Abstract
Objective: To determine whether the SP-B deficiency and gene mutations in exon 4 is associated with neonatal RDS in China Han ethnic population. Methods: The study population consisted of 40 neonates with RDS and 40 neonates with other diseases as control in China Han ethnic population. We Compared SP-B expression in lung tissue and bronchoalveolar lavage fluid with immunoblotting, and analyzed mutations in the SP-B gene with polymerase chain reaction (PCR) and gene sequencing. Results: In RDS group, low mature Surfactant protein B was found in both lung tissue and bronchoalveolar lavage fluid in 8 neonates. In control group, only 4 neonates with low mature Surfactant protein B in both lung tissue and bronchoalveolar lavage fluid. In RDS group, 20 neonates were found to have mutations in exon 4, 12 homozygous mutations with C/C genotype and 8 heterozygous mutations with C/T genotype in surfactant protein B gene+1580 polymorphism. There were 8 cases mutations in control group, 1 in C/C and 7 in C/T genotype. The frequency of homozygotes with C/C genotype was 0.3 and frequency of heterozygotes with C/T genotype was 0.02 in RDS group. In control group, frequency of homozygotes with C/C genotype was 0.025 and frequency of heterozygote with C/T genotype was 0.175. Conclusion: Low mature Surfactant protein B is associated with the pathogenesis of neonatal respiratory distress syndrome (RDS) in China Han ethnic population. Mutations in exon 4 of the surfactant protein B gene demonstrate an association between homozygous mutations with C/C genotype in SP-B gene and neonatal RDS.
Keywords: Polymorphisms, respiratory distress syndrome, surfactant protein B, neonates
Introduction
Pulmonary surfactant is a lipid-protein complex essential for normal lung function, responsible for reducing the superficial tension of the airliquid interface of the alveoli, thus preventing lung collapse at the end of expiration [1,2]. Avery showed that pulmonary surfactant deficiency is a major factor in the pathophysiology of respiratory distress syndrome (RDS). Surfactant proteins play an important role in function of pulmonary surfactant [3]. Surfactant protein B gene polymorphism are associated with respiratory distress syndrome in preterm babies [4]. A developmentally regulated, quantitative deficiency of pulmonary surfactant phospholipids results in surfactant dysfunction and respiratory distress syndrome (RDS) in premature newborns with increasing risk and severity associated with decreasing gestational age [5,6]. Clinical, epidemiological and biochemical evidence has strongly suggested that RDS is a multifactorial and multigenic disease, and the surfactant proteins might be implicated in this genetic variation [7-9]. The study of the genetic variation of surfactant proteins can help understand individual variability in the susceptibility to the development of pulmonary pathologies. These genetic variants can be valuable markers in the mapping of several pathologies, particularly for the respiratory distress syndrome.
To characterize developmental and genetic regulation of SP-B expression in China Han ethnic group neonates with RDS, we compared the mature SP-B expression and SP-B gene polymorphisms in 40 neonates with RDS and 40 other diseases as control.
Materials and methods
Patient populations
The study population consisted of 40 neonates with RDS and 40 neonates with other diseases as control in China Han ethnic population. The RDS group consisted of 40 neonates from unrelated families; 20 (50%) were females and 20 (50%) males; 20 (50%) were preterm newborns (gestational age (GA) was 34-36 weeks) and 20 (50%) were full term newborns (gestational age ≥ 37weeks). The control group consisted of 40 neonates with non-RDS diseases such as congenital heart disease, persistent pulmonary hypertension and shock. Both groups matched with Baseline demographic data like GA and sex.
The diagnosis of RDS was made on the basis of clinical and radiological criteria: presence of signs and symptoms of respiratory distress (grunting, intercostal retractions, nasal flaring, cyanosis, and tachypnea), a chest radiograph with a diffuse reticulogranular pattern, and air bronchograms. All neonates with RDS were treated with bovine surfactant at a dose of 200mg/kg and mechanical ventilation. Additional doses (each, 100mg/kg) were given if the neonate had an a/A PO2<20, 12 and 24 h later. All of the RDS neonates died in 10-14 days.
Laboratory tests
Blood sample collection
Blood collection was performed at the hospital at the same time as other routine exams. Blood samples were placed in tubes containing EDTA and kept at 4°C until DNA extraction.
