Abstract
Mature B-cell lymphomas with both BCL2 and MYC translocations are known as “double hit” lymphomas. These lymphomas are aggressive and show high proliferation rate due to the growth advantages provided by MYC and BCL2 translocation and overexpression. Mantle cell lymphoma (MCL) is a neoplasm of mature B-lymphocytes with characteristic t(11;14) and subsequent Cyclin D1 overexpression. Secondary cytogenetic changes are frequent in MCL, but MYC translocation has only been rarely reported. In this study, we report four cases of MCL with MYC translocation or MYC gene amplification detected by conventional cytogenetics, fluorescence in situ hybridization and whole genome single nucleotide polymorphism (SNP) array, and determined the clinicopathologic features. Our study provides further evidence supporting the concept of “double hit” MCL with co-involvement of MYC gene rearrangement and/or amplification and CCND1 gene rearrangement.
Keywords: Mantle cell lymphoma, double hit lymphoma, MYC, 8q24, translocation, amplification
Introduction
Mantle cell lymphoma (MCL) is an aggressive and incurable mature B-cell lymphoma characterized by t(11;14)(q13;q32) and overexpression of cyclin D1 [1,2]. While patients with MCL typically follow a relentless progressive clinical course, there is some heterogeneity in the morphology, cytogenetics and even clinical behaviors among patients with MCL. Some cases of MCL have more indolent clinical course and are characterized by non-nodal leukemia disease [3,4]. On the other hand, blastoid and pleomorphic MCLs usually represent more advanced and aggressive disease [5]. The difference in clinical behavior depends on secondary cytogenetic abnormalities in addition to the translocation of the cyclin D1 gene locus. Cyclin D1 is essential for cell cycle progression through the G1-S checkpoint. Deregulated expression of cyclin D1 is considered an initiating event in MCL lymphomagenesis, but not sufficient for the development of MCL [6,7]. High numbers of secondary chromosomal aberrations are detected in MCL and the genes involved are likely related to tumor progression [8,9]. Abnormalities of the MYC gene locus in MCL are rare but associated with aggressive clinical behavior and blastoid morphology [10-13].
Here we report four cases of blastoid mantle cell lymphoma with secondary cytogenetic abnormalities involving the MYC gene locus. All cases had stage IV highly aggressive disease with systemic involvement. Fluorescence in situ hybridization (FISH) and single nucleotide polymorphism (SNP) microarray analysis showed either MYC translocation or gene amplification. The MYC translocation and gene amplification resulted in overexpression of MYC protein. We also reviewed cases with concurrent cyclin D1 gene rearrangement and MYC abnormalities in the literature and summarized their clinical and pathological features. Similar to double hit diffuse large B-cell lymphoma with both BCL-2 and MYC gene rearrangements, the “double hit” MCL with MYC abnormalities represent a transformed and highly aggressive disease.
Materials and methods
Case selection
Four cases of mantle cell lymphoma with MYC gene locus abnormalities were retrieved from the files of the Department of Hematopathology and Laboratory Medicine, H Lee Moffitt Cancer Center and Research Institute, Tampa, FL. Clinical information and follow-up were obtained from chart review. Relevant clinical information was summarized in Table 1. The study was approved by the Institutional Review Board of the University of South Florida.
Table 1.
Summary of clinical and laboratory findings in 4 patients
| Case 1 | Case 2 | Case 3 | Case 4 | |
|---|---|---|---|---|
| Age/Sex | 73/F | 61/M | 69/M | 81/F |
| Stage | IV | IV | IV | IV |
| CBC | HGB 11.9 g/dL WBC 96.2 ×109/L Platelet 121 ×109/L | HGB 6.6 g/dL WBC 438.5 ×109/L Platelet 59 ×109/L | HGB 9.1 g/dL WBC 21.2 ×109/L Platelet 18 ×109/L | HGB 9.4 g/dL WBC 84.7 ×109/L Platelet 141 ×109/L |
| Absolute lymphocyte count in PB | 86 ×109/L | 425 ×109/L | 14 ×109/L | 51 ×109/L |
| Bone marrow involvement | ND | ND | Yes | Yes |
| Lymphadenopathy | Yes, generalized | No | No | No |
| Splenomegaly | Yes, 15 cm by CT | Yes, 18 cm by CT | Yes, 20 cm by CT | Yes, 1 cm below left costal margin |
| Serum LDH | 688 U/L | 17870 U/L | 800 U/L | 3094 U/L |
| Therapy | R-CHOP Bendamustine and Rituxan | EPOCH Hyper-CVAD | CHOP Hyper-CVAD Autologous HSCT | None |
| Outcome, months | CR, 17 | CR, 4 | Persistent disease, 13 | In hospice, 3 |
M, male; F, female; CBC, Complete Blood Count; PB, peripheral blood; ND, not determined; CT, computerized axial tomography; CR, complete remission; R, Rituximab; CHOP, Cytoxan, adriamycin, vincristine, and prednisone; EPOCH, etoposide, prednisone, vincristine, cytoxan, and doxorubicin; Hyper-CVAD, cytoxan, adriamycin, vincristine, prednisone alternating with high dose cytarabine and methotrexate; HSCT, hematopoietic stem cell transplant.
Histology and immunohistochemistry
The morphology and immunohistochemistry were routinely studied on peripheral blood smear, and formalin-fixed and paraffin-embedded tissue sections. Immunohistochemical stains were performed using an automated immunostainer (Ventana Medical Systems, Inc. Tucson, AZ) according to established protocols. Antigen retrieval was performed using CC1, high pH buffer. Antibodies included CD20 (L26, predilute; Ventana), CD3 (2GV6, predilute, Ventana), CD5 (4C7, 1:10, Leica), CD23 (1B12, 1:20, Leica), Cyclin D1 (SP4, predilute, Ventana), p53 (BP53-11, predilute, Ventana), and MYC (Y69, 1:10, Biocare). Positive and negative controls were performed with all cases and showed appropriate reactivity and specificity.
Whole genome Single Nucleotide Polymorphism (SNP) array
SNP microarray analysis was performed using the Affymetrix Cytoscan HD platform which uses over 743,000 SNP probes and 1,953,000 non-polymorphic copy probes with a median spacing of 0.88 kb within genes. 250 ng of total genomic DNA extracted was digested with Nsp1 and then ligated to Nsp1 adaptors, and amplified using Titanium Taq with a GeneAmp PCR system 9700. PCR products were purified using AMPure beads and quantified using NanoDrop 8000. Purified DNA was fragmented and biotin labeled and hybridized to the Affymetrix cytoscan HD GeneChip. Data was analyzed using the Chromosome Analysis Suite. The analysis was based on GRCh37/hg 19 assembly.
