Abstract
Transcription-factor-directed reprogramming from somatic cells to induced pluripotent stem cells (iPSCs) is by nature an epigenetic process of cell fate change. Previous studies have demonstrated that this inefficient process can be facilitated by the inclusion of additional factors. To gain insight into the reprogramming mechanism, we aimed to identify epigenetic enzymes capable of promoting iPSC generation. Here we show that Kdm2b, a histone H3 Lys 36 dimethyl (H3K36me2)-specific demethylase, has the capacity to promote iPSC generation. This capacity depends on its demethylase and DNA-binding activities, but is largely independent of its role in antagonizing senescence. Kdm2b functions at the beginning of the reprogramming process and enhances activation of early responsive genes in reprogramming. Kdm2b contributes to gene activation by binding to and demethylating the gene promoters. Our studies not only identify an important epigenetic factor for iPSC generation, but also reveal the molecular mechanism underlying how Kdm2b contributes to reprogramming.
Direct reprogramming from somatic cells to a pluripotent state can be achieved by the introduction of defined transcription factors, such as Oct4, Sox2, Klf4 and c-Myc (ref. 1). The resultant iPSCs are molecularly and functionally similar to embryonic stem cells (ESCs) derived from the inner cell mass of a blastocyst2,3. However, the process of iPSC generation is highly inefficient with a low frequency and a long latency before the establishment of pluripotency2,3. A number of factors have been shown to affect reprogramming efficiency through cell-cycle-dependent or -independent mechanisms2,4. For example, inhibition of the p53–p21 pathway and the Ink4a/Arf locus increases the reprogramming efficiency and accelerates the reprogramming dynamics by affecting cell proliferation5–10; whereas, ectopic expression of Nanog enhances reprogramming presumably through an epigenetic mechanism without changing the proliferation status6.
A recent study indicates that reprogramming from fibroblasts to iPSCs involves a series of transcriptional changes11. At the beginning, epithelial genes that alter the morphology of fibroblasts to an ESC-like state are first activated, followed by the activation of Nanog and other pluripotent factors. After these waves of activation, mesenchymal genes are repressed, followed by the activation of ‘mature’ pluripotent genes11. Consistent with these sequential molecular events, factors facilitating mesenchymal-to-epithelial transition, such as BMPs, Tgf-β inhibitors and microRNA-200s, have been shown to promote iPSC generation11–14, indicating that activation of an epithelial transcription program early in reprogramming is crucial for the establishment of pluripotency. However, how such a program is activated is unknown at present.
Given that cell fate reprogramming is essentially a resetting of epigenetic states15, it is not surprising that chemical inhibitors of epigenetic enzymes16–19, and certain chromatin-remodelling factors, are capable of promoting iPSC generation20. Through studying epigenetic factors specifically enriched in ESCs, we found that Kdm2b (also known as Jhdm1b and Fbxl10), an H3K36me2-specific demethylase21,22, is able to facilitate iPSC generation. This property is independent of its effect on cell proliferation, but relies on its demethylase and DNA-binding activities that contribute to the activation of the reprogramming transcription program in the early stage.
RESULTS
Kdm2b promotes iPSC generation
To identify epigenetic factors that facilitate iPSC generation, we focused on the epigenetic factors enriched in ESCs. We found that isoform 1 (IF1) of Kdm2b (Supplementary Fig. S1) is highly expressed in ESCs (Fig. 1a) and progressively upregulated during reprogramming by Oct4, Sox2 and Klf4 (OSK; Supplementary Fig. S2a). To explore its potential role in iPSC generation, we constructed a doxycycline-inducible lentiviral plasmid expressing carboxy-terminal Flag-tagged Kdm2b-IF1 (hereafter referred to as Kdm2b). On doxycyclin induction, Kdm2b is expressed more than 50 times above the ESC level (Supplementary Fig. S2b). When introduced into Oct4–IRES–GFP/Rosa26-M2rtTA mouse embryonic fibroblasts (MEFs) together with the retroviral factors OSK, it can increase Oct4–GFP+ colony numbers by 4–6-fold (Fig. 1b). Furthermore, when c-Myc is included in the factor cocktail (OSKM), Kdm2b also increases Oct4–GFP+ colony numbers (Fig. 1c). Together, these data show that Kdm2b is capable of enhancing OSK- and OSKM-mediated iPSC generation.
Figure 1.
Kdm2b promotes iPSC generation. (a) RT–qPCR analysis of the expression levels of Kdm2a and different isoforms of Kdm2b (IF1, IF2 and IF3) in mouse ESCs and MEFs. Kdm2b-IF1 is highly expressed in mouse ESCs. The qPCR results were normalized to Gapdh and compared with the expression level in MEFs. (b) Kdm2b increases the efficiency of iPSC generation when co-introduced with OSK. The efficiency is represented by the number of Oct4–GFP+ colonies counted at post-transduction days 12 and 16 from 1 × 105 starting MEFs. n = 3. Error bars, s.e.m. (c) Kdm2b increases the reprogramming efficiency in the presence of c-Myc. Shown are numbers of Oct4–GFP+ colonies at day 12 reprogrammed by OSKM in the presence or absence of Kdm2b. n = 3. Error bars, s.e.m. (d) The reprogramming kinetics from day 6 to day 16 using different combinations of reprogramming factors. n = 3. Error bars, s.e.m. (e) RT–qPCR analysis of the knockdown efficiency of Kdm2b (Kdm2b-i). The results were normalized to the Gapdh level and are shown relative to the control shRNA treatment (Control-i). (f) Knockdown of Kdm2b reduced OSK-mediated reprogramming efficiency. Oct4–GFP+ colony numbers at days 14 and 18 are shown for OSK reprogramming with control or Kdm2b knockdown. Data in a, e and f represent the mean of two independent experiments.
By following the reprogramming kinetics, we found that although Kdm2b promotes iPSC generation, it does not significantly shorten the latency time, as the first Oct4–GFP+ colony appears around day 10 (OSK) or day 8 (OSKM) of reprogramming regardless of the presence of Kdm2b (Fig. 1d). This observation differs from the documented cell-cycle-based enhancement of iPSC generation, including depletion of p53 and p21 (refs 6,8,9) and inhibition of Ink4a/Arf (refs 5,7), which significantly shorten the latency time until the first appearance of iPSCs, but it is similar to the cell-cycle-independent enhancement by Nanog overexpression6.