DNA extraction and PCR amplification
The genomic DNA of the newborns was purified from total blood using the Wizard Genomic DNA Purification Kit® (Promega, USA) according to manufacturer instructions. DNA from patient and healthy newborn blood samples was amplified by PCR amplification protocols, as described by Lin et al. [9].
10 exons of SP-B genes were amplified using the Expand Long Template PCR system (Roche, Germany). The primers for 10 exons of SP-B genes were design by Primer 5 software. All primers used are listed in Table 1. The PCR mixture (total volume of 50 μL) consisted of 100 ng/μL DNA, 1X PCR buffer, 2.0 mM MgCl2, 1.5 mM deoxyribonucleotide triphosphates (dNTPs) (Promega®), 150 ng sense primer and anti-sense primer, and 0.75 μL of the Expand enzyme. PCR cycles consisted of one cycle at 95°C for 2 min, followed by 10 cycles at 95°C for 30 s, 58°C for 1 min and 72°C for 10 min and then by 20 cycles at 95°C for 30 s, 62°C for 10 min and 68°C for 12 min, with a final 20-min extension (68°C). The PCR products were identified on 2% agarose gels. The sequencing results of PCR products were analyzed with Chromas software.
Table 1.
Primers used in the present study
| SP-B exons | primers | sizes (bp) |
|---|---|---|
| 1 | 5’-AAGACAAACACTGAGGTCGC-3’ | 236 |
| 5’-CCTAGCACAAAGCAGTGCTC-3’ | ||
| 2 | 5’- ATGATGCCAGGTGTGTAGCC-3’ | 238 |
| 5’-AGCACCCTTCATTTCAGACC-3’ | ||
| 3 | 5’-AGAACCTCCCCATTGGAGC-3’ | 202 |
| 5’-CTGCTCTGTCCCTCATCTC-3’ | ||
| 4 | 5’- GGCCTTGTGTCCAGGGAC-3’ | 265 |
| 5’-TCCCCCATGGGTGGGCAC-3’ | ||
| 5 | 5’-TCCCTCCCAGACCCTAACAC-3’ | 272 |
| 5’-TCTGGATCTCCACTTTACTGG-3’ | ||
| 6 | 5’-GAGGACTCTTCTCCCAGC-3’ | 215 |
| 5’-GAGAGAGGTGGGAGCTGC-3’ | ||
| 7 | 5’-TGGGCAGAGAGTGGAGGCTTG-3’ | 288 |
| 5’-AGAGCGGGCATTGGGCTAAGG-3’ | ||
| 8 | 5’-ACAGAAACCAAACACCTCTG-3’ | 284 |
| 5’-CAAAGCCTTTCCTGGGCTC-3’ | ||
| 9 | 5’-CCTAGAGTGATCCAGAGATG-3’ | 259 |
| 5’-GAGGCCTCTGAGGATCAC-3’ | ||
| 10 | 5’-GGTGGAGTGAGTGCTGTTC-3’ | 227 |
| 5’-CCTGATGGTCAGGACCTG-3’ |
Western blotting
Tracheal aspirate sample acquisition and processing was described by Aaron et al. [10] Lung tissues were taken from dead neonates in 30 mins and kept at -80°C. Scraped frozen lung tissue were homogenized in extraction buffer (50 mM Tris·HCl, 0.1% SDS, 150 mM NaCl, 100 μg/ml phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1% NP-40 and 0.5% sodium orthovanadate), and incubated at 4°C for 20 min, centrifuged 20 min at 12000 g/min. Protein levels in the extracts were quantified using the BioRad DC protein assay. For western blot, whole cell extracts (50–100 μg/lane) were resolved in 8–12% SDS polyacrylamide gels, then transferred onto nitrocellulose membrane (0.45 μm, Millipore, USA) in 25 mM Tris-base, 190 mM glycine, and 20% methanol using a semi-dry blotter. Membranes were blocked with 8% fat-free milk and 0.1% Tween 20 in TBS. Rabbit antihuman SP-B antibody was used at the 1:3000 dilutions. Detection of polyclonal antibodies was performed using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulins and enhanced chemoluminescence (ECL).
