Abstract
XPG (Xeroderma pigmentosum group G complementing factor) is a protein associated with DNA repair and transcription. Point mutations in ERCC5, the gene coding for XPG, cause the cancer-prone disorder xeroderma pigmentosum (XP) while truncation mutations give rise to individuals with the combined clinical features of XP and Cockayne syndrome. Polymorphisms of ERCC5 or alterations in XPG mRNA expression were also associated to an increase risk of different cancers types and to prognosis of cancer patients. However, the expression of XPG protein in different normal or tumor human tissues is not well known. In the present work, we have validated an immunohistochemistry (IHC) assay for detection of expression levels of XPG protein in FFPE human tissue samples. We have also tested this IHC assay in different normal and tumor human tissues. On a microarray containing 28 normal cores, positive staining was observed in 60% of the samples. The highest staining was detected in adrenal gland, breast, colon, heart, kidney, thyroid and tongue. In tumors, positive staining was observed in 9 of 10 breast cancer samples and in all 5 ovarian cancer and 5 sarcomas samples. Subcellular localization was predominantly nuclear. The use of this validated methodology would help to interpret the role of XPG in tumorogenesis and its use as a possible prognostic or predictive factor.
Keywords: XPG, immunohistochemistry, tumor tissue
Introduction
The genome of eukaryotes is constantly being threatened by environmental agents (e.g. UV light, mutagenic chemicals and ionizing radiation) or endogenous cellular metabolites (e.g. reactive oxygen species) which can interact with and modify the DNA structure [1,2]. Some of these modifications affect the DNA bases while others can lead to the formation of single or double DNA strand breaks. These alterations in DNA can disrupt the replication and transcription processes or induce cellular dysfunctions, such as genetic instability and mutagenesis. The removal of these lesions is thus a vital process for every single cell. For this purpose, cells are equipped with complex and interconnected DNA repair pathways that detect and remove potentially lethal DNA lesions [1,3-5].
XPG (Xeroderma pigmentosum group G complementing factor) is a protein associated with the DNA repair machinery as it is a key member of the Nucleotide-Excision Repair (NER) pathway [6]. XPG, encoded by the Excision repair cross-complementation group 5 (ERCC5) gene, is a structure-specific 3′-endonuclease, member of the Fen-1 protein family [7-9]. In addition, XPG has been implicated in RNA transcription through its tight physical interaction with the transcription activator complex [11,12]. Moreover, in vitro data also suggests a role for XPG in the repair of oxidative DNA lesions by a NER-independent pathway mediated by its intrinsic single strand annealing activity that is independent of its nuclease activity [13]. Finally, it was recently published that XPG has a direct interaction with WRN protein, a 3’-5’ RECQ-like DNA helicase that participates in multiple DNA transactions during replication [13].
Point mutations in ERCC5 that inactivate the XPG endonuclease function cause the cancer-prone, sun-sensitive disorder xeroderma pigmentosum (XP) while truncation mutations in the ERCC5 gene give rise to individuals with the combined clinical features of XP and Cockayne syndrome (CS), a disorder associated with developmental and neurological abnormalities [14]. Alterations in XPG expression were also associated to an increase risk of different cancers types and to drug resistance to chemotherapeutic agents [15-20]. Most of these works determined ERCC5 expression at the DNA or mRNA level. However, the expression of XPG protein in different normal or tumor human tissues is not well known. Here we present the results concerning the validation and expression of XPG protein in human normal and tumor samples by immunohistochemistry (IHC).
Material and methods
Reagents
The XPG rabbit polyclonal antibody was purchased from Bethyl Laboratories (Montgomery, TX) and stored at 2-8°C in accordance with accompanying documentation. The rabbit IgG isotype control antibodies were purchased from BD Pharmingen (San Diego, CA) and from Dako (Carpinteria, CA). These antibodies were stored at 2-8°C in accordance with accompanying documentation. The Alexa Fluor 594 conjugated goat anti-rabbit IgG (H+L) antibodies were purchased from Molecular Probes. Hoechst was purchased from Sigma (St. Louis, MO, USA).
Cell lines and tissue samples
HeLa cells were obtained from ATCC (CCL-2) and are derived from an adenocarcinoma of the cervix. The XPG-specific knockdown cell line, XPG HeLa SilenciX® (HeLa/XPG), was purchased from Tebu-Bio (cat#01-00133). Both cell lines were maintained in D-MEM supplemented with 10% FCS and 100Uml-1 penicillin and streptomycin at 37°C and 5% CO2. For IHC, the viably frozen stock from HeLa and HeLa/XPG were thawed, cultured, fixed, processed, and embedded into paraffin in accordance with Mosaic Laboratories’ Standard Operating Procedures (SOPs).
Human normal and human cancer formalin-fixed, paraffin embedded (FFPE) material was provided by Mosaic Laboratories. Tissues were procured under an IRB-reviewed protocol (MOS001) that allows for use of remnant, deidentified, or anonymized human samples for in vitro analysis under the guidelines defining Exemption from Human Subject Research as defined by the Office of Human Research Protection (U.S.A).
Western blotting
After washing the cells in PBS, lysis buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% (v/v) Nonidet P-40, 2 mM EDTA, 1 mM PMSF, 10 mg/ml aprotinin, and 10 mg/ml leupeptin) was added and plates kept on ice for 15 min. Cell extracts were collected and cleared by microcentrifugation at 14000 g for 15 min at 4°C. Equal amount of proteins were resolved in SDS-PAGE and electroblotted to PVDF membranes (Immobilon-P, Millipore) following standard techniques. Membranes were sequentially probed with primary and appropriate secondary (horseradish-peroxidase-conjugated) antibodies following the manufacturer’s instructions. Antibody-antigen complexes were detected using the ECL system (Amersham Pharmacia Biotech).
Immunofluorescence
Cells were fixed (4% paraformaldehyde), permeabilized (0.1% Triton X-100) and incubated with the primary anti-XPG rabbit polyclonal antibody (dilution 1/150) for 1 h at 37°C. Then, cells were washed and incubated with the secondary anti-rabbit antibody conjugated with AlexaFluor 594 (dilution 1/100). Finally, the slides were incubated with Hoechst 33342 1 μg/ml and mounted with Mowiol mounting medium. Pictures were taken with a Leica DM IRM fluorescence microscope equipped with a 100x oil immersion objective and a DFC 340 FX digital camera (Leica, Wetzlar, Germany).