To further confirm the role of Kdm2b in reprogramming, we depleted Kdm2b by small hairpin RNA (shRNA; ref. 21; Fig. 1e) and induced reprogramming with OSK using the Kdm2b-depleted cells. Kdm2b depletion greatly reduces the reprogramming efficiency (Fig. 1f), indicating that Kdm2b is required for optimal induction of iPSCs. Collectively, these studies demonstrate that Kdm2b facilitates iPSC generation.
iPSCs generated in the presence of Kdm2b are pluripotent
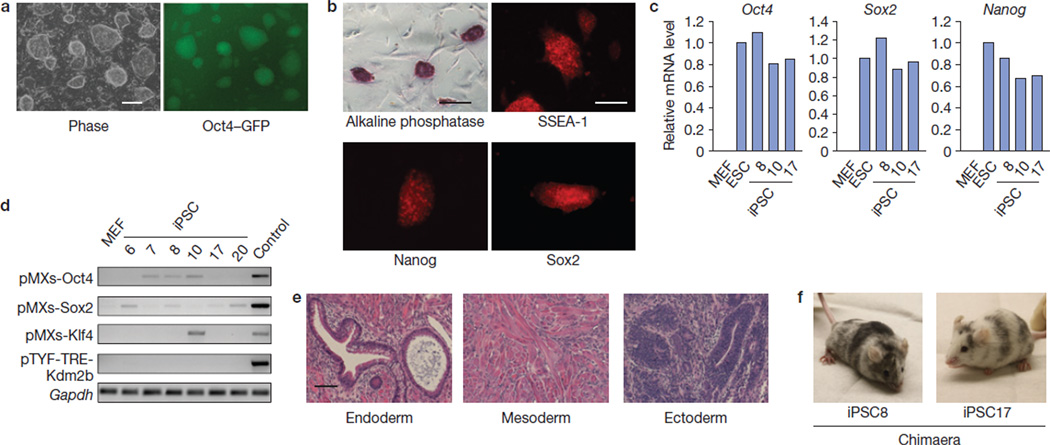
Next, we set out to characterize the iPSCs generated with OSK in the presence of exogenous Kdm2b. After 16–18 days of induction, Oct4–GFP+ colonies were picked up and propagated in the absence of doxycyclin. The resulting cell lines exhibit typical ESC morphology with Oct4–GFP expression (Fig. 2a), show alkaline phosphatase activity and express ESC markers SSEA-1 and pluripotent transcription factors Nanog and Sox2 (Fig. 2b). The endogenous loci of Oct4, Sox2 and Nanog in the iPSCs are activated to a level similar to that in ESCs (Fig. 2c); whereas the transgenes are mostly silenced (Fig. 2d). When implanted into immunodeficient mice, these iPSCs generate teratomas with tissues belonging to three germ layers (Fig. 2e). Importantly, they are competent for chimaera generation when injected into blastocysts (Fig. 2f). These data support that the iPSCs generated in the presence of exogenous Kdm2b are pluripotent.
Figure 2.
iPSCs generated by enforced expression of OSK plus Kdm2b are pluripotent. (a) The morphology and Oct4–GFP expression of a representative iPSC line derived from Kdm2b-assisted reprogramming. Scale bar, 200 µm. (b) Alkaline phosphatase activity and immunostaining of ESC markers (SSEA-1, Nanog and Sox2) of a selected iPSC line. Scale bar, 200 µm. (c) Activation of endogenous Oct4, Sox2 and Nanog in iPSCs derived from reprogramming by OSK plus Kdm2b. RT–qPCRs were carried out on RNA samples collected from MEFs, ESCs and selected iPSC lines (8, 10 and 17). (d) Silencing of retroviral transgenes pMXs-Oct4, -Sox2 and -Klf4 and lentiviral-inducible pTYF-TRE-Kdm2b in the absence of doxycyclin in selected iPSC lines (6, 7, 8, 10, 17 and 20). The RNA sample from doxycyclin-treated Oct4–IRES–GFP/Rosa26-M2rtTA MEFs introduced with the four transgenes serves as a positive control (Control). (e) Representative micrographs showing the presence of gut-like cavities (endoderm), muscle tissues (mesoderm) and neural rosettes (ectoderm) in teratoma derived from iPSCs reprogrammed by OSK plus Kdm2b. Scale bar, 100 µm. (f) iPSCs generated in the presence of Kdm2b are competent in chimaeric mice generation. Shown are chimaeric mice derived from two representative iPSC lines (8 and 17).
Kdm2b facilitates iPSC generation in an enzymatic-activity-dependent and a cell-proliferation-independent manner
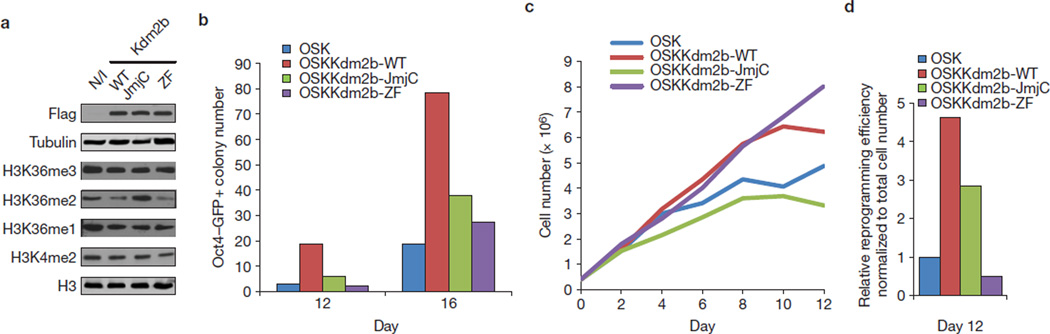
To understand how Kdm2b facilitates iPSC generation, we attempted to determine the domains of Kdm2b important for this property. To this end, we carried out reprogramming with point mutations in the catalytic JmjC domain21,22 and the DNA-binding CXXC-type zinc finger (ZF) domain23,24. We first confirmed that both the mutants and wild-type (WT) Kdm2b are expressed at similar levels on induction (Fig. 3a). Consistent with the enzymatic activity of Kdm2b, overexpression of the WT Kdm2b and the ZF mutant, but not the catalytic mutant, leads to a specific decrease of the H3K36me2 level (Fig. 3a). Importantly, mutations in either JmjC or ZF abrogate the capacity of Kdm2b in promoting iPSC generation (Fig. 3b), indicating that both the demethylase and DNA-binding activities of Kdm2b are essential for reprogramming enhancement.
Figure 3.
Kdm2b facilitates iPSC generation in a JmjC- and ZF-domain-dependent manner. (a) Western blot analysis demonstrates equal expression of wild-type and mutant Kdm2b with differential effects on global H3K36me2 levels. Lentiviruses carrying doxycyclin-inducible wild-type and mutant Kdm2b were respectively transduced into MEFs that constitutively express the transactivator rtTA. Expression of the constructs was induced by the addition of doxycyclin (1 µg ml−1). Western blotting shows the equal expression levels of the various Kdm2b forms as well as their effects on histone modifications. Tubulin and histone H3 serve as loading controls. Full scans of blots are shown in Supplementary Fig. S6a. (b) Mutations on the JmjC or ZF domain of Kdm2b abrogate its effect on iPSC generation. Shown are Oct4–GFP+ colony numbers counted at days 12 and 16 reprogrammed using OSK, OSK plus wild-type Kdm2b and the JmjC and ZF mutants. (c) The effect of Kdm2b on proliferation is dependent on its enzymatic activity, but independent of its ZF domain. Cell numbers were monitored for 12 days of reprogramming by OSK, and OSK plus WT and mutant Kdm2b. (d) Relative reprogramming efficiency independent of the cell proliferation effect. The Oct4–GFP+ colony numbers in b were divided by the total cell numbers in panel c, and the results were all compared with OSK reprogramming and are shown as the relative reprogramming efficiency. Data in b–d represent the mean of two independent experiments.
