Abstract
Skeletal muscle is a large and complex system that is crucial for structural support, movement and function. When injured, the repair of skeletal muscle undergoes three phases: inflammation and degeneration, regeneration and fibrosis formation in severe injuries. During fibrosis formation, muscle healing is impaired because of the accumulation of excess collagen. A group of zinc-dependent endopeptidases that have been found to aid in the repair of skeletal muscle are matrix metalloproteinases (MMPs). MMPs are able to assist in tissue remodeling through the regulation of extracellular matrix (ECM) components, as well as contributing to cell migration, proliferation, differentiation and angiogenesis. In the present study, the effect of GM6001, a broad-spectrum MMP inhibitor, on muscle-derived stem cells (MDSCs) is investigated. We find that MMP inhibition negatively impacts skeletal muscle healing by impairing MDSCs in migratory and multiple differentiation abilities. These results indicate that MMP signaling plays an essential role in the wound healing of muscle tissue because their inhibition is detrimental to stem cells residing in skeletal muscle.
Keywords: Skeletal muscle stem cells, MMPs, differentiation, cell migration
Introduction
Skeletal muscle is a dynamic tissue system that is essential for respiration, structural support and movement among other things, and is therefore highly vascularized and innervated [1-3]. However, as a consequence of its large size, it is susceptible to many types of injuries that include contusions, blunt force trauma, ischemia reperfusion, lacerations and burns [1,4-6]. Understanding the complexity of the cellular and molecular events during the healing process is vital to improve the quality of life for those afflicted with skeletal muscle injuries. The phases that occur in response to muscle injury are inflammation and degeneration, regeneration and fibrosis formation in severe injuries [3,7]. Inflammation is initiated first, resulting in a calcium influx that is associated with the disruption of the sarcolemma, and leads to calcium dependent proteolysis of the myofibrils, and thus degeneration [2]. Disruption of the blood vessels encompassing the damaged area stimulates the migration of neutrophils and macrophages to the site of injury, where neutrophils arrive first through chemotaxis. Macrophages arrive shortly after them and are responsible for the ingesting bacteria and cellular debris through phagocytosis as well as releasing chemokines including matrix metalloproteinases (MMPs) in order to recruit additional inflammatory cells and amplify this immune response [2,8-10]. The arrival of a second wave of non-phagocytic macrophages to the injury site begins to reduce the inflammatory response and initiate myogenesis [8,11].
In the following phases, regeneration is driven predominantly through the skeletal muscle precursors including muscle satellite cells and stem cells. These cells are normally quiescent in healthy muscular tissue and reside in between the basal lamina and the attached myofibers; however, an injury will activate them such as when the myofibers are disrupted from the basal lamina [2,12]. These precursor cells will migrate toward the bordering myofibers if the basal lamina is damaged or migrate under it to the site of injury [2,3]. These activated cells will enter the cell cycle by up-regulating MyoD and Myf5, which induces proliferation and differentiation into myoblasts [1,2,13,14]. After numerous successive rounds of proliferation, a small quantity of satellite cells will migrate beneath the basal lamina to maintain a quiescent pool of stem cells, thus ensuring their capacity for self-renewal [1,15,16]. The down regulation of Pax3 and Pax7 leads to an increased expression of myogenin and MyoD which helps facilitate myogenesis, and the formation of new myofibers to restore the original structure of the muscular tissue [2,14,16].
Sometimes, skeletal muscle healing results in fibrosis formation after severe injury, where elevated levels of collagen deposition are present during tissue remodeling. Fibroblasts and myofibroblasts, at the site of injury, can be stimulated to secrete extracellular matrix (ECM) components such as collagen type I, III, and fibronectin [17]. An overproduction of these ECM materials are necessary to aid in cell recruitment and migration to the site of injury to facilitate muscle healing and regeneration [17]. Unfortunately, an excessive secretion and inefficient turnover of collagen during this remodeling period will result in an unwanted accumulation, thus the formation of scar tissue [2,17]. Oversight of the ECM remodeling process is attributed to MMPs and the tissue inhibitors of metalloproteinases (TIMPs) [7,18,19].
MMPs are zinc-dependent endopeptidases that aid in tissue remodeling by promoting cell proliferation and migration as well as degrading various elements of the ECM in many tissues [7,18-20]. They participate during all stages of tissue repair, from secretion by leukocytes during inflammation, to ECM degradation and cell migration [3,12,20,21]. Collectively, there are 24 known MMPs that are classified into different families based on their small differences in structure and the substrate they act upon. MMP2 and MMP9, both gelatinases, play key roles in remodeling the architecture of injured and diseased skeletal muscle tissue by myoblast fusion/elongation and degradation of necrotic tissue, respectively [22,24]. Other MMPs, such as MMP1 serve in similar roles by degrading collagens I and III to stimulate cell migration and differentiation during injury, while MMP14, a membrane-type MMP, has been observed to aid in tissue remodeling by activating MMP2 and MMP9 [12,25-30]. The actions of MMPs are regulated by four TIMPs, which modify the action of MMPs during wound healing [31,32]. These enzymes modulate cellular processes by restricting cell behaviors or can even activate certain MMPs, such as MMP2 and MMP3 by TIMP2 [7,20,23,31]. The development of fibrotic scar tissue is dependent upon the interactions between the MMPs and the TIMPs, where an unfavorable ratio toward MMPs will result in fibrosis, and thus impair the processes of tissue remodeling and regeneration [10,17].
In the present study, the inhibition of MMP signaling on stem cells derived from skeletal muscle tissue of mice and C2C12 myoblasts was investigated. A broad spectrum MMP inhibitor, GM6001, which is known to inhibit MMP1, MMP2, MMP3, MMP8 and MMP9, was administered to both cell types. MMP inhibition had a negative impact on muscle-derived stem cells (MDSCs) characteristics by impairing stem cell marker expression. In turn, this affected MDSC migration and their ability to properly aid in the repair by causing erroneous skeletal muscle healing when GM6001 was injected into a laceration injury model of mice. These findings suggest that MMP signaling plays an essential role in tissue remodeling as well as in stem cell behavior, where MMP inhibition can have a tumultuous impact on wound healing.
Methods
Isolation and culture of MDSCs and C2C12 myoblasts
MDSCs were isolated from the gastrocnemius muscles (GMs) of 3-week-old C57BL/10J mice using a modified preplate technique [33,34]. C2C12 myoblasts were purchased from ATCC (American Type Culture Collection, Manassas, Virginia). Both cell types were cultured in proliferation media (PM) containing phenol red Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10% horse serum (HS), 1 % penicillin/streptomycin (P/S), and 0.5% chick embryo extract (CEE) at 5% CO2 and 37°C.
Wound migration assay
MDSCs and C2C12 myoblasts were grown to near confluence in a non-coated 12, 24, etc. multiwell plate? Prior to creating an artificial wound, several wells were treated with 25 μM of GM6001 (diluted with DMSO, Millipore) for 3 and 6 hours. After the given amount of GM6001 pretreatment, an artificial wound was created by disrupting the cell monolayer with a sterile plastic pipette tip. Cellular debris was aspirated and PM was added to the GM6001 pretreated cell groups. Another 2 groups of C2C12 myoblasts and MDSCs were treated with 2.5 and 25 μM GM6001 after creating an artificial wound. A live automated cell imager consisting of an inverted Nikon Eclipse TE 2000U microscope and Photometrics ES Cool Snap CCD camera was used to take images of cell migration into the artificial wound at 10 minute intervals for 6 hours as previously described [12]. Cell migration was measured in microns (μm) by the distance traveled into wound site at 1, 3 and 6 hours.
Single cell migration
A live automated cell imager consisting of an inverted Nikon Eclipse TE 2000U microscope and Photometrics ES Cool Snap CCD camera was used to take images of single cell migration for 2 hours at 3 minute intervals. Proper environmental conditions were maintained in a microincubator at 37°C and at 5% CO2. Similar treatment groups were used as in the wound migration assay, but only for MDSCs. All data analysis was assessed as previously described [12,35,36]. A series of images were analyzed using NIH ImageJ analysis software to track the centroid positions (x,y) of cell nuclei (which were assumed to be the representations of cell-bodies). Net translocation distance was measured as the distance between the starting point and the end point of cells after 2 hours. Migration speed was calculated as total length of the migration path during the 2 hour period. The directional persistency index was calculated as the ratio of the net translocation distance to the cumulative length of the migration path. The change of the direction of cellular centroid movement between consecutive images (i-1, i, i+1) was calculated as Δθ = θi,i+1 – θi,i-1, where (Equation 1)
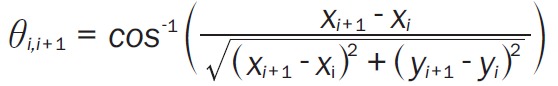 |
and from these values, the standard deviation of Δθ was determined.