Immunohistochemistry
IHC was performed at Mosaic Laboratories. The procedure for IHC analysis of XPG with DAB was performed using manual detection at room temperature (RT). Tissues were sectioned at 4 micron thickness, mounted onto positive-charged glass slides, dried, baked, deparaffinized, and rehydrated. Following rehydration, tissue sections were incubated in Envision Peroxidase (Dako) for 5 minutes to quench endogenous peroxidase. Tissue sections then underwent pretreatment using High Tide (Mosaic Laboratories, Lake Forest, CA) for 40 minutes in a water bath set to 95°C followed by a rinse in Splash-T Buffer (Mosaic Laboratories). Slides were incubated with anti-XPG antibody diluted to 2.5 μg/mL in antibody diluent (Dako) for 30 minutes. Slides were then rinsed twice in Splash-T Buffer for 5 minutes each followed by detection using the Envision+ Rabbit HRP Detection Kit (Dako) for 30 minutes. Slides were rinsed twice with Splash-T Buffer for 5 minutes each followed by incubation with DAB (Dako) for 5 minutes. Slides were rinsed with water, counterstained with hematoxylin (Dako), blued in ammonia water, dehydrated through graded alcohols, cleared in xylene, and coverslipped.
Data analysis
Staining of human tissue was evaluated by a pathologist and evaluation of reactivity involved a combination of the following: cellular localization of staining, staining intensity, subcellular localization, and percentage of cells staining in the primary component of the tissue type of interest. The XPG assay was evaluated on a semi-quantitative scale, and the percentage of cells staining at each of the following four levels was recorded: 0 (unstained), 1+ (weak staining), 2+ (moderate staining) and 3+ (strong staining). Slides were scanned using an Aperio ScanScope CS system (Aperio) to produce whole slide images. Representative 20X image of staining are shown in the different figures. Staining of HeLa cell lines was evaluated by image analysis using ImageScope software (Aperio). The nuclear algorithm (Aperio, Vista, CA) was used to enumerate the number of cells that stained positive and the intensity at which they stained. This algorithm serves as an approximate method to quantify the number of stained cells and report results as the percent of total cells within the region of interest that are stained at each intensity level. An H-score was calculated based on the summation of the product of percent of cells stained at each intensity using the following equation: (3 x % cells staining at 3+) + (2 x % cells staining at 2+) + (1 x % cells staining at 1+).
Results
XPG immunohistochemistry validation assay in human tumor cell lines
We firstly characterized the protein expression of XPG in HeLa cells and its XPG-specific knockdown derivative, HeLa/XPG, by western blot. As shown in Figure 1A, and as expected, HeLa/XPG cells presented a decreased expression of XPG compared to HeLa cells. This was later corroborated by immunoflourescence (Figure 1B).
Figure 1.
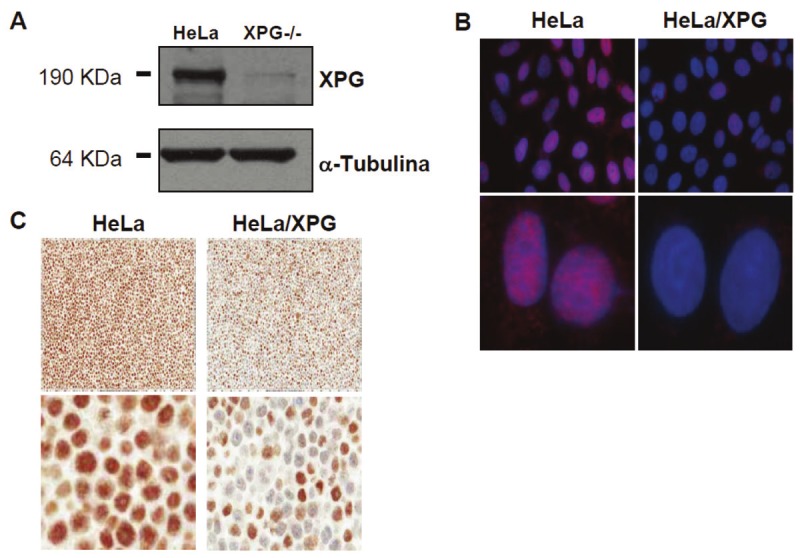
Expression of XPG protein in HeLa and HeLa/XPG cell lines by western blot (A), immunofluorescence (B) and immunohistochemistry (C). For A: cell lines were washed, lysed and electroblotted to PVDF membranes. Membranes were sequentially probed with primary and appropriate secondary antibodies. For B: cells were fixed and immunostained for XPG (red fluorescence); nuclei were visualized with Hoechst 33342 (blue fluorescence). For C: cells were fixed, processed, and embedded into paraffin.
IHC was then performed using these 2 characterized HeLa cell lines. These were evaluated by image analysis using a nuclear algorithm. The IHC image analysis results matched expectations. Photomicrographs of both cell lines are provided in Figure 1C. The percentage of positive HeLa control cells stained for XPG was 94.9. In this cell line, staining intensity was observed for 5.1%, 22%, 45.2% and 27.7% of 0, 1+, 2+ and 3+, respectively. These represented an H-score of 195.6. The percentage of positive HeLa/XPG cells stained for XPG was 51.8. In this cell line, staining intensity was observed for 48.2%, 18.2%, 23.3% and 10.3% of 0, 1+, 2+ and 3+, respectively. These represented an H-score of 95.6.
XPG immunohistochemistry validation assay in human samples
An antigen retrieval or pretreatment analysis of XPG was performed using a breast cancer specimen with various epitope retrieval methods. The procedure was performed with the following pretreatments: 1) no pretreatment; 2) Proteinase K (Dako) for 5 minutes at RT; 3) HIER using High Tide buffer for 40 minutes at 95°C in a water bath; 4) HIER using Red Tide buffer (Mosaic Laboratories) for 40 minutes at 95°C in a water bath; 5) HIER using Rip Tide buffer for 40 minutes at 95°C in a water bath; 6) HIER using Green Tide buffer (Mosaic Laboratories) for 40 minutes at 95°C in a water bath; 7) HIER using Diva buffer (Biocare Medical, Concord, CA) for 40 minutes at 95°C in a water bath; 8) HIER using Diva buffer with a decloaker set for 30 seconds at 125°C; and 9) Offshore Buffer (Mosaic Laboratories) for 10 minutes at RT. Untreated, proteinase K, Riptide and Offshore Buffer showed less than 10% staining and were discarded. Of the others, the greatest staining intensity and percent positive were observed with High Tide for 40 minutes at 95°C in a waterbath, and thus, this was selected for further experiments (Table 1). An antibody titration analysis was then performed using the same breast cancer specimen with serial dilutions of XPG antibody from 5.0 μg/mL to 0.3125 μg/mL (Table 2). Photomicrographs of titration analysis are provided in Figure 2. Based on these results, 2.5 μg/mL was selected for further experiments due to maximum signal to noise.
