Abstract
Purpose
Early events in colorectal tumorigenesis include mutation of the adenomatous polyposis coli (APC) gene and epigenetic hypermethylation with transcriptional silencing of the O6-methylguanine DNA methyltransferase (MGMT), human mut L homologue 1 (hMLH1), and P16/CDKN2A genes. Epigenetic alterations affect genetic events: Loss of MGMT via hypermethylation reportedly predisposes to guanine-to-adenine or cytosine-to-thymine (G:C→A:T) transition mutations in KRAS and P53, and silencing of hMLH1 leads to high levels of microsatellite instability (MSI-H)/mutator phenotype, suggesting that epigenetic-genetic subtypes exist.
Experimental Design
We evaluated the relationships of aberrant methylation of APC, MGMT, hMLH1, P16, N33, and five MINTs to mutations in APC, KRAS, BRAF, and P53 in 208 colorectal carcinomas.
Results
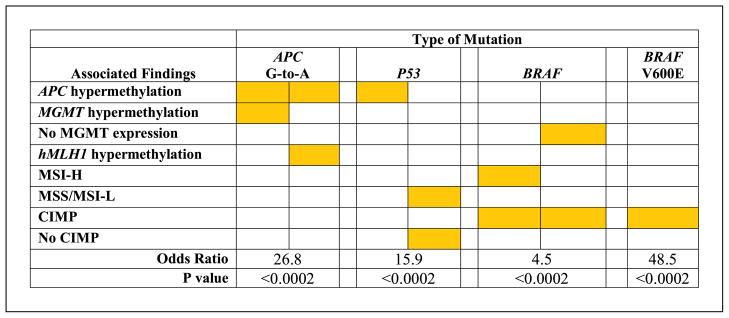
We found that APC hypermethylation was age related (P = 0.04), in contrast to the other genes, and did not cluster with CpG island methylator phenotype (CIMP) markers. Hypermethylation of APC concurrently with either MGMT or hMLH1 was strongly associated with occurrence of G-to-A transitions in APC [odds ratio (OR), 26.8; P < 0.0002 from multivariable logic regression model], but C-to-T transitions had no associations. There was no relationship of hypermethylation of any gene, including MGMT, with G-to-A or C-to-T transitions in KRAS or P53, although APC hypermethylation was associated with P53 mutation (P < 0.0002). CIMP with MSI-H due to hMLH1 hypermethylation, or CIMP with loss of MGMT expression in non – MSI-H tumors, was associated with BRAF mutation (OR, 4.5; P <0.0002). CIMP was also associated with BRAF V600E T-to-A transversion (OR, 48.5; P < 0.0002).
Conclusions
Our findings suggest that the heterogeneous epigenetic dysregulation of promoter methylation in various genes is interrelated with the occurrence of mutations, as manifested in epigenetic-genetic subgroups of tumors.
The development of most colorectal carcinomas (CRC) is thought to be initiated by inactivation of the APC/β-catenin/Wnt signaling pathway, usually by mutation of one copy of the APC gene followed by a second event that inactivates the other allele (1–4). The second inactivating event is usually described as allelic deletion or mutation but can be epigenetic methylation of cytosines in CpG islands of the promoter region of APC that results in transcriptional silencing (5, 6). Altered APC has wide-ranging downstream effects on cell-cell adhesion, transcriptional regulation, chromosomal instability, cell migration, proliferation and cell cycle control, differentiation, and apoptosis (1–4).
Somatic APC mutation is associated with the development of intraepithelial neoplasia (dysplasia) in aberrant crypt foci and early adenomas (7). The initiating alterations in APC in these early lesions persist whereas additional genetic and epigenetic events accumulate and drive tumor progression. Somatic mutation in the APC gene is present in as many as 80% of sporadic colorectal adenomas and carcinomas (7–9), and a mutation cluster region in exon 15 (9) accounts for the majority of mutations. Most APC mutations alter function of the gene product via point mutation or frameshift mutation that results in a truncated protein lacking all of the axin-binding sites and all but one or two of the 20-amino-acid β-catenin binding sites (10). Mutations of the KRAS or BRAF gene in the RAS/RAF pathway and of the P53 gene are later genetic events in colorectal tumorigenesis (3).
Well-known mechanisms of spontaneous generation of point mutations in genes include deamination of cytosine and 5-methylcytosine to uracil and thymine, respectively; depurination; DNA polymerase infidelity; and oxidative damage from endogenously produced free radicals (11, 12). Epigenetic events have been hypothesized to influence specific genetic changes and to have a complementary role with genetic alterations in colorectal tumorigenesis (13), albeit with transient controversy about the existence of the hypermethylation pathway, termed the CpG island methylator phenotype (CIMP; refs. 14, 15). A few reports have addressed the influence of epigenetics on specific gene mutations. Inactivation via cytosine methylation of CpG islands in the promoter of the DNA repair gene O6-methylguanine DNA methyltransferase (MGMT) has been linked to subsequent G:C→A:T transition mutations in other genes, representing G-to-A mutations on the sense strand or C-to-T mutations if the G-to-A is on the antisense strand (16). An explanatory mechanism of failure to repair adducts has been proposed (17–19). The MGMT gene product removes mutagenic and cytotoxic methyl adducts from the O6 position of guanine. Hypermethylation of the promoter of MGMT has been associated with transcriptional silencing of the gene and loss of protein expression. Failure of removal of O6-methylguanine adducts results from absent MGMT enzyme activity, and mispairing with thymines occurs during replication so that the guanine-cytosine pair is converted to an adenine-thymine pair (16, 20). MGMT hypermethylation has been associated in some studies with G:C→A:T mutations in the KRAS proto-oncogene and the P53 suppressor gene that are frequently mutated during progression of colorectal neoplasms (18, 19). Other studies, however, have not identified this association nor a relationship of these specific mutations to reduced or absent expression of MGMT protein (21, 22).
Hypermethylation of the promoter of the hMLH1 mismatch repair gene leads to another important molecular pathway in colorectal tumorigenesis, that is, the microsatellite instability (MSI) pathway (23–25). Loss of functional hMLH1 protein results in inability of tumor cells to repair nucleotide mismatches. This deficiency is manifested as slippage mutations in repeated nucleotide sequences (microsatellites), resulting in high levels of MSI (MSI-H), as well as instability in both homopolymeric runs and base substitutions in coding sequences called mutator phenotype (25). Hypermethylation of the P16 gene can silence expression of this suppressor gene and is an early event in colorectal neoplasia that can occur in adenomas (26–28) and aberrant crypt foci (29).
The relationships among epigenetic alterations of various individual genes, concurrent hypermethylation of multiple genes in CIMP, and specific genetic mutations are incompletely described and discordant among different studies. Comprehensive characterization of these multiple pathways that are important in CRC will contribute to understanding the spectrum of intertumoral heterogeneity. We therefore examined the interrelationships among these genomic pathways in 208 CRCs. Our findings provide additional evidence for the occurrence of epigenetic-genetic interactions in tumorigenesis and have important implications for understanding the molecular pathogenesis of subgroups of CRC.
