Abstract
Hypoxia-inducible factor, a heterodimeric transcription complex, regulates cellular and systemic responses to low oxygen levels (hypoxia) during normal mammalian development or tumor progression. Here, we present evidence that a similar complex mediates response to hypoxia in Caenorhabditis elegans. This complex consists of HIF-1 and AHA-1, which are encoded by C. elegans homologs of the hypoxia-inducible factor (HIF) α and β subunits, respectively. hif-1 mutants exhibit no severe defects under standard laboratory conditions, but they are unable to adapt to hypoxia. Although wild-type animals can survive and reproduce in 1% oxygen, the majority of hif-1-defective animals die in these conditions. We show that the expression of an HIF-1:green fluorescent protein fusion protein is induced by hypoxia and is subsequently reduced upon reoxygenation. Both hif-1 and aha-1 are expressed in most cell types, and the gene products can be coimmunoprecipitated. We conclude that the mechanisms of hypoxia signaling are likely conserved among metazoans. Additionally, we find that nuclear localization of AHA-1 is disrupted in an hif-1 mutant. This finding suggests that heterodimerization may be a prerequisite for efficient nuclear translocation of AHA-1.
Mammals use both systemic and cellular strategies to adapt to decreased oxygen levels during normal development and homeostasis. Hypoxic tissues secrete growth factors to increase vascularization, and individual cells increase anaerobic metabolism to sustain basic cellular functions (1). Hypoxia also plays a central role in tumor biology, as a mass of cancerous cells must adapt to hypoxia and induce angiogenesis to grow and metastasize (2).
The majority of the transcriptional responses to hypoxia are mediated by hypoxia-inducible factor (HIF) complexes, which consist of α and β subunits. The HIFβ subunit is also termed ARNT (aryl hydrocarbon receptor nuclear translocator) (3–7). When cellular oxygen levels are high, the von Hippel–Lindau tumor suppressor protein (VHL) binds directly to the α subunit and targets it for ubiquitination and proteosomal degradation. However, in hypoxic conditions, degradation of HIFα is inhibited (8–11). This permits HIFα to translocate to the nucleus, dimerize with ARNT, and activate the expression of target genes, which act to increase oxygen delivery or implement metabolic adaptation to hypoxia (2). Despite intensive study, the mechanisms by which cellular oxygen levels are sensed are not well understood. Progress in this field has been limited by the lack of genetic approaches to this important problem.
Widely divergent organisms have the ability to adapt to variable oxygen concentrations, which suggests that mechanisms of hypoxic sensing and response may have been established early in evolutionary history. Here, we investigate the molecular mechanisms of hypoxia response in a powerful genetic model organism, the nematode Caenorhabditis elegans. The natural habitat of C. elegans is often hypoxic, and C. elegans can adapt to very low environmental oxygen levels (12). We find that the C. elegans hif-1 and aha-1 gene products form a complex that is similar to the mammalian hypoxia-inducible factor, and we find that hif-1 function is required for adaptation to hypoxic conditions.
Methods
C. elegans Culture.
C. elegans were propagated and maintained as described in ref. 13. To isolate the ia04 mutation, N2 worms were mutagenized with ethyl methyl sulfonate, and the second generation of self-progeny was screened for hif-1 deletion mutations by PCR (14). The isolated deletion mutant was backcrossed to Bristol strain N2 nine times. To score viability in hypoxic conditions, animals were cultured on NGM plates in a sealed Plexiglas chamber with constant gas flow at 22°C. Compressed air and pure N2 were mixed to achieve the appropriate oxygen level, which was monitored by an oxygen analyzer. The transgenic C. elegans described here were generated by using standard protocols, and in each case, the coinjection marker was rol-6 (su1006) (15).
Molecular Biology.