Proliferation kinetics
A live cell automated imager captured images of the number of MDSCs and C2C12 cells per field of view at 20 minute intervals for 3 days. The population doubling time (PDT) as represented in the exponential equation Ni = N 02ti/PDT was calculated by fitting an exponential trendline to several measurements of N over the 3 day period. The exponential regression method provides a fitted curve in the form of Ni = N 0 ekti , where k = In(2)/PDT and PDT = In(2)/k [37]. The Click-iT 5-ethynyl-2’-deoxyuridine (EdU) imaging kit (Invitrogen) was used to evaluate the cell proliferation as per the manufacturer’s instructions. Briefly, MDSCs and C2C12 myoblasts were seeded on a 12 multiwell collagen coated plate at 2.5 x 103 cells and grown in PM containing 0.1% EdU for 12 hours. Later, the cells were fixed and a secondary antibody was applied, Alexa Fluor 594 (Invitrogen, 1:400), was used for EdU detection. Hoechst 33342 (Invitrogen) was used as a counterstain to visualize the cell nuclei at a 1:2000 dilution.
RT-PCR
MDSCs were subjected to a 25 μM treatment of GM6001 for 3 and 6 hours. Total RNA was extracted from the cells using the RNeasy plus mini kit (Qiagen) and cDNA was generated using the iScript cDNA Synthesis kit (Bio-Rad). For RT-PCR analysis after myogenic differentiation, the total RNA was also extracted from MDSCs after treatment with 25 μM of GM6001 for 3 and 6 hours and then cultured in myogenic differentiation media (DMEM supplemented with 2% HS and 1% P/S) for 1 day. The sense and anti-sense primers for RT-PCR and their product sizes are found in the Table 1. The cycling parameter used for all reactions were as follows: 94°C for 5 minutes; 30 cycles of: denature for 45 seconds at 95°C, anneal for 30 seconds (53°C – 56°C) and extend for 45 seconds at 72°C. RT-PCR was performed using a Bio-Rad MyiQ thermal cycler (Bio-Rad).
Table 1.
Primers for RT-PCR. Product size is in base pairs.
| Gene | Primer Sequences | Product Size |
|---|---|---|
| MyoD | Sense: 5’-GGCTACGACACCGCCTACTA-3’ | 204 |
| Anti-sense: 5’-GTTCTGTGTCGCTTAGGGAT-3’ | ||
| Notch1 | Sense: 5’-GCCGCAAGAGGCTTGAGAT-3’ | 129 |
| Anti-sense: 5’-GGAGTCCTGGCATCGTTGG-3’ | ||
| M-Cad | Sense: 5’-TCGGGCTGCTTGCCCAGAG-3’ | 222 |
| Anti-sense: CCCTGGATGCTGTAGATGACACTGC-3’ | ||
| GAPDH | Sense: CCTCTGGAAAGCTGTGGCGT-3’ | 190 |
| Anti-sense: 5’-TTGGCAGGTTTCTCCAGGCG-3’ | ||
| Myf5 | Sense: 5’-CCTGTCTGGTCCCGAAAGAAC-3’ | 132 |
| Anti-sense: 5’-TAGACGTGATCCGATCCACAAT-3’ | ||
| Myf6 | Sense: 5’-GCACCGGCTGGATCAGCAAGAG-3’ | 191 |
| Anti-sense: 5’-CTGAGGCATCCACGTTTGCTCC-3’ | ||
| CD34 | Sense: 5’-TCTCTGCCTGATGAGTCTGCTGC-3’ | 193 |
| Anti-sense: 5’-TCTCTGAGATGGCTGGTGTGGTC-3’ | ||
| Sca1 | Sense: 5’-CCTACTGTGTGCAGAAAGAGC-3’ | 238 |
| Anti-sense: 5’-CAGGAAGTCTTCACGTTGACC-3’ |
Myogenic differentiation
MDSCs and C2C12 myoblasts were cultured in PM until they reached 50% and 75% confluence, respectively. Both cell types cells were pretreated with 25 μM of GM6001 in DMEM for 3 and 6 hours prior to the addition of myogenic differentiation media. An additional group of cells did not receive a pretreatment, but instead received 25 μM of GM6001 for the duration of myogenic differentiation. At 5 and 7 days, MDSCs were fixed with formalin and evaluated for the presence of skeletal fast myosin heavy chain (MHC) positive myotubes (1:300, Sigma) and counterstained with DAPI (1000 ng/mL, Sigma). Fluorescent images were captured on a Leica DMIRB microscope (Deerfield, IL) with a Retiga 1300 digital camera and acquired using Northern Eclipse software (version 6.0; Empix Imagining, Mississauga, ON, Canada). The fusion index was quantified by the ratio of the total number of nuclei in myotube fused cells with the total number of nuclei of the entire cell population [38].
Osteogenic differentiation
Osteogenic diffentiation was performed as previously described [39]. Myoblasts were plated in a dish (3.0 x 103 cells per cm2) and allowed to attach to the dish for 24 hours. Prior to osteogenic induction, MDSCs were treated with 25 μM of GM6001 in DMEM for 3 and 6 hours. After treatment, cells were cultured in osteogenic differentiation media (DMEM supplemented with β-glycerolephosphate (10mM, Sigma), dexamethasone (0.1 μM, Sigma), ascorbate-2-phosphate (50 μM, Sigma), BMP4 (25 ng/mL, R&D Systems), 10% FBS and 1% P/S). One group of MDSCs was treated continuously with 25 μM of GM6001 for the duration of osteogenic induction. Osteogenesis was assessed by observing alkaline phosphatase (ALP) activity 3 days after initial osteogenic induction using an alkaline phosphatase kit from Sigma (86C-1KT).
Adipogenic differentiation
Adipogenic differentiation was performed as previously described [39]. MDSCs were plated in a dish (2.0 x 103 cells per well) and allowed to attach to the dish for 24 hours. Cells were cultured in adipogenic differentiation media (DMEM supplemented with insulin (10 μM), dexamethasone (1 μM), isobutyl-methylxanthine (0.5 μM) and indomethacin (200 μM). Two groups of MDSCs received GM6001 at concentrations of 2.5 and 25 μM for the duration of differentiation. Cell cultures were maintained for 14 days, with media changed every 2 days. After 2 weeks, cell cultures were fixed with 10% formalin for 10 minutes and stained with Oil Red O (Sigma), which is an indication of intracellular lipid accumulation. After fixation with formalin (4%) solution, cells were rinsed with 60% isopropanol, then incubated in filtered Oil Red O working solution at room temperature for 15 minutes. Images were captured on a Leica DMIRB microscope (Deerfield, IL) with a Retiga 1300 digital camera and acquired using Northern Eclipse software (version 6.0; Empix Imagining, Mississauga, ON, Canada). Once images were acquired, the remaining Oil Red O was eluted by adding 100% isopropanol for 10 minutes and then transferred to a 96 multiwell plate, where the OD was measured at 500 nm for 0.5 seconds.
Laceration muscle injury model
The muscle laceration injury model was performed as previously described [28]. Normal healthy (C57BL/10J, 4 weeks of age, female, Jackson Laboratory) mice were anesthetized by isofluorane for the duration of the procedure. A posterior longitudinal skin incision was performed to permit exposure of the entire GM muscle. The GM muscle was cut with a surgical blade, through the lateral 50% of their widths and 100% thickness at the muscle’s thickest region. After controlling the bleeding by simple compression the skin was sutured with 4.0 silk thread. Both of the legs were similarly injured. After the injuries, animals were returned to their cages for recovery with commercial pellets and water ad libitum. The right leg GM muscle received 25 mg/kg body weight of GM6001, where the opposing left leg received an injection of similar DMSO solution as a control at 1 and 4 days following the laceration injury. Three mice were sacrificed at 7 and 12 days after laceration for analysis of fibrosis development and 2 mice were sacrificed at 5 days following laceration injury for examination of stem cell markers. At the time of sacrifice, the GM muscles were isolated, mounted, and snap frozen in liquid-nitrogen cooled 2-methylbutane. Samples were then serially sectioned at 10 μm widths at -28°C with a cryostat for histological analysis.