Positive evaluation criteria include: DNA copy gain/loss within or including a known clinically significant cancer related gene (530 in database) of 50 Kb or greater; DNA copy number loss of >500Kb or gain >1Mb outside known clinical oncology significant regions with at least one OMIM annotated gene of possible clinical significance; Contiguous allele homozygosity >8Mb through the telomere of a single chromosome is consistent with acquired uniparental disomy (aUPD). These regions designate clonal evolution associated with the acquisition of homozygosity for a gene mutation within the homozygotic stretch. Empiric studies have detected whole chromosome 22 mosaicism below 10.0%.
Flow cytometric Immunophenotyping
The specimen processing and antibody staining were performed according to manufacturers’ recommendations. The antibody panel included the following: CD3-PerCP, CD5-APC, CD10-PE, CD4-FITC, CD7-PE, CD8-APC, CD11c-APC, CD14-FITC, CD19-PerCP, CD19-APC, CD20-APC, CD103-FITC, CD23-PE, FMC7-FITC, CD25-PE, CD38-APC, CD45-PerCP, CD62L-FITC, GPA-PE, IgG1-PE, IgG1-PerCP, IgG1-APC, IgG2a-FITC, kappa-FITC, and lambda-PE. Specimens were acquired with 6-parameter 4-color flow cytometry and all the antibodies were from Becton Dickinson. We used the FACSCalibur flow cytometer by Becton-Dickinson. Data acquisition was done in CellQuest Pro (sensitivity of fluorescent detectors monitored using standardized beads according to the manufacturer’s recommendations). At least 5,000 lymphocytes were acquired per tube. For analysis, cell populations were gated based on their forward and side scatter characteristics, in conjunction with antigen back-gating on CD19+/CD5+ cells to determine the appropriateness of the gates. Normal lymphoid cells within the specimens served as internal controls for determination of antibody binding intensity.
Fluorescence in situ hybridization
FISH probes were acquired from Vysis, Inc. and included a multiprobe panel for chronic lymphocytic leukemia (Vysis LSI TP53, Vysis LSI ATM, Vysis LSI D13S319, Vysis LSI 13q34, and Vysis CEP12), Vysis LSI IGH/CCND1 Dual Color, Dual Fusion Translocation Probe, Vysis LSI BCL6 (ABR) Dual Color Break Apart Rearrangement Probe, and Vysis LSI MYC Dual Color Break Apart Rearrangement Probe. The specimen processing and hybridizaton were performed according to manufacturers’ recommendations. The samples were analyzed using standard fluorescence microscopy methods.
Results
Clinical findings
The clinical features of the 4 patients are summarized in Table 1. There were two males and two females with an age range from 61 to 81 years (median, 71 years). All patients had stage IV disease with leukemic infiltrate in the peripheral blood. The white blood cells were markedly increased with absolute lymphocytosis. The absolute lymphocyte counts ranged from 14 ×109/L to 425 ×109/L (median, 68.5 ×109/L). All patients had marked splenomegaly, and two patients had involvement of the bone marrow (cases 3 and 4). No bone marrow biopsies were performed on patients 1 and 2. One patient had generalized lymphadenopathy (case 1), while the other three patients had no lymphadenopathy at the time of diagnosis. Serum LDH level was increased in all four patients, ranging from 688 U/L to 17870 U/L (median, 1947 U/L). Patient 1 had a reported history of chronic lymphocytic leukemia (CLL) with minimal lymphadenopathy and slightly elevated white blood cell count for 8 years. The diagnosis of CLL was based on flow cytometry with the presence of CD5 and CD23 positive clonal B-cells in the peripheral blood. Fluorescence in situ hybridization (FISH) study was not performed to confirm the diagnosis and a possibility of mantle cell lymphoma cannot be excluded by the authors. Two of the patients had good response to chemotherapy with complete remission (Case 1 and 2). Patient 1 was initially treated with R-CHOP (rituximab, cytoxan, adriamycin, vincristine, and prednisone) with complete remission. She relapsed with circulating lymphoma cells, increasing lymphadenopathy and splenomegaly 8 months later. She was subsequently treated with bendamustine and rituximab with complete remission, and placed on Rituximab maintenance. Patient 2 was treated with EPOCH (etoposide, prednisone, vincristine, cytoxan, and doxorubicin) for one cycle and then hyper-CVAD (cytoxan, adriamycin, vincristine, prednisone alternating with high dose cytarabine and methotrexate) for three cycles with complete remission. Patient 3 was treated with CHOP and hyper-CVAD with partial response. He underwent autologous hematopoietic stem cells after high dose chemotherapy with BEAM (BCNU, etoposide, cytrabine and melphalan), but showed persistent disease post transplant. Patient 4 elected not to receive chemotherapy and enrolled in hospice.
Morphologic and immunophenotypic findings
The lymphoma cells showed similar morphology in the peripheral blood of all 4 patients. The neoplastic cells were large with round to irregular nuclei, blastoid chromatin, prominent nucleoli and moderate amounts of basophilic cytoplasm (Figure 1). Some of the cells had cytoplasmic vacuoles. The bone marrow showed extensive infiltrate by mantle cell lymphoma with approximately 95% (case 3) and 60% (case 4) involvement, respectively. The lymphoma demonstrated an interstitial, nodular and diffuse infiltration pattern in the bone marrow (Figure 2A). Mitotic figures were easily seen. By immunohistochemical stains, the lymphoma cells were positive for CD20, cyclin D1, and MYC (Figure 2B, 2C). Ki-67 showed a moderately high proliferation rate (>50%). In the aspirate smears, the lymphoma cells showed similar blastoid morphology as seen in the peripheral blood (Figure 2D).
Figure 1.
