Abstract
Acute myelogenous leukemia (AML) is an extremely heterogeneous neoplasm with several clinical, pathological, genetic and molecular subtypes. Combinations of various doses and schedules of cytarabine and different anthracyclines have been the mainstay of treatment for all forms of AMLs in adult patients. Although this combination, with the addition of an occasional third agent, remains effective for treatment of some young-adult patients with de novo AML, the prognosis of AML secondary to myelodysplastic syndromes or myeloproliferative neoplasms, treatment-related AML, relapsed or refractory AML, and AML that occurs in older populations remains grim. Taken into account the heterogeneity of AML, one size does not and should not be tried to fit all. In this article, the authors review currently understood, applicable and relevant findings related to cytarabine and anthracycline drug-metabolizing enzymes and drug transporters in adult patients with AML. To provide a prime-time example of clinical applicability of pharmacogenomics in distinguishing a subset of patients with AML who might be better responders to farnesyltransferase inhibitors, the authors also reviewed findings related to a two-gene transcript signature consisting of high RASGRP1 and low APTX, the ratio of which appears to positively predict clinical response in AML patients treated with farnesyltransferase inhibitors.
Keywords: acute myelogenous leukemia, AML, anthracycline, APTX, ara-C, cytarabine, farnesyltransferase, pharmacogenomic, RASGRP1
Acute myelogenous leukemia (AML) is one of the most heterogeneous malignancies with several clinical, pathological and genetic subtypes. For example, from a clinical standpoint, AML in younger adults is probably a different disease to AML that occurs in the older population. AML, secondary to other bone marrow disorders such as myelodysplatic syndromes or myeloproliferative neoplasms (MPN), behaves differently compared to de novo AML. Previously untreated AML in general has a better prognosis than relapsed or refractory AML.
Karyotype or cytogenetic abnormalities represent the strongest pretreatment predictor of the rate of complete remission (CR), response duration and overall survival (OS) in adult patients with AML. On the basis of cytogenetics, AML is classified into three categories: favorable, with approximately 65–70% likelihood of cure with chemotherapy alone; intermediate, with 30–40% chance of long-term survival; and unfavorable, with less than 5–10% long-term survival without allogeneic stem cell transplantation. Intermediate-risk AML includes approximately 60% of patients and itself is comprised of a heterogeneous group with diverse structural and numerical chromosomal alterations. Cytogenetically normal AML belongs to the intermediate-risk category; however, discovery of several specific gene mutations such as FLT-3[1,2], NPM1 [3], CEBPA [4], DMNT3A [5], IDH1/2[6,7], KIT[8], WT-1 [9] and others, in patients with normal cytogenetic AML has provided further and sometimes independent prognostic insight. Nevertheless, different combinations of these genetic alterations in an individual person sometimes are too uncommon to be reliably allocated a prognostic value.
In the last four decades, combination of cytarabine (ara-C) and various doses of different anthracyclines has been the mainstay of treatment for all forms of AMLs in adult patients. Although this combination chemotherapy regimen, with addition of an occasional third agent, remains effective for treatment of some AML patients, it is far from ideal. Taken into consideration the heterogeneity of AML, one size does not and should not be tried to fit all.
Pharmacogenomics deals with the impact of genetic dissimilarity on pharmacokinetics, including absorption, distribution, metabolism and excretion of drugs as well as pharmacodynamics including efficacy and toxicity of drugs. Variations in drug-metabolizing enzymes, drug transporters and drug targets are the most practical aspects of pharmacogenomics.
In this article, the authors review currently understood, applicable and relevant pharmacogenomic findings in adult patients with AML that may suggest better strategies for the use of current chemotherapeutic agents. This includes choice of drugs, their dose intensities and schedules of their administration. The authors will also report on the clinical relevance of recent pharmacogenomic discoveries regarding new targets or new drug modifiers that may distinguish a subset of patients with AML who might be ‘better responders’ to novel agents under development for AML therapy. To provide a prime time example of clinical applicability of pharmacogenomics in distinguishing a subset of patients with AML who might be better responders to farnesyltransferase inhibitors, the authors specifically reviewed findings related to a two-gene transcript signature consisting of high RASGRP1 and low APTX, the ratio of which appears to positively predict clinical response in AML patients treated with farnesyltransferase inhibitors.
This review does not intend to review epidemiology, etiology, pathophysiology, diagnosis and management of AML. Further information on these aspects of AML are reviewed elsewhere [10–12].
Genes involved in metabolic pathways & transporters of drugs used for AML treatment
Ara-C
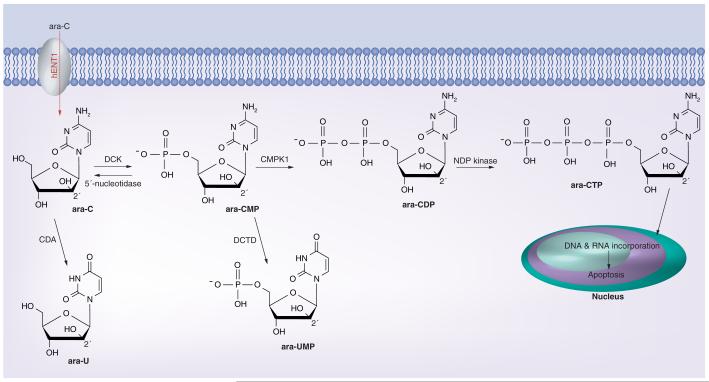
Ara-C was synthesized in 1959 [13] and since then it has been the most effective and universally used chemotherapeutic agent in the treatment of AML. Structurally, its arabinose sugar moiety is epimeric at the 2′-position with ribose. This difference, after conversion to the ara-C triphosphate (ara-CTP) nucleotide, causes it to prevent the transformation of cytidylate to 2′-deoxycytidylate [14]. Other mechanisms of action of ara-C include induction of miscoding after incorporation into DNA and RNA [15], and inhibition of DNA-dependent DNA polymerase [16]. Figure 1 illustrates the transporter and enzymes involved in ara-C metabolism.
Figure 1.
Ara-C metabolism. Ara-C: Cytarabine; Ara-CDP: Ara-C diphosphate; Ara-CMP: Ara-C monophosphate; Ara-CTP: Ara-C triphosphate; Ara-U: Ara-uracil; Ara-UMP: Ara-U monophosphate.
After entering the cells via hENT1, ara-C is phosphorylated in a stepwise fashion at the 5′ position of arabinoside. Phosphorylation is mediated initially by DCK to convert ara-C to ara-C monophosphate (ara-CMP), then by deoxycytidylate kinase to ara-C diphosphate and finally by nucleoside diphosphate kinase to the active metabolite, ara-CTP. Intracellular concentration of ara-CTP is directly correlated with the therapeutic effect of ara-C. Inactivation of ara-C can occur through ara-CMP dephosphorylation by 5′-nucleotidase back to ara-C. Deaminase enzymes can convert and inactivate ara-C and ara-CMP to ara-uracil (ara-U) and ara-U monophosphate, respectively.
The most important parameters in sensitivity or resistance to ara-C include: conversion to nucleotides by kinases; inactivation via de amination by deaminases; half-life of active form ara-CTP; and the magnitude of its incorporation into DNA.
Ara-C transporter: hENT1
Although at plasma concentrations greater than 10 μM, which is achieved with high-dose ara-C (2–3 g/m2 daily), the drug diffuses freely into the cell [17], at concentrations less than 1 μM, which is achieved with 100–200 mg/m2 daily, ara-C influx into the cells is strongly correlated with the number of nucleoside transporters per leukemic blast [18,19].
hENT1, localized on chromosome 6, is the transporter for ara-C and other nucleoside analogues such as gemcitabine and cladribine. Significant differences in the mRNA expression levels of hENT1 have been noted in patients with AML. AML patients with hENT1 deficiency at diagnosis had significantly shorter disease-free survival (DFS) and OS [20]. Hence, because ara-C is commonly used for treatment of adult AML, the expression level of its transporter, hENT1, can be considered as a predictive biomarker rather than a prognostic biomarker for patients with AML. The cytotoxicity of a single concentration of ara-C was closely correlated with the cell surface abundance of nucleoside transporter sites in a flow cytometry assay [21]. In a study to investigate variants of hENT1 in ethnically diverse DNA samples from 247 individuals, two nonsynonymous changes were identified in the coding region of hENT1 with no contributory effect on its transportation function. It has been suggested that variability in hENT1 expression is mediated by transcriptional regulation. Recently, it has been shown that activation or overexpression of the transcription factor PPARα resulted in higher hENT1 transport activity [22]. In the future, identification of genetic variations that affect hENT1 expression or interaction with transcription factors such as HIF-α [23] may provide practical insight for selection of nucleoside analogues in the treatment of AML and other malignancies such as pancreatic cancer.