Histology analysis
The sections of GM muscle from each mouse were washed in PBS and stained with Masson’s modified trichrome staining kit (IMEB, Sigma) according to the manufacturer’s specifications. This technique stains the muscle tissue red, collagen (or fibrous tissue) blue and the cell nuclei black. Five randomly selected high powered image fields of three sectioned slices of the injured area were obtained using a Nikon Eclipse 800 fitted with a Spot camera (Diagnostic Instruments). Images were analyzed using CellProfiler image analysis software to measure the percent area of the collagen of the tissue section [40]. Red and blue colors of each image were separated using the software program, where the area of the blue region was calculated and expressed as a percentage of the entire cross-sectional area of the muscle.
For immunohistochemistry, cross-sectional tissue sections of the GM muscle were fixed in 4% formalin. Afterward, the tissue slides were rinsed with PBS and 10% HS was used to block unspecific binding for 1 hour. Primary antibodies, dystrophin [1:200, Abcam] and Pax7 [1:100, DHSB]) were applied. For the Pax7 (produced in mice), the Vector Mouse on Mouse (MOM) kits (Vector Labs) were used to improve antibody specificity. Species specific secondary antibodies, Alexa Fluor 488 and 594 (1:400, Invitrogen), were used and the cell nuclei were counterstained with DAPI.
Measurement of results and statistical analysis
RT-PCR analysis was performed using ImageJ software (version 1.32j, National Institutes of Health, Bethesda, MD) where the integrated density (product of the area and the mean gray value) of bands was calculated. All molecular bands were represented as a percentage of a standard gene, GAPDH. All data are expressed as a mean ± SD. Comparisons of groups were completed using a one-way ANOVA, where significance levels were determined using Tukey HSD pairwise comparison. Statistical significance was determined if P < 0.05.
Results
Cell migration
Both the MMP inhibitor pretreatment time and concentration reduced migration distance into the artificial wound for both MDSCs and C2C12 myoblasts cells. Non-treated MDSCs (Figure 1D-F) migrated further 1, 3 and 6 hours after the artificial wound area was created in comparison to MDSCs that received 25μM of GM6001 as a pretreatment for 3 (Figure 1A-C) and 6 hours as well as MDSCs that were administered GM6001 at concentrations 2.5 and 25μM (Figure 1G-I) during the cell migration period. A difference in the migration distances of MDSCs was observed between the control and the GM6001 pretreatment groups (Figure 1J), as well as the MDSCS that received different concentrations of GM6001 (Figure 1K). MDSCs treated with 25μM of GM6001 migrated a shorter distance compared to the MDSCs treated with 2.5μM of GM6001. A similar pattern of cell migration is observed for C2C12 myoblasts, despite their shorter migration distances in comparison to the MDSCs. Pretreatment of the cells with GM6001 reduced C2C12 migration 1 and 3 hours after the artificial wound was created, but at 6 hours, migration distances of C2C12 cells pretreated for 3 hours were comparable to the control (Figure 1L). C2C12 myoblasts pretreated with GM6001 for 6 hours traveled significantly shorter distances than the untreated MDSCs, even at 6 hours after creating the artificial wound. C2C12 myoblast migration distances in response to different dosages of GM6001 were not nearly as significant compared to the MDSC migration results (Figure 1M). The cells that were treated with 2.5μM of GM6001 had reduced migration distances, yet the result was not significant from the control, suggesting that their cell migration is not dominated by MMP activity. At a ten-fold increase, 25μM GM6001 did significantly reduce the migration at 3 and 6 hours after beginning the wound assay.
Figure 1.
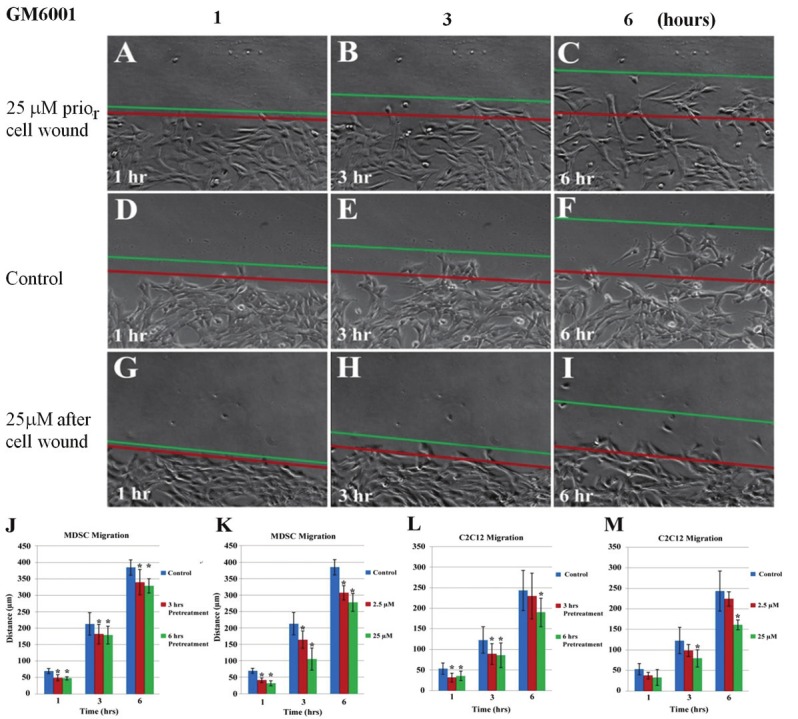
Cell migration is reduced with the administration of GM6001. A-I: phase contrast images taken of MDSCs using a live automated cell imager at 1 (A, D, G), 3 (B, E, H) and 6 (C, F, I) hours of MDSCs pretreated for 3 hours with 25μM GM6001 prior to an artificial wound (A, B, C), control group with no treatment (D, E, F), and 25μM GM6001 added to cell culture after artificial wound (G, H, I). The red line indicates the initial edge of the wound and the green line indicates the position of the cells after 1, 3 or 6 hours of migration into the wound area. Statistical analysis of MDSC (J, K) and C2C12 myoblast (L, M) migration based on pretreatment time with 25μM GM6001 and concentration (0, 2.5, 25μM) after the creation of an artificial wound. There is a significant difference (*P<0.05) from the non-treated group (control) at each time point.
To further investigate the effect of GM6001 on cell migration, time-lapse video microscopy was used to examine the migration pathways of MDSCs under different treatments. This experiment is representative of the in vivo conditions of MDSCs, whereby single MDSC would migrate in response to an injury, instead of a clustered group. Similar treatment groups of GM6001 were observed as before, where several plates of MDSCs treated with 25μM for 3 and 6 hours prior to time-lapse video microscopy. Two other groups were administered 2.5μM and 25μM of GM6001, and then immediately subjected to video imaging. All of the actual cell trajectories from each of the different groups were obtained from a 2 hour period where the data was pooled from 3 experiments (Figure 2A). The trajectories of MDSCs not treated with GM6001, migrated much further than the MDSCs that received any form of GM6001 treatment.
Figure 2.
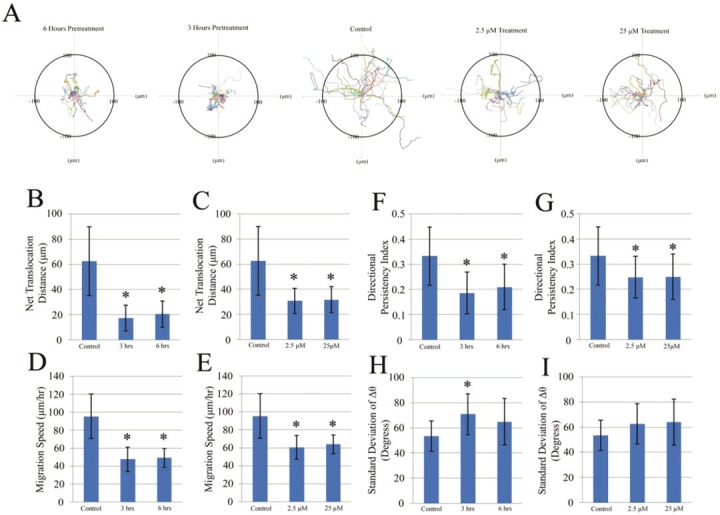
Tracking a single cell migration with GM6001 treatment. The migration paths of 20 individual MDSCs of different experimental groups captured in a time-lapse motility assay (A, data was pooled from three independent experiments). The net translocation distance (straight distance from the start to the end point) of each single MDSC over a 2 hour period is represented as the mean ± standard deviation of the paths of 20 randomly selected cells that were either pretreated with 25 μM of GM6001 prior to image capture (B) or treated with different concentrations at the start of image capture (C). The migration speed (total length of the migration path per hour) of each cell is shown as the mean ± standard deviation of 20 randomly selected cells that were either pretreated with 25 μM of GM6001 prior to image capture (D) or treated with different concentrations at the start of image capture (E). The directional persistency index (ratio of the net translocation distance to the cumulative length of the migration path) of each MDSC over a 2 hour period is represented at the mean ± standard deviation of the paths of 20 randomly selected cells that were either pretreated with 25 μM of GM6001 prior to image capture (F) or treated with different concentrations at the start of image capture (G). The centroid directional movement (a measure of the change in the direction of the centroid movement of a single cell) is shown as the mean ± standard deviation of 20 randomly selected cells that were either pretreated with 25 μM of GM6001 prior to image capture (H) or treated with different concentrations at the start of image capture (I). There is a significant difference (*P<0.05) from the non-treated group (control) at each time point.