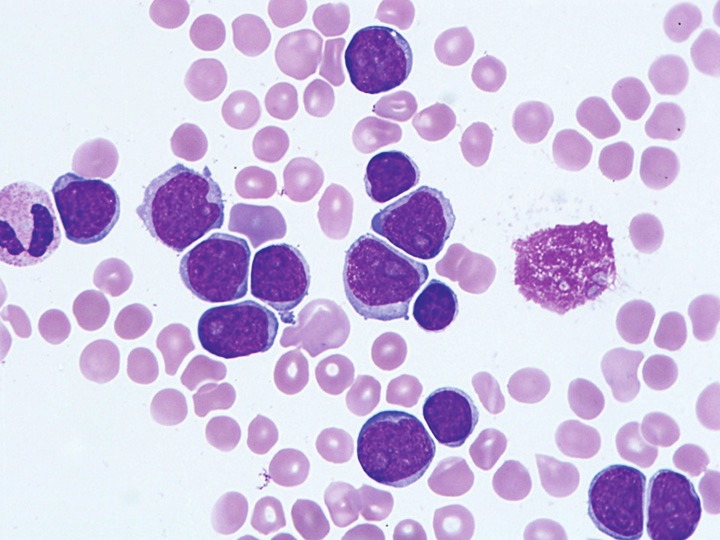
Representative image of cytological features of blastoid mantle cell lymphoma with MYC gene locus abnormalities in the peripheral blood smear (Case 2, Wright-Giemsa stain, magnification 1000x).
Figure 2.
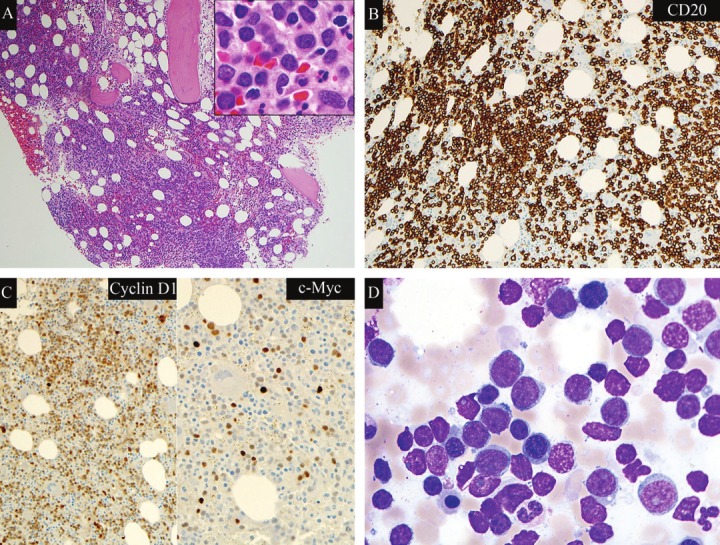
Hypercellular marrow with extensive infiltrate of blastoid mantle cell lymphoma. Case 4. A. hematoxylineosinstain, magnification 100x (Inlet, magnification 1000x). B. CD20 immunostain, magnification 200x. C. CyclinD1 immunostain, magnification 200x; MYC immunostain, magnification 400x. D. Bone marrow aspirate smear(Wright-Giemsa stain, magnification 1000x).
Flow cytometry analysis was performed on the peripheral blood of all four patients. Flow cytometry showed a predominance of B-cells with monotypic surface immunoglobulin light chain expression (2 kappa and 2 lambda). The neoplastic cells were positive for CD19 and CD20, but demonstrated immunophenotypic variations in all 4 cases. The lymphoma cells in three cases (1, 2 and 4) showed expression of CD5, but also weak to moderate expression of CD23 (Figure 3). Case 3 displayed either no expression or dim partial expression of CD5 in the lymphoma cells on different peripheral blood specimens (data not shown). CD23 was consistently negative in case 3.
Figure 3.
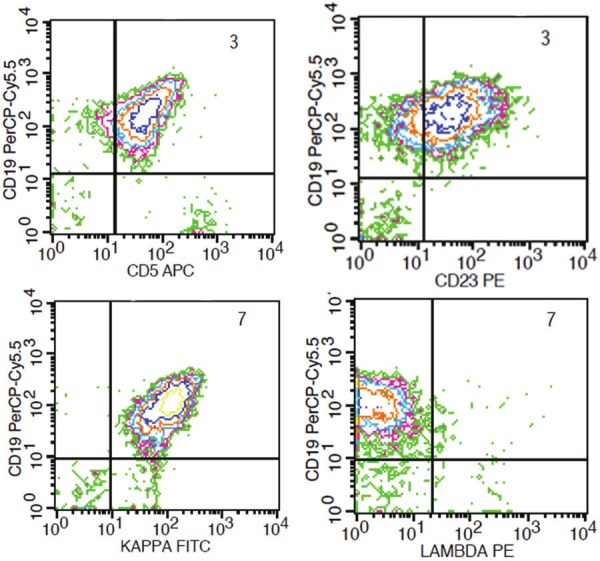
Blastoid mantle cell lymphoma cells with coexpression of CD5 and CD23 by flow cytometry analysis of peripheral blood. Case 2.
Cytogenetic, Fluorescence In Situ Hybridization (FISH) and whole genome Single Nucleotide Polymorphism (SNP) array findings
The cytogenetic and FISH findings are summarized in Table 2. Of the three cases with conventional cytogenetic analysis performed (cases 2, 3, 4), all showed complex karyotypes with abnormalities involving multiple chromosomes, in addition to the presence of t(11;14) (q13;q32). Chromosome 8q24 abnormalities were not detected by karyotype analysis, except the presence of trisomy 8 and isochromosome i(8)(q10) in case 3 (Figure 4C) FISH analysis using the CLL panel probes for 11q13/14q32 fusion, 11q22.3, 13q14.3, 17p13.1, and cen 12, along with BCL6 and MYC probes were performed in all four cases. The lymphoma cells in all cases showed CCND1/IGH fusion signals (case1, 73% of nuclei with an extra IGH signal; case 2, 65% of nuclei; case 3, 53.5% of nuclei; and case 4, 86% of nuclei with 2-3 cyclin D1/IGH gene fusion signals) (Figure 4A) ATM gene deletion at 11q22.3 was also detected in cases 1 and 4 (case 1, 85% of nuclei; case 4, 55% of nuclei). In case 1, FISH analysis using MYC dual color breakapart probe showed 73% of nuclei positive for MYC gene rearrangement, which highly suggested that the extra IGH signal noted in the CLL panel representing a second IGH gene rearrangement with MYC. In case 2, FISH analysis showed 98% of nuclei positive for MYC gene rearrangement and extra signals, and 92% of nuclei positive for extra BCL6 signals. The presence of three unsplit MYC and BCL6 signals were noted in most cells, consistent with trisomy of both 3q and 8q. There was no rearrangement of BCL6, although an extra 3’ portion of MYC was found in 98% of nuclei, consistent with additional rearrangement of MYC. In case 3, FISH showed 6.5% of nuclei positive for MYC gene rearrangement, 25.5% of nuclei positive for three MYC gene signals (Figure 4B), and 31% of nuclei positive for three BCL6 gene signals. In case 4, 60% of nuclei were positive for amplification of MYC gene without rearrangement.