The complementary DNAs for T01D3.2 and C15C8.2 were isolated from an embryonic cDNA expression library (16). The hif-1 cDNA plasmid pR8 was excised from the phage clone yk339c5 (from Y. Kohara, National Institute of Genetics, Shizuoka, Japan). The cDNA was modified to include a Kozak consensus translational start site (17) by amplifying the hif-1 cDNA fragment by using two primers: PASB6f, 5′-GCTCTAGAGCCAACCATGGAAGACAATCGGAAAAGAAAC-3′; and PASB2a, 5′-CAGTTGAAACTTTTGAATGAACCAGG-3′. The PCR product was then cut with XbaI to generate a 150-bp fragment, which was subsequently cloned into the two XbaI sites of pR8 to create pR33. To generate the hif-1:GFP expression construct pR19, a 9.0-kb KpnI fragment containing the hif-1 transcriptional start was excised from cosmid F38A6 and cloned into the KpnI site of pSP72. A PstI–SmaI fragment from the resulting plasmid was cloned into pPD95.75 (provided by A. Fire, Carnegie Institution of Washington). pHJ06 contains all of the hif-1 coding sequence fused in-frame to green fluorescent protein (GFP). pHJ06 was generated by inserting an hif-1 cDNA fragment into pR19. To create the aha-1:gfp plasmid pHJ02, 2.8 kb of 5′ regulatory sequence and the entire coding region of aha-1 was inserted into the pPD95.75 vector.
Antibodies, in Situ Staining, and Immunoprecipitation.
A bacterially expressed fusion protein containing AHA-1 (amino acids 8–57 and 353–451) and glutathione S-transferase was used to immunize mice and generate the 10H8 mAb. Immunostaining was performed as described (18). Images were captured with a Spot RT digital camera (Roche Diagnostics) on a Nikon E800 microscope. A commercially available GFP-specific mAb (Roche Diagnostics) was used to detect HIF-1:GFP on protein blots.
HIF-1 and AHA-1 were expressed in rabbit reticulocyte lysates (Promega TNT system) by using expression constructs pR33 and pJ343 (19). Radiolabeled methionine was included in the translation reaction, and the products were assayed by SDS/PAGE. A rabbit polyclonal antibody specific to AHA-1 (H.J., S. Wu, R.G., W. B. Wood, and J.A.P.-C., unpublished data) was used to immunoprecipitate AHA-1 and associated proteins. This process was performed as described in ref. 20, with an additional wash in 0.5 M LiCl/100 mM Tris⋅HCl, pH 7.4.
Results
Identification of hif-1.
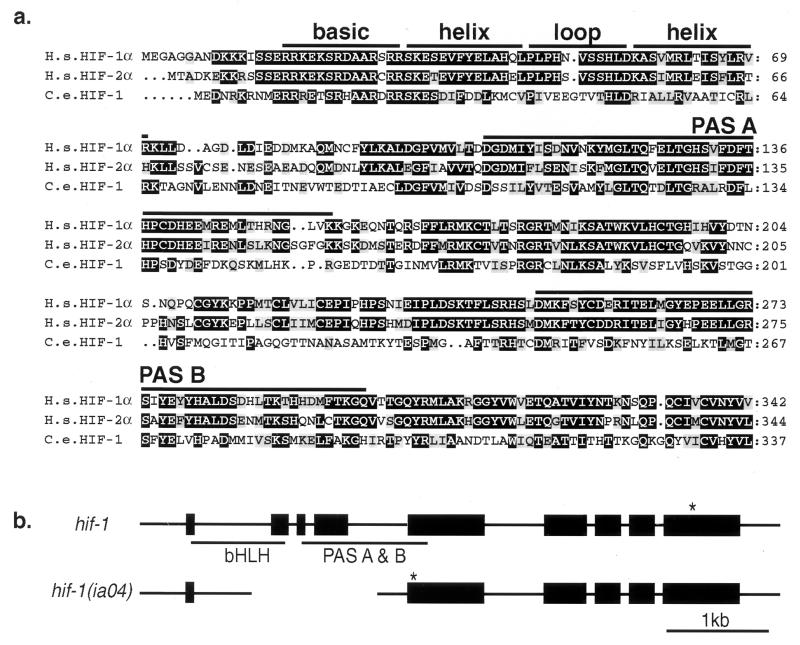
To determine whether the molecular mechanisms that regulate hypoxia response in C. elegans are similar to those described in mammals, we searched the C. elegans genome (21) for sequences encoding potential homologs of mammalian hypoxia-inducible factor α subunits (HIF-1α, HIF-2 α, and HIF-3α). The proteins that dimerize to form the hypoxia-inducible factor complex are members of a family of transcription factors containing basic helix–loop–helix (bHLH) and Per-ARNT-Sim (PAS) domains (3, 22). The bHLH domain is required for DNA binding and dimerization (23), and the PAS domain mediates interactions with other proteins and regulates dimerization specificity (23, 24). We isolated and sequenced complementary DNAs for the