Quantitative analysis of the single cell migration path revealed a significantly decreased net translocation distance (straight distance from the cell’s origin to the end point) (Figure 2B and C) and decreased migration speed (total length of the migration path per hour) (Figure 2D and E) for MDSCs that received any form of GM6001 treatment. Interestingly, there was no difference in the net translocation distance and migration speed between the different concentrations of GM6001. However since these cells were administered GM6001 at the initiation of the cell trajectory recording, these MDSCs were not exposed to GM6001 nearly as long as the pretreatment groups, thus their net translocation distance and migration speed were higher than the pretreatment groups, but still lower compared to the control. When the time-lapse video recording was started with the groups pretreated with GM6001, the media was replaced and then substituted with only DMEM. This data indicates that the effect of GM6001 exposure occurs quite quickly and when removed, it continues to have a residual impact on cell migration.
As a result, differences in the directional persistency index (ratio of the net translocation distance to the cumulative length of the migration path) (Figure 2F and G) and the centroid directional movement (a measure of the change in the direction of the centroid movement of a single cell) (Figure 2H and I) of MDSCs were observed. MDSCs not treated with GM6001 tended to migrate further from their starting location by maintaining the same direction of their pathway as demonstrated by a higher directional persistency index in comparison to MDSCs treated with any form of GM6001. These results corresponded moderately with the results of centroid directional movement. The change in centroid directional movement (Δθ) was calculated as a function of time, from which the standard deviation of Δθ was determined, in which, where a higher value indicates a larger fluctuation in the directionality of the cell’s movement. A lower directional persistency index value for the MDSCs that received any form of GM6001 treatment typically corresponded with a higher value of the standard deviation of Δθ, however only the MDSCs pretreated with 25μM of GM6001 for 3 hours was significantly different from the control. Non-treated MDSCs exhibited a higher directional persistency index with a lower standard deviation of Δθ.
Cell proliferation
The expression or secretion of MMPs can be associated with cell proliferation depending on cell type. In these experiments, MDSC proliferation was evaluated with a variety of GM6001 treatments and in comparison with GM6001 treated C2C12 myoblasts. MDSCs and C2C12 myoblasts were treated with 25μM of GM6001 for 3 and 6 hours. After the allotted time of treatment, the culture media supplemented with GM6001 was replaced with normal muscle cell PM and live cell imaging was used to capture images at fixed locations. Similarly, two groups of cells received muscle cell PM supplemented with either 2.5μM or 25μM of GM6001 and then imaged using a live cell imager. Once the data was collected over a period of 3 days, the number of cells per field of view was plotted as a function of time. The exponential regression line was fitted to the data using the equation, Ni = N 0 ekti, and the population doubling time (PDT) was calculated.
The results of this study indicated that there was no difference in the PDT of MDSCs nor C2C12 myoblasts treated with GM6001 for different time periods (Figure 3A) or that received different concentrations of GM6001 (Figure 3B). There was, however, a difference in the cell PDT between cell types. MDSCs had a PDT of approximately 30 hours, while the C2C12 myoblasts had a PDT of around 20 hours. During the artificial wound assay, the migration distances of the control group of C2C12 myoblasts were significantly less than the control group of the MDSCs. Specifically, more cell proliferation of the C2C12 myoblasts was observed at the edges of the artificially created wound assay during the 6 hour time period.
Figure 3.

GM6001 has no effect to muscle cell proliferation. The population doubling time of MDSCs and C2C12 myoblasts were unaffected based on pretreatment time (A) and different concentrations (B) of GM6001 administered to the cells. C2C12 cells (D) exhibit a greater expression of EdU (red) compared to MDSCs (C) regardless of MMP inhibition pretreatment (E) or different concentrations (F). All cells were stained with Hoechst 33342 (blue).
Cell proliferation of MDSCs and C2C12 myoblasts was confirmed using an EdU assay. EdU is a nucleoside analog of thymidine that is incorporated into DNA during active DNA synthesis and is detected by the copper-catalyzed covalent reaction between the azide and the alkyne. The percentage of MDSCs that incorporated EdU (Figure 3C) with or without GM6001 treatment was roughly 40%, much lower than the percentage of C2C12 myoblasts that incorporated EdU at around 80% (Figure 3D) over a 12 hour incubation period. Similarly, there was no difference in EdU incorporation for MDSCs or C2C12 myoblasts that were pretreated (Figure 3E) or received varying concentrations (Figure 3F) of GM6001 treatment compared to the control.
Stem cell characteristics and multiple differentiation
Since MMP1 stimulation influenced primary mouse myoblasts to exhibit stem cell characteristics, this prompted further investigation whether MMP inhibition had any detrimental effects of MDSC behavior besides impairing cell migration. As previously published, MDSCs characteristically express Sca1 and CD34 [41-43]. At the gene expression level, normal MDSCs express a high level of CD34 and Sca1 (Figure 4). When these cells are treated with 25 μM of GM6001 for 3 and 6 hours, their gene expression of Sca1 and CD34 is markedly reduced. The decrease in gene expression of Sca1 appears to depend on time, whereas, the gene expression of CD34 initially decreases at 3 hours, and then increases at 6 hours, but not to its original level.
Figure 4.

Stem cell characterizes’ change with GM6001 treatment. The gene expression of MDSCs treated with 25μM of GM6001 for 3 and 6 hours. These results indicate that MDSCs treated with an MMP inhibitor had reduced expression of stem cell markers, CD34 and Sca1. GAPDH was used as a loading control.
The multipotency of MDSCs has been observed by its capacity to undergo osteogenic differentiation in addition to myogenic differentiation [44-46]. MDSCs were plated at a moderate density and cultured in osteogenic differentiation media supplemented with a minimal amount of BMP4 (25 ng/mL) such that osteogenesis was driven by the MDSCs multipotency, not BMP4. After 3 days of osteogenic induction, MDSCs were fixed and stained for ALP expression to mark cell differentiation (Figure 5A). All forms of GM6001 treatment: 3 and 6 hours of pretreatment with 25μM of GM6001, and continuous treatment of 25μM of GM6001 negatively impacted their osteogenic differentiation potential compared to non-treated cells. Approximately 60% of the MDSCs that received some form of GM6001 treatment expressed ALP in comparison to 85% of the cells in the control (Figure 5B).
Figure 5.

GM6001 interacts muscle stem cells’ multiple differentiations. MDSC treated with GM6001 exhibited a reduced osteogenic differentiation potential based on staining for ALP, Von Kossa and Alizarin Red. The white arrows denote calcium deposition for Von Kossa stains and black arrows denote the red from Alizarin red stains. MDSCs treated with GM6001 (2.5μM and 25μM) exhibited reduced accumulation of lipids (red) within the cytoplasm after 2 weeks as shown with Oil Red O staining. The percentage of ALP positive MDSCs after 3 days of osteogenic differentiation is reduced with MMP inhibition (B). The percentage of MDSCs positive for EdU expression during osteogenic differentiation (C). The optical density was used to quantify adipogenesis (D). There is a significant difference (*P<0.05) between the control and other groups. Statistical significant between control and treated groups was determined if *P<0.05.
The osteogenic differentiation potential of MDSCs was further examined using to alternative stains to test osteogenesis, Von Kossa (Figure 5A) and alizarin red with similar treatment groups. The Von Kossa stain was used to assess mineralization of the MDSCs after 7 days in osteogenic differentiation media by the presence of a black color. Mineralization was observed in all of the test groups of MDSCs, however, the mineralization that occurred in the control groups appeared more frequently and larger. Since the Von Kossa stain alone is often not considered sufficient enough to detect in vitro mineralization because the silver ions in the staining solution are reacting on the phosphate in the cell culture, an alizarin red stain was also employed [47]. Alizarin red identifies the calcium-rich deposits in the MDSCs and the results of the staining yielded a similar outcome as the Von Kossa staining. MDSCs only began to exhibit alizarin red when they grew in a noticeably clustered manner. Within these clustering of cells, more alizarin red was found for the control group of MDSCs compared to MDSCs that were treated with any form of GM6001. Only one speck of alizarin red was detected on a cluster of MDSCs within the treatment group that received GM6001 for the duration of osteogenesis.