Kdm2b is able to promote cell proliferation by repressing senescence21,25,26. To determine whether the capacity of Kdm2b in enhancing reprogramming is mediated by promoting cell proliferation, we examined whether ectopic expression of Kdm2b affects cell proliferation in the context of OSK-induced reprogramming. We found that although the ZF domain of Kdm2b is critical for promoting iPSC generation (Fig. 3b), it is not required for promoting cell proliferation (Fig. 3c), indicating that the ability of Kdm2b to enhance proliferation is not sufficient for promoting iPSC generation. To separate its proliferation effect from the reprogramming effect, we normalized the reprogramming efficiency by dividing the Oct4–GFP+ colony numbers with the total cell numbers in the reprogramming populations. We found that exogenous WT Kdm2b increases the reprogramming efficiency by more than fourfold after normalization (Fig. 3d). These analyses not only confirm that the demethylase activity and DNA-binding capacity of Kdm2b are necessary for enhancing iPSC generation, but also indicate that cell proliferation stimulated by Kdm2b is not a major contributing factor for its role in promoting reprogramming.
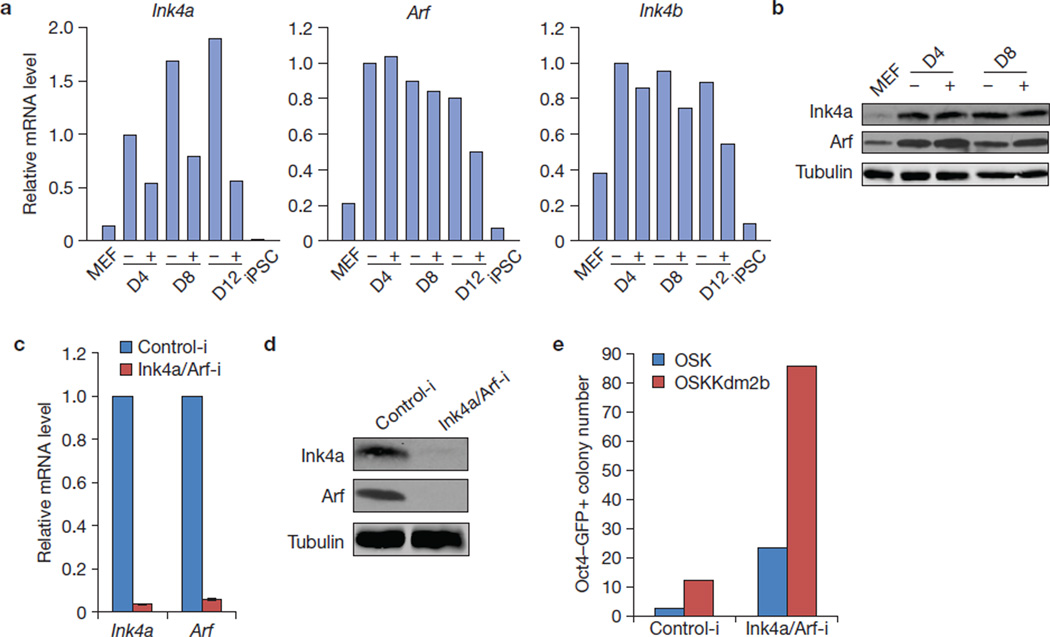
To further ascertain that Kdm2b-mediated suppression of senescence is not a major contributor to its role in reprogramming enhancement, we examined the transcript levels of Ink4a, Arf and Ink4b during reprogramming involving exogenous Kdm2b. We found that Ink4a, but not Arf or Ink4b, is significantly downregulated by Kdm2b in the first 12 days of OSK reprogramming (Fig. 4a). However, the protein level of Ink4a is less affected by Kdm2b (Fig. 4b), probably owing to the stability of this protein. To examine whether repression of this locus contributes to the effect of Kdm2b on reprogramming, we introduced shRNA that depletes Ink4a and Arf (Fig. 4c,d) to the reprogramming cells transduced with OSK and OSK plus Kdm2b. We found that Kdm2b was still able to increase the Oct4–GFP+ colony number following the depletion of Ink4a/Arf (Fig. 4e), indicating that the effect of Kdm2b on reprogramming is largely independent of its role in downregulating Ink4a/Arf. Consistent with previous findings that senescence suppression promotes reprogramming5,7,10, we also observed a great enhancement of reprogramming efficiency when Ink4a/Arf are knocked down regardless of whether Kdm2b is introduced (Fig. 4e). Collectively, our data indicate that the ability of Kdm2b to suppress cellular senescence is not the main reason for its role in enhancing reprogramming.
Figure 4.
Kdm2b promotes iPSC generation independently of Ink4a/Arf. (a) RT–qPCR analysis of the expression of Ink4a, Arf and Ink4b in Kdm2b-facilitated reprogramming. RNA samples were collected from MEFs, iPSCs and cells during reprogramming by OSK (−) and OSK plus Kdm2b (+) at days 4 (D4), 8 (D8) and 12 (D12). All of the qPCR results were normalized to the expression of Gapdh and compared with the sample in the absence of Kdm2b (−) at D4. Data represent the mean of two experiments. (b) Western blot analysis shows the protein level of Ink4a and Arf in Kdm2b-facilitated reprogramming at D4 and D8. Protein extracts from the reprogramming cell samples in a, as well as MEFs, were used. Tubulin serves as a loading control. (c) RT–qPCR analysis demonstrates depletion of Ink4a and Arf transcript by introduction of shRNA against Ink4a/Arf. n = 3. (d) Western blot analysis demonstrates efficient depletion of Ink4a and Arf by shRNA against Ink4a/Arf. (e) Kdm2b retains its capacity to enhance reprogramming following Ink4a and Arf depletion. Oct4–GFP+ colony number at day 12 for OSK or OSK plus Kdm2b reprogramming with or without Ink4a/Arf knockdown. Data represent the mean of two experiments. Full scans of blots are shown in Supplementary Fig. S6b. Data in a and e represent the mean of two independent experiments.