Table 1.
Anti-XPG expression using nine different antigen retrieval pretreatment conditions in a breast cancer specimen
| Pretreatment | % Cell staining at each Intensity | % Pos | Stromal staining | |||
|---|---|---|---|---|---|---|
|
| ||||||
| 0 | 1 | 2 | 3 | |||
| #1 | 100 | 0 | 0 | 0 | 0 | 1+ |
| #2 | 100 | 0 | 0 | 0 | 0 | 0 |
| #3 | 2 | 78 | 20 | 0 | 98 | 1+ |
| #4 | 10 | 90 | 0 | 0 | 90 | 0 |
| #5 | 92 | 5 | 0 | 3 | 8 | 2+ |
| #6 | 20 | 80 | 0 | 0 | 80 | 0 |
| #7 | 10 | 90 | 0 | 0 | 90 | 1+ |
| #8 | 3 | 94 | 3 | 0 | 97 | 1+ |
| #9 | 100 | 0 | 0 | 0 | 0 | 0 |
% pos, percentage of positive cells. #1: untreated; 2#: proteinase K (Dako) for 5 minutes at room temperature (RT); #3: HIER using High Tide buffer for 40 minutes at 95°C in a water bath; #4: HIER using Red Tide buffer (Mosaic Laboratories) for 40 minutes at 95°C in a water bath; #5: HIER using Rip Tide buffer for 40 minutes at 95°C in a water bath; #6: HIER using Green Tide buffer (Mosaic Laboratories) for 40 minutes at 95°C in a water bath; #7: HIER using Diva buffer (Biocare Medical, Concord, CA) for 40 minutes at 95°C in a water bath; #8: HIER using Diva buffer with a decloaker set for 30 seconds at 125°C; #9: offshore Buffer (Mosaic Laboratories) for 10 minutes at RT.
Table 2.
Anti-XPG titration analysis in a breast cancer specimen
| Titration | % Cell staining at each Intensity | Iso S | % Pos | Stroma | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 0 | 1 | 2 | 3 | ||||
| 5.0 μg/mL | 2 | 68 | 30 | 0 | 1+ | 98 | 1+ |
| 2.5 μg/mL | 8 | 92 | 0 | 0 | 0 | 92 | 1+ |
| 1.25 μg/mL | 40 | 60 | 0 | 0 | 0 | 60 | 0 |
| 0.62 μg/mL | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0.31 μg/mL | 10 | 0 | 0 | 0 | 0 | 0 | 0 |
Iso S, isotype staining; % pos, percentage of positive cells; Stroma, stroma staining.
Figure 2.
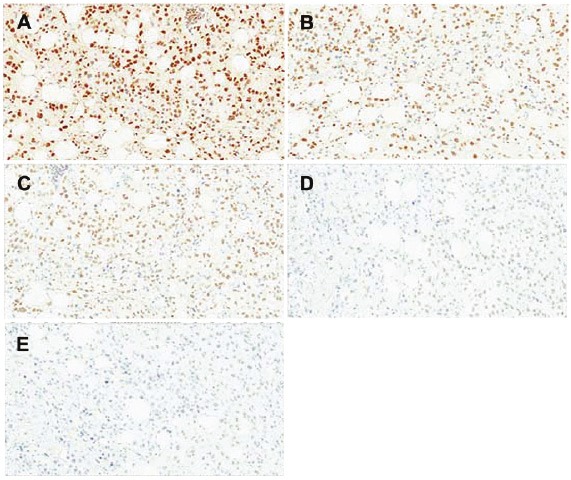
Photomicrographs of XPG titration analysis in a breast cancer specimen. A: concentration #1; B: concentration #2; C: concentration #3; D: concentration #4; E: concentration #5.
The precision of an immunohistochemistry assay is a combination of assay variance, tissue section and tissue architecture variance, and pathologist interpretation variance. Since the tissue varies from section to section, no two slides can be exactly alike. It is important to assess variance in the assay to determine the statistical significance of differences observed between tissues. A precision analysis was then performed using the optimized XPG IHC assay on 2 breast cancer, 1 ovarian cancer, and 1 sarcoma tissues on 5 separate days (Table 3). The mean percentage of staining, mean maximal intensity and mean H-Score of the 4 samples was 89.8±5.4%, 3±0 and 124±31, respectively. The average percent coefficient of variation (CV) was 2.1±1.8% for total positive staining, 0.00% for staining intensity, and 3.01±0.7% for H-Score.
Table 3.
Anti-XPG precision analysis in tumor tisues on 5 separate days
| Tumor type | % Cell staining at each Intensity | % Pos | Max SI | H-S | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 0 | 1 | 2 | 3 | ||||
| Day 1 | |||||||
| Breast | 5 | 10 | 84 | 1 | 95 | 3+ | 181 |
| Breast | 10 | 80 | 10 | 0 | 90 | 2+ | 100 |
| Ovary | 5 | 64 | 30 | 1 | 95 | 3+ | 127 |
| Sarcoma | 15 | 74 | 10 | 1 | 85 | 3+ | 97 |
| Day 2 | |||||||
| Breast | 6 | 20 | 74 | 0 | 94 | 2+ | 168 |
| Breast | 10 | 80 | 10 | 0 | 90 | 2+ | 100 |
| Ovary | 5 | 74 | 20 | 1 | 95 | 3+ | 117 |
| Sarcoma | 12 | 72 | 15 | 1 | 88 | 3+ | 105 |
| Day 3 | |||||||
| Breast | 4 | 10 | 85 | 1 | 96 | 3+ | 183 |
| Breast | 15 | 74 | 10 | 1 | 85 | 3+ | 97 |
| Ovary | 5 | 64 | 30 | 1 | 95 | 3+ | 127 |
| Sarcoma | 20 | 62 | 17 | 1 | 80 | 3+ | 99 |
| Day 4 | |||||||
| Breast | 5 | 15 | 79 | 1 | 95 | 3+ | 176 |
| Breast | 15 | 74 | 10 | 1 | 85 | 3+ | 97 |
| Ovary | 5 | 69 | 25 | 1 | 95 | 3+ | 122 |
| Sarcoma | 20 | 64 | 15 | 1 | 80 | 3+ | 97 |
| Day 5 | |||||||
| Breast | 5 | 15 | 79 | 1 | 95 | 3+ | 176 |
| Breast | 15 | 74 | 10 | 1 | 85 | 3+ | 97 |
| Ovary | 5 | 69 | 25 | 1 | 95 | 3+ | 122 |
| Sarcoma | 21 | 63 | 15 | 1 | 79 | 3+ | 96 |
% pos, percentage of positive cells; Max SI, maximal staining intensity; H-S, H-score.
