Abstract
Early diagnosis is the way to improve lung cancer survival rate and is almost impossible today due to the lack of molecular probes that recognize lung cancer cells sensitively and selectively. We have developed a new aptamer approach for the recognition of specific small cell lung cancer (SCLC) cell surface molecular markers. Our approach relies on cell based systematic evolution of ligands by exponential enrichment (cell-SELEX) to evolve aptamers for whole live cells that express a variety of surface markers representing molecular differences among cancer cells. When applied to different lung cancer cells including those from patient samples, these aptamers bind to SCLC cells with high affinity and specificity in different assay formats. When conjugated with magnetic and fluorescent nanoparticles, the aptamer nano-conjugates could effectively extract SCLC cells from mixed cell media for isolation, enrichment, and sensitive detection. These studies demonstrate the potential of the aptamer approach for early lung cancer detection.
Keywords: aptamer, cell-SELEX, detection, lung cancer, molecular marker
Introduction
As the leading cause of cancer mortality, lung cancer accounts for 29% of all cancer deaths in the United States with a 5-year survival rate less than 15%.[1] The main reason for the high death rate of lung cancer is that most lung cancer patients are diagnosed at an advanced stage where treatments are rarely successful.[2,3] Among all the lung cancer subtypes, small cell lung cancer (SCLC) has the highest tendency for early dissemination and the shortest median survival (7–12 months) as a clinically distinct entity.[4,5]
Survival of patients with lung cancer, especially SCLC, relies on early detection as well as effective treatment.[6] Although the previous clinical trial of early detection using sputum cytology and chest x-rays led to earlier diagnosis of lung cancers, no reduction in overall mortality could be justified thereafter.[7] The recent development of imaging based screening technologies such as spiral computed tomography (CT), optical coherent tomography, positron emission tomography (PET), virtual bronchoscopy, autofluorescence bronchoscopy, and confocal microscopy has not shown to improve this situation either.[7,8] The possible reason for this is that morphological criteria used in imaging approaches are not sensitive enough during the premalignant phase of lung cancer development,[9,10] especially SCLC, which arises without morphologically recognizable preneoplastic lesions.[11]
To further improve early detection, molecular approaches were exploited to complement imaging studies. Molecular abnormalities correlate with behavioral aspects of lung cancer, and therefore are more sensitive in detecting invasive malignant lesions in preclinical phase than tumor sizes revealed by imaging. Genetic and proteomic analysis are two major molecular-marker based early detection techniques.[12] Genetic changes can be detected reproducibly by PCR [13] and genomic hybridization,[14] but they do not always correlate with changes at protein level. Proteomic analysis using immunostaining [15] and mass spectrometry [16] is a new trend of early detection. However, there has been only limited success in developing sensitive and specific molecular probes for such analysis.[17]
Here, we describe a new nucleic acid probe based approach, in which a panel of DNA aptamers was purposely developed against the molecular differences among lung cancer cells to detect specific molecular markers on SCLC cell surfaces. Our approach relies on cell-SELEX (systematic evolution of ligands by exponential enrichment) to ensure the specificity and widespread availability of aptamer probes, which are missing in antibody based methods.[18–20]
With cell-SELEX, we have developed a panel of molecular aptamers for SCLC that is the most aggressive subtype of lung cancer with greater than 95% mortality in a few years, and has the shortest life expectancy among all lung cancer subtypes.[21,22] These aptamer probes were selected without prior knowledge about SCLC biomarkers and tested for their ability to specifically bind both cultured cells and clinical samples of SCLC in various assay formats. Aptamers were also exploited for detection and enrichment of SCLC cells, a critical step towards the goal of early detection where sensitive detection is needed.
Compared to other molecular recognition elements, aptamers used in this approach present several advantages for early detection.[23–25] While aptamers’ sensitivity leads to the detection of malignant cells, their specificity derived from cell-SELEX prevents cross reactivity with normal epithelial cells, resulting in fewer false positives. In practice, multiple aptamers can be readily developed for any cancer cells of interest without prior knowledge of cell surface marker proteins, and are more predictive of cancer progression than single probes used in previous studies. In addition, low-molecular weight aptamers can be easily synthesized and modified to recognize the target proteins at their native state on cell surfaces reproducibly.