Kdm2b functions early during reprogramming
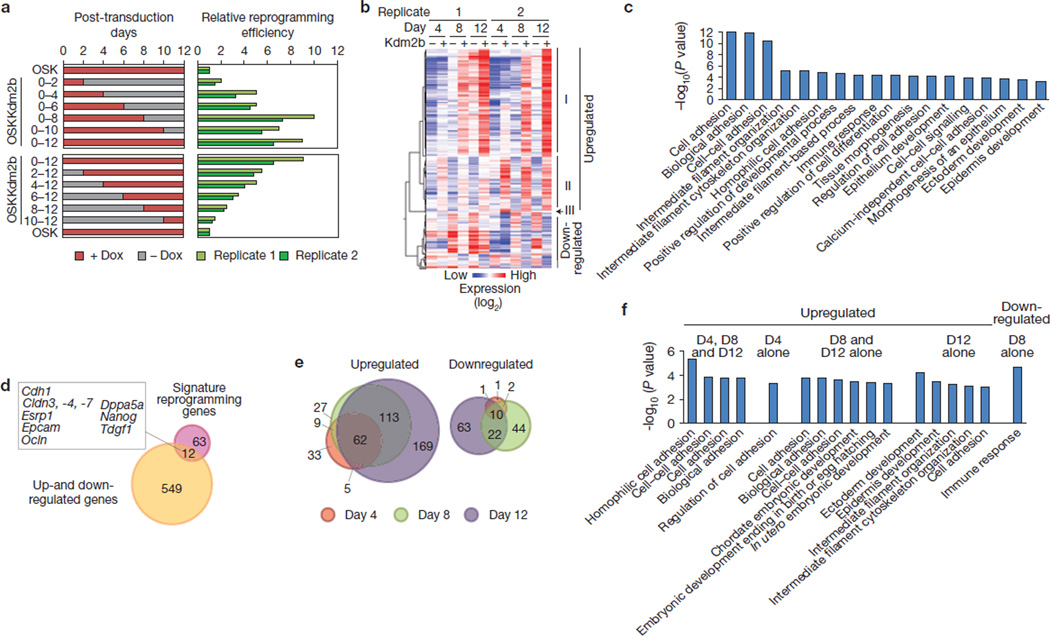
Given that the role of Kdm2b in suppressing senescence and/or promoting cell proliferation is not a main contributing factor, and that introduction of Kdm2b does not enhance expression of transduced reprogramming factors (Supplementary Fig. S3), we searched for alternative mechanisms. As reprogramming can be divided into stages with distinct molecular features4,11,27,28 and factors that promote iPSC generation can function at different stages during reprogramming14,29,30, we sought to determine the time window in which Kdm2b enhances iPSC generation. To this end, we induced Kdm2b expression for different durations and found that enforced expression of Kdm2b in the first 8 days constantly enhanced the reprogramming efficiency (Fig. 5a, upper panels). As a complement, we also induced Kdm2b expression beginning at different days after the initiation of reprogramming and found that the increase in reprogramming efficiency is largely proportional to the length of the doxycyclin treatment (Fig. 5a, lower panels). Together, these studies indicate that Kdm2b promotes iPSC generation by functioning early during the reprogramming process.
Figure 5.
Kdm2b exerts its effect from the beginning of the reprogramming process and facilitates gene activation early in reprogramming. (a) Kdm2b functions to promote iPSC generation from the beginning of the reprogramming process. Oct4–IRES–GFP/Rosa26-rtTAM2 MEFs were transduced with retroviral OSK and lentiviral Kdm2b driven by a doxycyclin-inducible promoter. After transduction, doxycyclin (Dox) was applied to (red bars, left panels) or omitted from (grey bars) the cell cultures during the initial 12 days of reprogramming. Numbers on the side of the left panel indicate the durations of doxycyclin treatments. Oct4–GFP+ cells were counted at day 12 and reprogramming efficiencies (green bars, right panels) are compared with that reprogrammed by OSK, which is set to 1. Two independent replicates are shown. (b) Hierarchical clustering of genes whose expression are significantly affected by the addition of Kdm2b into the reprogramming cocktail. The expression changes of the selected probes revealed by microarray analysis are at least twofold, and the changes occur in at least one of the three time points (days 4, 8 and 12). The heat map was derived using the Pearson correlation coefficient from data of two biological replicates. Genes upregulated or downregulated by Kdm2b constitute two distinct clusters, with the upregulated genes subdivided into three groups (I, II and III) on the basis of their time of activation. (c) Gene ontology analysis for all of the genes identified in b. All of the presented terms are above the P value of 1 × 10−3. For P values, see Supplementary Table S2. (d) Venn diagram showing the common genes of Kdm2b-affected genes (a) and the previously identified signature reprogramming genes. Ten genes, which are represented by 12 probes in the microarray, are shown in the rectangle. (e) Area-proportional Venn diagram showing the distribution of Kdm2b-affected probes at different times of reprogramming. (f) Gene ontology analysis for biological processes for different sections in e. The sections without any term enriched above the P value of 1 × 10−3 are omitted. For P values, see Supplementary Table S3.
Kdm2b amplifies early gene activation during reprogramming
To understand the effect of Kdm2b during reprogramming at the molecular level, we performed microarray analyses on cells collected at days 4, 8 and 12 during the OSK reprogramming in the presence or absence of Kdm2b. These analyses revealed that 418 probes are upregulated at least twofold in at least one of the three time points, and 143 probes show at least twofold downregulation (Supplementary Table S1). Hierarchical analysis revealed that the Kdm2b-upregulated genes can be grouped into three distinct clusters (I, II and III) on the basis of their activation timings (Fig. 5b), whereas Kdm2b-downregulated genes are distributed more ambiguously in terms of affected timings.
Gene ontology analysis revealed that Kdm2b-affected genes are exceptionally enriched for adhesion processes (P value = 10−10 ~ 10−12). Other terms with a significant P value (<10−3) include those related to cell morphology, development and epithelium-related processes (Fig. 5c and Supplementary Table S2). By comparing Kdm2b-affected genes (Fig. 5b) with a list of ‘signature’ reprogramming genes that are dynamically regulated in reprogramming11, we found that the former include early-activated epithelial genes (Cdh1, Cldn3, −4 and −7, Epcam, Esrp1 and Ocln) and pluripotent genes (Nanog, Dppa5a and Tdgf1; Fig. 5d). Indeed, all of the previously identified early-activated genes11 have enhanced expression in the presence of Kdm2b, whereas the expression of late-activated genes or repressed genes is not significantly altered (Supplementary Fig. S4). Combined with the fact that Kdm2b functions from the beginning of reprogramming (Fig. 5a), Kdm2b seems to facilitate activation of early responsive genes during reprogramming.
To further dissect the expression changes, we grouped the Kdm2b-affected genes on the basis of the timings of their expression changes (Supplementary Table S1). We found that most of the day 4 or day 8 affected genes are also affected at later time points, whereas a substantial portion of day 8 and/or day 12 affected genes are not affected at an earlier time point (Fig. 5e). Such a distribution pattern prompts us to reason that the introduction of Kdm2b probably amplifies a reprogramming transcriptional cascade. Moreover, gene ontology analyses revealed that genes upregulated from day 4 through 12 are enriched in adhesion molecules, whereas those exclusively upregulated at one or two time points seem to undergo a functional transition from cell adhesion to development processes (Fig. 5f and Supplementary Table S3). This observation indicates that a developmental-related transcription program is triggered sometime after an initial adhesion-related program, and these two programs may constitute a transcription cascade. Given that the developmental program seems to start at day 8 (Fig. 5f) and Kdm2b exerts its effect from the beginning of reprogramming (Fig. 5a), it is likely that Kdm2b directly contributes to the earlier adhesion-related program, whereas its effect on the later developmental program is the result of an amplified cascade.