XPG expression in human tissues as detected by immunohistochemistry
XPG expression in human tissues was firstly determined on a microarray containing 28 normal cores. Results are shown in Table 4 while representative photomicrographs are provided in Figure 3. Globally, expression of XPG ranged from 0% to 85% with an average of 19±27%. Maximal staining intensity was 2+. Mean H-Score was 20.9±30. Subcellular localization was predominantly nuclear, although cytoplasmic staining was observed in 5 specimens (adrenal gland, heart, kidney, cerebellum and cerebrum).
Table 4.
XPG IHC expression in 28 human normal tissues
| Tissue type | % Cell staining at each Intensity | % Pos | Max SI | H-S | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 0 | 1 | 2 | 3 | ||||
| Adrenal G | 25 | 70 | 5 | 0 | 75 | 2+ | 80 |
| Breast | 15 | 85 | 0 | 0 | 85 | 1+ | 85 |
| Cerebellum | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Cerebrum | 99 | 1 | 0 | 0 | 1 | 1+ | 1 |
| Cervix | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Colon | 35 | 65 | 0 | 0 | 65 | 1+ | 65 |
| Esophagus | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Heart | 40 | 30 | 30 | 0 | 60 | 2+ | 90 |
| Kidney | 65 | 20 | 15 | 0 | 35 | 2+ | 50 |
| Liver | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lung | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Skeletal M | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Smooth M | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ovary | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pancreas | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Parathyroid | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pitituary | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Prostate | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Salivary G | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Skin | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Spleen | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Stomach | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Testis | 85 | 15 | 0 | 0 | 15 | 1+ | 15 |
| Thymus | 95 | 5 | 0 | 0 | 5 | 1+ | 5 |
| Thyroid | 35 | 65 | 0 | 0 | 65 | 1+ | 65 |
| Tongue | 40 | 60 | 0 | 0 | 60 | 1+ | 60 |
| Tonsil | 85 | 15 | 0 | 0 | 15 | 1+ | 15 |
| Uterus | 95 | 5 | 0 | 0 | 5 | 1+ | 5 |
% pos, percentage of positive cells; Max SI, maximal staining intensity; H-S, H-score; adrenal G, adrenal gland; skeletal m, skeletal muscle.
Figure 3.
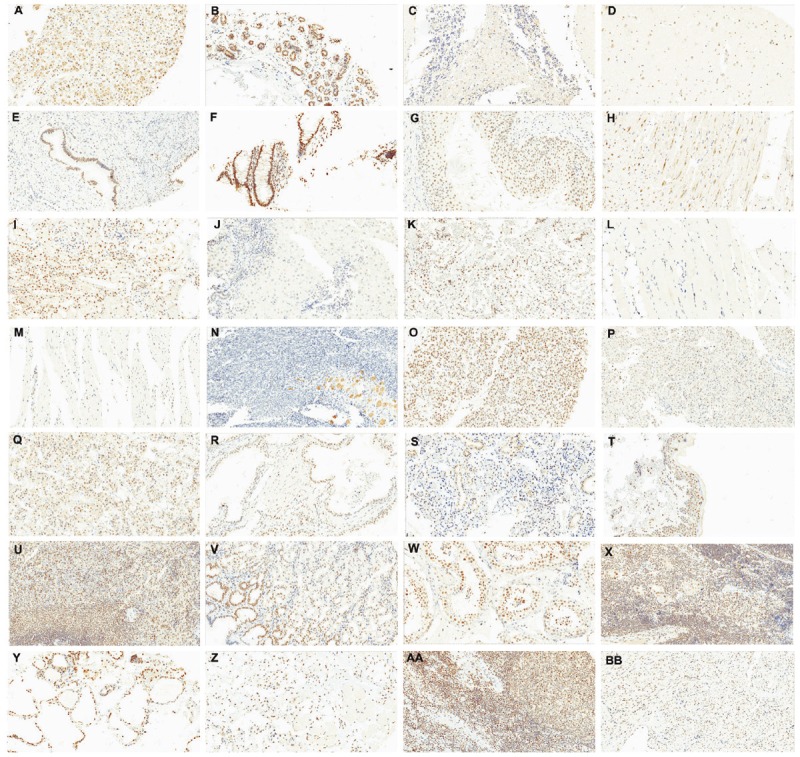
Expression of XPG protein in human normal tissues. A: adrenal gland; B: breast; C: cerebellum; D: cerebrum; E: cervix; F: colon; G: esophagus; H: heart; I: kidney; J: liver; K: lung; L: skeletal muscle; M: smooth muscle; N: ovary; O: pancreas; P: parathyroid; Q: pituitary; R: prostate; S: salivary gland; T: skin; U: spleen; V: stomach; W: testis; X: thymus; Y: thyroid; Z: tongue; AA: tonsil; BB: uterus. Selected photomicrographs were performed at 20X. Staining was performed on a tissue microarray.
The following tissues presented a moderate to high XPG expression, represented as an H-Score ≥50: adrenal gland, breast, colon, heart, kidney, thyroid and tongue. Interestingly, adrenal gland, heart and kidney samples showed nuclear and cytoplasmic staining intensity of 2+. In the colon and tongue samples, macrophages showed 1+ nuclear staining. In these samples, endothelial cells, smooth muscle, fibroblasts and stroma were not stained and considered as showing negative expression.