Ara-C kinases: DCK, CMPK1 & NDPK
DCK catalyzes the rate limiting first phosphorylation step in activation of many nucleoside analogues including ara-C. The enzyme attaches one phosphate group to carbon at 5′ position of the arabinose sugar moiety. The gene for DCK is located on chromosome 4. A greater than 30-fold variation in DCK mRNA expression was reported among AML cells, [24,25] and it appears that higher mRNA level correlates with longer event-free survival [20]. In a study involving approximately 120 Chinese patients with AML, two regulatory SNPs created two major haplotypes with lower mRNA level and poorer 2-year event-free survival associated with one compound genotype (-360CC/-201CC) [26]. Interestingly, no coding SNPs were detected in this population. These regulatory SNPs were reported with much less frequency in European patients [27]. Several other genetic polymorphisms related to DCK have been discovered with yet-to-be proven clinical relevance and significance [28].
CMPK1 or deoxycytidylate kinase catalyzes the second phosphorylation step, converting ara-CMP to ara-C diphosphate. The final phosphorylation step is catalyzed by nucleoside diphosphate kinase to generate the active ara-CTP. Major genetic polymorphisms in these two enzymes with clinical application have not been identified.
An increased sensitivity to ara-C in AML cell lines was reported when the intracellular level of deoxy cytidine 5′-triphosphate (dCTP) is diminished. The enzyme CTP synthetase catalyzes the conversion of UTP to CTP. Inhibition of this enzyme depletes cellular dCTP pool and promotes phosphorylation of ara-C by diminishing inhibitory feedback of dCTP on DCK [29]. In addition to feedback inhibition of DCK and reduced activation of ara-C [30], dCTP influences the ara-C treatment outcome by competing with ara-CTP for incorporation into DNA [31] and by allosterically activating ara-C deactivating the enzyme CDA [32]. Higher level of dCTP results in more resistant blasts to ara-C[32].
Another example that underscores the consequences of change in intracellular dCTP pool on ara-C, therapeutic effect deals with enzyme ribonucleotide reductase (RR). This enzyme is composed of a dimerized large (RRM1) and small (RRM2) subunits. The variation in the expression of an individual or both subunit genes can influence response to ara-C [33]. RR catalyzes the reduction of ribonucleotides to deoxy ribonucleotides for DNA synthesis [33]. RR regulates dCTP and other deoxyribonucleotide levels inside the cells, hence its activity is directly associated with ara-C sensitivity or resistance. Inhibition of RR by novel agents can culminate in accumulation of ara-CTP. Combination of RR inhibitor 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (Triapine) with ara-C and gemcitabine resulted in accumulation of metabolites of both agents in non-small-cell lung cancer cell lines [34]. We have tested the combination of triapine followed by the adenosine analogue fludarabine in adults with refractory acute leukemias and aggressive MPN in a Phase I clinical trial and demonstrated that this combination is active in refractory leukemias and warrants continuing study for patients with aggressive MPN [35].
Ara-CMP dephosphorylation: 5′-nucleotidase
5′-nucleotidases are enzymes with catalytic dephosphorylation function of deoxyribo- and ribo-nucleoside phosphates. Seven different members in 5′-nucleotidase family have been identified. NT5C2 reverses the function of CDK by removing 5′ phosphate from ara-CMP[36]. Increased expression of cytosolic 5′-nucleotidase II has been correlated with resistance to nucleoside-based chemotherapies including ara-C, gemcitabine and cladribine [37] and a lower rate of OS in approximately 100 patients with AML who were treated with ara-C [38].
Recently, sequencing the gene for cytosolic 5′-nucleotidase II identified 41 genetic variants (one insertion–deletion and 40 SNPs), including three nonsynonymous SNPs. Twenty-five of these SNPs were new [39]. Future studies to identify the clinical relevance of these genetic polymorphisms of cytosolic 5′-nucleotidase II are required.
Ara-C & ara-CMP deaminases
CDA inactivates ara-C by removing the amine group from its cytosine and converting it to ara-U. In patients with AML, elevated levels of CDA have been directly correlated with relapse and lower levels of CDA with prolonged remission [40,41]. In one study, CDA activity was the most sensitive parameter to predict adequate blast cell clearance [40]. Activity of CDA in previously untreated AML and in patients with CR is significantly lower than in refractory AML blasts and in persistent blast after induction chemotherapy, respectively [41]. Genetic evaluation of CDA in African–Americans, Caucasians [42] and Japanese [43] resulted in identification of new nonsynonymous coding polymorphisms, which need further evaluation to demonstrate clinical significance [44]. For example, A79C in the CDA gene is a common polymorphism, which changes a lysine residue to glutamine resulting in reduced enzyme activity. CDA A79C genotypes were found in approximately 450 children with AML. It was reported that postinduction treatment-related mortality was significantly higher in children with the CC geno type [45]. Nevertheless, the actual impact of the A79C polymorphism on CDA activity and clinical outcome with nucleosidic analogues even tually remains controversial [46–48], and so are the other SNPs commonly described in the CDA gene (e.g., C437T and G208A, with apparently a strong influence of ethnicity for the latter one). Hence, single genotype-based studies should be interpreted cautiously and that broader strategies are probably necessary to better picture the role CDA plays.
DCTD deaminates ara-CMP to ara-UMP. The clinical effects of genetic polymorphisms in this enzyme are uncertain and demand further evaluations [49,50].
Anthracyclines
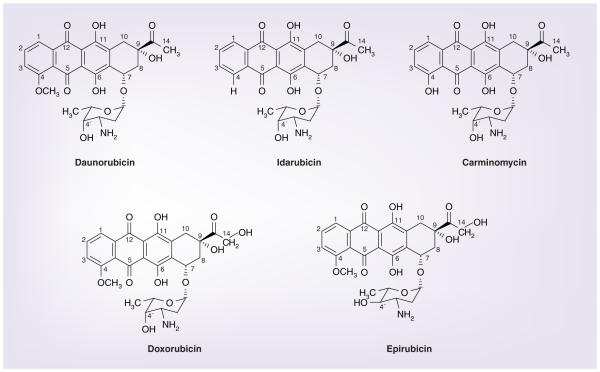
Anthracycline antibiotics (Figure 2) are another most important class of chemotherapeutic agents that have been widely used in the treatment of AML since the 1960s. In the 1950s, daunorubicin was isolated from soil-based bacteria by two independent research groups from Italy and France [51]. Minor modification in the structure of an anthracycline results in alteration of biological activity including potency of the compound. Idarubicin is the 4-demethoxy analogue of daunorubicin with five- to six-times higher antitumor potency. The 4-hydroxy analogue of daunorubicin, carminomycin, has been evaluated in Russia. Hydroxylation of the carbon 14 atom in daunorubicin results in another active chemotherapy, doxorubicin or adriamycin. Changing the spatial orientation of the hydroxyl group at the 4′ position of the sugar molecule of doxorubicin generates another anthracycline named epirubicin with presumed faster elimination from the body.
Figure 2.
Chemical structures of anthracyclines.
The major proposed mechanisms of action of anthracyclines include DNA intercalation, prevention of DNA replication by stabilization and inhibition of enzyme topoisomerase II, and production of reactive oxygen species generated by redox cycling of quinone moiety of the molecule [52–55].
Anthracyclines transporters: ATP-binding cassettes
One of the most clinically relevant pharmacogenomic aspects of anthracyclines is variation in drug transporters. The ATP-binding cassettes (ABCs) are membrane proteins that transport different types of molecules across the cellular membrane (influx, efflux or both)[56]. Forty nine ABC proteins are encoded by the human genome and classified into seven subfamilies with several of them not yet fully characterized [57,58]. The role of ABC efflux transporters in resistance to antineoplastic drug has been investigated in the last three decades[59,60]. Three ABC proteins appear to be responsible for most of multidrug resistance (MDR) in human; P-gp/ABCB1/MDR1, MRP1/ABCC1 and BCRP/ABCG2/ABCP/MXR[61]. Anthracyclines are among the chemotherapeutic agents that interact with these three ABC transporters [62]. Polymorphisms in the ABC drug transporters have been investigated extensively to better understand the significant variability in response to chemotherapies including anthracyclines.