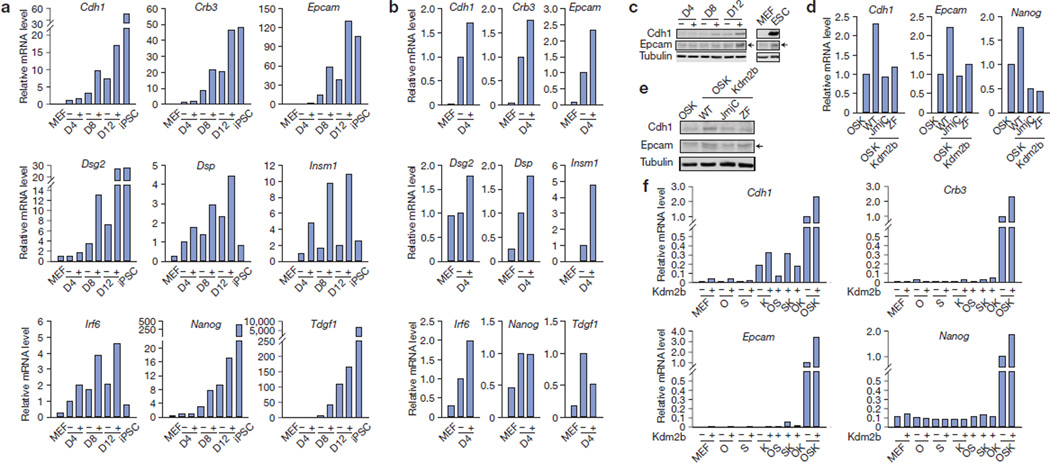
Quantitative PCR with reverse transcription (RT–qPCR) analysis confirmed the upregulation of genes encoding epithelial markers, such as Cdh1 (also known as E-cadherin), Crb3 and Epcam, as well as desmosomal components Dsg2 and Dsp (Fig. 6a–c). Enhanced expression of these genes starts at day 4 (Fig. 6b), and is affected more obviously at days 8 and 12 (Fig. 6a,c). We also note that genes activated earliest at day 4 include those encoding transcription factors implicated in cell adhesion and development, including Irf6, which is important for cleft palate development31,32, and Insm1, which is important for pancreatic and neuronal development33–35. Interestingly, these genes are upregulated by OSK and introduction of Kdm2b further augments their activations (Fig. 6a,b). The potential function of these early responsive genes in reprogramming remains to be analysed. Furthermore, the expression levels of pluripotent genes, such as Nanog and Tdgf1, are also enhanced by Kdm2b at days 8 and 12, but not at day 4 (Fig. 6a,b), indicating that their activation might be an indirect effect of Kdm2b.
Figure 6.
Kdm2b activates early responsive genes in reprogramming. (a) RT–qPCR analysis verifying genes upregulated at different time points in Kdm2b-facilitated reprogramming. RNA samples were extracted from MEFs, iPSCs and cells at day 4 (D4), 8 (D8) and 12 (D12) during reprogramming by OSK (−) and OSK plus Kdm2b (+). The qPCR results were normalized to Gapdh and compared with the samples in the absence of Kdm2b (−) at D4. (b) Epithelial gene expression amplified by Kdm2b at the earliest time point. Expression data from a were rescaled to manifest the enhancement by introducing Kdm2b into the OSK cocktail (−) at day 4 (D4). The expression of OSK reprogramming (+) is set to 1. (c) Western blot analysis demonstrates upregulation of Cdh1 and Epcam by Kdm2b. Protein extracts from the reprogramming cell samples in a, as well as MEFs and ESCs, were used. Tubulin serves as a loading control. (d) RT–qPCR analysis demonstrates that Kdm2b-enhanced activation of Cdh1, Epcam and Nanog depends on its JmjC and ZF domains. The analysis is performed at day 12 of reprogramming with cells reprogrammed by OSK, or OSK plus wild-type Kdm2b (WT), its JmjC or ZF mutants. All of the qPCR results were normalized to Gapdh and compared with the OSK reprogramming. (e) Western blot analysis of Cdh1, Epcam and tubulin with protein extract from the reprogramming cell samples in c. (f) Kdm2b cooperates with Oct4, Sox2 and/or Klf4 in transcription activation. RT–qPCR analysis of the expression of Cdh1, Crb3, Epcam and Nanog in MEFs transduced with individual factors of Kdm2b, Oct4 (O), Sox2 (S) and Klf4 (K), as well as different factor combinations. RNA samples were collected at post-transduction day 12. The qPCR results were first normalized to Gapdh and then compared with the sample transduced by OSK. Data in a, b, d and f represent the mean of two independent experiments. Full scans of blots are shown in Supplementary Fig. S6c.
We also examined the expression of mesenchymal-specific transcription factors, which are shown to be downregulated following the activation of Nanog and other pluripotent genes11. We observed a mild downregulation of Snai1, Snai2, Zeb1 and Zeb2 at days 8 and/or day 12 (Supplementary Fig. S5a), although their downregulation is less than twofold and consequently not picked up by the microarray analyses (Fig. 5d). Finally, we did not observe activation of endogenous Oct4 and Sox2 during the first 12 days of reprogramming (Supplementary Fig. S5b), consistent with the fact that even in the case of Kdm2b-assisted reprogramming, only a very small fraction of the starting cells gained pluripotency in the initial 12 days of reprogramming (Fig. 1b). Collectively, the data presented above indicate that Kdm2b facilitates activation of genes related to epithelial adhesion, which may in turn activate downstream genes including pluripotent genes. Thus, it seems sensible that Kdm2b promotes iPSC generation by facilitating the initiation of a putative transcription cascade.
As mutations in the JmjC domain and the ZF domain abrogate the capacity of Kdm2b in promoting iPSC generation, we investigated whether these mutations affect the capacity of Kdm2b to augment the gene activation in reprogramming. RT–qPCR and western blot analysis demonstrate that both mutations abrogate the ability of Kdm2b to activate Cdh1 and Epcam (Fig. 6d,e), as well as the subsequent activation of Nanog (Fig. 6d), consistent with the notion that Kdm2b enhances iPSC generation by facilitating early gene activation.
Kdm2b acts in concert with the key reprogramming factors in early gene activation
To understand how Kdm2b facilitates early gene activation in reprogramming, we investigated whether Kdm2b activates these genes alone or in concert with the key reprogramming factors. To this end, we introduced individual factors and different factor combinations into MEFs and examined their effects on the expression of early responsive epithelial genes, such as Cdh1, Crb3 and Epcam, as well as the later activated Nanog. We found that, overall when any of the OSK factors is omitted, activation of these genes is either greatly compromised or completely abolished despite the presence of Kdm2b (Fig. 6f). This observation is consistent with the fact that Kdm2b cannot replace any of the OSK in iPSC generation (data not shown). However, we note that Kdm2b alone does exhibit an about fivefold activation on the Cdh1 gene when compared with non-transduced MEF cells. Nevertheless, this activation seems to be minor (~5%) when compared with the activation by OSK (Fig. 6f). Consistent with a previous report13, transduction of Klf4 alone can partially activate Cdh1 when compared with OSK transduction. Its activation can be further boosted when Kdm2b is combined with Klf4 (Fig. 6f). For Crb3, Epcam and Nanog, Kdm2b barely activates them in the absence of any of the OSK (Fig. 6f). On the basis of these results, we conclude that Kdm2b acts in concert with the key reprogramming factors OSK to upregulate early responsive genes.