Materials and Methods
Specimens
We evaluated CRC specimens from 208 patients who were included in an ongoing study that compares the molecular pathology of CRC in Hong Kong and the United States. (The results of the international comparison of molecular characteristics are the subject of another report.) The patients had their resection between 1991 and 2003 at Queen Mary Hospital in Hong Kong (n = 90) or the Texas Medical Center in Houston at The University of Texas M. D. Anderson Cancer Center (n = 70) or St. Luke’s Episcopal Hospital (n = 48). The mean age of the patients was 61.6 years; 56.3% were male; 73.6% of the tumors were in the left colon or rectum; and 9.1% were stage I, 37.5% were stage II, 36.1% were stage III, and 17.3% were stage IV (30). No patients received preoperative neoadjuvant therapy before the surgical or biopsy procedure that produced the specimen used for our evaluation. The institutional review board at each collaborating institution approved the study.
Microdissection and DNA extraction
Routine formalin-fixed, paraffin-embedded or frozen archived resection or biopsy specimens were analyzed. DNA was prepared from tumor and from nonneoplastic mucosa after manual microdissection of 5-μm histopathologic sections, as described previously (31).
Hypermethylation of APC, MGMT, hMLH1, P16, N33, and MINTs
The methylation status of the APC promoter 1A; the promoters of the MGMT, hMLH1, P16/CDKN2A, and N33 genes; and methylated in tumor (MINT) loci 1, 2, 25, 27, and 31 were determined by bisulfite treatment of DNA followed by methylation-specific PCR, as shown in Fig. 1. The methods were those described previously (5, 26–29, 32, 33) except for N33 and MINT27. The primers for the N33 were 5′-CGGAG-GGTTTAGTTAGCGGGTTTTC-3′ and 5′-GACAAAACAATATCTCCTC-CACGCG-3′ for the methylated allele, and 5′-TGGAGGGTTTAGTTAG-TGGGTTTTT-3′ and 5′-CAACAAAACAATATCTCCTCCACACA-3′ for the unmethylated allele. The primers used for MINT27 were 5′-GGAGT-TTTGTGTTAGACGCGGC-3′ and 5′-AAAACGCCAAAAACTCCCTACG-3′ for the methylated allele, and 5′-GTGTGGAGTTTTGTGTTAGATGTGGT-3′ and 5′-CAAAAACACCAAAAACTCCCTACA-3′ for the unmethylated allele. DNA from normal lymphocytes was used as the control for unmethylated genes and loci, except for N33, whereas placental DNA treated with SssI (CpG) methylase (New England Biolabs) was used as the positive control. DNA obtained from the cell line UMUC3 was used as the control for unmethylated N33. PCR products from methylated and unmethylated reactions were electrophoresed on 3% agarose gels and visualized by ethidium bromide staining. Each sample was analyzed in triplicate. The criterion for presence of hypermethylation was detection of a methylated band in all three independent assays.
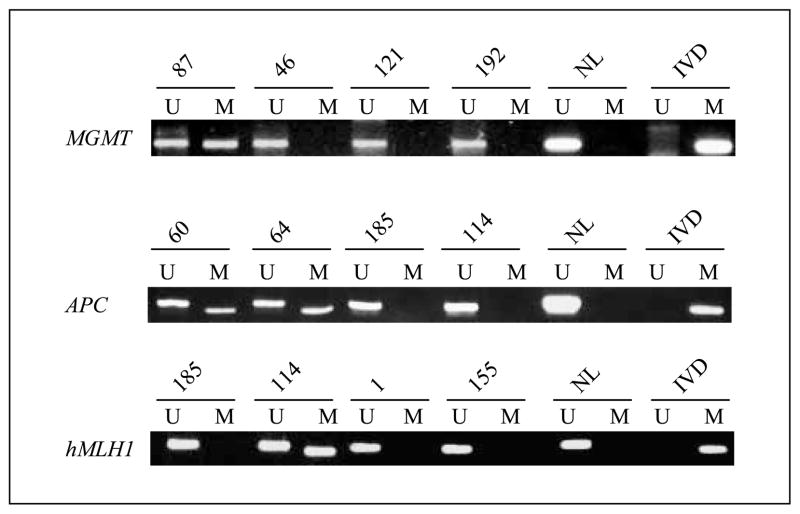
Fig. 1.
Examples of methylation assays of MGMT, APC, and hMLH1 gene promoters in CRCs. The presence of a visible PCR product from methylation-specific PCR in lanes labeled U indicates the presence of an unmethylated allele, and in lanes labeled M, the presence of a methylated allele. MGMT hypermethylation is evident in case 87; APC hypermethylation is evident in cases 60 and 64; and hMLH1 hypermethylation is evident in case 114. DNA from normal lymphocytes (NL) was used as a control for unmethylated MGMT and APC, and in vitro methylated DNA (IVD) from placenta was used as a control for the hypermethylated genes.
Hierarchical cluster analysis (34) of all tumors based on CpG island methylation in the 10 evaluated markers (Fig. 2) was used to identify the marker panel that defined CIMP status. The five markers identified in this fashion (MINT1, MINT2, MINT31, p16, and hMLH1) were found to be the same as those used in our previous studies (26, 32, 35). CRC were classified as having CIMP if three or more (≥60%) of the evaluated markers were methylated. Classification of CIMP on the basis of two or more methylated markers in the panel (≥40%) yielded similar results.
Fig. 2.
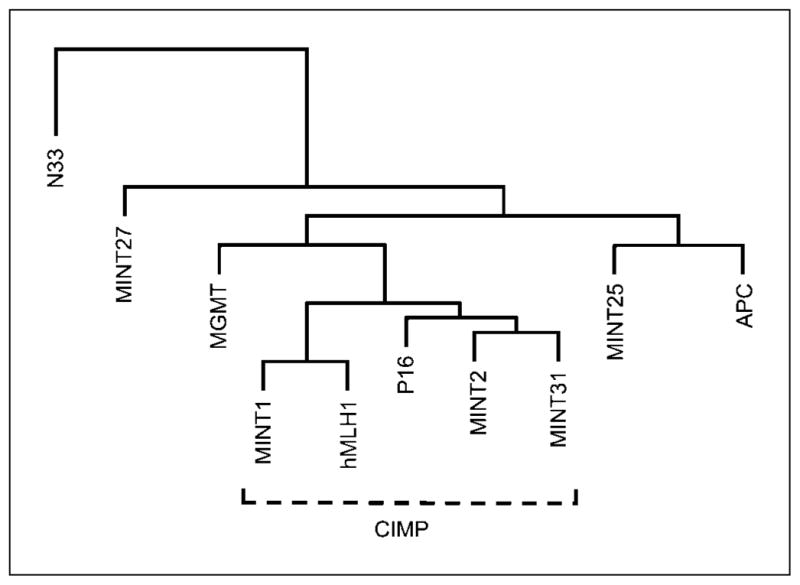
Hierarchical cluster analysis of the 10 methylation makers used. P16, hMLH1, and MINTs1, 2, and 31 clustered together and were used to define CIMP, as in our previous studies (26, 32, 35). Hypermethylation of the APC, MGMT, and N33 genes and of MINTs 25 and 27 did not cluster with the CIMP markers.