The function and expression of P-gp (ABCB1) is related to complete remission rate and drug resistance in AML [63]. Both ABCB1 expression and functional drug efflux increase with patient age, from 17% in patients less than 35 years old to 39% in patients aged 50 years or older [63]. Expression, function and genetic polymorphism of P-gp were studied in 817 AML samples. It was shown that the genetic polymorphism 3435TT (which results in unstable mRNA) had a significant effect on P-gp expression. However, this was only observed in 40% of cases in which mRNA and protein were detectable. Low white blood cell count, secondary AML and poor risk cytogenetics had a much greater impact on prognosis than genetic polymorphisms of P-gp expression in AML blasts [64].
P-gp can be modulated with different class of molecules including immunosuppressive agents (cyclosporine A [PSC-833] and tacrolimus [FK506]), calcium channel blockers (verapamil and nifedipine), tyrosine kinase inhibitors (imatinib and gefitinib), H2-receptor ant-agonists (cimetidine), and statins (lovastatin and simvastatin) [62]. On the basis of preclinical and preliminary studies that demonstrated an increased plasma concentration of an anthracycline when combined with cyclosporine A [65,66], several clinical trials tested the hypothesis of improvement in clinical benefit when combining P-gp modulators with different chemotherapy regimens. Unfortunately, these trials did not demonstrate significant clinical benefit including prolonged OS [67–72]. In some cases the combination of P-gp modulator with chemotherapy compared to chemotherapy alone resulted in worse toxicity profiles [67]. There have been several proposed explanations for the observed results including less frequent expression of the ABC transporters on AML blasts compared to normal hematopoietic stem/progenitor cells, abrogation of drug efflux completely by ABC modulators in normal cells but not in AML blasts, and different methods used to define MDR [73].
MRP1 (ABCC1) is expressed in approximately 10% of patients with AML with inverse correlation with age and with less significant relationship with chemotherapy response [63]. Neither ABCB1 nor ABCC1 expression were correlated with leukemia free survival or OS [63,74].
In a study aimed to investigate the correlation between the expression of other ABCC transporters including ABCC4 (MRP4), ABCC5 (MRP5) and ABCC11 (MRP8) on AML blasts with clinical outcomes, the researcher analyzed such expressions on blast samples from 50 patients with AML [75]. The results indicated that high expression of ABCC11 was inversely correlated with OS probability in 4 years (p = 0.03).
BCRP (ABCG2) was first discovered by Doyle and Ross at the University of Maryland in human MCF-7 breast cancer cell line that demonstrated an ATP-dependent reduction in the cellular levels of anthracyclines without overexpression of already indentified MDR transporters [76]. Compared to P-gp and MRP1, BCRP has a greater efflux transporting ability for mitoxantrone [77,78]. In a study involving blast cells from 20 AML patients, BCRP mRNA expression varied significantly (>1000-fold) among the samples with almost no expression in 50% of the samples and high expression in 33% [79]. The finding of high expression of BCRP in a subset of AML patients suggested further investigation of BCRP presence on AML blasts and its correlation with clinical outcome such as survival advantage. DNA from pretreatment bone marrow or blood samples from 261 adult patients with AML, who received ara-C and anthracycline-based chemotherapy was genotyped for eight nonsynonymous SNPs in the ABCB1, ABCC1 and ABCG2 genes. Heterozygous (AG) or homozygous (AA) variant genotypes for rs2231137 in the ABCG2 gene, compared to the wild-type (GG) genotype were associated with significantly improved survival, and increased toxicity [80]. In a study of 85 samples from patients with AML, P-gp, MRP3 and BCRP activities were inversely correlated with CR rate, DFS and OS. When samples expressed one or none of P-gp, MRP3 or BCRP proteins, patients had a better prognosis compared to the patients expressing two or three of these transporters [81].
Apart from the three major transporters, one study has demonstrated that low levels of ABCA3 transporter expression on AML blasts were statistically significantly associated with improved progression-free survival and OS compared to high ABCA3 levels [82].
Carcinogene & chemotherapy neutralizing enzymes: glutathione S-transferases
Glutathione S-transferase (GST) enzymes are one of the major cellular detoxifiers [83]. These dimeric isoenzymes catalyze reactions between the reduced form of glutathione, which carries sulfhydryl (-SH) group, and many endogenous or exogenous compounds [84] including epoxides, unsaturated aldehydes, peroxidized lipids[85], polyaromatic hydrocarbons [86–89], alkylating agent chemotherapies [90,91], and anthracyclines [92,93]. Generation of high levels of reactive oxygen species post anthracycline treatment induces GST to nullify cellular oxidative stress. GST family in mammalians comprises at least six classes, including alpha (α), mu (μ), omega (ω), pi (π), theta (τ) and zeta (ζ) [94]. GSTM1 and GSTT1 polymorphisms have been detected in their population distribution and associated with the development of many solid tumors [95] and hematologic malignancies including AML[96–98]. Interestingly, GSTM1 and GSTT1 null genotypes were not correlated with higher rates of treatment-related AML in children with acute lymphoblastic leukemia after chemotherapy [99].
In a study of 106 patients with AML, homozygous deletion resulting in null genotypes at GSTM1, GSTT1 or both were reported in 42, 28 and 18% of patients, respectively. The presence of at least one GST deletion appeared to be an independent negative prognostic risk factor for response to induction chemotherapy and OS. Compared with patients with an intact GST gene, AML patients with deletions of GSTM1 or GSTT1 or both achieved lower CR rate after induction therapy (60 vs 80%) and had shorter OS (8 vs 15 months) [100]. Concordant with these results, a study of 153 patients with de novo AML reported a lower probability of DFS in the presence of GST deletions [101].
On the contrary, a study of 200 AML patients older than 56 years treated with ara-C and dauno rubicin as induction therapy in the SWOG clinical trials reported opposite results [102]. In this study no statistically significant associations between treatment outcomes including CR rate, OS or toxicity and any GST genotypes were detected. The authors attributed the absence of correlation between GST polymorphisms and clinical outcomes to the older age of patients enrolled in this study, which could have trumped the potential effects of polymorphisms. Another study aimed to investigate the prognostic role of poly morphisms in three GST genes (GSTM1/T1/P1) in approximately 250 patients of the German Austrian AML Study Group confirmed the latter results[103]. In this study, GSTM1 and GSTT1 geno-types had no significant impact on response to induction therapy, relapse-free survival or OS. However, GSTP1*105 Val allele was associated with quicker neutrophil and platelet recovery, and longer relapse-free survival and OS.
These conflicting results warrant further investigations perhaps in prospective randomized clinical trials with prespecified primary and secondary end points including correlative laboratory measurements.
After reviewing the above polymorphisms in the enzymes and transporters involved in ara-C and anthracycline metabolism, here the authors will review the implication of pharmacogenomics in modern drug discovery by providing an example that underscores the power of testing for genetic signature to distinguish patients with AML who may demonstrate a more profound response to a novel class of chemotherapeutic agent.
Pharmacogenomics contribution to personalized medicine: design of clinical trials incorporating farnesyltransferase inhibitors in the treatment of AML
Farnesyltransferase inhibitors (FTIs) are selective inhibitors of FTase, which catalyzes the transfer of a 15-carbon farnesyl moiety to different acceptors [104,105]. Some of the poly-peptides to which farnesyl is transferred include centromeric proteins promoting completion of mitosis, and guanosine triphosphate-binding polypeptides of the Ras, Rho and Rheb families[106,107]. Tipifarnib, an oral methylquinolinone FTI, showed activity in refractory AML [108,109] and in newly diagnosed, unfavorable-risk elderly adults with AML [110,111]. A Phase III study compared single agent tipifarnib with best supportive care, including hydroxyurea, in approximately 450 patients older than 70 years with newly diagnosed AML who were not candidates for conventional chemotherapy [111]. The results showed no statistically significant survival advantage in patients treated with tipifarnib.
In primary AML cells obtained from patients, tipifarnib inhibited signaling downstream of the farnesylated Rheb and synergistically enhanced etoposide-induced antiproliferative effects in vitro [112]. These results prompted the design and execution of a multicenter Phase I clinical trial of tipifarnib (300–600 mg twice daily for 14 or 21 days) plus oral etoposide (100–200 mg daily on days 1–3 and 8–10 for each cycle) in 84 adults older than 70 years who were not candidates for conventional cytotoxic induction chemotherapy. A CR rate of 25% across multiple dose levels of both drugs, with a median OS of 22 months for patients in CR was reported [112].