Kdm2b binds to and regulates the H3K36me2 level at the promoter of early-activated genes
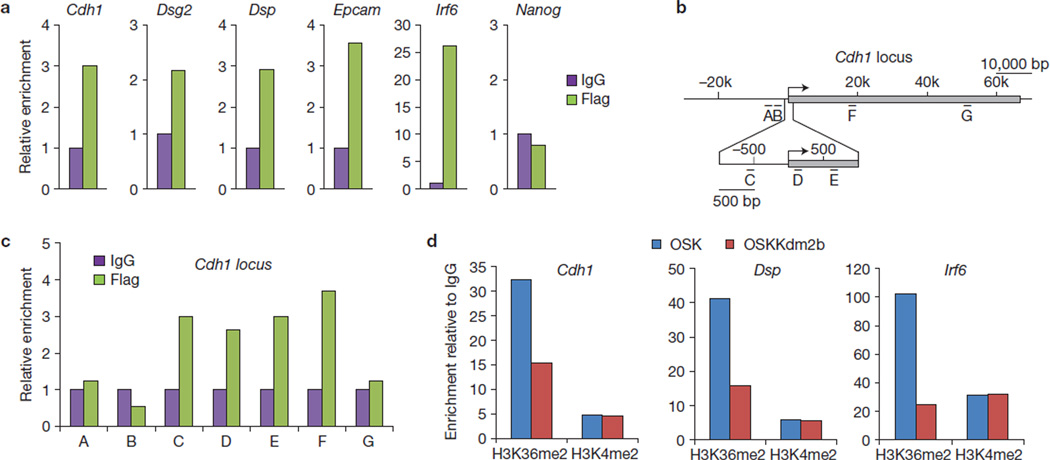
To determine whether Kdm2b directly contributes to the activation of the early responsive genes, we investigated whether Kdm2b binds to these genes. Taking advantage of the Flag epitope on the Kdm2b constructs, we performed chromatin immunoprecipitation (ChIP) analysis using cells at day 4 of reprogramming. This analysis demonstrates that Flag–Kdm2b is enriched in the promoter of the early-activated genes, including Cdh1, Epcam, Dsg2, Dsp and Irf6; however, no enrichment is detected at the Nanog promoter, consistent with the notion that Kdm2b contributes to Nanog activation indirectly (Fig. 7a). ChIP analysis also revealed the presence of exogenous Kdm2b at the promoter and part of the gene body of Cdh1 (Fig. 7b,c). Parallel ChIP experiments demonstrate that the level of H3K36me2 at the promoter of Cdh1, Dsp and Irf6 is decreased on the introduction of Kdm2b (Fig. 7d), consistent with the fact that Kdm2b preferentially removes H3K36me2 (ref. 21). These results support the notion that Kdm2b contributes to the activation of early responsive genes by binding to and demethylating H3K36me2 on their promoters.
Figure 7.
Kdm2b localizes on and demethylates the promoter region of early-activated genes. (a) ChIP analysis demonstrates that Kdm2b directly binds to the promoters of early-activated genes. ChIP analyses using anti-Flag antibody and IgG were performed, with cells collected at day 4 of reprogramming using OSK plus Flag-tagged Kdm2b. The qPCR results were compared with IgG controls. (b) Diagram of the Cdh1 locus illustrating the locations of amplicons (A–G) relative to the transcription start site (arrow) and the transcribed sequence (grey). (c) Kdm2b localizes from the promoter region to the middle of the gene body at the Cdh1 locus. ChIP was prepared and the results shown as a. Locations of amplicons A–G are shown in b. (d) ChIP analysis demonstrates that reduction of H3K36me2 level correlates with Kdm2b binding to the early-activated genes. ChIP analyses using anti-H3K36me2 and H3K4me2 antibody were performed with OSK or OSKKdm2b-introduced cells collected at day 4. The results for H3K36me2 and H3K4me2 were normalized to IgG. H3K4me2, a modification irrelevant to the enzymatic activity of Kdm2b, serves as a control. Data in a, c, d represent the mean of two independent experiments.
Enhanced early gene activation mediates the effect of Kdm2b in reprogramming
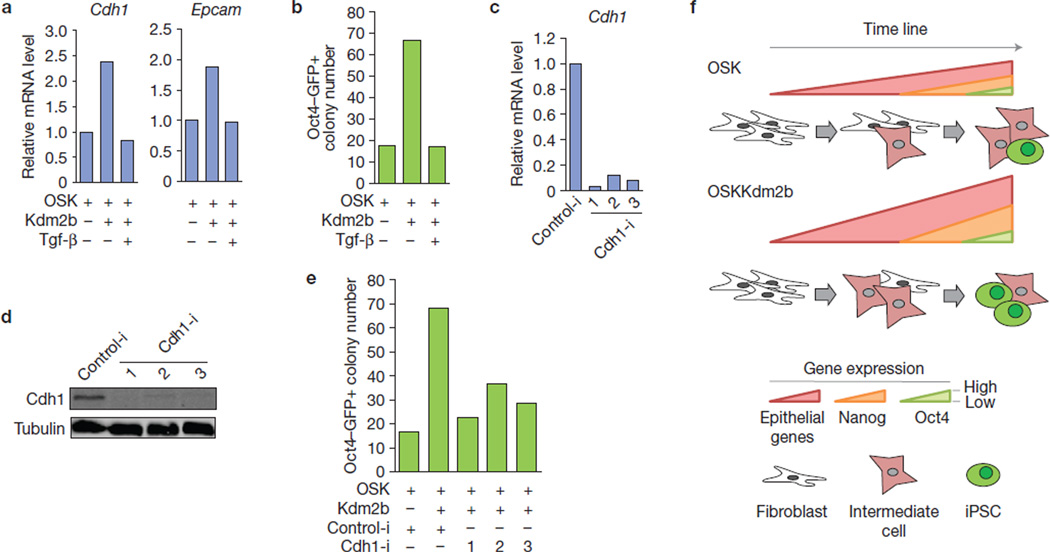
Epithelial genes, which are activated early11,13 and whose activation is enhanced by Kdm2b during reprogramming, are subjected to repression by Tgf-β signalling36. To demonstrate that increased expression of these genes contributes to Kdm2b-enhanced reprogramming, we performed reprogramming using OSK plus Kdm2b in the presence of Tgf-β. We found that, Kdm2b-mediated upregulation of epithelial genes, such as Cdh1 and Epcam, is abrogated on Tgf-β treatment (Fig. 8a). Importantly, Kdm2b-enhanced iPSC generation is also abrogated by Tgf-β (Fig. 8b), consistent with the notion that Kdm2b-enhanced expression of the epithelial genes contributes to its effect on reprogramming. To directly address the role of these epithelial genes in mediating Kdm2b-enhanced reprogramming, we focused on Cdh1, one of the epithelial genes important for reprogramming13,37,38 and directly regulated by Kdm2b. We found that depletion of Cdh1 by shRNAs (Fig. 8c,d) greatly compromised the capacity of Kdm2b to enhance OSK reprogramming (Fig. 8e), indicating that Cdh1 is one of the downstream targets that mediate the Kdm2b reprogramming effect.