five genes in the C. elegans genome that were predicted to contain both of these motifs (ahr-1, aha-1, F38A6.3, T01D3.2, and C15C8.2). F38A6.3 encodes the C. elegans gene product with the highest sequence similarity to the bHLH and PAS domains of mammalian HIF-1α, and we named the gene hif-1. The predicted hif-1 gene product has 42% identity and 67% similarity to human HIF-1α in the bHLH domain, and it has 30% identity and 55% similarity to HIF-1α in the two PAS core domains (Fig. 1a). C. elegans HIF-1 is similarly related to the mammalian HIF-2α and HIF-3α proteins within the bHLH-PAS regions. Phylogenetic analysis suggests that hif-1 diverged from the ancestral HIFα gene before the gene duplication events that gave rise to mammalian HIF-1α, HIF-2α, and HIF-3α.
Figure 1.
(a) Conserved N-terminal domains of the predicted HIF-1 protein, and the Lif-1 genomic structure. Alignment of the N-terminal amino acid sequences of human HIF-1α (H.s. HIF-1α; U22431), HIF-2α (H.s. HIF-2α; U81983), and C. elegans HIF-1 (C.e. HIF-1; AF364604). Identical and similar amino acids are highlighted in black and gray, respectively. (b) Genomic structure of hif-1 and the hif-1 (ia04) deletion mutant. Boxes represent exons. * indicate the first stop codon in the hif-1 ORF. The ia04 mutation is a 1,231-bp deletion of the second, third, and fourth exons. This introduces a frameshift and premature stop in the mutant mRNA.
We designed experiments to test the hypothesis that hif-1 had biochemical and functional similarities with mammalian HIFα subunits. This hypothesis predicted that (i) hif-1 mutants would be deficient in hypoxia response; (ii) the hif-1 gene product would interact with aha-1, the C. elegans ortholog of mammalian ARNT (19); (iii) HIF-1 protein would be more highly expressed in hypoxic conditions; and (iv) both aha-1 and hif-1 would be expressed in most, if not all, cells.
hif-1 Mediates Response to Hypoxia.
To determine whether hif-1 function was required for adaptation to hypoxia, we mutagenized C. elegans with ethyl methanesulfonate and screened for deletions in the hif-1 gene. The hif-1 (ia04) mutation is a 1,231-bp deletion of the second, third, and fourth exons. We analyzed hif-1 complementary DNA in the mutant and determined that the first exon splices to the fifth exon, resulting in deletion of most of the bHLH and PAS domains, a shift in frame, and a premature translational stop (Fig. 1b). This mutation likely results in complete loss of hif-1 function.
C. elegans adapt to environmental oxygen levels below 4% by decreasing their metabolic rate. At 1% oxygen, their metabolic rate is decreased by half (12). As shown in Table 1, wild-type C. elegans embryos hatch and grow to become fertile adults in 1% oxygen. However, 66% of hif-1 mutants do not survive embryogenesis when cultured in these hypoxic conditions. The requirement for hif-1 is alleviated when oxygen levels are raised to 2% (Table 1), and there is no critical developmental requirement for hif-1 when the animals are incubated in standard laboratory conditions (21% oxygen).