The following tissues showed a weak XPG expression, represented as an H-Score <50: cerebellum, cerebrum, salivary gland, skin, spleen, stomach, testis, thymus, tonsil and uterus. Of note, cerebellum and cerebrum showed nuclear and cytoplasmic expression. In the first case, stained cells were mostly in granular layer. In the salivary gland, thymus and tonsil samples, macrophages showed 1+ nuclear staining. In the uterus sample, smooth muscle cells showed 1+ nuclear staining. In thymus, 35% of cells of Hassall’s corpuscles had 1+ nuclear staining.
The following tissues were considered as presenting negative expression: cervix, esophagus, liver, lung, skeletal muscle, smooth muscle, ovary, pancreas, parathyroid, pituitary, and prostate. Interestingly, in the esophagus sample, smooth muscle cells showed 1+ positive staining. In the pancreatic sample, 100% of ductal cells had 1+ nuclear staining while 100% acini cells had a blush staining. Finally, in the lung and the ovary samples, stroma presented 1+ positive staining. Of note, in both samples macrophages showed 1+ cytoplasmic staining.
XPG expression in human tumor samples as detected by immunohistochemistry
XPG expression was also evaluated on 10 breast cancer, 5 ovarian cancer, and 5 sarcoma specimens. Results are shown in Table 5. Representative photomicrographs are provided in Figure 4. Globally, expression of XPG ranged from 0% to 99% with an average of 23.1±28%. Maximal staining intensity was 2+. Mean H-Score was 25.2±34. Subcellular localization was predominantly nuclear and 3 samples also demonstrated cytoplasmic staining (2 breast cancer and 1 sarcoma).
Table 5.
XPG IHC expression in tumor tissues
| Tumor type | % Cell staining at each Intensity | % Pos | Max SI | H-S | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 0 | 1 | 2 | 3 | ||||
| Breast | 1 | 66 | 33 | 0 | 99 | 2+ | 132 |
| Breast | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Breast | 75 | 25 | 0 | 0 | 25 | 1+ | 25 |
| Breast | 89 | 10 | 1 | 0 | 11 | 2+ | 12 |
| Breast | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Breast | 95 | 5 | 0 | 0 | 5 | 1+ | 5 |
| Breast | 95 | 5 | 0 | 0 | 5 | 1+ | 5 |
| Breast | 20 | 80 | 0 | 0 | 80 | 1+ | 80 |
| Breast | 94 | 6 | 0 | 0 | 6 | 1+ | 6 |
| Breast | 92 | 8 | 0 | 0 | 8 | 1+ | 8 |
| Ovary | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Ovary | 95 | 5 | 0 | 0 | 5 | 1+ | 5 |
| Ovary | 85 | 15 | 0 | 0 | 15 | 1+ | 15 |
| Ovary | 40 | 58 | 2 | 0 | 60 | 2+ | 62 |
| Ovary | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Sarcoma | 57 | 42 | 1 | 0 | 43 | 1+ | 44 |
| Sarcoma | 90 | 10 | 0 | 0 | 10 | 1+ | 10 |
| Sarcoma | 45 | 50 | 5 | 0 | 55 | 2+ | 60 |
| Sarcoma | 95 | 5 | 0 | 0 | 5 | 1+ | 5 |
| Sarcoma | 99 | 1 | 0 | 0 | 1 | 1+ | 1 |
% pos, percentage of positive cells; Max SI, maximal staining intensity; H-S, H-score.
Figure 4.
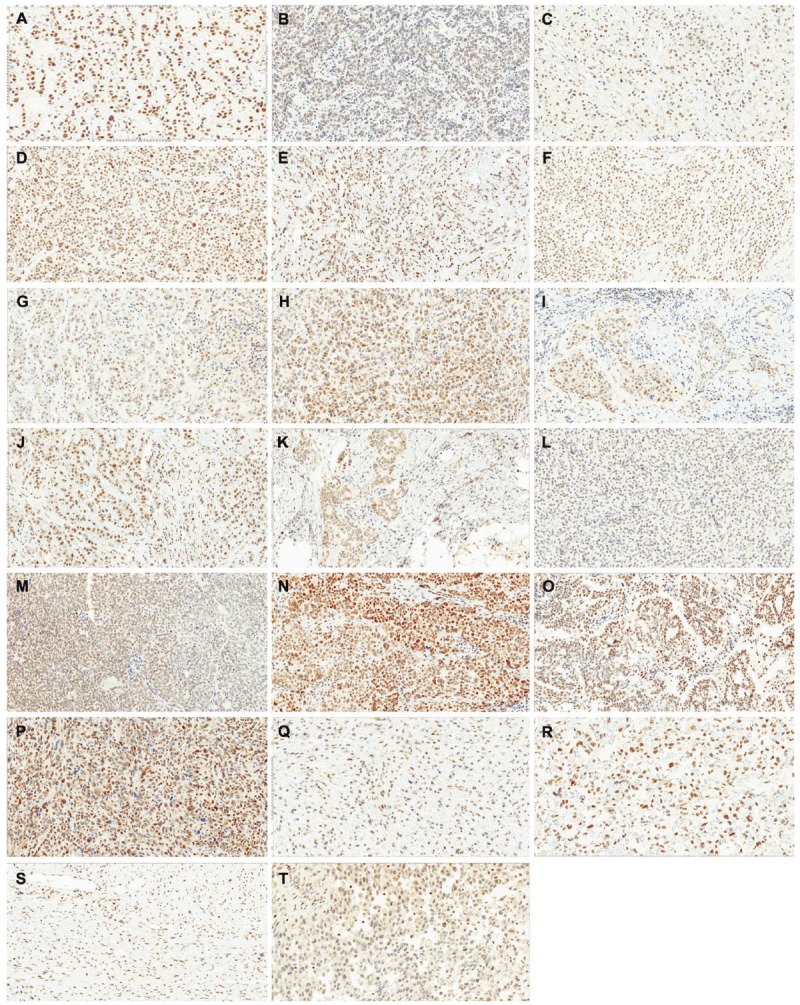
Expression of XPG protein in human tumor tissues. A-J: breast cancer samples; K-O: ovarian cancer samples; P-T: sarcoma samples. Staining was performed on whole tissue sections.