Figure 8.
Inhibition of the expression of early-activated genes compromises the capacity of Kdm2b to enhance iPSC generation. (a) Upregulation of early-activated genes in reprogramming by Kdm2b is abrogated by administration of Tgf-β. RNA samples were collected from reprogramming cells with indicated treatments at day 12. (b) The capacity of Kdm2b to increase iPSC generation efficiency is negated by Tgf-β. Oct4–GFP+ colony numbers were calculated at day 16 of reprogramming. (c,d) RT–qPCR (c) and western blotting (d) confirmation of Cdh1 depletion. RNA and protein extracts were collected from ESCs transduced with three individual shRNAs against Cdh1 (Cdh1-i, 1–3) or control shRNA (Control-i). Full scans of blots are shown in Supplementary Fig. S6d. (e) Inhibition of Cdh1 by shRNA compromises Kdm2b-mediated enhancement of iPSC generation. Shown are Oct4–GFP+ colony numbers at day 16 of reprogramming. (f) Transcription cascade model explaining how Kdm2b enhances iPSC generation. Introduction of Kdm2b into the reprogramming cocktail leads to upregulation of epithelial genes that are activated early during reprogramming, which in turn causes an amplified transcription cascade that enhances the activation of Nanog and the generation of iPSCs. Data in a, b and e represent the mean of two independent experiments.
DISCUSSION
Kdm2b promotes reprogramming independently of its effect on cell proliferation
Despite the fact that cellular senescence is a ‘roadblock’ for reprogramming5,7 and that Kdm2b is capable of suppressing senescence21,25, we demonstrate here that Kdm2b promotes iPSC generation largely independently of its role in senescence and/or proliferation. First, although Kdm2b increases cell proliferation during reprogramming (Fig. 3c), when the proliferation effect is subtracted, Kdm2b still exhibits a more than fourfold increase in reprogramming efficiency (Fig. 3d). Second, although an intact ZF domain is required for Kdm2b to promote iPSC generation, mutation on this domain does not alter its ability to stimulate cell proliferation (Fig. 3b–d), indicating that the roles of Kdm2b in promoting proliferation and iPSC generation are independent and separable. Third, when Ink4a/Arf, the key senescence regulators and documented targets of Kdm2b (ref. 25), are depleted, Kdm2b is still capable of promoting iPSC generation (Fig. 4e), indicating that antagonizing senescence is not a major contributor for Kdm2b-mediated reprogramming enhancement. Last, Kdm2b does not significantly shorten the latency time from induction until the first appearance of iPSCs (Fig. 1d), which is consistent with the reprogramming kinetics of cell-cycle-independent enhancement of reprogramming6.
While our manuscript was under review, a report was published demonstrating that Kdm2b greatly enhances iPSC generation in the presence of vitamin C and indicating that the effect of Kdm2b may be mediated through promoting cell cycle progression and activation of microRNA cluster 302/367 (ref. 39). However, as most of the experiments in this report were performed in the presence of both vitamin C and Kdm2b, the specific contribution of Kdm2b to iPSC generation is not addressed. Indeed, the same group previously found that vitamin C is able to alleviate the blockage of cell cycle progression imposed by p53–p21 (ref. 40). As vitamin C has a more pronounced effect on iPSC generation than Kdm2b (ref. 39), it is possible that the effect of vitamin C on cell cycle progression might mask the effect of Kdm2b and consequently the contribution of Kdm2b to iPSC generation might be overshadowed.
A transcription cascade model for gene regulation in reprogramming
By analysing the gene expression changes within or immediately after the functioning time window of Kdm2b, we found that exogenous Kdm2b enhances the expression of a set of early-activated genes during reprogramming (Fig. 5d and Supplementary Fig. S4). The enhanced expression of epithelial genes (Cdh1, Epcam and so on) and other uncharacterized genes (Irf6 and Insm1) takes place in the functioning time window of Kdm2b (Figs 5a and 6a,b). Following this first wave of activation, the expression of Nanog and other pluripotency factors is upregulated (Fig. 6a,b) concomitant with enrichment of developmental genes in the upregulated gene group (Fig. 5f). Meanwhile, some mesenchymal genes, whose downregulation follows Nanog activation11, begin to exhibit magnified downregulation in the presence of Kdm2b (Supplementary Fig. S5a). Soon after Nanog activation (day 8; Fig. 6a), the first Oct4–GFP+ colonies are observed at day 10 (Fig. 1a). The observation that Kdm2b amplifies these sequential transcription events prompted us to propose a transcription cascade model to explain how Kdm2b might contribute to reprogramming (Fig. 8e). We propose that Kdm2b facilitates initial gene activation that occurs on the epithelial genes, causing an amplified transcription cascade, which in turn enhances the activation of pluripotent genes such as Nanog, eventually resulting in an increase in reprogramming efficiency (Fig. 8e).
This model is consistent with previous observations that Tgf-β inhibitors induce Nanog expression and enhance reprogramming12,14, as Tgf-β signalling inhibits epithelial gene expression13,36 and suppression of Tgf-β potentially enhances the activation of epithelial genes. It also agrees with the notion that Nanog is not required for initiating reprogramming but plays a key role in driving the ‘pre-iPSCs’ to pluripotency30. Uncovering the potential links between transcription events is crucial for attesting the transcription cascade model.
Kdm2b facilitates early gene activation in reprogramming
The results of our study indicate that Kdm2b contributes to iPSC generation by facilitating activation of early responsive genes. We found that the ability of Kdm2b in promoting gene activation depends on OSK (Fig. 6f), supporting an axillary role for Kdm2b in enhancing gene expression. Furthermore, we demonstrate that Kdm2b binds to the promoters of early-activated genes and maintains a low level of H3K36me2 at these loci (Fig. 7a,c), indicating that Kdm2b directly participates in the regulation of these genes. A recent study indicates that depletion of H3K36me2 is a feature of CpG island promoters23. It is possible that Kdm2b-mediated removal of H3K36me2 at promoters facilitates OSK-directed gene activation by creating a chromatin environment favourable for cofactor recruitment. Such a scenario explains why Kdm2b can promote gene activation (Fig. 6) and facilitate reprogramming by OSK (Fig. 1) but fails to substitute for any of OSK (data not shown). Future studies should reveal the molecular mechanisms underlying how demethylation of H3K36me2 contributes to gene activation.