Table 2.
Cytogenetic and fluorescent in situ hybridization (FISH) results
| Case | Karyotype | FISH |
|---|---|---|
| 1 | ND | nuc ish 11q13(CCND1x3),14q32(IGHx4),(CCND1 con IGHx2) [146/200] nuc ish 8q24(CMYCx2)(5’CMYC sep 3’CMYCx1)[146/200] nuc ish 11q22.3(ATMx1)[170/200] |
| 2 | 45,X,-Y,t(3;15)(q21;q11.2),del(6)(q22), add(8)(p12),t(11;14) (q13;q32),add(17)(q25),+mar[cp13]/46,XY[7] | nuc ish 11q13(CCND1x3),14q32(IGHx3),(CCND1 con IGHx2) [136/200] nuc ish 8q24(5’C-MYCx3,3’C MYCx4)(5’C-MYC con 3’C-MYCx3) [98/100] nuc ish 6cen(CEP6x2),6q22(MYBx1)[151/200] nuc ish 14q32.3(IGHx3),18q21(BCL2x2)[139/200] nuc ish 3q27(BCL6x3)[120/150]/nuc ish 3q27(BCL6x4)[19/150] |
| 3 | 44,X,-Y,add(1)(q32),+8,i(8)(q10)x2, t(11;14)(q13;q32),-13,i(18)(q10),-19,-22,+mar[cp2]/46,XY[cp20] | nuc ish 11q13(CCND1x4),14q32(IGHx4)(CCND1 con IGHx3) [107/200] nuc ish 8q24(CMYCx3)[51/200]/(CMYCx2) (5’CMYC sep 3’CMYCx1)[13/200] nuc ish 3q27(BCL6x3 [62/200] |
| 4 | 42-43,XX,der(1)del(1)(p13.3p21)add(1)(q12),add(2)(p23),add(8)(p21), -9,add(9)(p21),add(10)(q22),t(11;14)(q13;q32),-13,del(13)(q12q22),-14, der(18)t(1;18)(q21;p11.2),-22,+mar[cp12]/46,XX[8] | nuc ish 11q13(CCND1x3),14q32(IGHx3),(CCND1 con IGHx2) [118/150] nuc ish 11q13(CCND1x4),14q32(IGHx4),(CCND1 con IGHx3)[11/150] nuc ish 8q24 (C-MYCx2,3’CMYCampx10-20)[60/100] nuc ish 11q22.3(ATMx1)[59/98] nuc ish 13q14.3(D13S319x1),13q34(LAMPx2)[38/150] |
ND, not done; FISH, fluorescent in situ hybridization.
Figure 4.
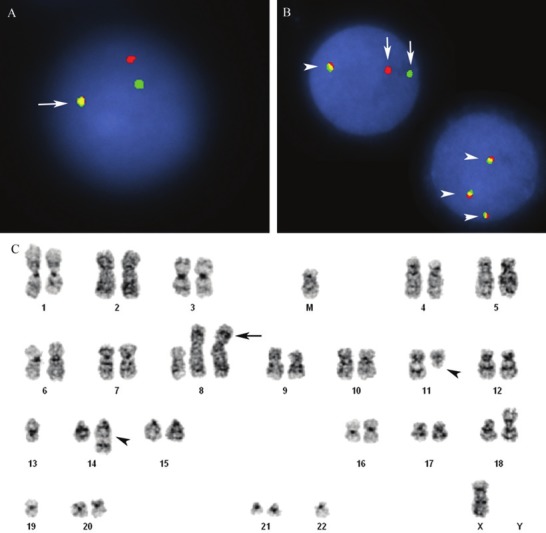
Double hit mantle cell lymphoma with t(11;14) and MYC gene rearrangement. Case 3. A. Interphase FISH using immunoglobulin heavy chain/CCND1 dual fusion probe for t(11;14). IGH/CCND1 translocation represented by fused green/red (yellow) signals (arrow), in addition to 1 red and 1 green signal. Normal signal pattern represented by 2 red and 2 green signals. B. Interphase FISH using MYC dual color breakapart probe. MYC gene rearrangement represented by separated 1 green and 1 red signals (arrow), normal MYC gene locus represented by fused green/red (yellow) signal (arrowheads). One cell has three yellow signals consistent with trisomy 8. C. Complex karyotype with a t(11;14) IGH/CCND1 gene rearrangement (arrowheads) and concurrent trisomy 8 and i(8)(q10) x2 [44,X,-Y,add(1)(q32),+8,i(8) (q10)x2, t(11;14)(q13;q32),-13,i 18)(q10),-19,-22,+mar].
The whole genome chromosome SNP/CN microarray copy number analysis was performed on case 4, and showed numerous duplications and deletions of chromosomal segments in chromosomes 1p, 1q, 6p, 8p, 8q, 9p, 10p, 11q, 13q, 14q, 18p, 18q, and 22q. The analysis showed clonal evolution: primary (50% of the cells) and secondary (30% of the cells). The primary changes include deletions of 1p, 8p and 8q, 9p, 10p, 11q, 13q, 14q, 18p, 22q (multiple), and duplications of 13q and 18p. The secondary changes include deletions of 1q, 13q, 18q and duplications of 6p and 22q. Also seen were amplifications of the 8q MYC gene and adjacent chromosomal segment (Figure 5) and 13q, as well as 8q and 9p (p16 gene) homozygous deletions. Chymotrypsis of chromosome 8q and 13q were delineated. Uniparental disomy for chromosome 22 was detected in approximately 50% of the cells. No other significant DNA copy number changes or copy neutral LOH were detected in the 1,695,000 region specific SNP/CNs.
Figure 5.

MYC gene amplification by whole genome single nucleotide polymorphism (SNP) array in case 4. CN, copy number.