In order to select patients, particularly elderly AML patients intolerable of conventional intensive chemotherapeutic regimens, who are most likely to benefit from tipifarnib-containing regimens, serial studies of gene-expression profiling of AML cell lines and primary AML marrow blasts were performed [113–115]. Studies in the newly diagnosed patients with AML revealed a two-gene transcript signature consisting of high RASGRP1 (which encodes the Ras-activating guanine nucleotide exchange factor RASGRP1) and low APTX (which encodes the DNA excision repair protein aprataxin), the ratio of which positively predicted clinical response [115].
RASGRP1 is a member of a family of proteins characterized by a Ras guanine nucleotide exchange factor domain. The protein as a diacylglycerol-regulated nucleotide exchange factor regulates activation of Ras via the exchange of bound guanosine diphosphate for guanosine triphosphate. RASGRP1 also regulates T-cells and B-cells development and differentiation as well as activates the Erk/MAP kinase pathway [116,117]. Mutations in APTX gene as a single-stranded DNA repair protein have been associated with ataxia-ocular apraxia [118].
Retrospectively, the two-gene signature correlation with clinical response in elderly patients with AML treated with tipifarnib and etoposide was confirmed [119]. Analysis of bone marrow aspirates of patients who were treated with tipifarnib and etoposide in the clinical trial validated the two-gene signature as a reproducible and reliable predictor of response to tipifarnib. This suggests the feasibility of a prospective trial to discriminate those patients with AML who are likely to respond to tipifarnib compared with those who are not.
The authors showed that AML patients treated with the combination of tipifarnib and etoposide with a RASGRP1/APTX ratio of 5.2 or greater had a CR rate of 78% compared with patients with a ratio of less than 5.2, who had a CR rate of 13%. Based on these results, negative and positive predictive values were 87 and 78%, respectively [119]. The negative predictive value in studies that used tipifarnib as single agent was approximately 95% [115].
To determine the specificity of the two-gene signature for tipifarnib responsiveness versus general chemotherapy sensitivity, bone marrow blasts from 41 adults with newly diagnosed AML undergoing intensive multiagent induction chemotherapy, including ara-C, anthra cyclines and etoposide were examined. The results demonstrated no association between the two-gene ratio and clinical outcome for the patients who were not treated with tipifarnib, hence, no significant value in predicting clinical response to chemotherapeutic regimens that did not contain tipifarnib [119].
The two-gene signature and correlation with response to tipifarnib just make sense with respect to the proposed mechanism of action of FTIs, which requires post-translational farnesylation of Ras proteins for Ras-dependent signal transduction signals [106,107,120]. It has been demonstrated that RASGRP1 can act as a guanine nucleotide exchange factor, which activates H-Ras and N-Ras and is associated with decreased chemotherapy sensitivity in leukemia cells [121].
Another conclusion that can be drawn from these results is correlation between low APTX expression and overall tipifarnib responsiveness. It can be speculated that when AML cells with low APTX expression are treated with tipifarnib, they have a decreased capability to undergo DNA excision repair. The evidence for the hypothesis that APTX downregulation confers a diminished ability to repair tipifarnib associated DNA damage was provided in the Phase I trial of tipifarnib and etoposide, [112] where DNA damage and apoptosis in bone marrow blasts were seen a week after initiation of tipifarnib.
Conclusion
Although, currently in 2012, outside clinical trials, it is not a routine practice to guide treatment plans for adult patients with AML based on pretreatment results of genetic tests deemed to identify polymorphisms in the enzymes and transporters, the use of pharmacogenomics to guide such optimal treatment of adult patients with AML appears to be ready for prime time. Continuing and future efforts are warranted to harmonize laboratories, to standardize assays and to apply these standardized assays prospectively in well-designed clinical trials. This concept is already exploited in clinical practice in childhood acute lymphoblastic leukemia, where polymorphisms in drug-metabolizing enzymes determine specific drug administration and dosing [122–125]. The current NCI 8977 trial, a Phase II trial of tipifarnib in previously untreated older adults with AML and baseline presence of a specific two-gene expression signature ratio, is a vanguard in ability to select AML patients based on their gene signatures [201].
Future perspective
In this article, we aimed to show that pharmacogenomic changes in drug-metabolizing enzymes, drug transporters and drug targets may have important clinical implications in the treatment of patients with AML. We are aware of the fact that the majority of these findings were identified retrospectively and on the basis of ‘responder analysis’. Responder analysis is subject to significant levels of biases. For this reason, to test the validity, reliability and clinical benefit of different pharmacogenomic changes, future trials of AML need to test prespecified hypotheses and prospectively incorporate them in randomized clinical trials. This is the only way that we can determine whether or not these changes are ready for prime time.
Executive summary.
■ Cytarabine and anthracyclines are the most commonly used chemotherapies for the treatment of acute myelogenous leukemia (AML). Several clinical trials have enrolled patients with AML to find the optimal dose and schedule of administration of these agents. The heterogeneous nature of AML explains, to a great extent, significant differences in response rate, response duration and survival of patients treated with combination of cytarabine and anthracycline. Another important factor in this equation of response to chemotherapies is interpatient variability irrespective of the AML type.
■ Genetic polymorphisms in enzymes responsible for intracellular metabolism of cytarabine (ara-C) and anthracyclines may be important factors in determination of response to chemotherapy. DCK, 5′-nucleotidase, CDA, cytidine-5′-triphosphate synthetase, ribonucleotide reductase and glutathione S-transferase are enzymes that are directly or indirectly affect response to ara-C and anthracyclines, and their pharmacogenomic changes have been suggested to carry clinical relevance.
■ Polymorphisms in drug transporters are other determinants of sensitivity or resistance to ara-C and anthracyclines. hENT1 for ara-C and P-gp, MRP1 and BCRP for anthracyclines are important drug transporters, polymorphisms of which appear to be relevant in the clinic. Modulation of P-gp with different class of agents has been tested in several prospective randomized clinical trials. To date, these trials have not demonstrated significant clinical benefit including prolonged overall survival by such modulation.
■ It appears that farnesyltransferase inhibitors have reasonable antileukemia activity. However, not all patients with AML respond favorably to these class of drugs. AML patients treated with the combination of tipifarnib and etoposide with a RASGRP1/APTX ratio of 5.2 or greater had a significantly higher CR rate compared with patients with a ratio of less than 5.2. Negative and positive predictive values of response for this two-gene signature were 87 and 78%, respectively. This example underscores the potential for pharmacogenomics to play an important role in selection of most likely beneficial chemotherapy combination for patients with particular genetic characteristics in the future.
