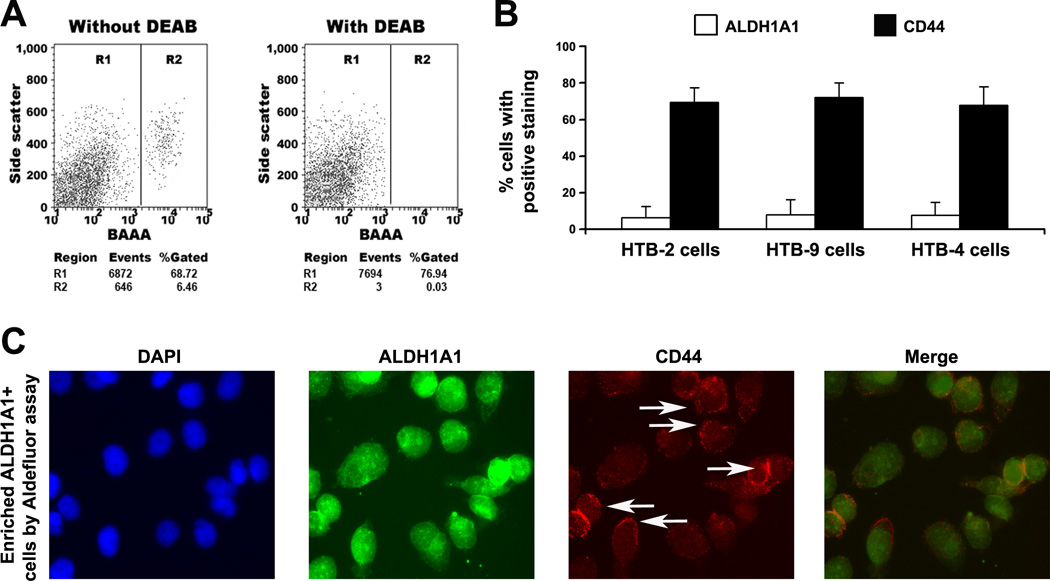
The three different cell lines displayed a small and similar size of ALDH1A1+ population: HTB-2 showed 6.4% (6.4 ± 1.1%), HTB-9 had 8.2% (8.2 ± 2.0%), and HTB-4 exhibited 7.9% (7.6 ± 1.9%) ALDH1A1+ cells, respectively (Fig. 1A). Percentage of CD44+ population in HTB-2, HTB-9, and HTB-4 cells was 69.4% (68.8± 5.3%), 72.1% (73.5 ± 6.3%), and 57.9% (57.3 ± 6.7%). Therefore, the ALDH1A1+ cell proportion (6.4–8.2%) in the bladder cancer cells was considerably lower than that of (57–72%) of CD44+ cells (P < 0.01) (Fig. 1B).
Fig. 1.
Identification of a small ALDH1A1+ population in bladder cancer cell lines. A. FACS analysis of cancer cells using the Aldefluor assay. The brightly fluorescent ALDH1A1-expressing cells (ALDH1A1+ cells) were detected in the green fluorescence channel. The cells incubated with DEAB were used to establish the baseline fluorescence of these cells (R1) and define the ALDEFLUOR (ALDH1A1)-positive region (R2). B. The ALDH1A1+ cell proportion in the bladder cancer cells was significantly lower than that of CD44+ cells. The percentage of ALDH1A1+ population in the celll lines was from 6.4% to 8.2%. However, the proportion of CD44+ cells was from 57% to 72%. C. Merged immunofluorescence imaging showed a high percentage of ALDH1A1+ cancer cells positive for CD44 staining. ALDH1A1+ cells isolated from HTB-2 cells were positive for ALDH1A1 antibody (green fluorescent staining of the cytoplasm). Fraction of the same cells was positively stained for CD44 as demonstrated by red fluorescent staining of the cell membrane (white arrows). 4',6-diamidino-2-phenylindole (DAPI) was used to stain nuclei. The experiments were undertaken on all bladder cancer cell lines and repeated three times. The Fig.1A and C only showed the result from HTB-2 cell line.
To evaluate proportion of cell population positive for both ALDH1A1 and CD44 in the three cell lines, we first enriched CD44+ cells by using FACS with CD44 antibody, from which, then isolated ALDH1A1+ cells by using Aldefluor assay. 16.5% (16.8±2.3%), 14.8% (13.1±3.1%), and 12.6% (12.8±2.9%) of CD44+ HTB-2, HTB-8, and HTB-4 cells were also positive for ALDH1A1, respectively. Vice versa, after serially enriched by Aldefluor assay followed by FACS with CD44 antibody, 79.2% (79.1±4.8%), 82.6% (82.3±5.2%), and 77.2% (77.3±5.6%) of ALDH1A1+ HTB-2, HTB-8, and HTB-4 cells were positive for CD44. The results indicate that the ALDH1A1+ population might be a subset of the CD44+ cells. The observation was also confirmed by merged immunofluorescence imaging that showed an average of 86.2 % (86.6±5.1%) ALDH1A1+ HTB-2 cancer cells were positive for CD44 staining (Fig. 1C).