Table 1.
hif-1 mutants have reduced viability in hypoxic conditions
| Oxygen levels, % | Wild type
|
hif-1 (ia04)
|
||||
|---|---|---|---|---|---|---|
| Embryonic lethality, % | Larval lethality, % | N | Embryonic lethality, % | Larval lethality, % | N | |
| 21 | 1.3 | 0.8 | 475 | 0.2 | 2 | 501 |
| 2 | ND | ND | — | 2.3 | 1.6 | 736 |
| 1 | 3.6 | 2.4 | 503 | 66.3 | 9.4 | 596 |
ND, not determined.
HIF-1 Binds AHA-1.
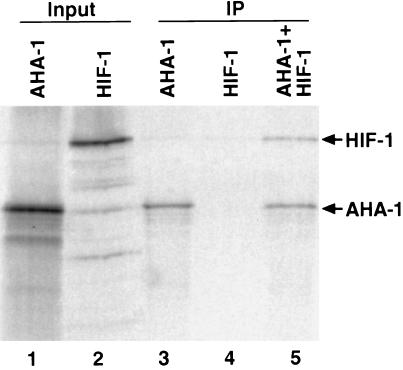
The mammalian hypoxia-inducible factors consist of two protein subunits: HIF-1α (or the related HIF-2α or HIF-3α) and ARNT (or ARNT2 or ARNT3) (2). To determine whether C. elegans HIF-1 forms a similar complex, we assayed its ability to bind AHA-1, the ortholog of mammalian ARNT (19). We expressed both gene products in rabbit reticulocyte lysates and immunoprecipitated the proteins with an antibody that specifically recognizes AHA-1. When HIF-1 is coincubated with AHA-1, both proteins are immunoprecipitated (Fig. 2, lane 5). Thus, HIF-1 binds AHA-1 in vitro.
Figure 2.
HIF-1 binds AHA-1. HIF-1 and AHA-1 were expressed in rabbit reticulocyte lysates in the presence of [35S]methionine (lanes 1 and 2), subjected to immunoprecipitation with an AHA-1-specific antibody (IP; lanes 3–5), and fractionated by SDS/PAGE. The antibody immunoprecipitates AHA-1 (lane 3) but not HIF-1 (lane 4). When AHA-1 and HIF-1 are coincubated, the antibody pulls down both proteins (lane 5). The molar ratio of HIF-1/AHA-1 in lane 5 is 1:2.5.
HIF-1:GFP Protein Levels Are Increased by Hypoxia.
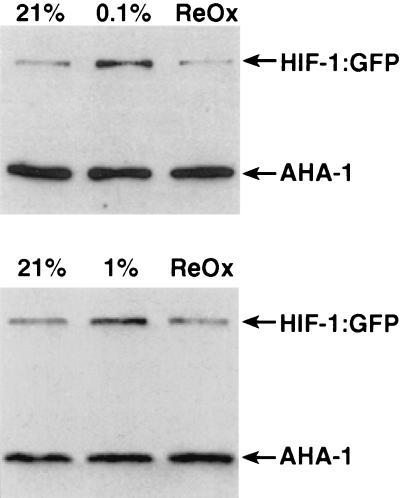
The degradation of mammalian HIFα protein is inhibited when oxygen levels are low. Therefore, we predicted that the expression of C. elegans HIF-1 protein would increase in hypoxic conditions. To test this, we generated transgenic animals carrying pHJ06, a reporter gene containing 2.7 kb of hif-1 5′ regulatory sequence and the entire hif-1 coding sequence fused to GFP (25). Transgenic worms were incubated in hypoxic conditions for 8 h. A fraction of the worms was then reoxygenated for 10 min. The expression of HIF-1:GFP was assayed by immunoblot by using a GFP-specific antibody. As shown in Fig. 3, HIF-1:GFP expression is increased by hypoxia treatment, and the protein is rapidly degraded upon reoxygenation. hif-1 mRNA is not substantially increased by hypoxia treatment (data not shown). The levels of AHA-1 protein remained constant, regardless of oxygen concentration (Fig. 3).