Footnotes
Financial & competing interests disclosure The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pnding, or royalties. No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■of interest
■■of considerable interest
- 1.Levis M, Murphy KM, Pham R, et al. Internal tandem duplications of the FLT3 gene are present in leukemia stem cells. Blood. 2005;106(2):673–680. doi: 10.1182/blood-2004-05-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levis M. FLT3/ITD AML and the law of unintended consequences. Blood. 2011;117(26):6987–6990. doi: 10.1182/blood-2011-03-340273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boissel N, Renneville A, Biggio V, et al. Prevalence, clinical profile, and prognosis of NPM mutations in AML with normal karyotype. Blood. 2005;106(10):3618–3620. doi: 10.1182/blood-2005-05-2174. [DOI] [PubMed] [Google Scholar]
- 4.Dufour A, Schneider F, Hoster E, et al. Monoallelic CEBPA mutations in normal karyotype acute myeloid leukemia: independent favorable prognostic factor within NPM1 mutated patients. Ann. Hematol. 2012;91(7):1051–1063. doi: 10.1007/s00277-012-1423-4. [DOI] [PubMed] [Google Scholar]
- 5.Hou HA, Kuo YY, Liu CY, et al. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012;119(2):559–568. doi: 10.1182/blood-2011-07-369934. [DOI] [PubMed] [Google Scholar]
- 6.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 2010;28(14):2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 2010;28(22):3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 8.Park SH, Chi HS, Min SK, Park BG, Jang S, Park CJ. Prognostic impact of c-KIT mutations in core binding factor acute myeloid leukemia. Leuk. Res. 2011;35(10):1376–1383. doi: 10.1016/j.leukres.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Paschka P, Marcucci G, Ruppert AS, et al. Wilms’ tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 2008;26(28):4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 11.Sanz MA, Grimwade D, Tallman MS, et al. Management of acute promyelocytic leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113(9):1875–1891. doi: 10.1182/blood-2008-04-150250. [DOI] [PubMed] [Google Scholar]
- 12.Estey EH. Acute myeloid leukemia: 2012 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2012;87(1):89–99. doi: 10.1002/ajh.22246. [DOI] [PubMed] [Google Scholar]
- 13.Cohen SS. Introduction to the biochemistry of d-arabinosyl nucleosides. Prog. Nucleic Acid Res. Mol. Biol. 1966;5:1–88. doi: 10.1016/s0079-6603(08)60231-7. [DOI] [PubMed] [Google Scholar]
- 14.Chu MY, Fischer GA. A proposed mechanism of action of 1-β-d-arabinofuranosyl-cytosine as an inhibitor of the growth of leukemic cells. Biochem. Pharmacol. 1962;11:423–430. doi: 10.1016/0006-2952(62)90225-3. [DOI] [PubMed] [Google Scholar]
- 15.Borun TW, Scharff MD, Robbins E. Rapidly labeled, polyribosome-associated RNA having the properties of histone messenger. Proc. Natl Acad. Sci. USA. 1967;58(5):1977–1983. doi: 10.1073/pnas.58.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creasey WA, Deconti RC, Kaplan SR. Biochemical studies with 1-β-d-arabinofuranosylcytosine in human leukemic leukocytes and normal bone marrow cells. Cancer Res. 1968;28(6):1074–1081. [PubMed] [Google Scholar]
- 17.Capizzi RL, Yang JL, Cheng E, et al. Alteration of the pharmacokinetics of high-dose ara-C by its metabolite, high ara-U in patients with acute leukemia. J. Clin. Oncol. 1983;1(12):763–771. doi: 10.1200/JCO.1983.1.12.763. [DOI] [PubMed] [Google Scholar]
- 18.Wiley JS, Taupin J, Jamieson GP, Snook M, Sawyer WH, Finch LR. Cytosine arabinoside transport and metabolism in acute leukemias and T cell lymphoblastic lymphoma. J. Clin. Invest. 1985;75(2):632–642. doi: 10.1172/JCI111741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundaram M, Yao SY, Ingram JC, et al. Topology of a human equilibrative, nitrobenzylthioinosine (NBMPR)-sensitive nucleoside transporter (hENT1) implicated in the cellular uptake of adenosine and anti-cancer drugs. J. Biol. Chem. 2001;276(48):45270–45275. doi: 10.1074/jbc.M107169200. [DOI] [PubMed] [Google Scholar]
- 20.Galmarini CM, Thomas X, Calvo F, et al. Potential mechanisms of resistance to cytarabine in AML patients. Leuk. Res. 2002;26(7):621–629. doi: 10.1016/s0145-2126(01)00184-9. ■Level of expression of many enzymes and transporters were measured in 77 acute myelogenous leukemia (AML) samples at diagnosis and correlation with clinical outcome was reported.
- 21.Gati WP, Paterson AR, Larratt LM, Turner AR, Belch AR. Sensitivity of acute leukemia cells to cytarabine is a correlate of cellular es nucleoside transporter site content measured by flow cytometry with SAENTA-fluorescein. Blood. 1997;90(1):346–353. [PubMed] [Google Scholar]
- 22.Montero TD, Racordon D, Bravo L, Owen GI, Bronfman ML, Leisewitz AV. PPARα and PPARγ regulate the nucleoside transporter hENT1. Biochem. Biophys. Res. Commun. 2012;419(2):405–411. doi: 10.1016/j.bbrc.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 23.Eltzschig HK, Abdulla P, Hoffman E, et al. HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J. Exp. Med. 2005;202(11):1493–1505. doi: 10.1084/jem.20050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakihara T, Fukuda T, Tanaka A, et al. Expression of deoxycytidine kinase (dCK) gene in leukemic cells in childhood: decreased expression of dCK gene in relapsed leukemia. Leuk. Lymphoma. 1998;31(3–4):405–409. doi: 10.3109/10428199809059234. [DOI] [PubMed] [Google Scholar]
- 25.Van Der Wilt CL, Kroep JR, Loves WJ, et al. Expression of deoxycytidine kinase in leukaemic cells compared with solid tumour cell lines, liver metastases and normal liver. Eur. J. Cancer. 2003;39(5):691–697. doi: 10.1016/s0959-8049(02)00813-4. [DOI] [PubMed] [Google Scholar]
- 26.Shi JY, Shi ZZ, Zhang SJ, et al. Association between single nucleotide polymorphisms in deoxycytidine kinase and treatment response among acute myeloid leukaemia patients. Pharmacogenetics. 2004;14(11):759–768. doi: 10.1097/00008571-200411000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Mahlknecht U, Dransfeld CL, Bulut N, et al. SNP analyses in cytarabine metabolizing enzymes in AML patients and their impact on treatment response and patient survival: identification of CDA SNP C-451T as an independent prognostic parameter for survival. Leukemia. 2009;23(10):1929–1932. doi: 10.1038/leu.2009.113. [DOI] [PubMed] [Google Scholar]
- 28.Lamba JK. Genetic factors influencing cytarabine therapy. Pharmacogenomics. 2009;10(10):1657–1674. doi: 10.2217/pgs.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verschuur AC, Van Gennip AH, Leen R, Voute PA, Van Kuilenburg AB. Cyclopentenyl cytosine increases the phosphorylation and incorporation into DNA of arabinofu-ranosyl cytosine in a myeloid leukemic cell-line. Adv. Exp. Med. Biol. 2000;486:311–317. doi: 10.1007/0-306-46843-3_61. [DOI] [PubMed] [Google Scholar]
- 30.Tattersall MH, Ganeshaguru K, Hoffbrand AV. Mechanisms of resistance of human acute leukaemia cells to cytosine arabinoside. Br. J. Haematol. 1974;27(1):39–46. doi: 10.1111/j.1365-2141.1974.tb06772.x. [DOI] [PubMed] [Google Scholar]
- 31.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp. Cell. Res. 1989;181(2):305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 32.Chiba P, Tihan T, Szekeres T, et al. Concordant changes of pyrimidine metabolism in blasts of two cases of acute myeloid leukemia after repeated treatment with ara-C in vivo. Leukemia. 1990;4(11):761–765. [PubMed] [Google Scholar]
- 33.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr. Cancer Drug Targets. 2006;6(5):409–431. doi: 10.2174/156800906777723949. ■Elegant review of ribonucleotide reductase chemistry, its tumorigenesis and its inhibition for cancer treatment.
- 34.Sigmond J, Kamphuis JA, Laan AC, Hoebe EK, Bergman AM, Peters GJ. The synergistic interaction of gemcitabine and cytosine arabinoside with the ribonucleotide reductase inhibitor triapine is schedule dependent. Biochem. Pharmacol. 2007;73(10):1548–1557. doi: 10.1016/j.bcp.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 35.Karp JE, Giles FJ, Gojo I, et al. A Phase I study of the novel ribonucleotide reductase inhibitor 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) in combination with the nucleoside analog fludarabine for patients with refractory acute leukemias and aggressive myeloproliferative disorders. Leuk. Res. 2008;32(1):71–77. doi: 10.1016/j.leukres.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dumontet C, Bauchu EC, Fabianowska K, et al. Common resistance mechanisms to nucleoside analogues in variants of the human erythroleukemic line K562. Adv. Exp. Med. Biol. 1999;457:571–577. doi: 10.1007/978-1-4615-4811-9_63. [DOI] [PubMed] [Google Scholar]
- 37.Hunsucker SA, Mitchell BS, Spychala J. The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol. Ther. 2005;107(1):1–30. doi: 10.1016/j.pharmthera.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Galmarini CM, Cros E, Thomas X, Jordheim L, Dumontet C. The prognostic value of cN-II and cN-III enzymes in adult acute myeloid leukemia. Haematologica. 2005;90(12):1699–1701. ■Expression of many enzymes in the metabolic pathway of cytarabine was analyzed and association with clinical outcome was made.
- 39.Mitra AK, Crews KR, Pounds S, et al. Genetic variants in cytosolic 5′-nucleotidase II are associated with its expression and cytarabine sensitivity in HapMap cell lines and in patients with acute myeloid leukemia. J. Pharmacol. Exp. Ther. 2011;339(1):9–23. doi: 10.1124/jpet.111.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jahns-Streubel G, Reuter C, Auf Der Landwehr U, et al. Activity of thymidine kinase and of polymerase α as well as activity and gene expression of deoxycytidine deaminase in leukemic blasts are correlated with clinical response in the setting of granulocyte-macrophage colony-stimulating factor-based priming before and during TAD-9 induction therapy in acute myeloid leukemia. Blood. 1997;90(5):1968–1976. ■Important study, which demonstrated a significant correlation between deoxycytidine deaminase expression and clinical outcome in AML patients who were treated with cytarabine.