To verify that the reduced osteogenic differentiation potential of the MDSCs was not the effect of GM6001 interacting with BMP4 to decrease cell proliferation, an EdU assay was employed. Using the identical treatment groups as before, MDSCs were cultured in osteogenic differentiation media for 5 days, with the EdU solution incubated during the final 12 hours of osteogenic induction. The results of the assay yielded no difference between any of the groups during osteogenic differentiation in the percentage of cells that incorporated EdU into their DNA (Figure 5C). Approximately 15% of the MDSCs across all groups were actively proliferating during osteogenic differentiation, which was considerably less than the 40% of MDSCs that were proliferating during a 12 hour period while in normal PM.
Finally, the ability of MDSCs to undergo adipogenesis in the presence of an MMP inhibitor was investigated. In this experiment, 2 groups of MDSCs were cultured in adipogenic differentiation media supplemented with either 2.5μM or 25μM of GM6001 and compared to control groups of MDSCs (Figure 5A). An Oil Red O staining was performed after 2 weeks, revealing the cytoplasmic lipid droplets, which are an indication of the adipogenic phenotype. The degree of lipid droplet accumulation was quantified by removing the total amount of Oil Red O within the MDSCs and measuring its OD at 500nm (Figure 5D). While there was no difference between the groups treated with GM6001, there was a significantly higher accumulation of lipids within the non-treated MDSCs. Unlike adipocytes where lipid droplets have a large appearance within the cell, the intracellular lipids in the MDSCs after 2 weeks of adipogenesis were small and in abundance within the cytoplasm.
Myogenic differentiation effects of MMP inhibition in vitro and in vivo
With apparent differences in the gene expression of markers associated with MDSCs, their attribute of multipotency was explored as presented above. MDSCs normally proliferate when cultured in growth media with serum, however, in serum free conditions; they exhibit a tendency to differentiate in multinucleated myotubes in culture. MMP inhibition was investigated to determine whether it would reduce myotube formation in vitro using a myogenic differentiation assay (Figure 6A). Four groups were assessed including: no treatment, 3 and 6 hours pretreatment with GM6001, and GM6001 treatment for the duration of the myogenic differentiation assay. After 5 and 7 days in myogenic differentiation media, the fusion index was quantified using fluorescent microscopy by the ratio of the number of cell nuclei within the myotube fused cells stained positive for MHC to the total number of nuclei of the entire cell population (Figure 6B). When comparing the different GM6001 treatment groups, there were no differences in the fusion index, however, all forms of GM6001 treatment negatively affected the myogenic differentiation potential of the MDSCs by producing a lower fusion index. There was no difference seen between groups subjected to 5 or 7 days of myogenic differentiation.
Figure 6.
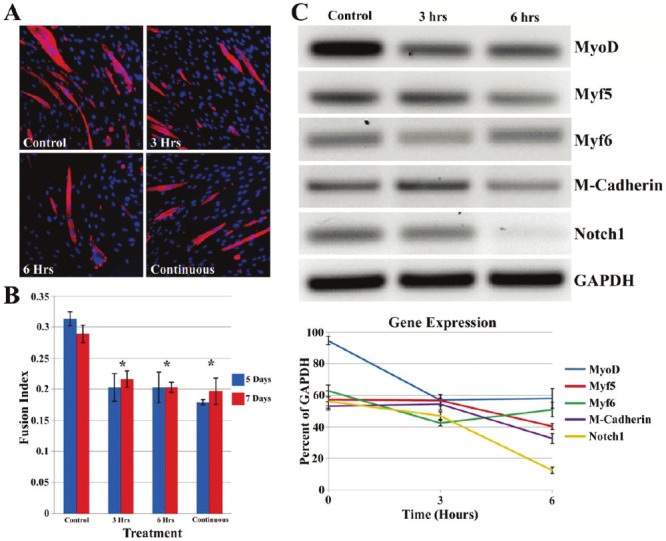
GM6001 inhibits myogenic differentiation in vitro. MMP inhibitor, GM6001, reduces muscle cell myotube formation (A). Immunostaining of MHC (red) in MDSC. There is a significant difference (*P<0.05) between GM6001 treated MDSCs and the control at each time point (B). MMP inhibition negatively impacts genes associated with myotube formation in MDSCs (C). Gene expression was examined for 4 genes related to myogenic differentiation (MyoD, Myf5, Myf6 and M-cadherin) and 1 signaling gene (Notch1) after 3 and 6 hours pretreatment with GM6001 and 1 day cultured with myogenic differentiation meda. GAPDH was used as loading control.
For gene expression analysis, MDSCs received GM6001 pretreatment for 3 and 6 hours prior to the addition myogenic differentiation media, and 1 day later, the mRNA was isolated for RT-PCR. Genes in the myogenic regulatory factor family, MyoD, Myf5 and Myf6 were observed (Figure 6C). The gene expression of MyoD was very strong in the initiation of myogenic differentiation of MDSCs, but the expression was severely affected with the administration of GM6001 for 3 and 6 hours. Myf5 and Myf6 exhibited similar trends, with a slight decrease in expression occurring at 6 hours for Myf5 and 3 hours for Myf6. M-cadherin, known as an adhesion molecule associated with terminal muscle cell differentiation in myotubes, showed a decrease in gene expression after 6 hours of GM6001 treatment, resembling an expression pattern of Myf5. One gene related to cell signaling and myogenesis of muscle cells, Notch1, was examined. Notch1 exhibited similar behavior as the gene expression as M-cadherin gene, where its gene expression was significantly reduced after 6 hours with GM6001 treatment. These data suggest that the use of GM6001 has a negative impact on myotube formation by altering the expression of genes related to myogenesis.
As demonstrated, the use of a broad spectrum MMP inhibitor, GM6001, has a number of negative effects on MDSCs as well as C2C12 myoblasts in vitro, which play important roles in the healing of skeletal muscle tissue in vivo. During skeletal muscle injury, MDSCs or muscle progenitor cells become activated from their residing location between the basal lamina and myofibers [48]. These cells can be detected by their expression of Pax7, which regulates the maintenance of these MDSCs by regulating other myogenic genes such as MyoD and Myf5 [49]. For in vivo experimentation, laceration injuries were performed on the GM muscle of several mice with injections of 25mg/kg GM6001 at 1 and 4 days following the injury. On day 5, the GM was harvested and histologically observed for Pax7 and dystrophin expression. A qualitative assessment revealed that mice which did not receive GM6001 treatment after a laceration injury expressed more Pax7 positive cells in comparison to mice which received GM6001 treatment, where Pax7 was not detected (Figure 7A). The expression of dystrophin in either case appeared to be relatively unaffected by GM6001 administration.
Figure 7.

GM6001 inhibits muscle repair processes in vivo. Pax7 expression (red) was reduced 5 days after laceration injury with GM6001 administration in comparison to non-treated laceration injuries (A). Dystrophin (green) and cell nuclei (blue) were also counterstained. GM6001 treatment of injured mouse skeletal muscle tissue exhibits elevated levels of collagen deposition 7 days post laceration injury than the non-treated skeletal muscle tissue (B). Masson’s modified trichrome staining was used to identify normal healthy skeletal muscle tissue (red) and collagen (blue). While an elevated level of collagen deposition was observed 7 and 12 days after laceration injury, there was no significant difference (C).
At later time points, the GM muscle was isolated and analyzed for collagen deposition during muscle recovery. The GM muscle that received GM6001 treatment at 7 days had an elevated level of collagen deposition compare to GM muscle that did not receive treatment (Figure 7B). Similar results were also observed at 12 days after the laceration injury, where the percentage of collagen deposition with GM6001 treatment was greater than the GM muscle that did not receive any form of treatment (Figure 7C). The percentage of collagen deposition did decrease with more recovery time, however, there was no significant difference between muscle tissue that did or did not receive GM6001 treatment.