Results and Discussion
SELEX for whole live cancer cells
To develop cell specific aptamer probes, live cancer cells were directly used as the target for cell-SELEX, an approach previously established in our lab. This approach was adapted in a few aspects to work with floating aggregates of SCLC and adherent monolayers of non-small cell lung cancer (NSCLC), which are two typical growth patterns of lung cancer culture. Because of their heterogeneous population and poor viability, it is more challenging to perform cell-SELEX with lung cancer than leukemia in previous study.[20] We chose to develop aptamer probes targeting SCLC (classic) in initial study as it possesses the worst clinical presentation among all lung cancer phenotypes. NSCLC (large cell) was adopted as a control for cell-SELEX to generate aptamers exclusive to the cell surface markers of SCLC. These cell surface markers are so exclusive to SCLC that normal lung epithelial cells are also not expected to bear them and cross-react with developed aptamers, as observed in previous studies with antibodies.[10] With counter-selection against control cells, the aptamers achieve great selectivity necessary for the reliable detection of lung cancer antigens.
In the actual selection (Figure 1A), a cultured SCLC cell line, NCI-H69, was first incubated with a 71-base synthetic single stranded DNA library. The DNA sequences that bound to target cells were then eluted after stringent washing. A cultured NSCLC cell line, NCI-H661, was now introduced as control cell to separate aptamers with affinity to both the target and control cells from those aptamers recognizing only target cells in the previously eluted DNA pool. The remaining target cell specific sequences from counter-selection were further PCR amplified to form the starting pool of next round of selection. A panel of aptamer probes eventually evolved to have great specificity and high affinity for SCLC along with the SELEX progress.
Figure 1. Scheme of Cell-SELEX for SCLC and Enrichment of Aptamers Along with the Progress of SELEX.


A) A number of DNA molecules from ssDNA library bind to SCLC cells after incubation, and are retained for counter-selection with NSCLC cells. The SCLC specific DNA molecules are subsequently PCR amplified for next round of selection, or for cloning and sequencing to identify individual aptamers in most selected pool.
B) Gradual evolution of SCLC specific aptamers along with the progress of SELEX. FITC-labeled ssDNA library and selected DNA pools were tested for binding to NCI-H69 (SCLC) and NCI-H661 (NSCLC) cells by flow cytometry. The binding ability of selected DNA pools gradually increased for SCLC, and no significant change was observed for NSCLC.
Enrichment of aptamers
We monitored the gradual enrichment of aptamers during the selection process by both flow cytometry (Figure 1B) and confocal microscopy (data not shown). The ability of DNA pools from each round of selection to bind target cells was assessed. The increase in the fluorescence intensity of the dye labeled DNA pools bound to target cells is gradual and steady along with the progress of selection, indicating a successful evolution of high affinity aptamers. By contrast, no significant change was observed in the response to the control cells during the selection process, demonstrating the specificity of selected DNA pools.
SCLC specific aptamers
After 25 rounds of selection, the binding ability of selected DNA pool reached a plateau, and cloning was performed to isolate individual aptamers in the most selected DNA pool. Results of subsequent sequencing were further analyzed by multiple sequence alignment software (Figure S1). We found that the majority of aptamers in the selected pool belong to several families based on the consensus sequences they have.
To deconvolute the selected DNA pool, those consensus sequences with high repeats were synthesized to test their ability to specifically bind SCLC cells. A few of them showed prominent binding ability for SCLC but not NSCLC (control cell), as illustrated by flow cytometry results (Figure 2A). The dominent peak refers to the binding of aptamer with SCLC cells. A second peak with high fluorescence signal was also noticed, which may represents the population of dead cells. According to confocal imaging results, fluorescent dye labeled aptamers only specifically bind to target SCLC cells (Figure 2B). In addition, individual aptamers were tested with saturation analysis as depicted in (Figure S2), and found to have high affinity with equilibrium dissociation constant in the nanomolar range (Table 1).
Figure 2. Molecular Aptamers Bind Specifically to SCLC Cells.