- 41.Schroder JK, Kirch C, Seeber S, Schutte J. Structural and functional analysis of the cytidine deaminase gene in patients with acute myeloid leukaemia. Br. J. Haematol. 1998;103(4):1096–1103. doi: 10.1046/j.1365-2141.1998.01084.x. [DOI] [PubMed] [Google Scholar]
- 42.Gilbert JA, Salavaggione OE, Ji Y, et al. Gemcitabine pharmacogenomics: cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin. Cancer Res. 2006;12(6):1794–1803. doi: 10.1158/1078-0432.CCR-05-1969. [DOI] [PubMed] [Google Scholar]
- 43.Yue L, Saikawa Y, Ota K, et al. A functional single-nucleotide polymorphism in the human cytidine deaminase gene contributing to ara-C sensitivity. Pharmacogenetics. 2003;13(1):29–38. doi: 10.1097/00008571-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 44.Fitzgerald SM, Goyal RK, Osborne WR, Roy JD, Wilson JW, Ferrell RE. Identification of functional single nucleotide polymorphism haplotypes in the cytidine deaminase promoter. Hum. Genet. 2006;119(3):276–283. doi: 10.1007/s00439-006-0142-0. [DOI] [PubMed] [Google Scholar]
- 45.Bhatla D, Gerbing RB, Alonzo TA, et al. Cytidine deaminase genotype and toxicity of cytosine arabinoside therapy in children with acute myeloid leukemia. Br. J. Haematol. 2009;144(3):388–394. doi: 10.1111/j.1365-2141.2008.07461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciccolini J, Dahan L, Andre N, et al. Cytidine deaminase residual activity in serum is a predictive marker of early severe toxicities in adults after gemcitabine-based chemotherapies. J. Clin. Oncol. 2010;28(1):160–165. doi: 10.1200/JCO.2009.24.4491. [DOI] [PubMed] [Google Scholar]
- 47.Giovannetti E, Tibaldi C, Falcone A, Danesi R, Peters GJ. Impact of cytidine deaminase polymorphisms on toxicity after gemcitabine: the question is still ongoing. J. Clin. Oncol. 2010;28(14):E221–E225. doi: 10.1200/JCO.2009.27.4928. [DOI] [PubMed] [Google Scholar]
- 48.Banklau C, Jindadamrongwech S, Sawangpanich R, et al. Effect of genetic alterations of cytarabine- metabolizing enzymes in childhood acute lymphoblastic leukemia. Hematol. Oncol. Stem Cell. Ther. 2010;3(3):103–108. doi: 10.1016/s1658-3876(10)50019-0. [DOI] [PubMed] [Google Scholar]
- 49.Capizzi RL, White JC, Powell BL, Perrino F. Effect of dose on the pharmacokinetic and pharmacodynamic effects of cytarabine. Semin. Hematol. 1991;28(3 Suppl. 4):S54–S69. [PubMed] [Google Scholar]
- 50.Fridland A, Verhoef V. Mechanism for ara-CTP catabolism in human leukemic cells and effect of deaminase inhibitors on this process. Semin. Oncol. 1987;14(2 Suppl. 1):S262–S268. [PubMed] [Google Scholar]
- 51.Weiss RB. The anthracyclines: will we ever find a better doxorubicin? Semin. Oncol. 1992;19(6):670–686. [PubMed] [Google Scholar]
- 52.Momparler RL, Karon M, Siegel SE, Avila F. Effect of adriamycin on DNA, RNA, and protein synthesis in cell-free systems and intact cells. Cancer Res. 1976;36(8):2891–2895. [PubMed] [Google Scholar]
- 53.Frederick CA, Williams LD, Ughetto G, et al. Structural comparison of anticancer drug– DNA complexes: adriamycin and daunomycin. Biochemistry. 1990;29(10):2538–2549. [PubMed] [Google Scholar]
- 54.Pigram WJ, Fuller W, Hamilton LD. Stereochemistry of intercalation: interaction of daunomycin with DNA. Nat. New Biol. 1972;235(53):17–19. doi: 10.1038/newbio235017a0. [DOI] [PubMed] [Google Scholar]
- 55.Lown JW, Chen HH, Plambeck JA, Acton EM. Diminished superoxide anion generation by reduced 5-iminodaunorubicin relative to daunorubicin and the relationship to cardiotoxicity of the anthracycline antitumor agents. Biochem. Pharmacol. 1979;28(17):2563–2568. doi: 10.1016/0006-2952(79)90027-3. [DOI] [PubMed] [Google Scholar]
- 56.Dassa E, Bouige P. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 2001;152(3–4):211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 57.Dean M, Allikmets R. Complete characterization of the human ABC gene family. J. Bioenerg. Biomembr. 2001;33(6):475–479. doi: 10.1023/a:1012823120935. [DOI] [PubMed] [Google Scholar]
- 58.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11(7):1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 59.Gottesman MM, Ling V. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett. 2006;580(4):998–1009. doi: 10.1016/j.febslet.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 60.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006;5(3):219–234. doi: 10.1038/nrd1984. ■■Comprehensive and elegant review of ATP-binding cassette transporters and multidrug resistance in different malignancies.
- 61.Litman T, Druley TE, Stein WD, Bates SE. From MDR to MXR: new understanding of multidrug resistance systems, their properties and clinical significance. Cell Mol. Life Sci. 2001;58(7):931–959. doi: 10.1007/PL00000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharom FJ. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9(1):105–127. doi: 10.2217/14622416.9.1.105. [DOI] [PubMed] [Google Scholar]
- 63.Leith CP, Kopecky KJ, Chen IM, et al. Frequency and clinical significance of the expression of the multidrug resistance proteins MDR1/P-glycoprotein, MRP1, and LRP in acute myeloid leukemia: a Southwest Oncology Group Study. Blood. 1999;94(3):1086–1099. ■Seminal article on the frequency and role of MDR proteins in AML.
- 64.Seedhouse CH, Grundy M, White P, et al. Sequential influences of leukemia-specific and genetic factors on p-glycoprotein expression in blasts from 817 patients entered into the National Cancer Research Network acute myeloid leukemia 14 and 15 trials. Clin. Cancer Res. 2007;13(23):7059–7066. doi: 10.1158/1078-0432.CCR-07-1484. [DOI] [PubMed] [Google Scholar]
- 65.List AF, Kopecky KJ, Willman CL, et al. Benefit of cyclosporine modulation of drug resistance in patients with poor-risk acute myeloid leukemia: a Southwest Oncology Group study. Blood. 2001;98(12):3212–3220. doi: 10.1182/blood.v98.12.3212. [DOI] [PubMed] [Google Scholar]
- 66.Kolitz JE, George SL, Dodge RK, et al. Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: final induction results of Cancer and Leukemia Group B Study 9621. J. Clin. Oncol. 2004;22(21):4290–4301. doi: 10.1200/JCO.2004.11.106. [DOI] [PubMed] [Google Scholar]
- 67.Baer MR, George SL, Dodge RK, et al. Phase 3 study of the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age and older with acute myeloid leukemia: Cancer and Leukemia Group B Study 9720. Blood. 2002;100(4):1224–1232. [PubMed] [Google Scholar]
- 68.Liu Yin JA, Wheatley K, Rees JK, Burnett AK. Comparison of ‘sequential’ versus ‘standard’ chemotherapy as reinduction treatment, with or without cyclosporine, in refractory/relapsed acute myeloid leukaemia (AML): results of the UK Medical Research Council AML-R trial. Br. J. Haematol. 2001;113(3):713–726. doi: 10.1046/j.1365-2141.2001.02785.x. [DOI] [PubMed] [Google Scholar]
- 69.Van Der Holt B, Lowenberg B, Burnett AK, et al. The value of the MDR1 reversal agent PSC-833 in addition to daunorubicin and cytarabine in the treatment of elderly patients with previously untreated acute myeloid leukemia (AML), in relation to MDR1 status at diagnosis. Blood. 2005;106(8):2646–2654. doi: 10.1182/blood-2005-04-1395. [DOI] [PubMed] [Google Scholar]
- 70.Sonneveld P, Suciu S, Weijermans P, et al. Cyclosporin A combined with vincristine, doxorubicin and dexamethasone (VAD) compared with VAD alone in patients with advanced refractory multiple myeloma: an EORTC-HOVON randomized Phase III study (06914) Br. J. Haematol. 2001;115(4):895–902. doi: 10.1046/j.1365-2141.2001.03171.x. [DOI] [PubMed] [Google Scholar]
- 71.Solary E, Drenou B, Campos L, et al. Quinine as a multidrug resistance inhibitor: a Phase 3 multicentric randomized study in adult de novo acute myelogenous leukemia. Blood. 2003;102(4):1202–1210. doi: 10.1182/blood-2002-11-3419. [DOI] [PubMed] [Google Scholar]
- 72.Solary E, Witz B, Caillot D, et al. Combination of quinine as a potential reversing agent with mitoxantrone and cytarabine for the treatment of acute leukemias: a randomized multicenter study. Blood. 1996;88(4):1198–1205. [PubMed] [Google Scholar]
- 73.Raaijmakers MH, De Grouw EP, Van Der Reijden BA, De Witte TJ, Jansen JH, Raymakers RA. ABCB1 modulation does not circumvent drug extrusion from primitive leukemic progenitor cells and may preferentially target residual normal cells in acute myelogenous leukemia. Clin. Cancer Res. 2006;12(11 Pt 1):3452–3458. doi: 10.1158/1078-0432.CCR-05-1945. [DOI] [PubMed] [Google Scholar]
- 74.Damiani D, Tiribelli M, Calistri E, et al. The prognostic value of P-glycoprotein (ABCB) and breast cancer resistance protein (ABCG2) in adults with de novo acute myeloid leukemia with normal karyotype. Haematologica. 2006;91(6):825–828. [PubMed] [Google Scholar]
- 75.Guo Y, Kock K, Ritter CA, et al. Expression of ABCC-type nucleotide exporters in blasts of adult acute myeloid leukemia: relation to long-term survival. Clin. Cancer Res. 2009;15(5):1762–1769. doi: 10.1158/1078-0432.CCR-08-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doyle LA, Yang W, Abruzzo LV, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl Acad. Sci. USA. 1998;95(26):15665–15670. doi: 10.1073/pnas.95.26.15665. ■Original article on the discovery of BCRP.