Statistical analysis and human studies committee approval
Statistical differences were evaluated by oneway analysis of variance (ANOVA) followed by LSD multiple comparisons tests through using statistical SPSS software package (SPSS Inc, Chicago, USA),and the difference was considered significant when P<0.05. The study protocol was approved by the Ethics Committees of Neonatal Intensive Care Unit of the Children’s Institute, 81 children hospital, in Beijing.
Results
More SP-B deficiency in RDS group
In RDS group, low mature Surfactant protein B was found in both lung tissue and bronchoalveolar lavage fluid in 8 neonates. And in other 7 RDS cases, low mature SP-B was found in only bronchoalveolar lavage fluid. In control group, low mature Surfactant protein B was found in both lung tissue and bronchoalveolar lavage fluid in 4 neonates. And in other 3control cases, low mature SP-B was found only in bronchoalveolar lavage fluid. (Figures 1 and 2)
Figure 1.
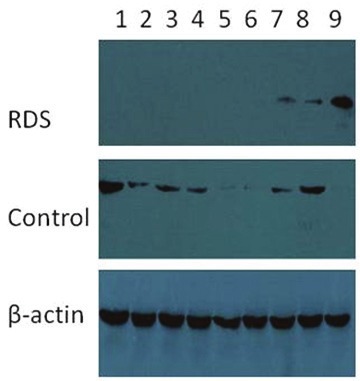
SP-B expression in lung tissue. Low mature surfactant protein B in lung tissue in 1-8 lanes in RDS group, while SP-B deficiency in lung tissue only in 4 lanes (5,6,7,9) in control group.
Figure 2.
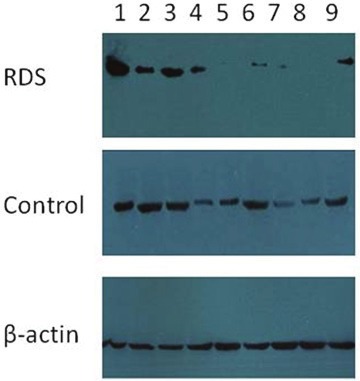
SP-B expression in bronchoalveolar lavage fluid. Low mature surfactant B in bronchoalveolar lavage fluid in 2, 4-9 lanes in RDS group, while SP-B deficiency in bronchoalveolar fluid only in 3 lanes (4, 7, 9) in the control group.
More mutation of exon 4 in SP-B gene in RDS group
In RDS group, 20 neonates were found to have mutations in exon 4, 12 homozygous mutations with C/C genotype and 8 heterozygous mutations with C/T genotype in surfactant protein B gene+1580 polymorphism. There were 8 cases mutations in control group, 1 in C/C and 7 in C/T genotype. To further assess the relationship between SP-B expression and gene mutation, genotype of SP-B gene+1580 polymorphism was analyzed in low SP-B expression neonates both in RDS and control groups. In 15 RDS neonates with low SP-B expression,12 in C/C and 2 in C/T genotype in surfactant protein B gene+1580 polymorphism. In 8 control neonates with low Surfactant protein expression, 1 in C/C and 7 in C/T genotype was found. To analyze further, 12 homozygous mutations with C/C genotype in SP-B gene led to SP-B deficiency in lung while 8 heterozygous mutations with C/T genotype in SP- B gene did not led to SP-B deficiency in lung.
SP-B gene polymorphisms associated with RDS
Overall, there were statistically significant differences among genotypes between groups. Our data showed that the frequency of homozygotes with C/C genotype was 0.3 and frequency of heterozygotes with C/T genotype was 0.02 in RDS group. In control group, frequency of homozygotes with C/C genotype was 0.025 and frequency of heterozygote with C/T genotype was 0.175.The homozygotes with C/C genotype was more frequently found in RDS neonates. (X2 =12.144, P<0.01) (Table 2, Figure 3).
Table 2.
Frequencies of the SP-B+1580 genotypes
| Groups | Patients(n) | Genotypes(n/%) | ||
|---|---|---|---|---|
|
| ||||
| C/C | C/T | T/T | ||
| RDS | 40 | 12(30)* | 8(20) | 20(50) |
| Control | 40 | 1(2.5) | 7(17.5) | 32(80) |
X2 = 12.144, P<0.01.
Figure 3.