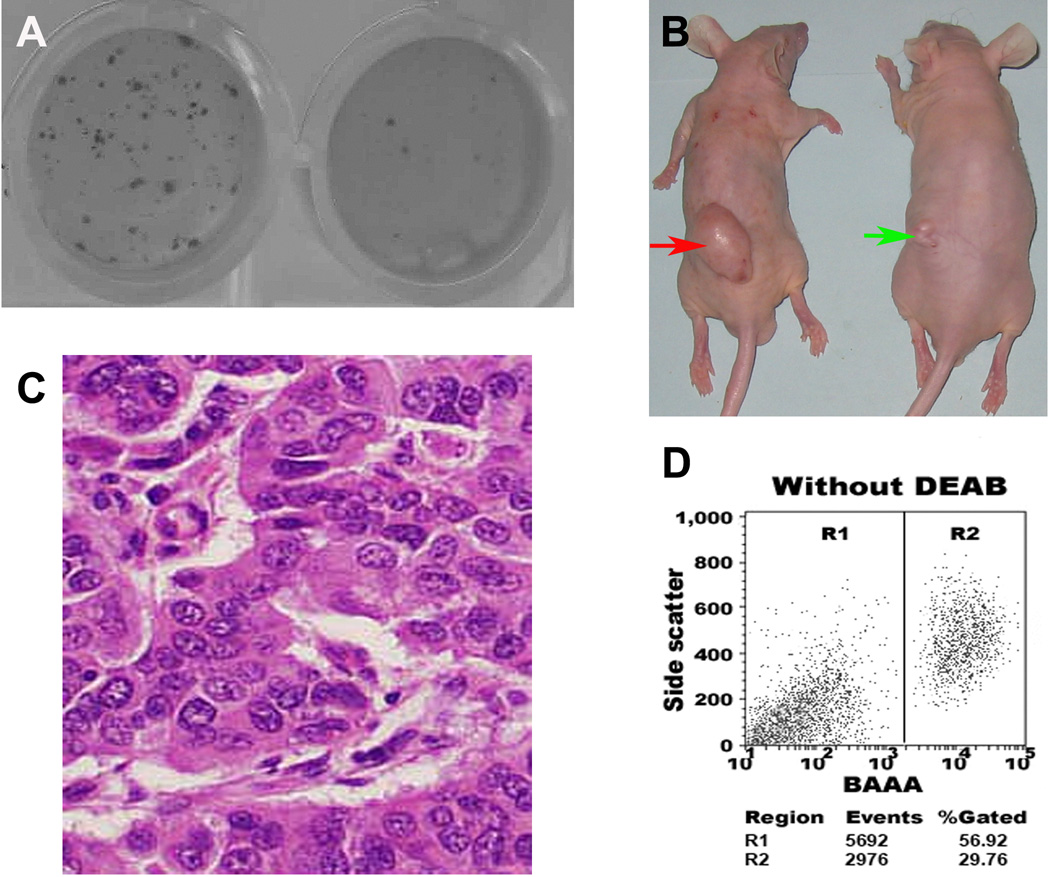
The ALDH1A1+ HTB-2, HTB-9, and HTB-4 cells displayed significantly higher colony-forming efficiency by forming larger and more clones (P < 0.0001) compared with the ALDH1A1− isogenic cells at the end of the clonal assays. The data indicated that ALDH1A1+ cancer cells could possess high proliferative capacity. We further tested the ability of the ALDH1A1+ and ALDH1A1− bladder cancer cells to produce colonies by using soft agar assay. As shown in Fig. 2A, the ALDH1A1+ HTB-2 cells generated at least 10 times as many colonies as the ALDH1A1− HTB-2 cells. Furthermore, the colonies from the ALDH1A1+ HTB-2 cells were larger in size compared with ones from the ALDH1A1− cells (all p<0.01). Similarly, the ALDH1A1+ HTB-9 and HTB-4 cells also resulted in more than 10 times as many colonies as did the ALDH1A1− cells. ALDH1A1+/CD44+ cells showed similar clonogenic formation efficiency as did ALDH1A1+ cells. Furthermore, although diaplaying higher clonogenicity compared with CD44− cells, the CD44+ cells had lower clonogenicity than did ALDH1A1+ cells and ALDH1A1+/CD44+ cells. In addition, there was no considerable difference of CD44− and unsorted bladder cancer cells to form colonies in the soft agar assay (P>0.05). The results implied that ALDH1A1+ cells had high clonogeniety and further enriching cancer cells using CD44+ could not increase clonogenic formation efficiency. Anchorage-independent growth in the soft agar assay has been suggested as an approximation of tumorigenesis, and CSCs are thought to be the tumor-initiating cells [28–9]. Therefore, these in vitro observations indicate that ALDH1A1+ bladder cancer cells present higher survival, proliferation, and tumorigenicity compared with the isogenic ALDH1A1− and CD44+ cells.