Mutation of APC, KRAS, BRAF, and P53 genes
DNA sequencing was used to evaluate mutations in the APC, KRAS, BRAF, and P53 genes. For the APC gene, four sets of oligonucleotide primers were used to amplify the mutation cluster region in exon 15 from codons 1260 to 1592 as described previously (36), except for redesigned primers C1 and C2. For codons 1416 to 1510, C1 was 5′-AAGCCCCAGT-GATCTTCCA-3′ and C2 was 5′-CAGAGCACTCAGGCTGGAT-3′. The primers for exon 11 and exon 15 of BRAF and exon 2 of KRAS used in the current study were described previously (37).
For the P53 gene, five sets of oligonucleotide primers were used to amplify exons 5 to 8 (exon 5 was amplified in two overlapping segments). Exon 5a-forward was 5′-CTTTCAACTCTGTCTCCTTCCTC-3′ and exon 5a-reverse was 5′-GCTGTGACTGCTTGTAGATGG-3′. Exon 5b-forward was 5′-GCCAAGACCTGCCCTGTG-3′ and exon 5b-reverse was 5′-CAACCAGCCCTGTCGTCTCT-3′. Exon 6-forward was 5′-GGCC-TCTGATTCCTCACTGA-3′ and exon 6-reverse was 5′-CTCCTCCCAGAG-ACCCCAGT-3′. Exon 7-forward was 5′-CCTCATCTTGGGCCTGTGTT-3′ and exon 7-reverse was 5′-GCAGGGTGGCAAGTGGCTCC-3′. Exon 8-forward was 5′-CCTTACTGCCTCTTGCTTCTCT-3′ and exon 8-reverse was 5′-CTCCACCGCTTCTTGTCCTG-3′.
For each primer pair, PCR products were generated from two independent PCR reactions for sequencing of the forward and reverse products. The PCR products were purified by use of shrimp alkaline phosphatase and exonuclease I (Amersham) in accordance with the manufacturer’s instructions. The purified PCR products were sequenced on an ABI Prism 3730 DNA Analyzer with the ABI Prism Big Dye Terminator Cycle Sequencing Kit version 1.1 (Applied Biosystems). The primers used for amplification were also used for sequencing. The sequencing results were analyzed using DNA Sequencing Analysis Software Version 5.0 (Applied Biosystems). All mutations were confirmed by analysis in both the forward and reverse directions.
Microsatellite instability
MSI was evaluated in genomic DNA extracted from microdissected tumor tissue and nonneoplastic mucosa as described previously (38). A panel of seven microsatellites was evaluated: four mononucleotide repeats (BAT25, BAT26, BAT40, and TGFβRII) and three dinucleotide repeats (D2S123, D5S346, and D17S250). Tumors were categorized according to criteria from the National Cancer Institute conference on MSI (39) as microsatellite-stable (MSS) if no marker had altered size, MSI-low (MSI-L) if at least one marker but <40% of total markers were altered, and MSI-high (MSI-H) if at least 40% of markers were altered.
MGMT and hMLH1 gene product expression
Histologic sections of formalin-fixed paraffin-embedded tissue cut 5 μm thick were evaluated for immunohistochemical staining of tumor cell nuclei with commercially available mouse anti-MGMT monoclonal antibody (Chemicon) or anti-hMLH1 monoclonal antibody (PharMingen) and routine methods, as described previously (40, 41). Nuclear expression was classified as present or absent relative to staining of nuclei of nonneoplastic cells that provided a positive internal control for each slide.
Statistical analysis
Each assay was done and interpreted independently of all other assays, and the results were then entered into a database for statistical analysis. Hierarchical cluster analysis was done to assess the association among the 10 evaluated CpG methylation sites (34). Descriptive statistics were calculated. Associations between categorical variables were evaluated with two-tailed Fisher’s exact tests. The odds ratios (OR) and 95% confidence intervals were estimated for each association. P values <0.05 were considered statistically significant.
In addition, a multivariable logic regression model (42) was fit for the binary outcome of presence or absence of mutation of the APC, Ki-ras, BRAF, and P53 genes and for the subsets of G-to-A, C-to-T, and G:C→A:T transition mutations, and of BRAF V600E nucleotide 1799 T-to-A transversion. Methylation, gene product expression, and microsatellite instability were used as predictors. Logic regression is one type of logistic regression model that uses the logic or Boolean combinations of binary covariates as potential predictors. A subset of cases with all available results was created. Through implementing a “greedy search” algorithm (42), we then found the optimal logic regression model that minimized the score function. The greedy search algorithm first identified a best single predictor among all candidate single predictors. Then, it searched among the remaining predictors to find the best combination that lowered the score function. This process continued until the score function could no longer be reduced. To account for multiple testing inherent in the logic regression procedure, permutation tests were carried out using 5,000 permutations to compute the P values. The ORs were adjusted by tumor stage. All statistical analyses were carried out in Splus.
Results
The epigenetic, genetic, and associated phenotypic alterations in the 208 CRCs we studied are summarized in Table 1 and Fig. 3.
Table 1.