Discussion
The wound healing process of skeletal muscle is a complex series of events that aim to reestablish the original architecture of the afflicted tissue [2,50]. This regenerative process is aided by MMPs that initially degrade components of the ECM, allowing greater cell mobility to the injury site for proliferation and repair of the damaged tissue structure [20]. While inflammatory cells such as macrophages and neutrophils are some of the first responders to the damaged tissue, ultimately muscle precursors, including satellite cell and stem cells, are the forerunners leading the recovery. Unfortunately, most efforts to repair the skeletal muscle tissue are impeded by the development of fibrous scar tissue several weeks after the initial injury [51]. Our previous studies have indicated administering MMP1 could improve muscle healing with less scar tissue formation [28,29,52-54]. However, the direct role of MMP on muscle precursors, a major group of cells involved in muscle fiber regeneration, is unclear.
As observed using an artificial wound assay, the use of a MMP inhibitor, GM6001, severely reduced the migration distance of MDSCs based on pretreatment time and dosage. The migration of myoblasts was also impaired; however at a lower concentration (2.5μM), cell migration was not significantly affected. In a similar study, a 10μM dose of GM6001 did not produce a lower migration distance in comparison to the control, suggesting that migration of the myoblasts is not solely guided by the MMPs [53]. While the results of the wound migration assay demonstrated a significant reduction in the migration distance of MDSCs using GM6001, it may not be the most appropriate model. In the in vivo setting, MDSCs are not located in large clusters; therefore a more ideal model is observing single cell migration. Likewise, using the same treatment groups, MMP inhibition was found to reduce both cell migration, speed and its directional persistency index over a 2 hour period. These results were comparable to those conducted in another study where two different types of MMP inhibitors were found to decrease the migration speed of C2C12 myoblasts over a 24 hour period, yet increase the directional persistency index [55]. Moreover, a number of publications provide evidence which suggested the importance of MMPs in skeletal muscle healing by actively promoting myoblasts migration and demonstrated the opposite outcome when they are inhibited [12,56-58].
MMP inhibition does not influence the rate of cell proliferation for either MDSCs or C2C12 myoblasts. These results were confirmed using both a population doubling assay by live cell imaging and an EdU assay. As observed, the cell proliferation occurred much faster for the myoblasts than the MDSCs, while MDSCs tended to migrate further even when comparing either control groups or GM6001 treatments. Similar to in vivo studies, muscle stem cells are one of the early responders, in addition to inflammatory cells, that actively participate in the myoblasts proliferation and differentiation to facilitate skeletal muscle healing [1-3,13,14,16]. This data suggests that cell proliferation may have an inverse relationship with distance migrating cells travel. Qualitatively, cell division appeared quite frequently at the edges of the artificial wound created for the wound assay for C2C12 myoblasts, which corresponds with their shorter population doubling time and reduced cell migration distance. Because of their behavior in response to injuries, muscle stem cells are better suited to aid in the wound healing process.
The most unique characteristic of MDSCs during tissue-related healing is their ability to undergo multiple differentiations into various cell lineages [4,37,43,46,59-66]. Their multipotency has been demonstrated in a plethora of investigations through differentiating into cells types such as osteogenic, myogenic, chondrogenic, adipogenic, neuro-like cells, urinary bladder cells and hepatocyte-like cells. When a broad spectrum MMP1 inhibitor is administered to MDSCs, it is observed that their multiple differentiation capacity into myogenic, osteogenic and adipogenic cells are reduced. Similar findings were made in other investigations where the myogenic and adipogenic differentiation capacity of C2C12 myoblasts and adipocytes, respectively, were significantly impaired through the use of the MMP inhibitor, batimastat [25,67,68]. This decrease in cell differentiation can be attributed to the decrease in the stem cell markers, Sca1 and CD34, of MDSCs as evaluated through gene expression analysis. While MMP inhibition significantly represses the multipotency of MDSCs, it is discernable that MMPs are not the sole regulators of cell differentiation.
During the repair phase of skeletal muscle healing, myogenic factors, MyoD, Myf5, and Myf6, as well as M-cadherin becomes highly upregulated during injury [2,14]. This characteristic response was observed for MDSCs after 1 day in myogenic differentiation in vitro, however, the gene expression profiles of MyoD and Myf5 were noticeably reduced with GM6001 treatment. Myf6 was downregulated with 3 hours of GM6001 treatment, but returned almost to the original level after 6 hours, suggesting that MMP inhibition is ineffective in adversely impacting all myogenic differentiation genes. The most perplexing outcome of the genes associated with early MDSC myogenic differentiation is the decrease in Notch1 expression. Notch signaling is commonly identified for its involvement with proliferation of satellite muscle cell progeny, where inhibition of Notch signaling prevents the expansion of these cells and therefore inhibits effective muscle regeneration [69,70]. For MDSCs, the Notch1 gene was observed to decrease with a 6 hour pretreatment of GM6001 before initiating myogenic differentiation, while a 3 hour pretreatment with GM6001 exhibited levels comparable to the control. While a decrease in the gene expression of the Notch1 receptor may suggest reduced MDSC cell proliferation, thus inhibiting myogenic differentiation, ultimately there are a total of four Notch receptors interacting with 3 delta-like (DLL) and 2 Jagged (JAG) ligands to influence a downstream transcription factor of the CBF1/Su(H)/LAG1 (CSL) family [71]. Further experimentation would be necessary to elucidate how or if MMP inhibition has any effect on the actual Notch signaling pathway, other than decreasing the gene expression of Notch1. Instead, the Notch1 receptor may be more closely linked to Notch signaling influencing cell differentiation by potential adhesion forces between DLL ligands and itself as its gene expression parallels that of the adhesion marker, M-cadherin.
In vivo experiments using mice were performed to examine the effect of GM6001 administration during skeletal muscle healing. Pax7 is commonly used to identify MDSCs or muscle satellite cells that help orchestrate the wound healing process [48,49]. Through qualitative inspection, the presence of Pax7 cells were observed 5 days post laceration injury in comparison to GM6001 treated laceration injuries, where Pax7 positive cells were undetectable. At later time points when fibrosis develops and impedes muscle regeneration, lacerated muscles at 7 and 12 days post injury exhibited an increase in the amount of collagen content compared to the control [3,72]. While the result in collagen deposition between both groups at the two time points was not significantly different, this outcome can potentially be attributed to the administering 25mg/kg body weight of GM6001, which was half the amount of the recommended dosage (50-100mg/kg body weight).
In general, the results of this study demonstrate the importance of MMP signaling in skeletal muscle healing. It has the abilities not only to reduce fibrosis formation and promote myoblast migration, but also influence muscle precursor cells, such as muscle stem cells, for the purpose of aiding in myogenic differentiation and recovery of the injured tissue. Because MMPs are sometimes regarded with a negative connotation in terms of cancer, these studies provide meaningful results that they may benefit individuals affected by skeletal muscle injuries or diseases [73].
Acknowledgement
Source of support: Partially from NIH (R21AR055725) and DOD (23RYX9132N613) to YL; Publish cost pay by the Texas Emerging Technology Fund and the Department of Pediatric Surgery of UT Health.