Figure 3.
HIF-1:GFP protein levels are increased by hypoxia. The transgenic C. elegans strain pHJ06 Ex6, which expresses the complete HIF-1 protein fused to GFP, was harvested before (21% O2) and after incubation in 0.1% oxygen (Upper) or 1% oxygen (Lower) for 8 h. In each experiment, a fraction of the hypoxia-treated worms were reoxygenated for 10 min (ReOx). Worms were lysed by boiling in loading buffer, and equal amounts of total protein were loaded in each lane. The expression of HIF-1:GFP and AHA-1 was assayed by probing the immunoblot with a GFP-specific antibody and the AHA-1-specific mAb 10H8.
aha-1 and hif-1 Are Broadly Expressed.
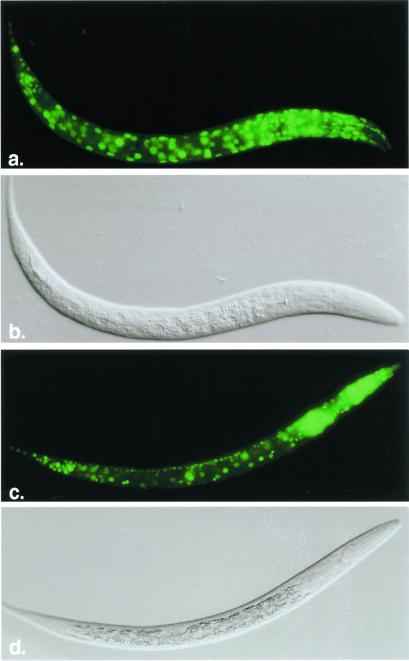
C. elegans do not have a complex circulatory system. Instead, they rely on diffusion for gas exchange. Therefore, individual cells must sense and respond to hypoxic conditions. We expected that most, if not all, cells would express hif-1 and aha-1. To test this hypothesis, we assayed the expression of reporter constructs in which hif-1 or aha-1 regulatory sequence directed the transcription of GFP. The hif-1:gfp fusion gene encoded by the pHJ06 plasmid was not appropriate for this study because it includes the region(s) of hif-1 that mediate oxygen-dependent degradation. Instead, we used the pR19 plasmid, which contains 2.7 kb of hif-1 5′ regulatory sequence and the genomic sequence encoding the first 45 aa of hif-1 fused to GFP. Transgenic animals carrying this reporter express GFP at high levels in every somatic cell (Fig. 4a). The aha-1:gfp fusion gene contains 2.8 kb of aha-1 5′ regulatory sequence and the complete aha-1 coding region. aha-1:gfp is expressed in many cell types throughout development, including hypodermal cells, intestinal cells, pharyngeal cells, and neurons (Fig. 4c). To further analyze AHA-1 expression, we generated the 10H8 mAb, which recognizes AHA-1 on protein blots and in situ. 10H8 recognizes AHA-1 in every cell of the C. elegans embryo (data not shown). Thus, aha-1 and hif-1 are both expressed in most, if not all, cells.
Figure 4.
hif-1 and aha-1 promoters direct expression of GFP reporter genes in most cell types. Fluorescent images and Nomarski images of a C. elegans first-stage larvae expressing hif-1:gfp (a and b) and aha-1:gfp (c and d).
Nuclear Localization of AHA-1 Depends on hif-1 Function.