Fig. 2.
ALDH1A1+ bladder cancer cells had high in vitro and in vivo tumorigenic potential. A. ALDH1A1+ cancer cells (left) possessed significantly higher colony-forming efficiency compared with ALDH1A1− populations (right). ALDH1A1+ and ALDH1A1− cells were plated in six-well dishes coated with a thin layer of agar. Three weeks after plating, ALDH1A1+ cells formatted larger and more colonies as compared with the ALDH1A1− cells. B. Tumor formation ability of ALDH1A1+ cancer cells was greater than that of isogenic ALDH1A1− cells. ALDH1A1+ and ALDH1A1− HTB-2 cells were implanted into flanks of nude mice. After four weeks, the dose of 1 × 105 ALDH1A1+ cells yielded larger tumors (red arrow) in all 10 mice with diameters of 32±2.6 mm3, whereas the same dose of ALDH1A1− cells only generated a small tumor mass (5.5 mm3) (green arrow) in only one mouse. The animal experiments were done by using all cell lines. The Fig. 2B only showed the result from HTB-2 cell line. C. Histopathologic examination of the engrafted tumors formed by the ALDH1A1+ HTB-9 cancer cells revealed a highly cellular mass with characteristics of bladder transitional cell carcinoma. D. ALDH1A1+ cancer cells created bladder tumors with heterogeneity in vivo. Reanalyzing cells of the engrafted tumors generated from the ALDH1A1+ HTB-9 by using the Aldefluor assay showed that the xenograft tumors produced 56.9% ALDH1A1+ cells and 29.7% ALDH1A1− cells. The experiments were undertaken on all bladder cancer cell lines and repeated three times. The Fig. 2D only showed the result from HTB-9 cell line.
ALDH1A1 knock-down on tumorigenicity/clonal growth
The ALDH1A1+ HTB-2 cells transfected with lentiviral-ALDH1A1-siRNA showed reduced ALDH1A1 at least 75%. Furthermore, this down-regulation was specific because there was no significant change of ALDH1A1 in the mock cells and the cells infected with the scrambled siRNAs (control cells). There was also no change in the expression of the other members of ALDH family in the cells treated with lentiviral-ALDH1A1-siRNA.
The ALDH1A1+ HTB-2 cells, after their ALDH1A1 was reduced, exhibited considerably lower colony-forming efficiency by forming smaller and less clones than did the ALDH1A1+ HTB-2 cells whose ALDH1A1 was not knocked down (P < 0.0001). Furthermore, soft agar assay showed that the ALDH1A1+ HTB-2 cells, after the ALDH1A1 was reduced, generated much less colonies compared with the enriched ALDH1A1+ HTB-2 cells without reduced ALDH1A1 expression (P < 0.0001). This observation implied that the increased tumorigenicity of ALDH1A1 cells was attributed to expression of this molecule. Altogether, the data demonstrates that high ALDH1A1 expression could directly contribute to tumorigenicity in bladder cancer cells.
Tumor formation of ALDH1A1+ bladder cancer cells in vivo
We implanted ALDH1A1+, ALDH1A1−, ALDH1A1+/CD44+, ALDH1A1+/CD44−, ALDH1A1−/CD44+, CD44+, CD44−, and unsorted HTB-2 cells into 10 mice, respectively. In vivo tumorigenicity was measured by tumor incidence and size at week four. As shown in Table 1, 1 × 103 ALDH1A1+ cells yielded tumors with an average of 19±1.6 mm3 in 8 of the mice after four weeks, while the same number of ALDH1A1− cells did not produce tumor in any mouse. Furthermore, 1 × 105 ALDH1A1+ HTB-2 cells generated much larger tumors in all mice with an average of 32±2.6 mm3, whereas the same amount of ALDH1A1− bladder cancer cells produced a small tumor mass (5.5 mm3) in only one of the mice (Fig. 2B). Therefore, the ALDH1A1+ bladder cancer cells were 100 times more potential in inducing in vivo tumorigenicity compared with the ALDH1A1− cells.