In breast cancer samples, staining ranged from 0% to 99% with an average of 25±34%. Positive staining was present in 9/10 samples. Of these positive samples, only two samples presented an H-Score ≥50 (Table 5). One of these samples showed a 2+ positive nuclear staining. Of note, in all positive samples, macrophages showed 2+, or even, 3+ nuclear staining. Other non-tumor cells showing a positive staining were endothelial cells (4/9 positive samples), smooth muscle (1/9 positive samples) and fibroblasts (7/9 positive samples). This last cell subtype showed 1+ (4 cases) or 2+ staining (3 cases). The negative sample did not show any staining in non-tumor cells.
In ovarian cancer samples, staining ranged from 5% to 60% with an average of 20±22%. Although positive staining was present in all samples, only one presented an H-Score ≥50. Of the analyzed non-tumor cell types, macrophages presented the higher expression (2+ and 3+) while fibroblasts showed moderate (1+ and 2+) and endothelial cells weak (1+) expression. In sarcoma samples, staining ranged from 1% to 55% with an average of 23±24%. Although positive staining was present in all samples, only one presented an H-Score ≥50. As in breast and ovarian cancer samples, macrophages presented the higher expression (1+ and 2+) of other non-tumor cell types. Endothelial cells and fibroblasts showed 1+ staining.
Discussion
The purpose of this study was to validate an immunohistochemical assay for XPG protein using a commercially available antibody for use in FFPE human tissues and to analyse its expression in human normal and cancer samples. The optimal pretreatment procedure to maximize staining was determined by testing 9 different antigen retrieval techniques. The optimal primary antibody titer was selected to maximize signal to noise through analysis of a breast cancer tissue while assay precision was determined in 2 breast cancer, 1 ovarian cancer, and 1 sarcoma tissues on 5 different days. These experiments established that the best IHC assay was: pretreament of samples with High Tide for 40 minutes at 95°C in a water-bath and staining using a 2.5 μg/mL antibody dilution. The optimized assay was then validated by staining 2 tumor cell lines. Once validated, the differential expression of XPG was determined in 28 normal tissues and 20 tumor tissues consisting of 10 breast, 5 ovarian, and 5 sarcoma cancer samples.
XPG is one of multiple proteins that are members of the NER system in mammalian cells [6]. The NER pathway is involved in DNA repair and allows tumor cells to survive DNA damage caused by ultraviolet light or different genotoxins such as anticancer therapeutic agents [21,22]. Briefly, after DNA damage recognition, the sequential action of NER helicases and endonucleases open the double helix and cleave the damaged strand few bases away from the lesion. This is followed by the removal of the DNA segment containing the lesion, DNA gap filling using the intact strand as template and restoration of the chromatin structure [1,12,23]. During this process, XPG functions as a structure-specific endonuclease that cleaves DNA bubbles and flaps near the junctions of single-stranded and double-stranded DNA with specific polarity [10,24,25]. XPG endonuclease activity has no preference for the sequencing of DNA damage. At the site of injury, NER complex proteins create a DNA bubble of an approximate length of 25 nucleotides. XPG cuts the DNA damage between 0-2 nucleotides below 3’ of the double-fork single-stranded chain. The cells that do not express ERCC5/XPG are inefficient in repairing damage to DNA.
XPG also binds strongly to various structured DNAs that it does not cleave, implying separate biological functions for its binding and incision activities [13,26,27]. For example, XPG has a non-enzymatic scaffolding role in several steps of NER, including coordination of incision with the resynthesis step [28,29]. In addition, XPG forms a complex with the transcription and repair factor TFIIH being this important for stable association of the CAK kinase subunit with TFIIH [12]. Thus, XPG alleles with C-terminal mutations or truncations are unable to bind TFIIH, resulting in impaired activated RNA polymerase II-mediated transcription [26,30]. More recently, it was shown that XPG has an intrinsic single-strand annealing activity that is independent of its nuclease activity but requires intact N- and C-terminal domains [29]. This activity is performed in cooperation with WRN protein through a direct physical interaction between both proteins and is observed during the late S-phase of the cell cycle [13]. Finally, XPG has a role in the early steps of base excision repair (BER) of oxidative DNA damage through direct interaction and stimulation of different DNA glycosylases [13].
As the incision activity of XPG is essential for removing bulky DNA adducts by NER, point mutations that inactivate the XPG endonuclease function cause the cancer-prone, sun-sensitive disorder xeroderma pigmentosum (XP) in XP-G patients [14,31] University Medical Centre (CMU. Moreover, patients with rare truncating mutations in XPG have the combined diseases of XP with Cockayne syndrome (XP-G/CS) [32-34]. XP-G/CS presents as severe primarily postnatal neurological and developmental dysfunction with mental retardation, wasting, greatly accelerated symptoms of segmental aging and death in early childhood. Mutations and polymorphisms of the ERCC5 gene were also associated to an increase risk of different cancers types such as endometrial, melanoma, prostate, bladder, gastric, cervix or lung cancer [15,16,35-38]. Polymorphisms of ERCC5 gene or differential expression levels of its mRNA were correlated with patient prognosis in different tumor pathologies. For example, the Asp1104His or His46His polymorphisms were reported to be an independent prognostic factor in ovarian cancer, sarcoma and cutaneous melanoma, and non-small cell lung cancer (NSCLC), respectively [19,39-41]. On the other hand, high expression of XPG mRNA was associated as an independent prognostic factor in NSCLC and sarcoma patients and to significantly correlate with increased response to chemotherapeutic agents such as oxaliplatin, fluoropyrimidin or trabectedin [42-44]. Thus, XPG seems to be an important molecular factor involved in tumorigenesis and prognosis of cancer patients. However, all these studies were performed using DNA sequencing or real-time PCR techniques that are quite cumbersome to be used in a routine pathology laboratory.