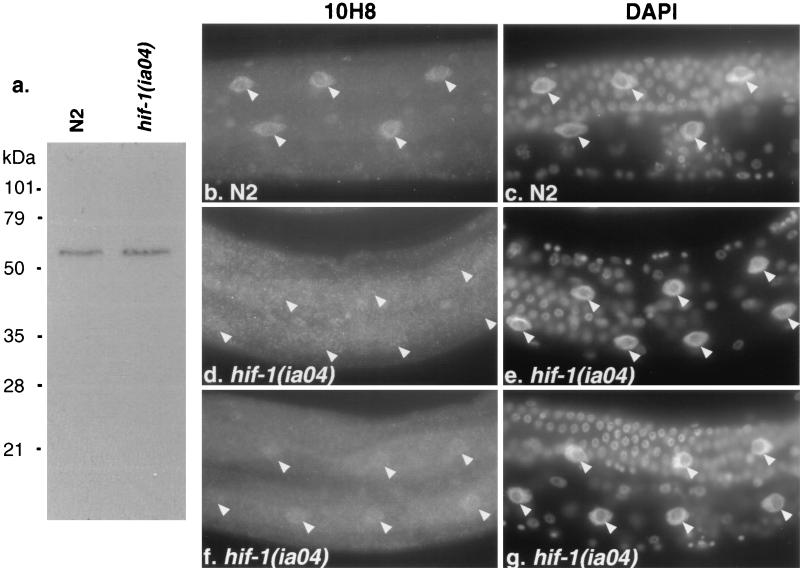
We used the 10H8 antibody to compare the subcellular localization of AHA-1 in wild-type and hif-1-defective animals. We reasoned that the interactions between HIF-1 and AHA-1 might be more readily detectable in cells that did not express other bHLH-PAS proteins, which might dimerize with AHA-1 and/or HIF-1. Therefore, we focused on the intestinal cells, which do not express any of the other C. elegans genes that include bHLH and PAS motifs (H.J., S. Wu, R.G., W. B. Wood, and J.A.P.-C., unpublished data). In the intestinal cells of wild-type animals, AHA-1 localization is predominantly nuclear (Fig. 5b). In the hif-1 mutants, this expression pattern is disrupted. In most cases, the staining is either slightly stronger in the nucleus or diffuse throughout the entire intestine (Fig. 5 d and f). These data suggest that AHA-1 is not efficiently localized to the intestinal nuclei in the hif-1 mutant. It is also possible that AHA-1 may be destabilized in the absence of hif-1. This requirement for hif-1 is cell type-specific, as AHA-1 is localized to the nuclei of some neuronal cells in the hif-1 mutant (data not shown). Total AHA-1 levels are equivalent in wild-type and hif-1 mutant animals (Fig. 5a).
Figure 5.
Subcellular localization of AHA-1 is disrupted in the hif-1 (ia04) mutant. (a) Equal amounts of total protein from wild-type (N2) or hif-1 (ia04) were size-fractionated by SDS/PAGE, blotted, and probed with the AHA-1-specific antibody 10H8. (b, d, and f) Immunostaining of intestinal cells with 10H8 in wild-type (N2) (b) or hif-1 mutant adults (d and f). (c, e, and g) The 4′,6-diamidino-2-phenylindole staining of the cells that are shown in b, d, and f, respectively. The arrowheads label the positions of intestinal cell nuclei.
Discussion
These data demonstrate that C. elegans hif-1 is a functional homolog of mammalian HIFα. First, hif-1 mutants cannot adapt to 1% oxygen. Second, the level of a HIF-1:GFP fusion protein is regulated by oxygen concentration. Third, HIF-1 forms a complex with AHA-1, the C. elegans ortholog of ARNT (HIF-1β) in vitro. The gene products also interact in vivo, as the nuclear localization of AHA-1 depends on hif-1 function in intestinal cells. Fourth, HIF-1:GFP and AHA-1 are expressed in most, if not all, cells.
Physiological Role of hif-1.
Although mammalian HIF-1α has an essential role in embryonic development, hif-1 defective C. elegans are viable and fertile when cultured in standard laboratory conditions. This reflects the relatively simple physiology of C. elegans. A mammalian embryo relies on hypoxia-induced angiogenesis to oxygenate tissues. Hif-1α−/− mice die by E9.0 with severe vascular defects (4, 5). In contrast, an adult C. elegans hermaphrodite has no apparent need for specialized respiratory structures or a complex circulatory system. Any cell in the organism is only a few cell widths from the outer surface of the worm or the intestinal lumen (26).