1 × 103 ALDH1A1+/CD44+ HTB-2 bladder cancer cells and ALDH1A1+/CD44− HTB-2 bladder cancer cells created tumors of average 20±1.3 mm3 and 19±1.1 mm3, respectively, in 8 of the mice, which had no statistical difference from the tumors generated by ALDH1A1+ cells alone (P>0.05). At a dose of 2 × 102, none of ALDH1A1+ cells, ALDH1A1+/CD44+ cells, and ALDH1A1+/CD44− cells could produce tumor. The findings suggest that CD44, when used serially with ALDH1A1 to sort bladder cancer cells, do not improve enrichment of tumor-initiating cells over ALDH1A1+ cells.
1×103 CD44+ HTB-2 cells produced 3 tumors in 10 mice. At a dose of 1×105, the CD44+ cancer cells generated 8 tumors. However, 1×103 CD44− HTB-2 cells did not create any tumor. 1×105 CD44− HTB-2 cells were able to produce 2 tumors. The observation suggest that although CD44+ cancer cells have higher in vivo tumorigenity compared with CD44− cancer cells, they are inferior to ALDH1A1+ cancer cells in producing xenograft tumors. Interestingly, unlike CD44+ HTB-2 cells, 1×103 ALDH1A1−/CD44+ cancer cells did not produce any tumor. 1×105 ALDH1A1−/CD44+ cells created only 4 tumors. The data imply that the CD44+ cells that have tumorigenity, when lacking ALDH1A1+ in the population, will have reduced potential to create xenograft tumors. In other words, although being a small number, ALDH1A1+ cancer cells in CD44+ cell population may have stronger tumorigenic capacity compared with the rest of CD44+ cells that are negative for ALDH1A1 staining.
We repeated the experiments using HTB-9 and HTB-4 bladder cancer cells and obtained similar results. These data, taken together with above in vitro findings, suggest that ALDH1A1+ cancer cells had higher tumorigenic potential compared with ALDH1A1−, CD44+, and unsorted populations.
Histopathologic examination revealed a highly cellular mass with characteristics of bladder transitional cell carcinoma in the engrafts from the ALDH1A1+ bladder cancer cells (Fig. 2C). Because the gold standard in determining CSCs is whether the testing cells can preferentially initiate tumor development in animal models [28–9], the observation could provide functional evidence that the ALDH1A1+ cancer cells have the property of in vivo self-renewal.
Furthermore, serial transplantation experiments showed that bladder tumors developed from ALDH1A1+ cells could be regenerated for 3 cycles until we prepared the paper. To elucidate whether ALDH1A1+ cancer cells could create bladder tumors with heterogeneity in vivo, Aldefluor analysis was performed on disassociated cells of the engrafts. The xenograft tumors in the first passage xenografts gave rise to 56.9% (58.7±4.6%) ALDH1A1+ cells and 29.7% (29.3±1.9%) ALDH1A1− cells (Fig. 2D). The percentage of the ALDH1A1+ and ALDH1A1− population in the third passage xenografts were 44.9% (45.1±3.2%), 45.4% (46.2±2.3%), respectively. The result again supports that the ALDH1A1+ cancer cells have the property of self-renewal in animal model. Furthermore, it also indicates that ALDH1A1+ cancer cells could differentiate into ALDH1A1− cells, and thus create bladder tumors with heterogeneity in vivo.
ALDH1A1 expression in human bladder tissue specimens
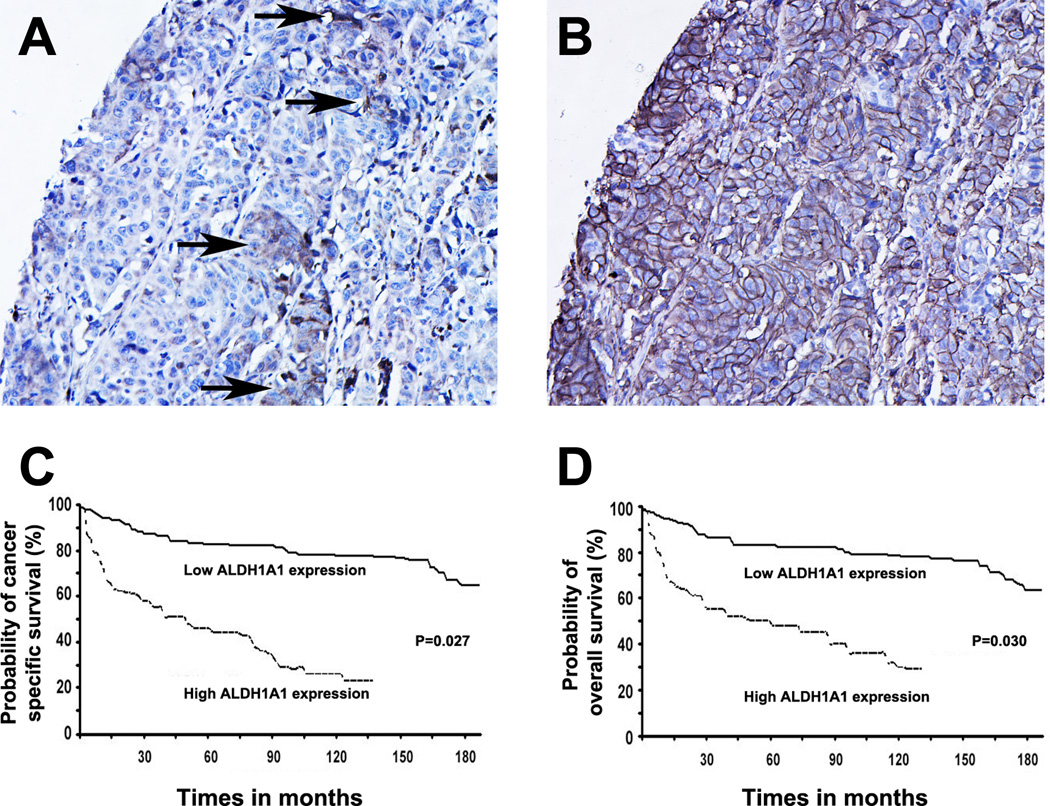