In the present work, we have validated an IHC assay for detection of expression levels of XPG protein in FFPE human tissue samples. We have also tested this IHC assay in different normal and tumor human tissues. XPG expression was firstly determined on a microarray containing 28 normal tissue cores. Positive staining was observed in 60% of the normal samples. The highest staining was detected in adrenal gland, breast, colon, heart, kidney, thyroid and tongue. A weaker staining was observed in cerebellum, cerebrum, salivary gland, skin, spleen, stomach, testis, thymus, tonsil and uterus. Finally, cervix, esophagus, liver, lung, skeletal muscle, smooth muscle, ovary, pancreas, parathyroid, pituitary, and prostate samples were considered as negative. Subcellular localization was predominantly nuclear, although cytoplasmic staining was also observed in 5 specimens (adrenal gland, heart, kidney, cerebellum and cerebrum). Interestingly, a weak to moderate staining intensity was also correlated with a cytoplasmic staining in adrenal gland, heart and kidney samples while both central nervous system samples presented cytoplasmic staining even in the case of presenting a weak nuclear staining. Another interesting observation concerned the staining of macrophages present on these normal tissues. In fact, these inflammatory cells showed a weak nuclear staining in positive and negative samples. However, while in positive samples (colon, tongue, salivary gland, thymus and tonsil) it was mainly nuclear, in negative tissues (lung and ovary), it was cytoplasmic. Other interesting observations were the expression of XPG in the Hassall’s corpuscles of the thymus or smooth muscle cells in esophagus and uterus.
In tumors, positive staining was observed in 90% of breast cancer samples and in all ovarian cancer and sarcomas samples. XPG expression in breast and ovary cancer samples was expected as breast and uterus normal samples already expressed weak to moderate levels of the protein. Subcellular localization was predominantly nuclear. Of note, in all positive samples (19/20 analyzed samples), inflammatory cells showed 2+, or even, 3+ nuclear staining. These contrasted with that observed in normal tissues where macrophage nuclear staining was seen only in 17% (5/28) of analyzed tissues. The biological significance of this XPG overexpression in tumor-associated macrophages (TAMs) is not well understood. One possibility is that this over-expression is related to chronic hypoxia or inflammation in tissue microenvironments that will favour higher levels of DNA lesions that need to be repaired. A second possibility would be that TAMs would need high levels of XPG to support high levels of transcription in order to maintain elevated inflammatory cytokine secretions as observed in tumor microenvironments. Other non-tumor cells showing a positive staining were endothelial cells, smooth muscle and fibroblasts.
In summary, we have validated an immunohistochemical assay for XPG protein detection using a commercially available antibody for use in formalin-fixed, FFPE human tissues. To our knowledge, this is the first time that XPG expression was evaluated at the protein level in human samples. The use of this validated methodology would help to interpret the role of XPG in tumorogenesis and its use as a possible prognostic or predictive factor.
References
- 1.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt PC. Paradigms for the three rs: DNA replication, recombination, and repair. Mol Cell. 2007;28:702–707. doi: 10.1016/j.molcel.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Jiang G, Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol Cell Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ljungman M. Activation of DNA damage signaling. Mutat Res. 2005;577:203–216. doi: 10.1016/j.mrfmmm.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Friedberg EC, Wood RD. New insights into the combined Cockayne/xeroderma pigmentosum complex: human XPG protein can function in transcription factor stability. Mol Cell. 2007;26:162–164. doi: 10.1016/j.molcel.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Costa RM, Chigancas V, Galhardo Rda S, Carvalho H, Menck CF. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Berneburg M, Lehmann AR. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv Genet. 2001;43:71–102. doi: 10.1016/s0065-2660(01)43004-5. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 10.O’Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3’ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 11.Lee SK, Yu SL, Prakash L, Prakash S. Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription. implications for Cockayne syndrome. Cell. 2002;109:823–834. doi: 10.1016/s0092-8674(02)00795-x. [DOI] [PubMed] [Google Scholar]
- 12.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 13.Trego KS, Chernikova SB, Davalos AR, Perry JJ, Finger LD, Ng C, Tsai MS, Yannone SM, Tainer JA, Campisi J, Cooper PK. The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle. 2011;10:1998–2007. doi: 10.4161/cc.10.12.15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nouspikel T, Lalle P, Leadon SA, Cooper PK, Clarkson SG. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc Natl Acad Sci USA. 1997;94:3116–3121. doi: 10.1073/pnas.94.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Berhane N, Sobti RC, Mahdi SA. DNA repair genes polymorphism (XPG and XRCC1) and association of prostate cancer in a north Indian population. Mol Biol Rep. 2012;39:2471–2479. doi: 10.1007/s11033-011-0998-5. [DOI] [PubMed] [Google Scholar]
- 16.He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP, Wang YN, Sun MH, Zhou XY, Yang YJ, Wang JC, Jin L, Wei QY, Li J. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235–1244. doi: 10.1007/s00439-012-1152-8. [DOI] [PubMed] [Google Scholar]