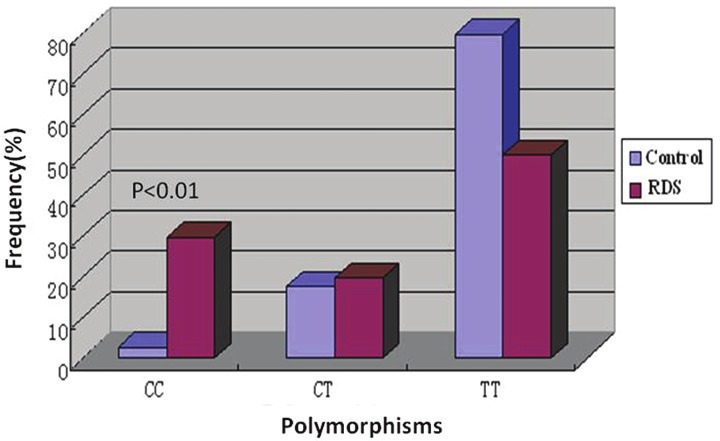
Frequencies of the SP-B+1580 genotypes. The relative frequencies of the SP-B genotypes (T/T and C/C and C/T) are shown for the two groups under study in Table 2. The homozygotes with C/C genotype was more frequently found in RDS neonates. (X2=12.144, P<0.01).
Discussion
RDS represents the most frequent form of respiratory insufficiency in the preterm newborn, and is still a major cause of morbidity and mortality in this group of patients [11,12]. Pulmonary morbidity has been attributed to oxygen toxicity, barotrauma, lung immaturity, and nutritional deficiencies. However, significant variations in pulmonary outcomes of similar infants with comparable exposures to oxygen, mechanical ventilation and nutritional deficiencies suggest that genetic factors also contribute to pulmonary outcomes [13]. RDS is the result of complex interactions between several environmental and genetic factors associated with prematurity, gender, race, and maternal diseases [14]. Some genetic variants of surfactant proteins, particularly SP-A and SP-B, are risk or protective factors in RDS etiology [15,16].
The development of lung disease in infants who made reduced amounts of mature SP-B supports the notion that a critical level of SP-B production is needed to prevent lung disease, and it suggests that individuals with even one SP-B mutation limiting SP-B production may be at risk for lung disease if other factors further decrease SP-B production. This may account for the findings in a full-term infant with transient SP-B deficiency that had a missense mutation on one allele [17]. In support of this hypothesis, genetically engineered mice heterozygous for one null SP-B allele had half-normal levels of SP-B mRNA and protein, exhibited abnormal lung compliance, and were more susceptible to hyperoxic lung injury when compared with homozygous wild-type litter mates [18,19]. In our early work, we found that there was SP-B deficiency in lung [20] and gene deficiency in intron 4 [21].
We have found low mature Surfactant protein B in both lung tissue and bronchoalveolar lavage fluid in 8 In RDS group. And in other 7 cases, low mature SP-B was found in only bronchoalveolar lavage. But the cases were 4 and 3 in the control group, and it is possible that such reduction of SP-B could be associated with RDS. The mechanism for SP-B deficiency remains unclear.
The present study analyzed mutations in exon 4 of SP-B gene (C/T at nucleotide +1580) in China Han ethnic group neonates with respiratory distress with RDS. The C/T polymorphism at nucleotide +1580 is located at the end of exon 4, at nucleotide +1580 and can alter the translation of amino acid 131 through a substitution from threonine (ACT) to isoleucine (ATT) [22]. This change eliminates a potential Linked glycosylation site but the real consequences of this alteration are not known [23,24]. In the literature, most studies suggest that the C/C genotype might be associated with a greater risk of pulmonary disease. In our results, 20 neonates were found to have mutations in exon 4, 12 in C/C and 8 in C/T genotype in surfactant protein B gene+1580 polymorphism in RDS group. There were 8 cases mutations in control group, 1 in C/C and 7 in C/T genotype. The C/C genotype was more frequently found in RDS neonates, To analyze further, 12 homozygous mutations with C/C genotype in SP-B gene led to SP-B deficiency in lung while 8 heterozygous mutations with C/T genotype in SP- B gene did not led to SP-B deficiency in lung. This suggests that homozygous mutations with C/C genotype may participate in development of neonatal RDS. That results agreed with the etiology of RDS, which was multifactorial and multigenic, and genetics may play a role in its pathogenesis. However, Lyra et al [25] have reported that no significantly differences in the frequencies of genotypes CC, CT and TT when the healthy term group and the RDS group were compared. This might be attributed either to ethnic differences between the studied population and individuals analyzed in other countries, or to sample size or geographical environment. Liu et al. [26] have reported that ethnic background is an important risk factor to be considered in analytical studies of allele and genotype frequencies. These investigators analyzed the similarity of genetic markers among populations of three different ethnic groups (Caucasian, Black, and Hispanic) in order to determine whether individuals of different races or ethnic groups could be grouped together in linkage studies. The results showed that the allele and genotype frequencies can differ among distinct ethnic groups, especially between ethnic groups of different races.