References
- 1.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–38. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 2.Grefte S, Kuijpers-Jagtman AM, Torensma R, Von den Hoff JW. Skeletal muscle development and regeneration. Stem Cells Dev. 2007;16:857–68. doi: 10.1089/scd.2007.0058. [DOI] [PubMed] [Google Scholar]
- 3.Huard J, Li Y, Fu FH. Muscle injuries and repair: current trends in research. J Bone Joint Surg Am. 2002;84-A:822–32. [PubMed] [Google Scholar]
- 4.Nozaki M, Li Y, Zhu J, Ambrosio F, Uehara K, Fu FH, Huard J. Improved muscle healing after contusion injury by the inhibitory effect of suramin on myostatin, a negative regulator of muscle growth. Am J Sports Med. 2008;36:2354–62. doi: 10.1177/0363546508322886. [DOI] [PubMed] [Google Scholar]
- 5.Ritenour AE, Christy RJ, Roe JL, Baer DG, Dubick MA, Wade CE, Holcomb JB, Walters TJ. The Effect of a Hypobaric, Hypoxic Environment on Acute Skeletal Muscle Edema after Ischemia-Reperfusion Injury in Rats. Journal of Surgical Research. 2010;160:253–259. doi: 10.1016/j.jss.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, Wolf SE, Walters TJ. Muscle contractile properties in severely burned rats. Burns. 2010;36:905–11. doi: 10.1016/j.burns.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellayr I, Mu X, Li Y. Biochemical insights into the role of matrix metalloproteinases in regeneration: challenges and recent developments. Future Med Chem. 2009;1:1095–1111. doi: 10.4155/fmc.09.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chazaud B, Brigitte M, Yacoub-Youssef H, Arnold L, Gherardi R, Sonnet C, Lafuste P, Chretien F. Dual and beneficial roles of macrophages during skeletal muscle regeneration. Exerc Sport Sci Rev. 2009;37:18–22. doi: 10.1097/JES.0b013e318190ebdb. [DOI] [PubMed] [Google Scholar]
- 9.Chazaud B, Sonnet C, Lafuste P, Bassez G, Rimaniol AC, Poron F, Authier FJ, Dreyfus PA, Gherardi RK. Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. J Cell Biol. 2003;163:1133–43. doi: 10.1083/jcb.200212046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Li Y. Role of matrix metalloproteinases in skeletal muscle: migration, differentiation, regeneration and fibrosis. Cell Adh Migr. 2009;3:337–41. doi: 10.4161/cam.3.4.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunelli S, Rovere-Querini P. The immune system and the repair of skeletal muscle. Pharmacol Res. 2008;58:117–21. doi: 10.1016/j.phrs.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Pan H, Murray K, Jefferson BS, Li Y. Matrix metalloproteinase-1 promotes muscle cell migration and differentiation. Am J Pathol. 2009;174:541–9. doi: 10.2353/ajpath.2009.080509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrhardt J, Morgan J. Regenerative capacity of skeletal muscle. Curr Opin Neurol. 2005;18:548–53. doi: 10.1097/01.wco.0000177382.62156.82. [DOI] [PubMed] [Google Scholar]
- 14.Ten Broek RW, Grefte S, Von den Hoff JW. Regulatory factors and cell populations involved in skeletal muscle regeneration. J Cell Physiol. 2010;224:7–16. doi: 10.1002/jcp.22127. [DOI] [PubMed] [Google Scholar]
- 15.Kuang S, Gillespie MA, Rudnicki MA. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell. 2008;2:22–31. doi: 10.1016/j.stem.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Boonen KJ, Post MJ. The muscle stem cell niche: regulation of satellite cells during regeneration. Tissue Eng Part B Rev. 2008;14:419–31. doi: 10.1089/ten.teb.2008.0045. [DOI] [PubMed] [Google Scholar]
- 17.Serrano AL, Munoz-Canoves P. Regulation and dysregulation of fibrosis in skeletal muscle. Exp Cell Res. 2010;316:3050–8. doi: 10.1016/j.yexcr.2010.05.035. [DOI] [PubMed] [Google Scholar]
- 18.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–39. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 19.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–29. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 20.Gill SE, Parks WC. Metalloproteinases and their inhibitors: regulators of wound healing. Int J Biochem Cell Biol. 2008;40:1334–47. doi: 10.1016/j.biocel.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–33. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bani C, Lagrota-Candido J, Pinheiro DF, Leite PE, Salimena MC, Henriques-Pons A, Quirico-Santos T. Pattern of metalloprotease activity and myofiber regeneration in skeletal muscles of mdx mice. Muscle Nerve. 2008;37:583–92. doi: 10.1002/mus.20970. [DOI] [PubMed] [Google Scholar]
- 23.Fukushima K, Nakamura A, Ueda H, Yuasa K, Yoshida K, Takeda S, Ikeda S. Activation and localization of matrix metalloproteinase-2 and -9 in the skeletal muscle of the muscular dystrophy dog (CXMDJ) BMC Musculoskelet Disord. 2007;8:54. doi: 10.1186/1471-2474-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimowska M, Brzoska E, Swierczynska M, Streminska W, Moraczewski J. Distinct patterns of MMP-9 and MMP-2 activity in slow and fast twitch skeletal muscle regeneration in vivo. Int J Dev Biol. 2008;52:307–14. doi: 10.1387/ijdb.072331mz. [DOI] [PubMed] [Google Scholar]
- 25.Ohtake Y, Tojo H, Seiki M. Multifunctional roles of MT1-MMP in myofiber formation and morphostatic maintenance of skeletal muscle. J Cell Sci. 2006;119:3822–32. doi: 10.1242/jcs.03158. [DOI] [PubMed] [Google Scholar]
- 26.Soini Y, Satta J, Maatta M, Autio-Harmainen H. Expression of MMP2, MMP9, MT1-MMP, TIMP1, and TIMP2 mRNA in valvular lesions of the heart. J Pathol. 2001;194:225–31. doi: 10.1002/path.850. [DOI] [PubMed] [Google Scholar]
- 27.Toth M, Chvyrkova I, Bernardo MM, Hernandez-Barrantes S, Fridman R. Pro-MMP-9 activation by the MT1-MMP/MMP-2 axis and MMP-3: role of TIMP-2 and plasma membranes. Biochem Biophys Res Commun. 2003;308:386–95. doi: 10.1016/s0006-291x(03)01405-0. [DOI] [PubMed] [Google Scholar]
- 28.Bedair H, Liu TT, Kaar JL, Badlani S, Russell AJ, Li Y, Huard J. Matrix metalloproteinase-1 therapy improves muscle healing. J Appl Physiol. 2007;102:2338–45. doi: 10.1152/japplphysiol.00670.2006. [DOI] [PubMed] [Google Scholar]
- 29.Kaar JL, Li Y, Blair HC, Asche G, Koepsel RR, Huard J, Russell AJ. Matrix metalloproteinase-1 treatment of muscle fibrosis. Acta Biomater. 2008;4:1411–20. doi: 10.1016/j.actbio.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Han YP, Yan C, Zhou L, Qin L, Tsukamoto H. A matrix metalloproteinase-9 activation cascade by hepatic stellate cells in trans-differentiation in the three-dimensional extracellular matrix. J Biol Chem. 2007;282:12928–39. doi: 10.1074/jbc.M700554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stetler-Stevenson WG. Tissue inhibitors of metalloproteinases in cell signaling: metalloproteinase-independent biological activities. Sci Signal. 2008;1:re6. doi: 10.1126/scisignal.127re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melendez-Zajgla J, Del Pozo L, Ceballos G, Maldonado V. Tissue inhibitor of metalloproteinases-4. The road less traveled. Mol Cancer. 2008;7:85. doi: 10.1186/1476-4598-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gharaibeh B, Lu A, Tebbets J, Zheng B, Feduska J, Crisan M, Peault B, Cummins J, Huard J. Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nat Protoc. 2008;3:1501–9. doi: 10.1038/nprot.2008.142. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Pan H, Huard J. Isolating stem cells from soft musculoskeletal tissues. J Vis Exp. 2010 doi: 10.3791/2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae YH, Ding Z, Zou L, Wells A, Gertler F, Roy P. Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J Cell Physiol. 2009;219:354–64. doi: 10.1002/jcp.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J Cell Biol. 2005;171:349–59. doi: 10.1083/jcb.200504029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deasy BM, Gharaibeh BM, Pollett JB, Jones MM, Lucas MA, Kanda Y, Huard J. Long-term self-renewal of postnatal muscle-derived stem cells. Mol Biol Cell. 2005;16:3323–33. doi: 10.1091/mbc.E05-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urish KL, Vella JB, Okada M, Deasy BM, Tobita K, Keller BB, Cao B, Piganelli JD, Huard J. Antioxidant levels represent a major determinant in the regenerative capacity of muscle stem cells. Mol Biol Cell. 2009;20:509–20. doi: 10.1091/mbc.E08-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng B, Cao B, Li G, Huard J. Mouse adipose-derived stem cells undergo multilineage differentiation in vitro but primarily osteogenic and chondrogenic differentiation in vivo. Tissue Eng. 2006;12:1891–901. doi: 10.1089/ten.2006.12.1891. [DOI] [PubMed] [Google Scholar]