Oxygen sensing and hif-1 function is likely to be very important in the soil environment inhabited naturally by C. elegans. In the laboratory, C. elegans are cultured on top of an agar-based medium, and the ambient oxygen concentrations are relatively high. However, high concentrations of bacteria, the C. elegans food source, can deplete oxygen in a soil microenvironment. The nematodes must be able to sense and adapt to these hypoxic conditions.
Regulation of AHA-1 Nuclear Translocation.
We find that AHA-1 translocation to the nuclei of intestinal cells is inefficient in hif-1 mutants. This result was not predicted by the prevalent models for hypoxia signaling. In the mammalian cell lines commonly used to study hypoxia-inducible factor or aryl hydrocarbon receptor (AHR) signaling, ARNT is localized to the nucleus constitutively. After HIF-1α or activated AHR translocates to the nucleus, it forms a dimer with ARNT (2). However, in the Drosophila embryo, Drosophila ARNT (encoded by the tango gene) apparently remains cytoplasmic until a bHLH-PAS dimerization partner is expressed (27, 28). We conclude that the role of AHA-1 in the formation and nuclear localization of an active transcriptional complex may depend on cell type-specific factors, such as the expression of other bHLH-PAS proteins. We will explore this hypothesis by examining the expression of other bHLH-PAS genes in those cells that localize AHA-1 to the nucleus in the absence of hif-1.
Importance of a Genetic Model System for the Study of Hypoxia.
A thorough understanding of the molecular mechanisms by which cells sense and respond to hypoxia is essential to the treatment of many human diseases, including cancer, stroke, and cardiac failure. However, several critical questions remain. The oxygen sensors are undiscovered, and it will be important to determine how they regulate HIFα stability. Insulin signaling has also been shown to activate the hypoxia-inducible factor (29–31), but the roles of this interaction during hypoxia response, cellular differentiation, or tumor progression are not understood. Other signal transduction pathways may also modify hypoxia-inducible factor to modulate cellular response to low oxygen levels (32).
C. elegans is an increasingly powerful model system for the dissection of signal transduction pathways, and further genetic analysis of hif-1 and its interacting genes will likely reveal mechanisms of hypoxia signaling and response that are conserved throughout the animal kingdom. In support of this hypothesis, a putative homolog of VHL has recently been identified in C. elegans (33), and the cul-2 gene encodes another evolutionarily conserved component of the ubiquitin–proteasome proteolytic mechanism (34).
While this manuscript was in review, two studies were published that demonstrated that mammalian HIFα is targeted for VHL-mediated degradation by proline hydroxylation (35, 36). The modified proline and an essential leucine have been identified in HIFα, and these residues are conserved in C. elegans HIF-1 (35).
Acknowledgments
We are grateful to David Vleck and Mark Hargrove, who provided equipment and advice that made the hypoxia experiments possible. We thank Andy Fire, Peter Okkema, and Yuji Kohara for reporter constructs, cDNA libraries, and the yk339c5 cDNA, respectively. Xun Gu and Gavin Naylor provided useful advice regarding phylogenetic analysis. Some C. elegans strains were provided by the Caenorhabditis Genetic Stock Center, which is supported by the National Institutes of Health National Center for Research Resources. We thank Clark Coffman and members of the Powell–Coffman lab for helpful discussions and critical reading of the manuscript. This work was supported by National Science Foundation Grant 9874456 (to J.A.P.-C.).