In summary, this study showed low mature Surfactant protein B is associated with the pathogenesis of neonatal RDS in China Han ethnic population. Mutations in exon 4 of the SP- B gene demonstrate an association between homozygous mutations in SP-B gene with RDS. And further SP-B gene Polymorphisms study suggests that homozygotes with genotype C/C in SP- B gene+1580 might be associated with neonatal RDS China Han ethnic population and reinforces the need for other studies to address this issue.
Acknowledgment
This study was funded by the National Natural Science Foundation of China (30871397).
References
- 1.Avery ME, Mead J. Surface properties in relation to atelectasis and hyaline membrane disease. AMA J Dis Child. 1959;97:517–523. doi: 10.1001/archpedi.1959.02070010519001. [DOI] [PubMed] [Google Scholar]
- 2.Chroneos ZC, Sever-Chroneos Z, Shepherd VL. Pulmonary surfactant: An immunological perspective. Cell Physiol Biochem. 2010;25:13–26. doi: 10.1159/000272047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orgeig S, Hiemstra PS, Veldhuizen EJ, Casals C, Clark HW, Haczku A, Knudsen L, Possmayer F. Recent advances in alveolar biology: Evolution and function of alveolar proteins. Respiratory Physiology & Neurobiology. 2010;173S:S43–S54. doi: 10.1016/j.resp.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyra PP, Diniz EM, Abe-Sandes K, Angelo AL, Machado TM, Cardeal M. Surfactant protein B gene polymorphism in preterm babies with respiratory distress syndrome. Braz J Med Biol Res. 2011;44:66–72. doi: 10.1590/s0100-879x2010007500147. [DOI] [PubMed] [Google Scholar]
- 5.Mazela J, Merritt TA, Gadzinowski J, Sinha S. Evolution of pulmonary surfactants for the treatment of neonatal respiratory distress syndrome and pediatric lung diseases. Acta Paediatr. 2006;95:1036–48. doi: 10.1080/08035250600615168. [DOI] [PubMed] [Google Scholar]
- 6.Kramer B. The respiratory distress syndrome in preterm infants: physiology, prophylaxis, and new therapeutic approaches. Intensivmed. 2007;44:403–408. [Google Scholar]
- 7.Floros J, Kala P. Surfactant proteins: molecular genetics of neonatal pulmonary diseases. Annu Rev Physiol. 1998;60:365–384. doi: 10.1146/annurev.physiol.60.1.365. [DOI] [PubMed] [Google Scholar]
- 8.Haataja R, Hallman M. Surfactant proteins as genetic determinants of multifactorial pulmonary diseases. Ann Med. 2002;34:324–333. doi: 10.1080/078538902320772089. [DOI] [PubMed] [Google Scholar]
- 9.Lin Z, deMello DE, Wallot M, Floros J. An SP-B gene mutation responsible for SP-B deficiency in fatal congenital alveolar proteinosis: evidence for a mutation hotspot in exon 4. Mol Genet Metab. 1998;64:25–35. doi: 10.1006/mgme.1998.2702. [DOI] [PubMed] [Google Scholar]
- 10.Hamvas A, Heins HB, Guttentag SH, Wegner DJ, Trusgnich MA, Bennet KW, Yang P, Carlson CS, An P, Cole FS. Developmental and genetic regulation of human surfactant protein-B in vivo. Neonatology. 2009;95:117–124. doi: 10.1159/000153095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curstedt T, Johansson J. New synthetic surfactants-basic science. Biol Neonate. 2005;87:332–337. doi: 10.1159/000084881. [DOI] [PubMed] [Google Scholar]
- 12.Jobe AH. Pulmonary surfactant therapy. N Engl J Med. 1993;328:861–868. doi: 10.1056/NEJM199303253281208. [DOI] [PubMed] [Google Scholar]
- 13.Cole FS, Hamvas A, Nogee LM. Genetic disorders of neonatal respiratory function. Pediatr Res. 2001;50:157–162. doi: 10.1203/00006450-200108000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Haataja R, Hallman M. Surfactant proteins as genetic determinants of multifactorial pulmonary diseases. Ann Med. 2002;34:324–333. doi: 10.1080/078538902320772089. [DOI] [PubMed] [Google Scholar]