- 77.Ross DD, Yang W, Abruzzo LV, et al. Atypical multidrug resistance: breast cancer resistance protein messenger RNA expression in mitoxantrone-selected cell lines. J. Natl Cancer Inst. 1999;91(5):429–433. doi: 10.1093/jnci/91.5.429. [DOI] [PubMed] [Google Scholar]
- 78.Lemos C, Jansen G, Peters GJ. Drug transporters: recent advances concerning BCRP and tyrosine kinase inhibitors. Br. J. Cancer. 2008;98(5):857–862. doi: 10.1038/sj.bjc.6604213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ross DD, Karp JE, Chen TT, Doyle LA. Expression of breast cancer resistance protein in blast cells from patients with acute leukemia. Blood. 2000;96(1):365–368. [PubMed] [Google Scholar]
- 80.Hampras SS, Sucheston L, Weiss J, et al. Genetic polymorphisms of ATP-binding cassette (ABC) proteins, overall survival and drug toxicity in patients with acute myeloid leukemia. Int. J. Mol. Epidemiol. Genet. 2010;1(3):201–207. [PMC free article] [PubMed] [Google Scholar]
- 81.Benderra Z, Faussat AM, Sayada L, et al. MRP3, BCRP, and P-glycoprotein activities are prognostic factors in adult acute myeloid leukemia. Clin. Cancer Res. 2005;11(21):7764–7772. doi: 10.1158/1078-0432.CCR-04-1895. ■Important article, which demonstrated the value of different MDR proteins in AML as clinical prognosticators.
- 82.Chapuy B, Koch R, Radunski U, et al. Intracellular ABC transporter A3 confers multidrug resistance in leukemia cells by lysosomal drug sequestration. Leukemia. 2008;22(8):1576–1586. doi: 10.1038/leu.2008.103. [DOI] [PubMed] [Google Scholar]
- 83.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001;360(Pt 1):1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Douglas KT. Mechanism of action of glutathione-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 1987;59:103–167. doi: 10.1002/9780470123058.ch3. [DOI] [PubMed] [Google Scholar]
- 85.Tsunada S, Iwakiri R, Noda T, et al. Chronic exposure to subtoxic levels of peroxidized lipids suppresses mucosal cell turnover in rat small intestine and reversal by glutathione. Dig. Dis. Sci. 2003;48(1):210–222. doi: 10.1023/a:1021775524062. [DOI] [PubMed] [Google Scholar]
- 86.Santella RM, Perera FP, Young TL, et al. Polycyclic aromatic hydrocarbon–DNA and protein adducts in coal tar treated patients and controls and their relationship to glutathione S-transferase genotype. Mutat. Res. 1995;334(2):117–124. doi: 10.1016/0165-1161(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 87.Whyatt RM, Perera FP, Jedrychowski W, Santella RM, Garte S, Bell DA. Association between polycyclic aromatic hydrocarbon– DNA adduct levels in maternal and newborn white blood cells and glutathione S-transferase P1 and CYP1A1 polymorphisms. Cancer Epidemiol. Biomarkers Prev. 2000;9(2):207–212. [PubMed] [Google Scholar]
- 88.Weiserbs KF, Jacobson JS, Begg MD, et al. A cross-sectional study of polycyclic aromatic hydrocarbon–DNA adducts and polymorphism of glutathione S-transferases among heavy smokers by race/ethnicity. Biomarkers. 2003;8(2):142–155. doi: 10.1080/1354750031000086269. [DOI] [PubMed] [Google Scholar]
- 89.Obolenskaya MY, Teplyuk NM, Divi RL, et al. Human placental glutathione S-transferase activity and polycyclic aromatic hydrocarbon DNA adducts as biomarkers for environmental oxidative stress in placentas from pregnant women living in radioactivity- and chemically-polluted regions. Toxicol. Lett. 2010;196(2):80–86. doi: 10.1016/j.toxlet.2010.03.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Colvin OM, Friedman HS, Gamcsik MP, Fenselau C, Hilton J. Role of glutathione in cellular resistance to alkylating agents. Adv. Enzyme Regul. 1993;33:19–26. doi: 10.1016/0065-2571(93)90006-y. [DOI] [PubMed] [Google Scholar]
- 91.Biroccio A, Benassi B, Fiorentino F, Zupi G. Glutathione depletion induced by c-Myc downregulation triggers apoptosis on treatment with alkylating agents. Neoplasia. 2004;6(3):195–206. doi: 10.1593/neo.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lutzky J, Astor MB, Taub RN, et al. Role of glutathione and dependent enzymes in anthracycline-resistant HL60/AR cells. Cancer Res. 1989;49(15):4120–4125. [PubMed] [Google Scholar]
- 93.L’Ecuyer T, Allebban Z, Thomas R, Vander Heide R. Glutathione S-transferase overexpression protects against anthracycline-induced H9C2 cell death. Am. J. Physiol. Heart Circ. Physiol. 2004;286(6):H2057–H2064. doi: 10.1152/ajpheart.00778.2003. [DOI] [PubMed] [Google Scholar]
- 94.Wilce MC, Parker MW. Structure and function of glutathione S-transferases. Biochim. Biophys. Acta. 1994;1205(1):1–18. doi: 10.1016/0167-4838(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 95.Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 1997;6(9):733–743. [PubMed] [Google Scholar]
- 96.Basu T, Gale RE, Langabeer S, Linch DC. Glutathione S-transferase theta 1 (GSTT1) gene defect in myelodysplasia and acute myeloid leukaemia. Lancet. 1997;349(9063):1450. doi: 10.1016/s0140-6736(05)63726-9. [DOI] [PubMed] [Google Scholar]
- 97.Rollinson S, Roddam P, Kane E, et al. Polymorphic variation within the glutathione S-transferase genes and risk of adult acute leukaemia. Carcinogenesis. 2000;21(1):43–47. doi: 10.1093/carcin/21.1.43. [DOI] [PubMed] [Google Scholar]
- 98.Crump C, Chen C, Appelbaum FR, et al. Glutathione S-transferase θ 1 gene deletion and risk of acute myeloid leukemia. Cancer Epidemiol. Biomarkers Prev. 2000;9(5):457–460. [PubMed] [Google Scholar]
- 99.Woo MH, Shuster JJ, Chen C, et al. Glutathione S-transferase genotypes in children who develop treatment-related acute myeloid malignancies. Leukemia. 2000;14(2):232–237. doi: 10.1038/sj.leu.2401660. [DOI] [PubMed] [Google Scholar]
- 100.Voso MT, D’alo F, Putzulu R, et al. Negative prognostic value of glutathione S-transferase (GSTM1 and GSTT1) deletions in adult acute myeloid leukemia. Blood. 2002;100(8):2703–2707. doi: 10.1182/blood.V100.8.2703. [DOI] [PubMed] [Google Scholar]
- 101.Barragan E, Collado M, Cervera J, et al. The GST deletions and NQO1*2 polymorphism confers interindividual variability of response to treatment in patients with acute myeloid leukemia. Leuk. Res. 2007;31(7):947–953. doi: 10.1016/j.leukres.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 102.Weiss JR, Kopecky KJ, Godwin J, et al. Glutathione S-transferase (GSTM1, GSTT1 and GSTA1) polymorphisms and outcomes after treatment for acute myeloid leukemia: pharmacogenetics in Southwest Oncology Group (SWOG) clinical trials. Leukemia. 2006;20(12):2169–2171. doi: 10.1038/sj.leu.2404421. [DOI] [PubMed] [Google Scholar]