Summary of epigenetic, genetic, and associated phenotypic alterations
| Site and type of alteration | Percentage of tumors | No. tumors | Total tumors evaluated |
|---|---|---|---|
| APC gene | |||
| APC gene alteration identified | 70.2 | 146 | 208 |
| APC promoter hypermethylation | 31.7 | 64 | 202 |
| Mutation in APC mutation cluster region | 62.0 | 129 | 208 |
| Point mutation | — | 87 (136)* | — |
| G-to-A transition | — | 19 (25)* | — |
| C-to-T transition | — | 30 (32)* | |
| Frameshift mutation | — | 57 (59)* | — |
| MGMT gene and MGMT expression | |||
| MGMT promoter hypermethylation | 27.4 | 57 | 208 |
| Loss of MGMT protein expression | 24.5 | 51 | 208 |
| Concordance of MGMT methylation status with MGMT protein expression status | 82.7 | 172 | 208 |
| hMLH1 gene and microsatellite instability | |||
| hMLH1 promoter hypermethylation† | 9.2 | 19 | 207 |
| Loss of hMLH1 protein expression | 9.8 | 20 | 204 |
| Concordance of hMLH1 methylation status with hMLH1 protein expression status | 94.6 | 192 | 203 |
| High levels of microsatellite instability (MSI-H) | 10.6 | 22 | 208 |
| Concordance of hMLH1 methylation status with MSI status | 92.6 | 187 | 202 |
| CpG island methylation of additional markers | |||
| Frequency of hypermethylation | |||
| P16 gene† | 19.5 | 39 | 200 |
| N33 gene | 93.8 | 180 | 192 |
| MINT1† | 10.1 | 21 | 208 |
| MINT2† | 22.6 | 47 | 208 |
| MINT25 | 28.3 | 51 | 180 |
| MINT27 | 39.6 | 67 | 169 |
| MINT31† | 20.3 | 42 | 207 |
| CIMP with three markers (≥ 60%) | 10.1 | 21 | 208 |
| KRAS proto-oncogene/BRAF gene | |||
| KRAS codon 12 or 13 mutation | 39.2 | 80 | 204 |
| KRAS codon 12 or 13 G-to-A transition | — | 56 | — |
| Codon 12a G-to-A | — | 5 | — |
| Codon 12b G-to-A | — | 26 | — |
| Codon 13b G-to-A | — | 25 | — |
| BRAF mutation | 6.5 | 13 | 199 |
| BRAF V600E nt 1799 T-to-A transversion | 4.5 | 9 | 199 |
| Discordant KRAS and BRAF V600E mutation status | 100 | 9 | 9 |
| P53 gene | |||
| Exon 5–8 mutation | 55.2 | 112 | 203 |
| G-to-A transition | — | 40 | — |
| C-to-T transition | — | 31 | — |
Number in parenthesis is the number of APC mutations. The total number of APC mutations exceeds the number of tumors due to the occurrence of multiple mutations in 41 tumors. The total number of point mutations includes the number of G-to-A transition mutations.
Five markers used to define CIMP based on cluster analysis (see Fig. 2).
Fig. 3.
Diagram of epigenetic and genetic alterations in 208 CRCs arranged by methylation and mutation status. The CIMP markers from Fig. 2 are indicated by asterisk in the column headings. Shading, the presence of the molecular alteration; X, unavailable data; open cell, absence of the alteration. Numbers in the APC columns indicate when two or more mutations are present, and numbers in the BRAF column indicate the type of or nucleotide with mutation. The intertumoral heterogeneity of the alterations and the epigenetic-genetic associations are apparent. The strong relationship between hypermethylation of APC with hypermethylation of MGMT or hMLH1 and G-to-A transition mutation in APC is evident in cases 1 to 11.
Promoter hypermethylation and mutation of APC gene
An alteration of APC was found in 70.2% of the CRC; 31.7% had APC hypermethylation, and this alteration was age-related (43), occurring more frequently in older patients (mean age 64.6 years as contrasted with 60.2 years, P = 0.04; 45.8% of CRC with APC hypermethylation in patients in the oldest quartile >79 years of age but only 23.5% in the youngest quartile <51 years old). No other hypermethylation site we evaluated had an association with age. By contrast with the tumors, nonneoplastic colorectal mucosa rarely had APC hypermethylation (0.8%).
Sixty-two percent of the tumors had mutation of APC in the mutation cluster region between codons 1260 and 1596. Point mutation was the most common sequence alteration observed. Twenty-five G-to-A transition point mutations were found in 19 tumors (Table 2). Two tumors had two G-to-A transitions (cases 7 and 18), and two tumors had three (cases 3 and 16). Only one of the G-to-A mutations occurred in a CpG site (case 3). Twenty-four percent of the G-to-A mutations were silent, and the remainder resulted in amino acid substitutions. A C-to-T transition was found in 30 tumors. Thus, there was no evidence of strand bias in G:C→A:T mutations (25 G-to-A and 30 C-to-T on sense strand).
Table 2.
Summary of G-to-A transition mutations in APC gene
| Case | Codon | G in nucleotide sequence | Protein sequence change |
|---|---|---|---|
| 1 | 1578b | GAA TGT ATT | Cys→Tyr |
| 2 | 1550c | CAA GAG AAA | Glu→no change |
| 3 | 1400c | CGT TCG ATT | Ser→no change |
| 1513c | GAT GAG CCA | Glu→no change | |
| 1561c | GAA AAG GAC | Lys→no change | |
| 4 | 1530a | CAG GAA AAT | Glu→Lys |
| 5 | 1540c | TCA GAG CAG | Glu→no change |
| 6 | 1466b | AGT GGA CCT | Gly→Glu |
| 7 | 1357a | TCA GGA GCG | Gly→Arg |
| 1534a | AAT GGG AAT | Gly→Arg | |
| 8 | 1312a | ATT GGA ACT | Gly→Arg |
| 9 | 1338c | CTG CAG GGT | Gln→no change |
| 10 | 1470a | CAA GCT GCA | Ala→Thr |
| 11 | 1377a | TAT GTT CAG | Val→Ile |
| 12 | 1321b | GTG AGC GAA | Ser→Asn |
| 13 | 1426b | GAT AGC CCT | Ser→Asn |
| 14 | 1374a | CCT GAA CAC | Glu→Lys |
| 15 | 1416b | AGT GGC ATT | Gly→Asp |
| 16 | 1348b | GCC AGG CAC | Arg→Lys |
| 1460a | ACT GCT GAA | Ala→Thr | |
| 1475a | GCT GCA GTT | Ala→Thr | |
| 17 | 1435b | AGC AGA AGT | Arg→Lys |
| 18 | 1385b | TTT AGC AGA | Ser→Asn |
| 1521a | GTG GAA TTA | Glu→Lys | |
| 19 | 1317a | GCT GAA GAT | Glu→Lys |
Promoter hypermethylation of MGMT gene and loss of MGMT protein expression
Of the CRCs, 27.4% had hypermethylation of MGMT, and 24.5% had loss of MGMT protein by immunohistochemistry. Methylation status of the MGMT gene and MGMT gene product expression status were highly concordant, with the expected inverse relationship in 82.7% of the carcinomas (P < 0.001).
Promoter hypermethylation of hMLH1 gene, loss of hMLH1 protein expression, and microsatellite instability
Hypermethylation of the hMLH1 gene was found in 9.2% of the CRC, and loss of hMLH1 protein expression was found in 9.8%, with the expected inverse relationship in 94.6% the carcinomas. Hypermethylation of hMLH1 also had the expected strong association with high levels of MSI (MSI-H), with methylation status of hMLH1 and MSI status concordant in 92.7% of tumors (P < 0.0001).
Concurrent CpG island hypermethylation
Hierarchical cluster analysis showed that the methylation status of the 10 markers had distinct relationships (Fig. 2). MINT1, MINT2, MINT31, P16, and hMLH1 clustered together, as we found in our previous studies (26, 32, 35). These five markers therefore were used to define CIMP based on hypermethylation of three or more markers (≥60%) and identified 10.1% of the CRC as having CIMP. The methylation status of the APC, MGMT, and hMLH1 genes were not significantly associated (OR, 0.5–1.1; 95% confidence interval, 0.1–1.7 to 0.6–2.1), as reported in previous studies (5, 44).