ALDH1A1 was not observed in the normal bladder specimens, however,was found in bladder tumor tissues as illustrated by strong cytoplasm staining (Fig. 3A). Furthermore, IHC analysis of serial tumor sections displayed that ALDH1A1+ cells existed within and were restricted to tumor cells that had positive CD44 staining (Fig. B). The ALDH1A1+ cancer cells, therefore, accounted for a fraction of the CD44+ tumor cell population. The observation is consistent with finding in the above in vitro experiments, in which, the ALDH1A1+ population is a subset of the CD44+ bladder cancer cells.
Fig. 3.
High ALDH1A1 expression in human bladder cancer specimens was associated with a poor prognosis for the patients. A and B. ALDH1A1+ cancer cells comprised a fraction of the CD44+ tumor cell population. Immunohistochemical study of adjacent sections of a bladder tumor for the expression of ALDH1A1 and CD44 showed that a small number of ALDH1A1+ cells (arrows in A) were present within the CD44 immunopositive tumor areas (B). C. Survival analysis of 216 patients with bladder urothelial carcinomas based on high ALDH1A1 expression status. C. Cancer-specific survival time; D. Overall survival time. P values were calculated using the log-rank test.
To assess clinical significance of ALDH1A1 expression in bladder cancer patients, we used a cut-off value to determine whether a tumor had low or high ALDH1A1 expression as previously described [22], that was, a tumor specimen with more than 10% overall score was defined as one with high ALDH1A1 expression. High expression of ALDH1A1 was found to be present in 26% (56/216) of the total bladder tumor samples. There was a stepwise increase in the prevalence of ALDH1A1 expression with the grades and stages of the bladder carcinomas (Table 2): 14% (12 of 84), 23% (19 of 82), and 50% (25 of 50) in grade 1, 2, and 3, respectively (p = 0.01). Elevated ALDH1A1 expression was observed in 21% (33 of 160) of noninvasive urothelial carcinomas (12% Ta and 31% T1/pTis), 41% (24 of 46) advanced bladder carcinomas (T2–4), and 68% (13 of 19) lymph node metastases (p = 0.02). However, there was no significant association between ALDH1A1 expression and other clinicopathological features such as patient’s age and gender and tumor size, and multiplicity (Table 2). Therefore, ALDH1A1 expression status was closely correlated with important histopathologic characteristics (grades and stages) of bladder urothelial carcinomas.
High ALDH1A1 expression was related to recurrence and progression of bladder urothelial carcinomas and poor prognosis of the patients
In the follow-up period, 32% (45 of 140) of tumors with high ALDH1A1 expression recurred compared with 15% (11 of 76) tumors with low ALDH1A1 expression having recurrence (P = 0.007) (Table 2). Furthermore, 55% (38 of 69) of tumors with high ALDH1A1 expression showed progression, as compared with only 12% (18 of 147) of tumors with low ALDH1A1 expression having progression (P = 0.001). Therefore, overexpression of ALDH1A1 was positively associated with the incidence of recurrence and progression of bladder urothelial carcinomas.
Kaplan-Meier plots and log-rank tests showed that the bladder cancer patients with high ALDH1A1 expression in their tumor tissues had statistically significant shorter cancer-specific survival, as compared with those whose tumors had low ALDH1A1 expression (P = 0.027; Fig. 3, C). Moreover, patients with high ALDH1A1 positive staining in their tumors had statistically significantly shorter overall survival rate compared with those whose tumors had low ALDH1A1 expression (P = 0.030; Fig. 3, D). Therefore, ALDH1A1 expression in bladder urothelial carcinomas was inversely associated with poor prognosis of the patients.