- 40.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jankowski RJ, Deasy BM, Huard J. Muscle-derived stem cells. Gene Ther. 2002;9:642–7. doi: 10.1038/sj.gt.3301719. [DOI] [PubMed] [Google Scholar]
- 42.Jankowski RJ, Haluszczak C, Trucco M, Huard J. Flow cytometric characterization of myogenic cell populations obtained via the preplate technique: potential for rapid isolation of muscle-derived stem cells. Hum Gene Ther. 2001;12:619–28. doi: 10.1089/104303401300057306. [DOI] [PubMed] [Google Scholar]
- 43.Lee JY, Qu-Petersen Z, Cao B, Kimura S, Jankowski R, Cummins J, Usas A, Gates C, Robbins P, Wernig A, Huard J. Clonal isolation of muscle-derived cells capable of enhancing muscle regeneration and bone healing. J Cell Biol. 2000;150:1085–100. doi: 10.1083/jcb.150.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corsi KA, Pollett JB, Phillippi JA, Usas A, Li G, Huard J. Osteogenic potential of postnatal skeletal muscle-derived stem cells is influenced by donor sex. J Bone Miner Res. 2007;22:1592–602. doi: 10.1359/jbmr.070702. [DOI] [PubMed] [Google Scholar]
- 45.Kim KS, Lee JH, Ahn HH, Lee JY, Khang G, Lee B, Lee HB, Kim MS. The osteogenic differentiation of rat muscle-derived stem cells in vivo within in situ-forming chitosan scaffolds. Biomaterials. 2008;29:4420–8. doi: 10.1016/j.biomaterials.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 46.Payne KA, Meszaros LB, Phillippi JA, Huard J. Effect of phosphatidyl inositol 3-kinase, extracellular signal-regulated kinases 1/2, and p38 mitogen-activated protein kinase inhibition on osteogenic differentiation of muscle-derived stem cells. Tissue Eng Part A. 2010;16:3647–55. doi: 10.1089/ten.tea.2009.0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonewald LF, Harris SE, Rosser J, Dallas MR, Dallas SL, Camacho NP, Boyan B, Boskey A. von Kossa staining alone is not sufficient to confirm that mineralization in vitro represents bone formation. Calcif Tissue Int. 2003;72:537–47. doi: 10.1007/s00223-002-1057-y. [DOI] [PubMed] [Google Scholar]
- 48.Peault B, Rudnicki M, Torrente Y, Cossu G, Tremblay JP, Partridge T, Gussoni E, Kunkel LM, Huard J. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol Ther. 2007;15:867–77. doi: 10.1038/mt.sj.6300145. [DOI] [PubMed] [Google Scholar]
- 49.Pawlikowski B, Lee L, Zuo J, Kramer RH. Analysis of human muscle stem cells reveals a differentiation-resistant progenitor cell population expressing Pax7 capable of self-renewal. Dev Dyn. 2009;238:138–49. doi: 10.1002/dvdy.21833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurme T, Kalimo H, Lehto M, Jarvinen M. Healing of skeletal muscle injury: an ultrastructural and immunohistochemical study. Med Sci Sports Exerc. 1991;23:801–10. [PubMed] [Google Scholar]
- 51.Huard J, Gharaibeh B, Usas A. Regenerative Medicine Based on Muscle-Derived Stem Cells. Oper Tech Orthop. 2010;20:119–126. [Google Scholar]
- 52.Wang XX, Li YC, Ma XP, Han ZC. [Effects of electroacupuncture at Jiaji (EX-B 2) on extracellular matrix MMP-1, MMP-3 and TIMP-1 levels in the degenerated cervical intervertebral disc in rats] . Zhen Ci Yan Jiu. 2009;34:248–51. [PubMed] [Google Scholar]
- 53.Mu X, Urso ML, Murray K, Fu F, Li Y. Relaxin regulates MMP expression and promotes satellite cell mobilization during muscle healing in both young and aged mice. Am J Pathol. 2010;177:2399–410. doi: 10.2353/ajpath.2010.091121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bellayr IH, Mu X, Li Y. Biochemical insights into the role of matrix metalloproteinases in regeneration: challenges and recent developments. Future Med Chem. 2009;1:1095–1111. doi: 10.4155/fmc.09.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishimura T, Nakamura K, Kishioka Y, Kato-Mori Y, Wakamatsu J, Hattori A. Inhibition of matrix metalloproteinases suppresses the migration of skeletal muscle cells. J Muscle Res Cell Motil. 2008;29:37–44. doi: 10.1007/s10974-008-9140-2. [DOI] [PubMed] [Google Scholar]
- 56.Allen DL, Teitelbaum DH, Kurachi K. Growth factor stimulation of matrix metalloproteinase expression and myoblast migration and invasion in vitro. Am J Physiol Cell Physiol. 2003;284:C805–15. doi: 10.1152/ajpcell.00215.2002. [DOI] [PubMed] [Google Scholar]
- 57.Barnes BR, Szelenyi ER, Warren GL, Urso ML. Alterations in mRNA and protein levels of metalloproteinases-2, -9, and -14 and tissue inhibitor of metalloproteinase-2 responses to traumatic skeletal muscle injury. Am J Physiol Cell Physiol. 2009;297:C1501–8. doi: 10.1152/ajpcell.00217.2009. [DOI] [PubMed] [Google Scholar]
- 58.El Fahime E, Torrente Y, Caron NJ, Bresolin MD, Tremblay JP. In vivo migration of transplanted myoblasts requires matrix metalloproteinase activity. Exp Cell Res. 2000;258:279–87. doi: 10.1006/excr.2000.4962. [DOI] [PubMed] [Google Scholar]
- 59.Bellayr IH, Gharaibeh B, Huard J, Li Y. Skeletal muscle-derived stem cells differentiate into hepatocyte-like cells and aid in liver regeneration. Int J Clin Exp Pathol. 2010;3:681–90. [PMC free article] [PubMed] [Google Scholar]
- 60.Cao B, Zheng B, Jankowski RJ, Kimura S, Ikezawa M, Deasy B, Cummins J, Epperly M, Qu-Petersen Z, Huard J. Muscle stem cells differentiate into haematopoietic lineages but retain myogenic potential. Nat Cell Biol. 2003;5:640–6. doi: 10.1038/ncb1008. [DOI] [PubMed] [Google Scholar]
- 61.Kubo S, Cooper GM, Matsumoto T, Phillippi JA, Corsi KA, Usas A, Li G, Fu FH, Huard J. Blocking vascular endothelial growth factor with soluble Flt-1 improves the chondrogenic potential of mouse skeletal muscle-derived stem cells. Arthritis Rheum. 2009;60:155–65. doi: 10.1002/art.24153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lavasani M, Lu A, Peng H, Cummins J, Huard J. Nerve growth factor improves the muscle regeneration capacity of muscle stem cells in dystrophic muscle. Hum Gene Ther. 2006;17:80–92. doi: 10.1089/hum.2006.17.180. [DOI] [PubMed] [Google Scholar]
- 63.Li G, Corsi-Payne K, Zheng B, Usas A, Peng H, Huard J. The dose of growth factors influences the synergistic effect of vascular endothelial growth factor on bone morphogenetic protein 4-induced ectopic bone formation. Tissue Eng Part A. 2009;15:2123–33. doi: 10.1089/ten.tea.2008.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsumoto T, Kubo S, Meszaros LB, Corsi KA, Cooper GM, Li G, Usas A, Osawa A, Fu FH, Huard J. The influence of sex on the chondrogenic potential of muscle-derived stem cells: implications for cartilage regeneration and repair. Arthritis Rheum. 2008;58:3809–19. doi: 10.1002/art.24125. [DOI] [PubMed] [Google Scholar]
- 65.Romero-Ramos M, Vourc’h P, Young HE, Lucas PA, Wu Y, Chivatakarn O, Zaman R, Dunkelman N, el-Kalay MA, Chesselet MF. Neuronal differentiation of stem cells isolated from adult muscle. J Neurosci Res. 2002;69:894–907. doi: 10.1002/jnr.10374. [DOI] [PubMed] [Google Scholar]
- 66.Smaldone MC, Chancellor MB. Muscle derived stem cell therapy for stress urinary incontinence. World J Urol. 2008;26:327–32. doi: 10.1007/s00345-008-0269-9. [DOI] [PubMed] [Google Scholar]
- 67.Bouloumie A, Sengenes C, Portolan G, Galitzky J, Lafontan M. Adipocyte produces matrix metalloproteinases 2 and 9: involvement in adipose differentiation. Diabetes. 2001;50:2080–6. doi: 10.2337/diabetes.50.9.2080. [DOI] [PubMed] [Google Scholar]
- 68.Chavey C, Mari B, Monthouel MN, Bonnafous S, Anglard P, Van Obberghen E, Tartare-Deckert S. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J Biol Chem. 2003;278:11888–96. doi: 10.1074/jbc.M209196200. [DOI] [PubMed] [Google Scholar]
- 69.Brack AS, Conboy IM, Conboy MJ, Shen J, Rando TA. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–9. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 70.Luo D, Renault VM, Rando TA. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin Cell Dev Biol. 2005;16:612–22. doi: 10.1016/j.semcdb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 71.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 72.Li Y, Foster W, Deasy BM, Chan Y, Prisk V, Tang Y, Cummins J, Huard J. Transforming growth factor-beta1 induces the differentiation of myogenic cells into fibrotic cells in injured skeletal muscle: a key event in muscle fibrogenesis. Am J Pathol. 2004;164:1007–19. doi: 10.1016/s0002-9440(10)63188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–39. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]