Discussion
Mantle cell lymphoma (MCL) is a well defined mature B-cell neoplasm with characteristic morphologic and immunophenotypic features and a specific chromosomal translocation juxtaposing IGH@ and cyclin D1 (CCND1) genes [1,2]. Despite these unifying features, MCL represents a heterogeneous disorder, ranging from indolent MCL with benign clinical course to highly aggressive blastoid or pleomorphic variants [3,4]. Most patients with MCL have a progressively aggressive clinical course and are characterized morphologically by small to medium sized lymphoma cells with aberrant expression of CD5 but not CD23 [1]. The t(11;14) (q13;q32) with deregulated cyclin D1 expression is the primary genomic alteration in MCL. But the t(11;14) (q13;q32) and overexpression of cyclin D1 are not sufficient for lymphomagenesis [6,14]. Additional genetic alterations are apparently required for the development of MCL. Cytogenetic changes of MCL roughly correlate to the clinical and morphologic groups. While indolent MCL usually shows stable genome without many secondary chromosomal abnormalities other than t(11;14) (q13;q32), blastoid and pleomorphic MCL typically show a complex karyotype with highly unstable genome [15]. The cytogenetic studies with comparative genomic hybridization microarray (CGH) have revealed frequent secondary gains and losses, involving genes important for genomic stability, proliferation, and apoptosis, such as TP53, ATM, MYC, BMI1, CDK4 and BCL2 et al [15,16]. Among the large numbers of genes implicated in the lymphomagenesis of MCL, MYC appears to play an important role in transforming the initial B-cell clone harboring t(11;14)(q13;q32). Studies with transgenic mice showed strong cooperation between the MYC gene and the transcriptionally activated cyclin D1 in oncogenic transformation of B-cell lymphomas [6].
MYC is a global transcription factor regulating up to 10% of the human genes [17]. MYC regulates essential cell functions such as cell growth, cell cycle progression, cell survival, cellular metabolism and biosynthesis. MYC expression is tightly regulated at the transcripitional and translational levels. Deregulated MYC expression in the B-cells in transgenic mice is oncogenic and results in the development of B-cell lymphomas [18]. MYC deregulation in lymphoma occurs in two scenarios, one as a primary event such as in Burkitt lymphoma, and the other as a secondary event in a variety of lymphomas, including diffuse large B-cell lymphoma, transformed follicular lymphoma, plasmablastic lymphoma, lymphoblastic lymphoma, B-cell lymphoma unclassifiable with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma and mantle cell lymphoma [19]. MYC deregulation typically results from chromosomal translocation in lymphoma. In Burkitt lymphoma, MYC activation is the result of MYC translocation at 8q24 to the IG heavy chain locus at 14q32, or less frequently to the IG kappa or lambda light chain loci at 2p12 and 22q11 respectively. In contrast to Burkitt lymphoma, secondary MYC translocation in other B-cell lymphomas usually occurs in an unstable genome with complex karyotype and juxtaposed to non-IG gene locus. The presence of MYC translocation in this setting is typically associated with an aggressive clinical course. A subset of aggressive B-cell lymphoma with both BCL2 and MYC gene rearrangements is defined as “double hit” lymphoma and shows very poor prognosis. In consistent with the pivotal role of MYC in cell proliferation and metabolism, the double hit lymphoma and other aggressive B-cell lymphomas with MYC gene rearrangement usually show extremely high proliferation rate in the neoplastic cells. MCL with 8q24 abnormalities involving MYC gene is rare and often associated with blastoid morphology and poor prognosis [10-13,20-27].
In this study we presented four cases of MCL with secondary cytogenetic abnormalities involving the MYC gene locus (three with MYC gene translocation and one with MYC amplification). The MYC translocation was not associated with IG heavy or light chain gene loci. All cases were blastoid MCL with markedly elevated white cell blood count, circulating large lymphoma cells, extensive involvement of bone marrow (in cases with bone marrow biopsy), splenomegaly and elevated LDH. In case 4 whole genome chromosome SNP/CN microarray (Reveal) analysis showed numerous duplications and deletions in various chromosomes along with uniparental disomy on chromosome 22. Review of the literature shows 26 cases of MCL with 8q24 and MYC gene translocation or amplification (Table 3) [10-13,20-27]. All of the cases were advanced stage IV disease with high mortality mostly within the first two years after diagnosis despite aggressive chemotherapy. The majority were blastoid or pleomorphic MCL, only three cases with classic lymphocytic MCL. Complex karyotype was invariably present in addition to t(11;14)(q13;q32). Conventional cytogenetic analysis and FISH studies showed 15 cases with MYC translocation and 11 cases with additional 8q24 or MYC amplification. Of the 15 cases with MYC translocation, 5 cases showed translocation with the IGH@ gene locus, 4 cases showed translocation with the IG light chains gene locus (3 IGK@, 1 IGL@), and 6 cases showed non-IG translocation with 8q24/MYC. Cryptic MYC gene rearrangement in two cases [22,24] and MYC gene amplification in one case [26] were detected by FISH only, indicating the importance of performing FISH studies in aggressive B-cell lymphomas. These cases and the four cases presented in this study constitute an unique group of highly aggressive MCL with similar clinical and pathologic features. It should be noted that two of the patients in the current study achieved complete remission following aggressive chemotherapy. However, the follow-up period was relatively short (4 and 17 months) for the development of relapsed disease, as seen in several previous studies.