Multivariate Cox proportional hazards regression analysis revealed that elevated ALDH1A1 expression in urothelial tumors was an independent predictor for cancer-specific and overall survival rates. Furthermore, tumor stage was associated with cancer-specific and overall survival rates (P = 0.002 and P = 0.027, respectively). Lymph node metastasis was also related with cancer-specific and overall survival rates of the patients (P = 0.013 and P = 0.029, respectively). In addition, treatments including BCG, radiation therapy, and systematic chemotherapy could serve as significant factors for cancer-specific survival (all P < 0.05), while age only was shown to be an independent prognostic factor for overall survival of bladder cancer patients (P = 0.039).
Discussion
Development of specific markers to isolate and characterize CSCs or stem-like cancer cells of urothelial bladder carcinoma would greatly advance our understanding of tumor biology of bladder. In this study, we first isolated ALDH1A1+ cells from bladder cancer cell lines by uisng Aldefluor assay, and then evaluated their CSC-associated function by in vitro and in vivo approaches. Important properties of CSCs include in vitro self-renewal, in vivo tumor initiation, and giving rise to a heterogeneous population of cancer cells. Several pieces of evidence support that ALDH1A1+ bladder cancer cells could be enriched in CSCs or stem-like cancer cells. First, the in vitro assays revealed that ALDH1A1+ cancer cells had higher clone formation efficiency than did ALDH1A1− cancer cells. Furthermore, ALDH1A1+ bladder cancer cells grew in an anchorage-independent manner. Therefore, ALDH1A1+ bladder cancer cells were highly in vitro colongenic and tumorigenic cells. Moreover, the ALDH1A1+ cancer cells, after ALDH1A1 expression was reduced, had lower tumorigenicity compared with the cells that had high expression of ALDH1A1, demonstrating that this molecule could be directly responsible for cancer cell growth. Second, our in vivo experiments showed that the ALDH1A1+ bladder cancer cells were at least 100 times more tumorigenic than ALDH1A1− cancer cells. Furthermore, the engrafted tumors illustrated histopathologic patterns similar to those of the primary bladder cancer cells, implying that the ALDH1A1+ population could resemble the characteristics of the tumor subtype and possess the capacity to self-renew. Third, the dissociated cells of the engraftments created from ALDH1A1+ bladder cancer cells presented an average of 29% ALDH1A1− cancer cells. The data demonstrated that ALDH1A1+ cells might give rise to a heterogeneous property of tumors of bladder. Fourth, importantly, ALDH1A1+ bladder cancer cells could be serially passaged in vivo. Considering that the most crucial standard for CSCs is their ability to re-initiate successively transplantable xenografts that resemble the original tumor histology and heterogeneity [5, 28, 32], the ALDH1A1+ bladder cancer cells could represent CSCs of bladder cancer.
CD44, a surface biomarker, has been applied to isolate CSCs from established bladder cancer cell lines [30]. Chan et al. [9] recently used CD44 to isolate CSCs from bladder tumors. Our finding from the established cancer cell lines is consistent with their observation that a small population of CSCs in deed exist in bladder cancer cells and CD44 can be used as marker to isolate the cancer stem cells. Furthermore, our and their data showed that CD44+ bladder cancer cells had higher in vitro and in vivo tumorigenity compared with CD44− cancer cells. In our study, however, we further compared the ALDH1A1+ bladder cancer cells and the CD44+ isogenic cells for their in vitro and in vivo tumorigenicity. We found that the CD44+ cells displayed lower clonogenicity than did ALDH1A1+ cells and ALDH1A1+/CD44+ cells. The in vitro observation is supported by subsequent in vivo findings, in which, although displaying higher tumorigenicity compared with CD44− cells, the CD44+ cells had lower potential to create tumor compared with ALDH1A1+ cells. Both in vitro and in vivo experiments also showed that ALDH1A1+/CD44+ did not exhibit higher tumor-initiating capability than did ALDH1A1+ and ALDH1A1+/CD44− cells. ALDH1A1+ cells could, therefore, be more specific for stemness than CD44+ population in bladder cancer. Additionally, of the CD44+ cancer cells, only small proportion (6.4–8.2%) of these cells are positive for ALDH1A1, however, up to 72.1% ALDH1A1+ cells had positive CD44 staining in the same cell lines. The data suggested that although CD44+ population contained CSCs of bladder cancer, only a subset of the population had CSC characteristics and were “real” CSCs, which might be ALDH1A1+ cells within the population. The possibility was further supported by a higher percentage of CD44+ population existed in clinical bladder tumors and a small number of ALDH1A1+ cells that were solely present within the CD44 immunopositive tumor areas. The result was also consistent with a recent study of colon carcinomas [23], in which, ALDH1A1+/CD44+ colon cancer cells did not display statistically better efficiency in generating tumors than did isogenic ALDH1A1+ cells.