Mutations of KRAS proto-oncogene, BRAF, and P53 suppressor gene
Mutation of the KRAS gene at codon 12 or 13 was found in 39.2% of the CRC. Among the 80 mutations, 70.0% were a G-to-A transition at codon 12a (GGT->AGT, n = 5), at 12b (GGT→GAT, n = 26), or at 13b (GGC→GAC, n = 25), but no tumor had a G-to-A transition at 13a (GGC→AGC). C-to-T transition at 13c does not occur, and therefore strand bias for G:C→A:T mutation is not a consideration. Among all of the molecular alterations we studied, KRAS mutation was associated with stage, occurring with higher frequency in more advanced tumors: 15.8% of stage I, 37.7% in stage II, 38.4% in stage III, and 57.1% in stage IV (P = 0.03).
BRAF mutation was identified in 6.5% of the tumors, with the V600E T-to-A transversion at nucleotide 1799 accounting for 9 of the 13 mutations. The other BRAF mutations were G469V G-to-T transversion (case 80), T599I C-to-T transition (case 35), F468S T-to-C transition (case 49), and N581S A-to-G transition (case 166). KRAS and BRAF V600E mutations, but not the other BRAF mutations, were mutually exclusive (Fig. 3), as expected (37, 38, 45, 46).
Mutation in exons 5 to 8 of the P53 gene was present in 55.2% of the carcinomas. Of the 112 P53 mutations, 35.7% were a G-to-A transition, and seven of these were located in a non-CpG site. C-to-T mutation occurred in 31 tumors. Thus, there was no evidence of strand bias in G:C→A:T mutations (40 G-to-A and 31 C-to-T on sense strand).
Associations among hypermethylation and gene mutations
Table 3 and Fig. 4 summarize the relationships among the epigenetic and genetic alterations identified in multivariable analysis. Hypermethylation of specific genes was associated with the pattern of gene mutations in subgroups of CRC.
Table 3.
Associations between composite binary predictor variables created through logic regression and mutation of APC, KRAS, BRAF, or P53 genes
| A. APC mutation
| |||||
|---|---|---|---|---|---|
| Created composite variable | APC mutation present | No APC mutation found | Total | OR* | P† |
| APC hypermethylation present or (neither MGMT protein expression nor CIMP found) | 65 | 22 | 87 | 2.9 | 0.11 |
| APC hypermethylation absent and (presence of MGMT protein expression or of CIMP) | 56 | 50 | 106 | ||
| Total | 121 | 72 | 193 | ||
| B. APC G-to-A transition
| |||||
|---|---|---|---|---|---|
| Created composite variable | APC G-to-A present | No APC G-to-A found | Total | OR* | P† |
| (Concurrent APC and MGMT hypermethylation) or (concurrent APC and hMLH1 hypermethylation) | 11 | 11 | 22 | 26.8 | <0.0002 |
| (Concurrent APC and MGMT hypermethylation not found) or (concurrent APC and hMLH1 hypermethylation not found) | 8 | 174 | 182 | ||
| Total | 19 | 185 | 204 | ||
| C. KRAS mutation
| |||||
|---|---|---|---|---|---|
| Created composite variable | KRAS mutation present | No KRAS mutation found | Total | OR* | P† |
| CIMP present | 4 | 16 | 20 | 3.3 | 0.25 |
| CIMP not found | 74 | 97 | 171 | ||
| Total | 78 | 113 | 191 | ||
| D. KRAS G-to-A transition
| |||||
|---|---|---|---|---|---|
| Created composite variable | KRAS G-to-A present | No KRAS G-to-A found | Total | OR* | P† |
| MSI-H present and CIMP not found | 7 | 5 | 12 | 4.4 | 0.35 |
| MSS/MSI-L or CIMP present | 48 | 131 | 179 | ||
| Total | 55 | 136 | 191 | ||
| E. BRAF mutation
| |||||
|---|---|---|---|---|---|
| Created composite variable | BRAF mutation present | No BRAF mutation found | Total | OR* | P† |
| (Concurrent CIMP and MSI-H) or (CIMP present and MGMT protein expression not found) | 8 | 13 | 21 | 4.5 | <0.0002 |
| CIMP not found or (concurrent MSS/MSI-L and MGMT protein expression) | 2 | 163 | 1657 | ||
| Total | 10 | 176 | 186 | ||
| F. BRAF V600E T-to-A transversion
| |||||
|---|---|---|---|---|---|
| Created composite variable | BRAF V600E mutation present | No BRAF V600E mutation found | Total | OR* | P† |
| CIMP present | 6 | 27 | 153 | 48.5 | <0.0002 |
| CIMP not found | 1 | 152 | 33 | ||
| Total | 7 | 179 | 186 | ||
| G. P53 mutation
| |||||
|---|---|---|---|---|---|
| Created composite variable | P53 mutation present | No P53 mutation found | Total | OR* | P† |
| APC hypermethylation present or (MSS/MSI-L present and CIMP not found) | 103 | 66 | 169 | 15.9 | <0.0002 |
| No APC hypermethylation found and (MSI-H or CIMP present) | 2 | 20 | 22 | ||
| Total | 105 | 86 | 191 | ||
| H. P53 G-to-A transition
| |||||
|---|---|---|---|---|---|
| Created composite variable | P53 G-to-A present | No P53 G-to-A found | Total | OR* | P† |
| MSS/MSI-L present and CIMP not found | 36 | 124 | 160 | 11.6 | 0.13 |
| MSI-H or CIMP present | 0 | 31 | 31 | ||
| Total | 36 | 155 | 191 | ||
ORs were adjusted by tumor stage.
All P values are based on 5,000 permutations of the original data set.
Fig. 4.
Diagram illustrating the subgroups of CRC with epigenetic-genetic interactions identified through multivariable logic regression model. Hypermethylation of APC concurrently with either MGMT or hMLH1 hypermethylation was associated with G-to-A transition mutation in APC, whereas APC hypermethylation or absence of CIMP in MSS/MSI-L tumors was associated with P53 mutation. CIMP and MSI-H or CIMP in the absence of MSI-H but with loss of MGMT expression was associated with BRAF mutation. CIMP was also associated with BRAF V600E. ORs were adjusted by tumor stage, and all P values are based on 5,000 permutations of the original data set.
Hypermethylation of APC in concert with either MGMT or hMLH1 was strongly associated with G-to-A transition mutation in APC (cases 1–19 in Fig. 3; OR, 26.8; P < 0.0002; Table 3B; Fig. 4). A G-to-A transition was found in 50.0% of 22 CRC with APC hypermethylation concurrent with either MGMT or hMLH1 hypermethylation, but in only 4.4% of 182 carcinomas without concurrent methylation of these gene pairs. APC hypermethylation was not associated, however, with APC mutations in general (Table 3A) or with APC mutations of other subtypes. C-to-T transitions representing G-to-A on the antisense strand as occurs with MGMT deficiency (16) as well as C-to-T transitions on the sense strand had no associations. Further evidence for the influence of specific, rather than generalized, methylation on mutation pattern was provided by the lack of association of hypermethylation of the other genes and markers with mutation of KRAS (Table 3C and D).