Table 3.
Mantle cell lymphoma with c-Myc translocation or amplification in the literature
| Reference | Cases | Morphology | Karyotype | c-Myc gene abnormality | Stage | Outcome |
|---|---|---|---|---|---|---|
| Au et al. [20] | 4 | Blastoid | 43-44,X,-Y,add(3)(p11),t(8;9)(q24;q13),der(11)t(11;14)(q13;q32), der(14)(14pter-->14q32::?::9q13 -->9qter), 15, der(17)t(3;17)(q13;p11)42-45,XY,add(2)(p25), del(6)(p21),del6(q15),der(8)ins(8;14)(q24;q24q32),del(9)(p22),t(11;14)(q13;q32), del(12)(q15),-14,-15,add(16)(q24)[cp8]/46,XX[8]64-65,XX,-X,-2,add(2)(p23),add(3)(p12)x2,-4,-4,+6,+8, add(8)(q24)x2, +9,del(10)(q24),+add(10)(q24),+11, t(11;14) (q13;q24),+12,-13x3,+14,-15,der(15)t(5;15)(p11;p11)x2,-16x3,+17,add(18)(q23),?idic(18)(q23), +dic(19)t(19;?)(p13;?),-20x3,-21,-22,+7mar[cp7]/46, XX[7]45,XY,dic(8;9)(q24;p24),-9,t(11;14)(q13;q32)[13]/46,XY[13] | ND | IV | Dead 1-15 m |
| Onciu et al. [21] | 3 | NA | 42-44,-X,Y,add(3)(q29),-5,add(8)(q24),add(9)(q34),add(10)(q26),t(11;14)(q13;q32),add(11)(p15),-13, add(13)(p13),-17,-18,add(22)(q13),del(22)(q11),+2,-6mar[cp13]/46,XY[7]45,X,-X,del(1)(p13p22),-2,der(8)t(2;8)(q13;q24),del(9) (p12),t(11;14)(q13;q32),t(12;18) (q13;q23),-13,add(15)(p15),add(21)(p13),+2mar[9]/46,XY[11]46,XY,del(7)(q22),add(8)(q24),t(11;14)(q13;q32)[4] | NA | NA | NA |
| Hao et al. [10] | 5 | 2 Blastoid 3 pleomorphic | 46-48,XX,add(8)(q24),add(9)(q34),t(11;14) (q13;q32), 13, add(13)(q22),del(17)(p11.2),+0-2 mar[cp28]46,XY,del(7)(q22),add(8)(q24),t(11;14)(q13;q32)[4]42-44,-X,Y,add(3)(q29),-5,add(8)(q24),add(9)(q34),add(10)(q26),t(11;14)(q13;q32), add(11)(p15),-13,add(13)(p13),-17,-18,add(22)(q13),del(22)(q11),+2,-6 mar[cp13]45,X,-X,del(1)(p13;p22),-2,der(8)t(2;8) (q13;q24),del(9) (p12),t(11;14)(q13;q32),t(12;18)(q13;q23),-13,add(15)(p15),add(21)(p13),+2mar[9]46,X,del(X)(q26),t(8;14)(q24;q32),t(11;14) (q13;q32), add(13)(q32)add(15)(q24),del(17)(p11;p13), add(18) (p11)[14] | Three c-Myc translocation by FISH Two c-Myc amplification by FISH | IV | 2 Dead, 22, 60 m 3 Alive, 3-21 m |
| Michaux et al. [22] | 1 | Pleomorphic | 46,XY,t(11;19;14)(q13;q13;q32),ins(12;?)(q21;?),del(13)(q14q22)[6]/46,idem,add(2)(p25)[3]/46,idem, add(3)(q29)[2]. | t(8;22) (q24;q11) by FISH Trisomy 8 by FISH | IV | Dead, 2 m |
| Felten et al. [11] | 1 | Blastoid | ND | c-Myc gene rearrangement by FISH | IV | Dead, 4 m |
| Zhang et al. [23] | 1 | Classic | 46,XY,t(8;17;21;14;17;21;14)(q24;p13;q22;q11.2;p13; q22;q32),del(10)(q24),t(11;14)(q13;q32) | c-Myc gene rearrangement by FISH | IV | Alive, 22 m |
| Seok et al. [24] | 1 | Blastoid | 45-47,XY,-9,-11,der(14)t(11;14)(q13;q32),-22,+2~3mar[cp10]/46,XY[2] | c-Myc/IGH translocation by FISH | IV | Dead, 1 m |
| Tirier et al. [25] | 1 | Classic | 45,XY,t(2;8)(p12;q24), -10,dic(10;15)(p10;p13),del(11), t(11;14)(q13;q32),del(11)(q14q22),-15[20]/45,idem, del(6)(q?15q?24)[7]/46,XY[6] | ND | IV | Dead, 4 m |
| Vaishampayan et al. [12] | 2 | Blastoid | 44,X,-Y,add(6)(p25),t(8;14)(q24;q32),del(9) (q12),t(11;14) (q13;q32),-12,der(19)t(1;19) (q21;p13)42,X,-Y,add(6)(q27),dup(8)(q24q13),t(11;14)(q13;q32), dup(12)(q13q15),der(14)t(14;19) (p11;q11),add (15) (p11),-15,-17,-19 | c-Myc/IGH translocation by FISH in case 1 3 copies of c-Myc by FISH in case 2 | IV | Dead, 4, 10 m |
| Mossafa et al. [26] | 3 | Blastoid | 40-43,XY,-3,dic(5;13)(q35;p11),der(6)(q?),-6,add(8)(q24), der(9)t(9;?)(q34;?), der(11),-12,der(14) (q?),del(15)(q13q24),der(16)t(16;?)(p11;?),der(17)t(8;17)(q11;q11), r(?),+mar1,+mar2 [CP13]45-46,XX,del(1)(p34),i(6)(p10),10,t(11;14) (q13;q32), +mar1[CP06]43,XY,-9,t(11;14)(q13;q32),-12,-15,-17,-21,-22,+2-3mar [CP10] | c-Myc amplification by FISH (5->7 copies) | IV | 2 Dead, 1-2 m 1 Alive, PR |
| Mossafa et al. [26] | 1 | Classic | 51-55,XY,+X,+5,+6,del(6)(q15q22),+8,+9,+11,t(11;14)(q13;q32),4xdm[CP13] | c-Myc amplification by FISH (4-5 copies) | NA | Alive, CR |
| Oliveira et al. [27] | 1 | Blastoid | 47,XY,t(2;8)(p12;q24)[21],+7[21],del(9)(q13)[21],t(11;14)(q13;q32)[21],del(17)(p11.2) [21]/46,XY[9][cp21] | c-Myc gene overexpression by Real-time PCR | IV | Dead, 9 m |
| Reddy et al. [13] | 2 | Blastoid | 44-45,XY,del(2)(q11.2q21),der(3;17)(p10;q10),der(5) t(3;5)(q12;q15),t(11;14)(q13;q32) [cp6]/46,XY[14]42-44,X,-Y,add(1)(p13),t(2;8)(p12;q24),der(2)t(2;15)(p25;q11.2),+3,del(9)(p22p24),þdel(9)(p22p24),-10, del(11)(q21q23),t(11;14)(q13;q32),-13,-15,-17,add(17)(p11.2)[cp7]/46,XY[17] | Myc/IGH and MYC translocation in case 1, and Myc/IGK translocation in case 2 by FISH | IV | NA |
M, male; F, female; ND, not done; NA, not available; FISH, fluorescent in situ hybridization; PCR, polymerase chain reaction; m, month; CR, complete remission; PR, partial remission.