The elevated ALDH1A1 expression was found in 26% (56 of 216) human bladder tumor tissues. The observation of a minority (26%) of cancer samples positive for ALDH1A1 was consistent with the findings in other types of human solid malignancies [21–3, 28, 31]. For example, increased ALDH1A1 expression occurred in 30% of 345 breast tumor specimens (21). We recently found that elevated ALDH1A1 expression existed in 29% human lung tumor tissues [22]. Especially, our current observation is in agreement with feature of CSCs that only account for small population of cancer cells within tumors [5, 31–2]. Therefore, the data further provides evidences supporting that the ALDH1A1+ cancer cells might be enriched in tumor-stem cells or CSCs of bladder urothelial cell carcinoma.
The development of molecular biomarkers that can predict bladder cancer cells to recur and progress would help to identify tumors that could benefit the most effective treatments, and is thus clinically important. We demonstrated that increased ALDH1A1 expression levels were correlated with advanced tumor grade and stage. Furthermore, patients with elevated ALDH1A1 expression in their tumors had higher recurrence and shorter survival rates compared with patients with low ALDH1A1 expression. In addition, the increased ALDH1A1 expression was an independent prognostic factor for both cancer-specific and overall survivals in the patients. The findings are potentially important in practice, because the IHC-staining for detecting ALDH1A1 changes is a simple and readily available assay, and will easily be performable in clinical settings. Moreover, the ALDH1A1 antibody specifically binds its target with very low or no background, reducing inconsistent or variable results of IHC. The future application of an automated instrument with standard protocol will diminish inter-observer and intra-observer variations in interpretation of IHC staining results [33]. Therefore, ALDH1A1 would potentially be added to the current prognostic methods to improve the accuracy of clinical outcome predictions and the choice of appropriate therapy for bladder cancer patients who might have disease recurrence and progression.
The ALDH1A1 gene is about 53 kb long and contains 13 exons, encoding a 501-residue protein [16]. Human ALDH1 isozyme is expressed at different levels in various tissues, with the highest level in the liver and the lowest or undetectable levels in the heart [17], implying the ALDH1 expression is tissue-specific. The molecular mechanism of its tissues-specific expression has been investigated [11, 17–8]. Yanagawa et al. [11] found that 5'-flanking region of ALDH1A1 contains a number of putative regulatory elements. These include NF-IL6, HNF-5, GATA binding sites, and putative response elements for interleukin-6, phenobarbital and androgen, a noncanonical TATA box (ATAAA), and a CCAAT box. The protein level in various types of human cells might be regulated at the transcriptional level [11, 18]. For example, they defined a minimal promoter region from −91 to −53 bp of human “liver” ALDH1, and observed a 65-fold increase in CAT activity in a hepatoma cell line, Hep3B. In contrast, only 6–7-fold increases in CAT activity were found in a human leukemia cell line, K562. Whether and how ALDH1A1 is regulated at the transcriptional level in human urinary bladder urothelial cells is currently investigated at our laboratory. The study would deep our understanding of ALDH1A1 function in malignant transformation of SCs in carcinogenesis of bladder.
In summary, we show that ALDH1A1+ bladder cancer cells are endowed with extensive tumorigenicity and self-renewal potential, being able to generate tumors that resemble the histopathologic characteristics and heterogeneity of the parental tumor cells, and thus have the properties of CSCs or stem-like cancer cells. Furthermore, ALDH1A1 could function as a prognostic factor for predicting outcome in patients with the challenge malignancy. Nevertheless, a longitudinal clinical study in large population to validate its prognostic value for improving treatment efficiencies of bladder cancer will be needed.
Acknowledgments
Grant support: This work was supported in part by American Cancer Society Research Scholar Grant, National Cancer Institute (NCI) Grants CA-135382, CA-137742, and CA-133956, Wendy Will Case Cancer Award, Associate Member Award from NCI-The Early Detection Research Network, and Clinical Innovator Award from Flight Attendant Medical Research Institute (F. J.).