The presence of APC hypermethylation or the presence of MSS or MSI-L in the absence of CIMP was associated with mutation of P53 (60.9%, 103 of 169, versus 9.1%, 2 of 22; OR, 15.9; P < 0.0002; Table 3G; Fig. 4). The inverse relationship of P53 mutation and CIMP has been reported in previous studies (35), as has the inverse relationship between P53 mutation and MSI-H (47–50), but the association of P53 mutation with APC hypermethylation has not been reported previously.
CIMP in concert with MSI-H due to hypermethylation of hMLH1 was associated with mutation of BRAF, as in previous studies (50–54). In addition, we identified the novel association of CIMP and loss of MGMT protein due to MGMT methylation with BRAF mutation (38.1%, 8 of 21, versus 1.2%, 2 of 165; OR, 4.5; P < 0.0002; Table 3E; Fig. 4). Only 2 of 13 CRC with BRAF mutation lacked CIMP, MSI-H, or loss of MGMT. CIMP was also strongly associated with BRAF V600E T-to-A transversion (18.2%, 6 of 33, versus 0.7%, 1 of 153; OR, 48.5; P < 0.0002; Table 3F; Fig. 4), as described previously (51–54). Only one of five CRCs with BRAF V600E mutation and MSI-H had methylation of MGMT and loss of MGMT protein expression, whereas all four tumors with BRAF V600E mutation that were MSS or MSI-L had lost MGMT expression (Fig. 3). These findings raise the possibility that MSI-H/mutator phenotype and MGMT expression have alternative relationships with BRAF mutation.
Discussion
Our study emphasizes the heterogeneity of both hypermethylation and epigenetic-genetic interactions in molecular subgroups of CRC. The characteristics of hypermethylation of APC, a key gene in CRC, differed from those of the other genes and markers we evaluated. APC hypermethylation was more frequent in older patients, but hypermethylation of other genes and markers in our panel was not age-related. APC hypermethylation did not cluster with methylation of the markers that defined the CIMP, as is evident in Fig. 2. Although age-related, APC hypermethylation was also tumor specific, occurring only rarely in colorectal mucosa, in contrast to other genes with age-related methylation that are frequently methylated in the mucosa as well (55). Although a recent study (56) suggested that APC hypermethylation had little effect on colorectal carcinogenesis, another study (44) reported that APC hypermethylation was discordant with methylation of other genes, as we found, and that the phenotype of tumors with APC hypermethylation was different from those with hypermethylation of other genes. These results emphasize the importance of identifying associations to clarify CIMP-positive and other subgroups of CRC that have hypermethylation of the type not involved in CIMP.
The influence of epigenetic alterations on genetic events has been the subject of speculation and hypotheses addressed at both tumor initiation and tumor progression, including the role of epigenetics in potentiation and modulation of the effects of genetic changes (13). Among the small number of published studies that have addressed directly the topic of epigenetic-genetic interactions (57), Esteller et al. reported an association between MGMT hypermethylation and G:C→A:T transitions in both KRAS and P53 (18, 19). Their studies led to the proposal that methylation-induced loss of MGMT protein expression, and therefore its intracellular function, influenced the mechanism and the type of subsequent mutations in these other genes involved in progression through failure of adduct repair that should have been accomplished by MGMT. Our study shows that hypermethylation of APC in concert with hypermethylation of either hMLH1 or MGMT is strongly associated with G-to-A transition in non-CpG sites of the APC gene, but not with C-to-T transition. To our knowledge, ours is the first report of the relationship in tumors of the epigenetic mechanism of concordant gene hypermethylation that includes APC to mutation type in that gene in the WNT signaling pathway. In addition, we found that APC hypermethylation was associated with P53 mutation, whereas CIMP was inversely related to P53 mutation, as in previous studies (35, 50, 58).
In contrast to the strong association of G-to-A mutation of APC with concurrent MGMT and APC hypermethylation, we found no relationship to loss of MGMT gene product expression. Other investigators also found no specific association with MGMT protein expression (22), although they found a weak relationship between reduced or absent MGMT expression by immunohistochemistry and G:C→A:T mutations in various genes. The methylation of MGMT may be more strongly associated with G-to-A transitions than is reduced protein expression because methylation indicates stable loss of expression, detects heterogeneous loss of expression, or indicates effects of expression level differences that are mechanistically important but not discernible by nonquantitative immunohistochemical methodology. In addition, G-to-A transitions among G:C→A:T mutations had an association with methylation in our study, whereas C-to-T transitions did not. In vitro studies of MGMT-deficient cells have shown that G-to-A transitions accounted for 94% of G:T→A:T transition hotspots after exposure to a methylating agent, and in T-lymphocytes of normal individuals, 72% of hotspots (16). Thus, G-to-A transition may be influenced preferentially by MGMT gene or MGMT protein alterations. We found in the small number of tumors with BRAF mutation in our study that MGMT protein expression was usually present in those with MSI-H but lost in those with MSS or MSI-L, raising the possibility of a role of MGMT in BRAF mutation occurrence or selection.
We found that MSI-H was inversely associated with P53 mutation, as in previous studies (47–50), and that hMLH1 methylation status and MSI-H status were strongly concordant (92.6% of our CRC, Table 1). Our sample size, however, was insufficient to distinguish among the possible effects on APC mutation of hypermethylation of multiple genes that include hMLH1 and the mutator phenotype/MSI-H that results from hMLH1 hypermethylation with loss of the gene product. We found a strong inverse relationship of P53 mutation to CIMP and an association of BRAF mutation with CIMP, MSI-H, and hypermethylation of hMLH1, as reported previously (35, 46, 53, 55, 58). There was no such relationship between CIMP and APC or KRAS mutation, also as reported previously (43, 44, 50, 59). Thus, the effect of CIMP status on gene mutation is quite different among genes, providing additional evidence that suggests specificity of some epigenetic-genetic interactions resulting in molecular subgroups of CRC.
In the current study, we found no association between MGMT hypermethylation and G-to-A or C-to-T mutations in KRAS and P53. Our results corroborate those of other investigators (21, 22), but contrast with the studies of Esteller et al. (18, 19) who first reported these associations. We also found a clear association of APC mutation type (i.e., G-to-A transition) with hypermethylation of APC concurrent with MGMT or hMLH1 hypermethylation. However, Esteller et al. (5) reported that APC promoter hypermethylation is biased toward CRC with genetically intact APC, whereas other investigators (21, 22) found no correlation between APC methylation status and APC mutations. The discordance in results of the reported studies may be explained by several factors. Technical issues such as the use of different assays could lead to different sensitivities in detecting the mutations, and target sequences for the detection of both mutations and methylation abnormalities differ among studies (51). Standardization of methylation markers and criteria for classification, as has been done for MSI (39), is not in place for hypermethylation studies. Alternatively, the discordance could reflect intertumoral differences in molecular mechanisms and clinical-pathologic characteristics, including stage of disease, in the patient populations studied. For example, correlation between dietary factors and specific mutations of KRAS (59–61) and APC (62) in CRC has been reported. Thus, differences in environmental factors among study populations may affect the methylation and mutation events that are found and reported.