METHODS
Plasmids and virus preparation
Mouse Kdm2b (isoform 1) was amplified from complementary DNA, fused with a C-terminal Flag tag and cloned into a doxycyclin-inducible lentiviral vector pTYF-TRE. The JmjC (H211A, D213A) and ZF (C573A C576A C579A) mutants were constructed by mutagenesis PCR and confirmed by sequencing. Target sequences of control shRNA and shRNAs against Kdm2b (ref. 21), Ink4a/Arf and Cdh1 are described in Supplementary Table S4. These shRNAs were expressed in lentiviral plasmid pTY-U6-Pgk-Puro. Lentivirus was prepared by co-transfection of pTY/pTYF plasmids with pHP, pHEF1α-VSVG and pCEP4-Tat into 293T cells, and collected at 24, 36 and 48 h after transfection. Viral supernatant was filtered through a 0:45-µm membrane and concentrated by a spin column before being applied to MEFs. Retroviral plasmids pMXs-Oct4, Sox2, Klf4 and c-Myc were obtained from Addgene, and retrovirus was prepared as previously described18.
MEF derivation and iPSC generation
MEFs for iPSC generation were prepared from E13.5 embryos of Oct4–IRES–GFP/Rosa26-M2rtTA double knock-in mice. To derive iPSCs, MEFs at the first 2 passages were seeded onto 6-well plates at a density of 1 × 105 cells per well, 16 h before viral infection. Two doses of retrovirus and/or one dose of lentivirus was applied within 48 h in the presence of Polybrene (10 µg ml−1). At 24 h after the second retroviral transduction, the virus supernatant was withdrawn and the day was designated as day 0 post-transduction. Subsequently, iPSCs were induced for 12–18 days in mouse ESC medium (DMEM with 15% FBS, non-essential amino acid, GlutaMax, sodium pyruvate, β-mercaptoethanol, penicillin/streptomycin and 1,000 U ml−1 leukaemia inhibitory factors) in the presence of doxycyclin (1 µg ml−1). If indicated, Tgf-β treatment was carried out by applying Tgf-β1 (R&D Systems) at 2 ng ml−1. Oct4–GFP+ colonies were counted on selected days from day 6 to day 18. Reprogramming efficiency was presented as the number of Oct4–GFP+ colonies derived from 1 × 105 MEFs. Relative reprogramming efficiency over a control induction is also used in some cases. At day 18, Oct4–GFP+ colonies were manually picked, trypsinized and seeded onto mitomycin-C-treated feeder MEF cells. The derived iPSC lines were propagated in mouse ESC medium in the absence of doxycyclin for at least 8 passages before being characterized.
Cell staining, teratoma assay and chimaera generation
For immunofluorescent staining, antibodies against SSEA-1 (Chemicon mAB4301, clone MC-480), Nanog (Bentyl, IHC-00205) and Sox2 (Millipore, AB5603) were applied at a concentration of 1:500, 1:250 and 1:1,000, respectively. Alkaline phosphatase staining was carried out with an alkaline phosphatase detection kit (Millipore). Teratoma analysis was performed as previously reported18. For generation of chimaeric mice, 12-week-old Albino B6 (C57Bl/6J-Tyr < c-2J >) female mice were stimulated to superovulation by injection with pregnant mare serum gonadotropin (2.5 IU) followed by administration of human chorionic gonadotropin (5 IU) 47 h later. The female mice were subsequently mated with Albino B6 stud males and blastocysts were collected on gestation day 3.5. On the day of microinjection, iPSCs (line 8 and 17) were rinsed twice with PBS, dissociated with 0.05% trypsin, washed once with and then resuspended in Knockout DMEM supplemented with 15% FBS. Each blastocyst was injected with 10–15 iPSCs using a piezo impact micromanipulator. Injected embryos were then implanted into the uterus of pseudopregnant Swiss Webster recipient females.
RT–PCR and western blotting
Quantitative and semi-quantitative RT–PCR was carried out using primers in Supplementary Table S5. Western blotting was performed using antibodies against Arf (Santa Cruz sc-32748, clone 5-C3-1, 1:200), Cdh1 (Cell Signaling 3195, clone 24E10, 1:1,000), Epcam (Abcam ab32392, clone E144, 1:500), Flag (Sigma F1804, clone M2, 1:5,000), histone H3 (Abcam ab1791, 1:5,000), H3K4me2 (Active Motif 39141, 1:1,000), H3K36me1 (Abcam ab9048, 1:1,000), H3K36me2 (ref. 22; 1:1,000), H3K36me3 (Abcam ab9050, 1:1,000), Ink4a (Santa Cruz sc-1207, 1:200) and α-tubulin (Sigma T6199, clone DM1A, 1:2,000).
Microarray analysis
RNA samples were extracted from cells transduced with OSK or OSK plus Kdm2b at post-transduction day 4, 8 and 12. The reverse transcription and hybridization procedure was carried out as previously described18. The microarray data were analysed with GeneSpring software and are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MEXP-3433.
ChIP
Cells transduced with OSK plus Flag-tagged Kdm2b were collected for ChIP at post-transduction day 4. ChIP was performed using Imprint ChIP Kit (Sigma) according to the manufacturer’s instruction. Chromatin was prepared by sonication at 4 °C on a Bioruptor 300 (Diagenode) with a high magnitude for 10 cycles with 30 s on and 30 s off. For each precipitation reaction, chromatin from 2 × 105 cells was applied to a Stripwell plate pre-bound with antibodies against Flag (Sigma F1804, clone M2), H3K36me2 (ref. 22), H3K4me2 (Active Motif 39141) or mouse IgG. If necessary, immunoprecipitated and purified DNA fragments were subjected to amplification using a Whole Genome Amplification Kit (Sigma). Immunoprecipitated or amplified DNA was analysed by qPCR using primers listed in Supplementary Table S6.
Supplementary Material
ACKNOWLEDGEMENTS
We thank UNC Animal Models Core for chimaera generation and UNC Functional Genomic Core for microarray analysis. We thank S. Yamaguchi and K-H. Hong for helpful discussions. This work is supported by U01DK089565 from the NIH. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website
AUTHOR CONTRIBUTIONS
G.L. and Y.Z. designed all of the experiments and wrote the manuscript. G.L. performed most of the experiments. J.H. constructed the Kdm2b plasmids and lentiviral doxycyclin inducible system.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plath K, Lowry WM. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Maherali N, Hochedlinger K. Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Ichida JK, et al. A small-molecule inhibitor of tgf-β signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huangfu D, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J. Biol. Chem. 2010;285:25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Singhal N, et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 21.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat. Struct. Mol. Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 23.Blackledge NP, et al. CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc. Natl Acad. Sci. USA. 2009;106:2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfau R, et al. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc. Natl Acad. Sci. USA. 2008;105:1907–1912. doi: 10.1073/pnas.0711865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sridharan R, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson RJ, Dixon J, Jiang R, Dixon MJ. Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum. Mol. Genet. 2009;18:2632–2642. doi: 10.1093/hmg/ddp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomason HA, et al. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J. Clin. Invest. 2010;120:1561–1569. doi: 10.1172/JCI40266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gierl MS, Karoulias N, Wende H, Strehle M, Birchmeier C. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 2006;20:2465–2478. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellitzer G, et al. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 2006;25:1344–1352. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farkas LM, et al. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron. 2008;60:40–55. doi: 10.1016/j.neuron.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 36.Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 2009;21:166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 37.Chen T, et al. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- 38.Redmer T, et al. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Esteban MA, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.