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5:713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 3.Habuchi T, Marberger M, Droller MJ, et al. Prognostic markers for bladder cancer: International Consensus Panel on bladder tumor markers. Urology. 2005;66:64–74. doi: 10.1016/j.urology.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 4.Malats N, Bustos A, Nascimento CM, et al. P53 as a prognosticmarker for bladder cancer: a meta-analysis and review. Lancet Oncol. 2005;6:678–686. doi: 10.1016/S1470-2045(05)70315-6. [DOI] [PubMed] [Google Scholar]
- 5.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;12:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 6.Chang CC, Shieh GS, Wu P, Lin CC, Shiau AL, Wu CL. Oct-3/4 expression reflects tumor progression and regulates motility of bladder cancer cells. Cancer Res. 2008;68:6281–6291. doi: 10.1158/0008-5472.CAN-08-0094. [DOI] [PubMed] [Google Scholar]
- 7.He X, Marchionni L, Hansel DE, et al. Differentiation of a highly tumorigenic basal cell compartment in urothelial carcinoma. Stem Cells. 2009;9:1487–1495. doi: 10.1002/stem.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan KS, Espinosa I, Chao M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci U S A. 2009;18:14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.She JJ, Zhang PG, Wang ZM, Gan WM, Che XM. Identification of side population cells from bladder cancer cells by dyecycle violet staining. Cancer Biol Ther. 2008;7:1664–1669. doi: 10.4161/cbt.7.10.6637. [DOI] [PubMed] [Google Scholar]
- 11.Yanagawa Y, Chen JC, Hsu LC, Yoshida A. The transcriptional regulation of human aldehyde dehydrogenase I gene. The structural and functional analysis of the promoter. J Biol Chem. 1995;29:17521–17527. doi: 10.1074/jbc.270.29.17521. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong L, Stojkovic M, Dimmick I, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- 13.Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess DA, Craft TP, Wirthlin L, et al. Widespread non-hematopoietic tissue distribution by transplanted human progenitor cells with high aldehyde dehydrogenase activity. Stem Cells. 2006;26:611–620. doi: 10.1634/stemcells.2007-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess DA, Wirthlin L, Craft TP, et al. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162–2169. doi: 10.1182/blood-2005-06-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu LC, Chang WC, Yoshida A. Genomic structure of the human cytosolic aldehyde dehydrogenase gene. Genomics. 1989;5:857–865. doi: 10.1016/0888-7543(89)90127-4. [DOI] [PubMed] [Google Scholar]
- 17.Harada S, Agarwal DP, Goedde HW. Electrophoretic and biochemical studies of human aldehyde dehydrogenase isozymes in various tissues. Life Sci. 1980;26:1773–1780. doi: 10.1016/0024-3205(80)90577-9. [DOI] [PubMed] [Google Scholar]
- 18.Elizondo G, Corchero J, Sterneck E, Gonzalez FJ. Feedback inhibition of the retinaldehyde dehydrogenase gene ALDH1 by retinoic acid through retinoic acid receptor alpha and CCAAT/enhancer-binding protein beta. J Biol Chem. 2000;15:39747–39753. doi: 10.1074/jbc.M004987200. [DOI] [PubMed] [Google Scholar]
- 19.Pearce DJ, Taussig D, Simpson C, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;6:752–760. doi: 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- 20.Balicki D. Moving forward in human mammary stem cell biology and breast cancer prognostication using ALDH1. Cell Stem Cell. 2007;15:485–487. doi: 10.1016/j.stem.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;15:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang F, Qiu Q, Khanna A, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Molecular Cancer Res. 2008;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;8:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreb JS, Baker HV, Chang LJ, et al. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer. 2008;7:87. doi: 10.1186/1476-4598-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong C, Tang DQ, Xie CQ, et al. Genetic engineering of human embryonic stem cells with lentiviral vectors. Stem Cells Dev. 2005;4:367–377. doi: 10.1089/scd.2005.14.367. [DOI] [PubMed] [Google Scholar]
- 26.Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A11 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–349. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 27.Miyake H, Eto H, Arakawa S, Kamidono S, Hara I. Over expression of CD44V8-10 in urinary exfoliated cells as an independent prognostic predictor in patients with urothelial cancer. J Urol. 2002;167:1282–1287. [PubMed] [Google Scholar]
- 28.Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res. 2007;67:6796–6805. doi: 10.1158/0008-5472.CAN-07-0490. [DOI] [PubMed] [Google Scholar]
- 29.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2007;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 30.Yang YM, Chang JW. Bladder cancer initiating cells (BCICs) are among EMA-CD44v6+ subset: novel methods for isolating undetermined cancer stem (initiating) cells. Cancer Invest. 2008;26:725–733. doi: 10.1080/07357900801941845. [DOI] [PubMed] [Google Scholar]
- 31.Chen YC, Chen YW, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385:307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 32.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;6859:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 33.Sitnikova L, Mendese G, Liu Q, et al. IMP3 predicts aggressive superficial urothelial carcinoma of the bladder. Clin Cancer Res. 2008;6:1701–1706. doi: 10.1158/1078-0432.CCR-07-2039. [DOI] [PubMed] [Google Scholar]