Just as aggressive B-cell lymphoma with both BCL2 and MYC rearrangements is defined as double hit lymphoma, MCL with both CCND1 and MYC gene rearrangements is specified as double hit MCL [28]. The effect of the second “hit” of MYC rearrangement on the morphologic features was well illustrated in an unusual case presented by Felten et al. [11]. In the case, the lymph node showed areas with classic MCL and areas resembling Burkitt lymphoma. The cells in the areas with Burkitt features showed non-IG associated MYC gene rearrangement by FISH. These cells also demonstrated acquisition of germinal center marker CD10, loss of BCL2, with persistent CCND1/IGH translocation and overexpression of cyclin D1. Since the MCL with MYC amplification in our study and others showed similar clinicopathologic features, these cases should also be classified as double hit MCL, even though the “second hit” by deregulated MYC is not mediated by chromosomal translocation. Gains of 8q24 do not necessarily correspond to MYC amplification. Tagawa et al. and Rubio-Moscardo et al. showed gains of 8q24 in 24% and 19% patients with MCL, respectively [29,30] The relatively high frequently of gains of 8q24 cannot explain the rarity of these double hit MCL. Beà et al. identified two cases of MCL with 8q24 amplification by CGH, one with typical morphology, and one with blastoid morphology [8]. However, only one of the two cases showed MYC gene amplification by Southern blot analysis. As MYC mRNA has a very short half life in normal cells, the mRNA level may not reflect the protein level and activation. In fact, Hernández et al showed 38% MCL had elevated MYC mRNA by Northern blot, with only slightly higher frequency in blastoid MCL [31]. In addition, Southern blot showed no MYC gene amplification in classic MCL, and only one of 13 blastoid MCL showed three fold MYC gene amplification. It appears the best methodology to detect MYC amplification is by FISH or high density SNP microarray. The SNP microarray allows the detection of exact gene copy number. The SNP microarray performed on case 4 of our study showed amplification of MYC gene, corresponding to the high copy number of MYC gene detected by FISH (Table 2). PCR studies may also be of use in determining the level of MYC gene amplification. Jardin et al. demonstrated that MYC gains were associated with high risk MCL with blastoid morphology by quantitative Multiplex PCR of Short Fluorescent Fragments (QMPSF) assays [32]. Recently, a specific antibody against MYC is available commercially for immunohistochemical staining on formalin fixed paraffin embedded tissue. The immunostain appeared promising for detection of MYC expression and activation, in correlation with MYC translocation [33]. However, there is still controversy as to the utility of immunostain for MYC in predicting MYC gene translocation in diffuse large B-cell lymphoma [34]. We showed variably strong MYC expression by immunostain in our cases, which seems to confirm the findings by Bajor-Dattilo et al. Immunostain for MYC has not been sufficiently evaluated in MCL.
In addition to MYC, other genes are also likely implicated in the aggressive behavior of lymphoma cells in double hit MCL. CDKN2A gene on chromosome 9 encodes two proteins by alternative splicing, INK4A and ARF, which are important in cell cycle control and stability of p53 respectively. SNP microarray in case 4 of our study showed homozygous deletion of the CDKN2A gene. Rubio-Moscardo et al. also demonstrated that deletions of INK4a gene were associated with poorer prognosis together with blastoid morphology, increased number of genomic gains, and TP53 gene deletion [30]. Assessment of the CDKN2A gene locus in double hit MCL pathogenesis will be important in future studies. The uniparental disomy is likely indicative of the presence of a homozygous mutation of a tumor suppressor gene. One such tumor suppressor on chromosome 22 is neurofibromatosis 2 (NF2), but its involvement in lymphoma is not established.
Both classic and blastoid/pleomorphic MCL typically show aberrant expression of CD5, but are negative for CD23. Three of the four cases in this study showed co-expression of CD5 and CD23, although the CD23 expression was only dim and partial in 2 cases. The aberrant expression of CD23 may be a common feature in the double hit MCL, although the cases are limited in our study.
In summary, we presented four cases of double hit MCL, defined as MYC gene rearrangement and/or amplification in addition to CCND1 gene rearrangement. Review of the literature shows that these cases represent a relatively unique group of MCL with highly aggressive clinical behavior. FISH, CGH or SNP microarray is recommended in blastoid mantle cell lymphoma with high circulating lymphoma cells and splenomegaly to determine MYC gene amplification, if conventional cytogenetics does not detect 8q24 translocation or is not available. Identification of such double hit MCL warrants aggressive Burkitt-type chemotherapy, as complete remission can be achieved, although the disease free survival is short as described in the literature. CDKN2A gene deletion may play a role in the pathogenesis of double hit MCL.
Acknowledgements
The authors would like to thank Helen S Molina in our immunohistology lab for her help with the immunostains.
References
- 1.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon: IARC Press; 2008. [Google Scholar]
- 2.Pileri SA, Falini B. Mantle cell lymphoma. Haematologica. 2009;94:1488–92. doi: 10.3324/haematol.2009.013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furtado M, Rule S. Indolent mantle cell lymphoma. Haematologica. 2011;96:1086–8. doi: 10.3324/haematol.2011.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ondrejka SL, Lai R, Smith SD, Hsi ED. Indolent mantle cell leukemia: a clinicopathological variant characterized by isolated lymphocytosis, interstitial bone marrow involvement, kappa light chain restriction, and good prognosis. Haematologica. 2011;96:1121–7. doi: 10.3324/haematol.2010.036277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raty R, Franssila K, Jansson SE, Joensuu H, Wartiovaara-Kautto U, Elonen E. Predictive factors for blastoid transformation in the common variant of mantle cell lymphoma. Eur J Cancer. 2003;39:321–9. doi: 10.1016/s0959-8049(02)00456-2. [DOI] [PubMed] [Google Scholar]
- 6.Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Cyclin D1/bcl-1 cooperates with MYC genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994;13:3487–95. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the MYC gene. EMBO J. 1994;13:2124–30. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bea S, Ribas M, Hernandez JM, Bosch F, Pinyol M, Hernandez L, Garcia JL, Flores T, Gonzalez M, Lopez-Guillermo A, Piris MA, Cardesa A, Montserrat E, Miro R, Campo E. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood. 1999;93:4365–74. [PubMed] [Google Scholar]