The mechanisms responsible for the epigenetic-genetic associations we found remain to be determined. Alkylation of DNA at the O6 position of guanine is an important step in the formation of G-to-A mutations in cancer. Human colorectal DNA has long been known to contain O6-methylguanine and N7-methylguanine adducts that arise from exposure to methylating factors and agents (16, 63, 64). The sources of these exposures are unknown but can potentially be dietary/lifestyle and occupational characteristics, endogenous alkylating agents, and in situ formation typically mediated by the bacterial or chemical nitrosation of amines (65–67).
Although APC mutation is considered to be the initiating alteration in the development of many CRC, this alteration is uncommon in small sporadic adenomas (68, 69). On the other hand, hypermethylation of MGMT and p16 are present at relatively high frequency even in small adenomas (17, 26, 28) and in aberrant crypt foci that are the earliest precursor lesion (29), whereas hypermethylation of hMLH1 is not frequent in these early lesions (48, 70). One recent hypothesis linking epigenetics and genetic alterations is that colon cancer cells with APC gene inactivation are protected from methylation-induced cytotoxicity (71) so that lack of APC function due to epigenetic silencing may prevent cell death after exposure to DNA-methylating agents. As a consequence, mutagenic adducts including O6-methylguanine could accumulate in the DNA, followed by development of G-to-A mutations. Hypermethylation of APC with MGMT or hMLH1 thus may produce synergistic effects on this specific type of APC mutation and may explain the association we found. On the other hand, APC may be merely a marker gene for the secondary effects of global hypermethylation on occurrence of mutation, because 24% (6 of 25) of the G-to-A mutations we found were silent and therefore would not affect APC function or provide selective advantage to the tumor cells through changes in that specific gene. The remainder of the G-to-A transitions in APC produced amino acid substitutions rather than truncation of the gene product. Additional studies to include the temporal order of the early epigenetic and genetic events are needed to determine the specific mechanisms by which the interactions affect the neoplastic process in large bowel mucosa and the premalignant lesions in which CRC develop.
Acknowledgments
Grant support: The Kadoorie Charitable Foundation and Cancer Center Support Grant P30 CA16672 from the National Cancer Institute, NIH. S.R. Hamilton is the recipient of the Frederick F. Becker Distinguished University Chair in Cancer Research from The University of Texas.
References
- 1.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 2.Behrens J. The role of the Wnt signalling pathway in colorectal tumorigenesis. Biochem Soc Trans. 2005;33:672–5. doi: 10.1042/BST0330672. [DOI] [PubMed] [Google Scholar]
- 3.Jaiswal AS, Balusu R, Narayan S. Involvement of adenomatous polyposis coli in colorectal tumorigenesis. Front Biosci. 2005;10:1118–34. doi: 10.2741/1605. [DOI] [PubMed] [Google Scholar]
- 4.Nathke IS. The adenomatous polyposis coli protein: the Achilles heel of the gut epithelium. Annu Rev Cell Dev Biol. 2004;20:337–66. doi: 10.1146/annurev.cellbio.20.012103.094541. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M, Sparks A, Toyota M, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–71. [PubMed] [Google Scholar]
- 6.Arnold CN, Goel A, Niedzwiecki D, et al. APC promoter hypermethylation contributes to the loss of APC expression in colorectal cancers with allelic loss in 5q. Cancer Biol Ther. 2004;3:960–64. doi: 10.4161/cbt.3.10.1113. [DOI] [PubMed] [Google Scholar]
- 7.Powell SM, Zilz N, Beazer-Barclay Y, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–7. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 8.Smith KJ, Johnson KA, Bryan TM. The APC gene product in normal and tumor cells. Proc Natl Acad Sci U S A. 1993;90:2846–50. doi: 10.1073/pnas.90.7.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyoshi Y, Nagase H, Ando H, et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–33. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 10.Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10:721–33. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 11.Hussain SP, Harris CC. Molecular epidemiology of human cancer: contribution of mutation spectra studies of tumor suppressor genes. Cancer Res. 1998;58:4023–37. [PubMed] [Google Scholar]
- 12.Strauss BS. The origin of point mutations in human tumor cells. Cancer Res. 1992;52:249–53. [PubMed] [Google Scholar]
- 13.Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14:427–32. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell. 2003;4:121–31. doi: 10.1016/s1535-6108(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 15.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–45. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Tomita-Mithcell A, Ling LL, Glover CL, Goodluck-Griffith J, Thilly WG. The mutational spectrum of the HPRT gene from human T cells in vivo shares a significant concordant set of hot spots with MNNG-treated human cells. Cancer Res. 2003;63:5793–8. [PubMed] [Google Scholar]
- 17.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–7. [PubMed] [Google Scholar]
- 18.Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–71. [PubMed] [Google Scholar]
- 19.Esteller M, Risques RA, Toyota M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–92. [PubMed] [Google Scholar]
- 20.Coulondre C, Miller JH. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977;117:577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- 21.Lind GE, Thorstensen L, Lovig T, et al. A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol Cancer. 2004;3:1–11. doi: 10.1186/1476-4598-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halford S, Rowan A, Sawyer E, Talbot I, Tomlinson I. O6-methylguanine methyltransferase in colorectal cancers: detection of mutations, loss of expression, and weak association with G:C>A:T transitions. Gut. 2005;54:797–802. doi: 10.1136/gut.2004.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jascur T, Boland CR. Structure and function of the components of the human DNA mismatch repair system. Int J Cancer. 2006;119:2030–5. doi: 10.1002/ijc.22023. [DOI] [PubMed] [Google Scholar]
- 24.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–46. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 25.Grady WM, Markowitz S. Genomic instability and colorectal cancer. Curr Opin Gastroenterol. 2000;16:62–7. doi: 10.1097/00001574-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 26.Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–35. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petko Z, Ghiassi M, Shuber A, et al. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res. 2005;11:1203–9. [PubMed] [Google Scholar]
- 28.Woodson K, Weisenberger DJ, Campan M, et al. Gene-specific methylation and subsequent risk of colorectal adenomas among participants of the polyp prevention trial. Cancer Epidemiol Biomarkers Prev. 2005;14:1219–23. doi: 10.1158/1055-9965.EPI-04-0726. [DOI] [PubMed] [Google Scholar]