- 103.Voso MT, Hohaus S, Guidi F, et al. Prognostic role of glutathione S-transferase polymorphisms in acute myeloid leukemia. Leukemia. 2008;22(9):1685–1691. doi: 10.1038/leu.2008.169. [DOI] [PubMed] [Google Scholar]
- 104.Karp JE, Lancet JE. Development of farnesyltransferase inhibitors for clinical cancer therapy: focus on hematologic malignancies. Cancer Invest. 2007;25(6):484–494. doi: 10.1080/07357900701359437. [DOI] [PubMed] [Google Scholar]
- 105.Sousa SF, Fernandes PA, Ramos MJ. Farnesyltransferase inhibitors: a detailed chemical view on an elusive biological problem. Curr. Med. Chem. 2008;15(15):1478–1492. doi: 10.2174/092986708784638825. [DOI] [PubMed] [Google Scholar]
- 106.Sebti SM, Adjei AA. Farnesyltransferase inhibitors. Semin. Oncol. 2004;31(1 Suppl. 1):S28–S39. doi: 10.1053/j.seminoncol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 107.Rowinsky EK, Windle JJ, Von Hoff DD. Ras protein farnesyltransferase: a strategic target for anticancer therapeutic development. J. Clin. Oncol. 1999;17(11):3631–3652. doi: 10.1200/JCO.1999.17.11.3631. [DOI] [PubMed] [Google Scholar]
- 108.Karp JE, Lancet JE, Kaufmann SH, et al. Clinical and biologic activity of the farnesyltransferase inhibitor R115777 in adults with refractory and relapsed acute leukemias: a Phase 1 clinical-laboratory correlative trial. Blood. 2001;97(11):3361–3369. doi: 10.1182/blood.v97.11.3361. [DOI] [PubMed] [Google Scholar]
- 109.Harousseau JL, Lancet JE, Reiffers J, et al. A Phase 2 study of the oral farnesyltransferase inhibitor tipifarnib in patients with refractory or relapsed acute myeloid leukemia. Blood. 2007;109(12):5151–5156. doi: 10.1182/blood-2006-09-046144. [DOI] [PubMed] [Google Scholar]
- 110.Lancet JE, Gojo I, Gotlib J, et al. A Phase 2 study of the farnesyltransferase inhibitor tipifarnib in poor-risk and elderly patients with previously untreated acute myelogenous leukemia. Blood. 2007;109(4):1387–1394. doi: 10.1182/blood-2006-04-014357. ■Phase II study, which demonstrated antileukemia activity of tipifarnib in patients with AML with unfavorable prognosis.
- 111.Harousseau JL, Martinelli G, Jedrzejczak WW, et al. A randomized Phase 3 study of tipifarnib compared with best supportive care, including hydroxyurea, in the treatment of newly diagnosed acute myeloid leukemia in patients 70 years or older. Blood. 2009;114(6):1166–1173. doi: 10.1182/blood-2009-01-198093. ■■Pivotal Phase III clinical trial of tipifarnib in newly diagnosed elderly AML.
- 112.Karp JE, Flatten K, Feldman EJ, et al. Active oral regimen for elderly adults with newly diagnosed acute myelogenous leukemia: a preclinical and Phase 1 trial of the farnesyltransferase inhibitor tipifarnib (R115777, Zarnestra) combined with etoposide. Blood. 2009;113(20):4841–4852. doi: 10.1182/blood-2008-08-172726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raponi M, Belly RT, Karp JE, Lancet JE, Atkins D, Wang Y. Microarray analysis reveals genetic pathways modulated by tipifarnib in acute myeloid leukemia. BMC Cancer. 2004;4:56. doi: 10.1186/1471-2407-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raponi M, Harousseau JL, Lancet JE, et al. Identification of molecular predictors of response in a study of tipifarnib treatment in relapsed and refractory acute myelogenous leukemia. Clin. Cancer Res. 2007;13(7):2254–2260. doi: 10.1158/1078-0432.CCR-06-2609. [DOI] [PubMed] [Google Scholar]
- 115.Raponi M, Lancet JE, Fan H, et al. A 2-gene classifier for predicting response to the farnesyltransferase inhibitor tipifarnib in acute myeloid leukemia. Blood. 2008;111(5):2589–2596. doi: 10.1182/blood-2007-09-112730. ■■Study that revealed the potential clinical importance of two-gene transcript signature for response to tipifarnib in older patients with AML.
- 116.Kosco KA, Cerignoli F, Williams S, Abraham RT, Mustelin T. SKAP55 modulates T cell antigen receptor-induced activation of the Ras-Erk-AP1 pathway by binding RasGRP1. Mol. Immunol. 2008;45(2):510–522. doi: 10.1016/j.molimm.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 117.Roose JP, Mollenauer M, Ho M, Kurosaki T, Weiss A. Unusual interplay of two types of Ras activators, RasGRP and SOS, establishes sensitive and robust Ras activation in lymphocytes. Mol. Cell. Biol. 2007;27(7):2732–2745. doi: 10.1128/MCB.01882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ferrarini M, Squintani G, Cavallaro T, Ferrari S, Rizzuto N, Fabrizi GM. A novel mutation of aprataxin associated with ataxia ocular apraxia type 1: phenotypical and genotypical characterization. J. Neurol. Sci. 2007;260(1–2):219–224. doi: 10.1016/j.jns.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 119.Karp JE, Vener TI, Raponi M, et al. Multi-institutional Phase 2 clinical and pharmacogenomic trial of tipifarnib plus etoposide for elderly adults with newly diagnosed acute myelogenous leukemia. Blood. 2012;119(1):55–63. doi: 10.1182/blood-2011-08-370825. ■■Study that validated the clinical relevance of two-gene transcript signature for response to tipifarnib in older patients with AML.
- 120.Basso AD, Kirschmeier P, Bishop WR. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J. Lipid Res. 2006;47(1):15–31. doi: 10.1194/jlr.R500012-JLR200. [DOI] [PubMed] [Google Scholar]
- 121.Caloca MJ, Zugaza JL, Bustelo XR. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J. Biol. Chem. 2003;278(35):33465–33473. doi: 10.1074/jbc.M302807200. [DOI] [PubMed] [Google Scholar]
- 122.Wall AM, Rubnitz JE. Pharmacogenomic effects on therapy for acute lymphoblastic leukemia in children. Pharmacogenomics J. 2003;3(3):128–135. doi: 10.1038/sj.tpj.6500174. [DOI] [PubMed] [Google Scholar]
- 123.Rocha JC, Cheng C, Liu W, et al. Pharmacogenetics of outcome in children with acute lymphoblastic leukemia. Blood. 2005;105(12):4752–4758. doi: 10.1182/blood-2004-11-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Holleman A, Den Boer ML, De Menezes RX, et al. The expression of 70 apoptosis genes in relation to lineage, genetic subtype, cellular drug resistance, and outcome in childhood acute lymphoblastic leukemia. Blood. 2006;107(2):769–776. doi: 10.1182/blood-2005-07-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang YL, Lin DT, Chang SK, et al. Pharmacogenomic variations in treatment protocols for childhood acute lymphoblastic leukemia. Pediatr. Blood Cancer. 2010;54(2):206–211. doi: 10.1002/pbc.22292. [DOI] [PubMed] [Google Scholar]
■ Website
- 201.Zarnestra in Newly Diagnosed Acute Myelogenous Leukemia (AML)With 2 Gene Expression Signature Ratio. http://clinicaltrials.gov/ct2/show/NCT01361464?term=NCI+8977&rank=1.