- 15.Floros J, Veletza SV, Kotikalapudi P, Krizkova L, Karinch AM, Friedman C, Buchter S, Marks K. Dinucleotide repeats in the human surfactant protein-B gene and respiratory-distress syndrome. Biochem J. 1995;305:583–590. doi: 10.1042/bj3050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kala P, Ten HT, Nielsen H, Dunn M, Floros J. Association of pulmonary surfactant protein A (SP-A) gene and respiratory distress syndrome: interaction with SP-B. Pediatr Res. 1998;43:169–177. doi: 10.1203/00006450-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Tokieda K, Iwamoto HS, Bachurski C, Wert SE, Hull WM, Ikeda K, Whitsett JA. Surfactant protein-B-deficient mice are susceptible to hyperoxic lung injury. Am J Respir Cell Mol Biol. 1999;21:463–472. doi: 10.1165/ajrcmb.21.4.3436. [DOI] [PubMed] [Google Scholar]
- 18.Klein JM, Thompson MW, Snyder JM, George TN, Whitsett JA, Bell EF, McCray PB Jr, Nogee LM. Transient surfactant protein B deficiency in a term infant with severe respiratory failure. J Pediatr. 1998;132:244–248. doi: 10.1016/s0022-3476(98)70439-1. [DOI] [PubMed] [Google Scholar]
- 19.Clark JC, Weaver TE, Iwamoto HS, Ikegami M, Jobe AH, Hull WM, Whitsett JA. Decreased lung compliance and air trapping in heterozygous SP-B-deficient mice. Am J Respir Cell Mol Biol. 1997;16:46–52. doi: 10.1165/ajrcmb.16.1.8998078. [DOI] [PubMed] [Google Scholar]
- 20.Yin XJ, Luo FP, Li AH, An Y, Feng ZC. Relationship between reduced expression of surfactant protein B and neonatal respiratory distress syndrome in twenty Han ethnic group neonates in China. Chin J Pediatr. 2008;46:9–12. (in Chinese) [PubMed] [Google Scholar]
- 21.Yin XJ, Li LH, Wang Y, Xi L, Feng ZC. A study on expression of surfactant protein B in neonatal respiratory distress syndrome. Chin J Neonatol. 2011;26:336–339. (in Chinese) [Google Scholar]
- 22.Lin Z, deMello DE, Wallot M, Floros J. An SP-B gene mutation responsible for SP-B deficiency in fatal congenital alveolar proteinosis: evidence for a mutation hotspot in exon 4. Mol Genet Metab. 1998;64:25–35. doi: 10.1006/mgme.1998.2702. [DOI] [PubMed] [Google Scholar]
- 23.Floros J, Fan R, Diangelo S, Guo X, Wert J, Luo J. Surfactant protein (SP) B associations and interactions with SP-A in white and black subjects with respiratory distress syndrome. Pediatr Int. 2001;43:567–576. doi: 10.1046/j.1442-200x.2001.01474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Christensen ND, Wigdahl B, Guttentag SH, Floros J. Differences in N-linked glycosylation between human surfactant protein-B variants of the C or T allele at the single-nucleotide polymorphism at position 1580: implications for disease. Biochem J. 2003;369:179–184. doi: 10.1042/BJ20021376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyra P, Vaz F, Moreir P. Comparison of surfactant protein B polymorphisms of healthy term newborns with preterm newborns having respiratory distress syndrome. Braz J Med Biol Res. 2007;40:779–786. doi: 10.1590/s0100-879x2006005000105. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Bentley CM, Floros J. Study of human SP-A, SP-B and SP-D loci: allele frequencies, linkage disequilibrium and heterozygosity in different races and ethnic groups. BMC Genet. 2003;4:13. doi: 10.1186/1471-2156-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