Abbreviations
- HIF
hypoxia-inducible factor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- GFP
green fluorescent protein
- bHLH
basic helix–loop–helix
Footnotes
References
- 1.Guillemin K, Krasnow M A. Cell. 1997;89:9–12. doi: 10.1016/s0092-8674(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 2.Semenza G L. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 3.Wang G L, Jiang B H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer N V, Kotch L E, Agani F, Leung S W, Laughner E, Wenger R H, Gassmann M, Gearhart J D, Lawler A M, Yu A Y, Semenza G L. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan H E, Lo J, Johnson R S. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maxwell P H, Dachs G U, Gleadle J M, Nicholls L G, Harris A L, Stratford I J, Hankinson O, Pugh C W, Ratcliffe P J. Proc Natl Acad Sci USA. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmeliet P, Dor Y, Herbert J M, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Nature (London) 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 8.Ohh M, Park C W, Ivan M, Hoffman M A, Kim T Y, Huang L E, Pavletich N, Chau V, Kaelin W G. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 9.Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. . (First Published September 5, 2000; 10.1073/pnas.190332597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maxwell P H, Wiesener M S, Chang G W, Clifford S C, Vaux E C, Cockman M E, Wykoff C C, Pugh C W, Maher E R, Ratcliffe P J. Nature (London) 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 11.Tanimoto K, Makino Y, Pereira T, Poellinger L. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Voorhies W A, Ward S. J Exp Biol. 2000;203:2467–2478. doi: 10.1242/jeb.203.16.2467. [DOI] [PubMed] [Google Scholar]
- 13.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen G, Hazendonk E, Thijssen K L, Plasterk R H. Nat Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- 15.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okkema P G, Fire A. Development (Cambridge, UK) 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 18.Finney M, Ruvkun G. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 19.Powell-Coffman J A, Bradfield C A, Wood W B. Proc Natl Acad Sci USA. 1998;95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukunaga B N, Probst M R, Reisz-Porszasz S, Hankinson O. J Biol Chem. 1995;270:29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 21.C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y Z, Hogenesch J B, Bradfield C A. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 23.Jiang B H, Rue E, Wang G L, Roe R, Semenza G L. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 24.Zelzer E, Wappner P, Shilo B Z. Genes Dev. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 26.Sulston J E, Schierenberg E, White J G, Thomson J N. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 27.Emmons R B, Duncan D, Estes P A, Kiefel P, Mosher J T, Sonnenfeld M, Ward M P, Duncan I, Crews S T. Development (Cambridge, UK) 1999;126:3937–3945. doi: 10.1242/dev.126.17.3937. [DOI] [PubMed] [Google Scholar]
- 28.Ward M P, Mosher J T, Crews S T. Development (Cambridge, UK) 1998;125:1599–1608. doi: 10.1242/dev.125.9.1599. [DOI] [PubMed] [Google Scholar]
- 29.Zelzer E, Levy Y, Kahana C, Shilo B Z, Rubinstein M, Cohen B. EMBO J. 1998;17:5085–5094. doi: 10.1093/emboj/17.17.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldser D, Agani F, Iyer N V, Pak B, Ferreira G, Semenza G L. Cancer Res. 1999;59:3915–3918. [PubMed] [Google Scholar]
- 31.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, Gottschalk A R, Ryan H E, Johnson R S, Jefferson A B, et al. Genes Dev. 2000;14:391–396. [PMC free article] [PubMed] [Google Scholar]
- 32.Agani F, Semenza G L. Mol Pharmacol. 1998;54:749–754. doi: 10.1124/mol.54.5.749. [DOI] [PubMed] [Google Scholar]
- 33.Woodward E R, Buchberger A, Clifford S C, Hurst L D, Affara N A, Maher E R. Genomics. 2000;65:253–265. doi: 10.1006/geno.2000.6144. [DOI] [PubMed] [Google Scholar]
- 34.Feng H, Zhong W, Punkosdy G, Gu S, Zhou L, Seabolt E, Kipreos E. Nat Cell Biol. 1999;1:486–492. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- 35.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara J M, Lane W S, Kaelin W G., Jr Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 36.Jaakkola P, Mole D R, Tian Y M, Wilson M I, Gielbert J, Gaskell S J, Kriegsheim Av A, Hebestreit H F, Mukherji M, Schofield C J, et al. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]