- 17.Graf N, Ang WH, Zhu G, Myint M, Lippard SJ. Role of endonucleases XPF and XPG in nucleotide excision repair of platinated DNA and cisplatin/oxaliplatin cytotoxicity. Chembiochem. 2011;12:1115–1123. doi: 10.1002/cbic.201000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabatino MA, Marabese M, Ganzinelli M, Caiola E, Geroni C, Broggini M. Down-regulation of the nucleotide excision repair gene XPG as a new mechanism of drug resistance in human and murine cancer cells. Mol Cancer. 2010;9:259. doi: 10.1186/1476-4598-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X, Li F, Sun N, Shukui Q, Baoan C, Jifeng F, Lu C, Zuhong L, Hongyan C, YuanDong C, Jiazhong J, Yingfeng Z. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer. 2009;65:230–236. doi: 10.1016/j.lungcan.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Takebayashi Y, Pourquier P, Zimonjic DB, Nakayama K, Emmert S, Ueda T, Urasaki Y, Kanzaki A, Akiyama SI, Popescu N, Kraemer KH, Pommier Y. Antiproliferative activity of ecteinascidin 743 is dependent upon transcription-coupled nucleotide-excision repair. Nat Med. 2001;7:961–966. doi: 10.1038/91008. [DOI] [PubMed] [Google Scholar]
- 21.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reardon JT, Vaisman A, Chaney SG, Sancar A. Efficient nucleotide excision repair of cisplatin, oxaliplatin, and Bis-aceto-ammine-dichloro-cyclohexylamine-platinum(IV) (JM216) platinum intrastrand DNA diadducts. Cancer Res. 1999;59:3968–3971. [PubMed] [Google Scholar]
- 23.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 24.O’Donovan A, Scherly D, Clarkson SG, Wood RD. Isolation of active recombinant XPG protein, a human DNA repair endonuclease. J Biol Chem. 1994;269:15965–15968. [PubMed] [Google Scholar]
- 25.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. Embo J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Hohl M, Thorel F, Clarkson SG, Scharer OD. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J Biol Chem. 2003;278:19500–19508. doi: 10.1074/jbc.M213155200. [DOI] [PubMed] [Google Scholar]
- 28.Mocquet V, Laine JP, Riedl T, Yajin Z, Lee MY, Egly JM. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. Embo J. 2008;27:155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Scharer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. Embo J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 31.O’Donovan A, Wood RD. Identical defects in DNA repair in xeroderma pigmentosum group G and rodent ERCC group 5. Nature. 1993;363:185–188. doi: 10.1038/363185a0. [DOI] [PubMed] [Google Scholar]
- 32.Emmert S, Slor H, Busch DB, Batko S, Albert RB, Coleman D, Khan SG, Abu-Libdeh B, Di-Giovanna JJ, Cunningham BB, Lee MM, Crollick J, Inui H, Ueda T, Hedayati M, Grossman L, Shahlavi T, Cleaver JE, Kraemer KH. Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J Invest Dermatol. 2002;118:972–982. doi: 10.1046/j.1523-1747.2002.01782.x. [DOI] [PubMed] [Google Scholar]
- 33.Lindenbaum Y, Dickson D, Rosenbaum P, Kraemer K, Robbins I, Rapin I. Xeroderma pigmentosum/cockayne syndrome complex: first neuropathological study and review of eight other cases. Eur J Paediatr Neurol. 2001;5:225–242. doi: 10.1053/ejpn.2001.0523. [DOI] [PubMed] [Google Scholar]
- 34.Scharer OD. XPG: its products and biological roles. Adv Exp Med Biol. 2008;637:83–92. doi: 10.1007/978-0-387-09599-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cincin ZB, Iyibozkurt AC, Kuran SB, Cakmakoglu B. DNA repair gene variants in endometrial carcinoma. Med Oncol. 2012 Jan 22;29:2949–54. doi: 10.1007/s12032-012-0162-7. [DOI] [PubMed] [Google Scholar]
- 36.Goncalves FT, Francisco G, de Souza SP, Luiz OC, Festa-Neto C, Sanches JA, Chammas R, Gattas GJ, Eluf-Neto J. European ancestry and polymorphisms in DNA repair genes modify the risk of melanoma: a case-control study in a high UV index region in Brazil. J Dermatol Sci. 2011;64:59–66. doi: 10.1016/j.jdermsci.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Rouissi K, Ouerhani S, Hamrita B, Bougatef K, Marrakchi R, Cherif M, Ben Slama MR, Bouzouita M, Chebil M, Ben Ammar Elgaaied A. Smoking and polymorphisms in xenobiotic metabolism and DNA repair genes are additive risk factors affecting bladder cancer in Northern Tunisia. Pathol Oncol Res. 2011;17:879–886. doi: 10.1007/s12253-011-9398-3. [DOI] [PubMed] [Google Scholar]
- 38.Jeon HS, Kim KM, Park SH, Lee SY, Choi JE, Lee GY, Kam S, Park RW, Kim IS, Kim CH, Jung TH, Park JY. Relationship between XPG codon 1104 polymorphism and risk of primary lung cancer. Carcinogenesis. 2003;24:1677–1681. doi: 10.1093/carcin/bgg120. [DOI] [PubMed] [Google Scholar]
- 39.Caiola E, Porcu L, Fruscio R, Giuliani D, Milani R, Torri V, Broggini M, Marabese M. DNA-damage response gene polymorphisms and therapeutic outcomes in ovarian cancer. Pharmacogenomics J. 2011 Dec 13; doi: 10.1038/tpj.2011.50. doi: 10.1038/tpj.2011.50. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 40.Italiano A, Laurand A, Laroche A, Casali P, Sanfilippo R, Le Cesne A, Judson I, Blay JY, Ray-Coquard I, Bui B, Coindre JM, Nieto A, Tercero JC, Jimeno J, Robert J, Pourquier P. ERCC5/XPG, ERCC1, and BRCA1 gene status and clinical benefit of trabectedin in patients with soft tissue sarcoma. Cancer. 2011;117:3445–3456. doi: 10.1002/cncr.25925. [DOI] [PubMed] [Google Scholar]
- 41.Schrama D, Scherer D, Schneider M, Zapatka M, Brocker EB, Schadendorf D, Ugurel S, Kumar R, Becker JC. ERCC5 p. Asp1104His and ERCC2 p.Lys751Gln polymorphisms are independent prognostic factors for the clinical course of melanoma. J Invest Dermatol. 2011;131:1280–1290. doi: 10.1038/jid.2011.35. [DOI] [PubMed] [Google Scholar]
- 42.Monzo M, Moreno I, Navarro A, Ibeas R, Artells R, Gel B, Martinez F, Moreno J, Hernandez R, Navarro-Vigo M. Single nucleotide polymorphisms in nucleotide excision repair genes XPA, XPD, XPG and ERCC1 in advanced colorectal cancer patients treated with firstline oxaliplatin/fluoropyrimidine. Oncology. 2007;72:364–370. doi: 10.1159/000113534. [DOI] [PubMed] [Google Scholar]
- 43.Bartolucci R, Wei J, Sanchez JJ, Perez-Roca L, Chaib I, Puma F, Farabi R, Mendez P, Roila F, Okamoto T, Taron M, Rosell R. XPG mRNA expression levels modulate prognosis in resected non-small-cell lung cancer in conjunction with BRCA1 and ERCC1 expression. Clin Lung Cancer. 2009;10:47–52. doi: 10.3816/CLC.2009.n.007. [DOI] [PubMed] [Google Scholar]
- 44.Schoffski P, Taron M, Jimeno J, Grosso F, Sanfilipio R, Casali PG, Le Cesne A, Jones RL, Blay JY, Poveda A, Maki RG, Nieto A, Tercero JC, Rosell R. Predictive impact of DNA repair functionality on clinical outcome of advanced sarcoma patients treated with trabectedin: a retrospective multicentric study. Eur J Cancer. 2011;47:1006–1012. doi: 10.1016/j.ejca.2011.01.016. [DOI] [PubMed] [Google Scholar]