- 29.Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823–30. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene FL, Page DL, Fleming ID, et al., editors. AJCC cancer staging manual. 6. New York: Springer; 2002. pp. 113–9. [Google Scholar]
- 31.Isola J, DeVries S, Chu L, Ghazvini S, Waldman F. Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol. 1994;145:1301–8. [PMC free article] [PubMed] [Google Scholar]
- 32.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–15. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol. 2003;162:815–22. doi: 10.1016/S0002-9440(10)63878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venables WN, Ripley BD. Modern applied statistics with S-plus. 3. New York: Springer; 1999. [Google Scholar]
- 35.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–5. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yashima K, Nakamori S, Murakami Y, et al. Mutations of the adenomatous polyposis coli gene in the mutation cluster region: comparison of human pancreatic and colorectal cancers. Int J Cancer. 1994;59:43–7. doi: 10.1002/ijc.2910590110. [DOI] [PubMed] [Google Scholar]
- 37.Chan TL, Zhao W, Leung SY, Yuen ST. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63:4878–81. [PubMed] [Google Scholar]
- 38.Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62:6451–5. [PubMed] [Google Scholar]
- 39.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen L, Kondo Y, Rosner GL, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–8. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 41.Leung SY, Yuen ST, Chung LP, Chu KM, Chan AS, Ho JC. hMLH1 promoter methylation and lack of hMLH1 expression in sporadic gastric carcinomas with high-frequency microsatellite instability. Cancer Res. 1999;59:159–64. [PubMed] [Google Scholar]
- 42.Ruczinski I, Kooperberg C, LeBlanc M. Logic regression. J Comput Graph Stat. 2003;12:475–511. [Google Scholar]
- 43.Rashid A, Issa JP. CpG island methylation in gastroenterologic neoplasia: a maturing field. Gastroenterology. 2004;127:1578–88. doi: 10.1053/j.gastro.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Iacopetta B, Grieu F, Li W, et al. APC gene methylation is inversely correlated with features of the CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2006;119:2272–8. doi: 10.1002/ijc.22237. [DOI] [PubMed] [Google Scholar]
- 45.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 46.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 47.Elsaleh H, Powell B, McCaul K, et al. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin Cancer Res. 2001;7:1343–9. [PubMed] [Google Scholar]
- 48.Konishi M, Kikuchi-Yanoshita R, Tanaka K, et al. Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology. 1996;111:307–17. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 49.Cottu PH, Muzeau F, Estreicher A, et al. Inverse correlation between RER+ status and p53 mutation in colorectal cancer cell lines. Oncogene. 1996;13:2727–30. [PubMed] [Google Scholar]
- 50.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis indentifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–9. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterized by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–6. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beach R, Chan AO, Wu TT, et al. BRAF mutations in aberrant crypt foci and hyperplastic polyposis. Am J Pathol. 2005;166:1069–75. doi: 10.1016/S0002-9440(10)62327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slattery ML, Curtin K, Sweeney C, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120:656–63. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 54.Koinuma K, Shitoh K, Miyakura Y, et al. Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas. Int J Cancer. 2004;108:237–42. doi: 10.1002/ijc.11523. [DOI] [PubMed] [Google Scholar]
- 55.Shen L, Issa JP. Epigenetics in colorectal cancer. Curr Opin Gastroenterol. 2002;18:68–73. doi: 10.1097/00001574-200201000-00012. [DOI] [PubMed] [Google Scholar]
- 56.Xu XL, Yu J, Zhang HY, et al. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol. 2004;10:3441–54. doi: 10.3748/wjg.v10.i23.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong C, Moorefield KS, Jun P, et al. Epigenome scans and cancer genome sequencing converge on WNK2, and a kinase-independent suppressor of cell growth. Proc Natl Acad Sci US A. 2007;104:10974–9. doi: 10.1073/pnas.0700683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51:797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagasaka T, Sasamoto H, Notohara K, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–94. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- 60.Giovannucci E, Stampfer MJ, Colditz GA, et al. Folate, methionine, and alcohol intake and risk of colorectal adenoma. J Natl Cancer Inst. 1993;85:875–84. doi: 10.1093/jnci/85.11.875. [DOI] [PubMed] [Google Scholar]
- 61.Slattery ML, Curtin K, Anderson K, et al. Associations between dietary intake and Ki-ras mutations in colon tumors: a population-based study. Cancer Res. 2000;60:6935–41. [PubMed] [Google Scholar]
- 62.Luchtenborg M, Weijenberg MP, de Goeij AF, et al. Meat and fish consumption, APC gene mutations and hMLH1 expression in colon and rectal cancer: a prospective cohort study (The Netherlands) Cancer Causes Control. 2005;16:1041–54. doi: 10.1007/s10552-005-0239-0. [DOI] [PubMed] [Google Scholar]
- 63.Hall CN, Badawi AF, O’Connor PJ, Saffhill R. The detection of alkylation damage in the DNA of human gastrointestinal tissues. Br J Cancer. 1991;64:59–63. doi: 10.1038/bjc.1991.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harrison KL, Wood M, Lees NP, Hall CN, Margison GP, Povey AC. Development and application of a sensitive and rapid immunoassay for the quantitation of N7-methyldeoxyguanosine in DNA samples. Chem Res Toxicol. 2001;14:295–301. doi: 10.1021/tx000071b. [DOI] [PubMed] [Google Scholar]
- 65.Povey AC, Hall CN, Badawi AF, Cooper DP, O’Connor PJ. Elevated levels of the pro-carcinogenic adduct, O(6)-methylguanine, in normal DNA from the cancer prone regions of the large bowel. Gut. 2000;47:362–5. doi: 10.1136/gut.47.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bartsch H, Montesano R. Relevance of nitros-amines to human cancer. Carcinogenesis. 1984;5:1381–93. doi: 10.1093/carcin/5.11.1381. [DOI] [PubMed] [Google Scholar]
- 67.Hotchkiss JH. A review of current literature on N-nitroso compounds in foods. Adv Food Res. 1987;31:53–115. doi: 10.1016/s0065-2628(08)60166-4. [DOI] [PubMed] [Google Scholar]
- 68.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 69.Thorstensen L, Lind GE, Lovig T, et al. Genetic and epigenetic changes of components affecting the WNT pathway in colorectal carcinomas stratified by microsatellite instability. Neoplasia. 2005;7:99–108. doi: 10.1593/neo.04448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samowitz WS, Slattery ML. Microsatellite instability in colorectal adenomas. Gastroenterology. 1997;112:1515–9. doi: 10.1016/s0016-5085(97)70032-5. [DOI] [PubMed] [Google Scholar]
- 71.Narayan S, Jaiswal AS, Balusu R. Tumor suppressor APC blocks DNA polymerase β-dependent strand displacement synthesis during long patch but not short patch base excision repair and increases sensitivity to methylmethane sulfonate. J Biol Chem. 2005;280:6942–9. doi: 10.1074/jbc.M409200200. [DOI] [PubMed] [Google Scholar]