- 9.Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–62. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- 10.Hao S, Sanger W, Onciu M, Lai R, Schlette EJ, Medeiros LJ. Mantle cell lymphoma with 8q24 chromosomal abnormalities: a report of 5 cases with blastoid features. Mod Pathol. 2002;15:1266–72. doi: 10.1097/01.MP.0000037310.82136.99. [DOI] [PubMed] [Google Scholar]
- 11.Felten CL, Stephenson CF, Ortiz RO, Hertzberg L. Burkitt transformation of mantle cell lymphoma. Leuk Lymphoma. 2004;45:2143–7. doi: 10.1080/10428190410001711479. [DOI] [PubMed] [Google Scholar]
- 12.Vaishampayan UN, Mohamed AN, Dugan MC, Bloom RE, Palutke M. Blastic mantle cell lymphoma associated with Burkitt-type translocation and hypodiploidy. Br J Haematol. 2001;115:66–8. doi: 10.1046/j.1365-2141.2001.03056.x. [DOI] [PubMed] [Google Scholar]
- 13.Reddy K, Ansari-Lari M, Dipasquale B. Blastic mantle cell lymphoma with a Burkitt translocation. Leuk Lymphoma. 2008;49:740–50. doi: 10.1080/10428190701852024. [DOI] [PubMed] [Google Scholar]
- 14.Hirt C, Schuler F, Dolken L, Schmidt CA, Dolken G. Low prevalence of circulating t(11;14) (q13;q32)-positive cells in the peripheral blood of healthy individuals as detected by real-time quantitative PCR. Blood. 2004;104:904–5. doi: 10.1182/blood-2004-02-0738. [DOI] [PubMed] [Google Scholar]
- 15.Royo C, Salaverria I, Hartmann EM, Rosenwald A, Campo E, Bea S. The complex landscape of genetic alterations in mantle cell lymphoma. Semin Cancer Biol. 2011;21:322–34. doi: 10.1016/j.semcancer.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Salaverria I, Zettl A, Bea S, Moreno V, Valls J, Hartmann E, Ott G, Wright G, Lopez-Guillermo A, Chan WC, Weisenburger DD, Gascoyne RD, Grogan TM, Delabie J, Jaffe ES, Montserrat E, Muller-Hermelink HK, Staudt LM, Rosenwald A, Campo E. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. J. Clin. Oncol. 2007;25:1216–22. doi: 10.1200/JCO.2006.08.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol. 2010;149:484–97. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- 18.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–8. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 19.Slack GW, Gascoyne RD. MYC and aggressive B-cell lymphomas. Adv Anat Pathol. 2011;18:219–28. doi: 10.1097/PAP.0b013e3182169948. [DOI] [PubMed] [Google Scholar]
- 20.Au WY, Horsman DE, Viswanatha DS, Connors JM, Klasa RJ, Gascoyne RD. 8q24 translocations in blastic transformation of mantle cell lymphoma. Haematologica. 2000;85:1225–7. [PubMed] [Google Scholar]
- 21.Onciu M, Schlette E, Medeiros LJ, Abruzzo LV, Keating M, Lai R. Cytogenetic findings in mantle cell lymphoma cases with a high level of peripheral blood involvement have a distinct pattern of abnormalities. Am J Clin Pathol. 2001;116:886–92. doi: 10.1309/JQMR-323G-71Y9-M7MB. [DOI] [PubMed] [Google Scholar]
- 22.Michaux L, Wlodarska I, Theate I, Stul M, Scheiff JM, Deneys V, Ferrant A, Hagemeijer A. Coexistence of BCL1/CCND1 and CMYC aberrations in blastoid mantle cell lymphoma: a rare finding associated with very poor outcome. Ann Hematol. 2004;83:578–83. doi: 10.1007/s00277-004-0879-2. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Kern WF, Yu Z, Mulvihill JJ, Li S. Cryptic and complex chromosomal rearrangements and the deletion of TP53 gene in a patient with leukemic mantle cell lymphoma. Cancer Genet Cytogenet. 2006;169:169–73. doi: 10.1016/j.cancergencyto.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Seok Y, Kim J, Choi JR, Kim YR, Park SJ, Kim SJ, Song J, Lee KA. CD5-negative blastoid variant mantle cell lymphoma with complex CCND1/IGH and MYC aberrations. Ann Lab Med. 2012;32:95–8. doi: 10.3343/alm.2012.32.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tirier C, Zhang Y, Plendl H, Weber-Matthiesen K, Langer W, Heit W, Schlegelberger B. Simultaneous presence of t(11;14) and a variant Burkitt’s translocation in the terminal phase of a mantle cell lymphoma. Leukemia. 1996;10:346–50. [PubMed] [Google Scholar]
- 26.Mossafa H, Damotte D, Jenabian A, Delarue R, Vincenneau A, Amouroux I, Jeandel R, Khoury E, Martelli JM, Samson T, Tapia S, Flandrin G, Troussard X. Non-Hodgkin’s lymphomas with Burkitt-like cells are associated with c-Myc amplification and poor prognosis. Leuk Lymphoma. 2006;47:1885–93. doi: 10.1080/10428190600687547. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira FM, Tone LG, Simoes BP, Rego EM, Araujo AG, Falcao RP. Blastoid mantle cell lymphoma with t(2;8) (p12;q24) Leuk Lymphoma. 2007;48:2079–82. doi: 10.1080/10428190701606834. [DOI] [PubMed] [Google Scholar]
- 28.Aukema SM, Siebert R, Schuuring E, van Imhoff GW, Kluin-Nelemans HC, Boerma EJ, Kluin PM. Double-hit B-cell lymphomas. Blood. 2011;117:2319–31. doi: 10.1182/blood-2010-09-297879. [DOI] [PubMed] [Google Scholar]
- 29.Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, Morishima Y, Nakamura S, Seto M. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–58. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 30.Rubio-Moscardo F, Climent J, Siebert R, Piris MA, Martin-Subero JI, Nielander I, Garcia-Conde J, Dyer MJ, Terol MJ, Pinkel D, Martinez-Climent JA. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105:4445–54. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez L, Hernandez S, Bea S, Pinyol M, Ferrer A, Bosch F, Nadal A, Fernandez PL, Palacin A, Montserrat E, Campo E. c-myc mRNA expression and genomic alterations in mantle cell lymphomas and other nodal non-Hodgkin’s lymphomas. Leukemia. 1999;13:2087–93. doi: 10.1038/sj.leu.2401599. [DOI] [PubMed] [Google Scholar]
- 32.Jardin F, Picquenot JM, Parmentier F, Ruminy P, Cornic M, Penther D, Bertrand P, Lanic H, Cassuto O, Humbrecht C, Lemasle E, Wautier A, Bastard C, Tilly H. Detection of gene copy number aberrations in mantle cell lymphoma by a single quantitative multiplex PCR assay: clinicopathological relevance and prognosis value. Br J Haematol. 2009;146:607–18. doi: 10.1111/j.1365-2141.2009.07791.x. [DOI] [PubMed] [Google Scholar]
- 33.Tapia G, Lopez R, Munoz-Marmol AM, Mate JL, Sanz C, Marginet R, Navarro JT, Ribera JM, Ariza A. Immunohistochemical detection of MYC protein correlates with MYC gene status in aggressive B cell lymphomas. Histopathology. 2011;59:672–8. doi: 10.1111/j.1365-2559.2011.03978.x. [DOI] [PubMed] [Google Scholar]
- 34.Correlation of MYC gene Translocation Status with MYC Protein Expression in Burkitt Lymphoma and Diffuse Large B Cell Lymphoma. United States & Canadian Academy of Pathology. 2012:2012. Modern Pathology. [Google Scholar]