A) SCLC cells, NCI-H69 (top panel) and NSCLC cells, NCI-H661 (bottom panel), were incubated with FITC-labeled aptamers and ssDNA library. Cell surface binding of aptamers was assessed by flow cytometry. Aptamer binding was restricted to SCLC cells. As the cells are different, different flow cytometry parameters were used. One can only compare the library and the aptamer for each type of cell. The dominent peak refers to the binding of aptamer with SCLC cells. A second peak with high fluorescence signal was also noticed, which may represents the population of dead cells.
B) SCLC cells, NCI-H69, were incubated with TAMRA-labeled ssDNA library (top panel) and HCH07 aptamer (bottom panel). The fluorescent dye labeled aptamers bind to the cell surface of SCLC cells as determined by confocal imaging at 60× magnification. Only background level fluorescence was observed on cells incubated with fluorescent dye labeled ssDNA library due to non-specific binding.
Table 1.
Equilibrium Dissociation Constant of Selected SCLC Aptamers
| Selected sequence name | Kd, nM |
|---|---|
| HCA12 | ~97nM |
| HCC03 | ~123nM |
| HCH07 | ~38nM |
| HCH01 | ~157nM |
Enzymatic treatment of cell surface markers
In addition to the prior studies of developed aptamers, their putative cell surface targets were examined by enzymatic treatment to further verify the binding of aptamers to SCLC cell surface markers. After the brief treatment of cells with trypsin or proteinase K, diminished binding of aptamers to SCLC cells was observed by flow cytometry in both cases (Figure S3A). We saw the same trend under confocal microscopy, only limited amount of fluorescent aptamers getting retained on enzyme treated cell surfaces (Figure S3B). These results suggested that selected aptamers indeed bind to cell membrane target molecules, and these discovered SCLC cell surface markers can be affected by protease.
Validation of aptamers with different cancer cells and assay formats
Before testing with clinical samples, we assessed the applicability of developed aptamer probes to other cultured SCLC cell lines (i.e., to validate the target molecules of developed aptamers as exclusive markers for SCLC). The panel of aptamers showed consistent binding pattern to NCI-H146 and NCI-H128 (Table 2), two SCLC cell lines that have similar cell characteristics as NCI-H69 (target cell used in cell-SELEX). In contrast to SCLC, three NSCLC cells lines including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma (the one used as control cell in cell-SELEX) did not respond to the selected aptamers except one case (aptamer HCH07 bound to NCI-H23) (Table 2). Moreover, other cancer types including two leukemia cell lines and two liver cancer cell lines were not recognized by these aptamers in most cases (Table 2). Interestingly, the aptamer that bound to adenocarcinoma NCI-H23 is also able to recognize liver cancer cell lines. Considering these cell lines were not used as control cell in cell-SELEX, there is still a chance for some of the selected aptamers to recognize them by the same target molecule SCLC has. It is expected to minimize this type of cross-reactivity by adding additional negative selection criteria.
Table 2.
Tests of Developed Aptamers with Cultured Cancer Cell Lines
| Cultured cancer cell lines | Receptors | HCA12 | HCC03 | HCH07 | HCH01 |
|---|---|---|---|---|---|
| NCI–H69 (small cell carcinoma) | IGF II | + | + | + | + |
| NCI–H146 (small cell carcinoma, bone marrow) | IGF II | − | + | + | + |
| NCI–H128 (small cell carcinoma, pleural effusion) | N/A | + | + | + | + |
| NCI–H661 (large cell carcinoma, lymph node) | N/A | − | − | − | − |
| NCI–H23 (adenocarcinoma) | PDGF; TGF; EGF | − | − | + | − |
| NCI–H1385 (squamous cell carcinoma, lymph node) | N/A | − | − | − | − |
| CCRF–CEM (T cell acute lymphoblastic leukemia) | N/A | − | − | − | − |
| Ramos (B cell human Burkitt’s lymphoma) | N/A | − | − | − | − |
| IMEA (liver cancer) | N/A | − | − | + | − |
| BNL (liver cancer) | N/A | − | − | + | − |
Besides the tests with live cancer cells, it is also interesting to see whether the aptamers developed from live cells can recognize fixed cells, which is the main assay format for retrospective analysis of preserved specimens such as sputum and biopsy in early detection studies. It will also be useful for histological examination in the clinical diagnosis of lung cancer. We processed formalin-fixed, paraffin-embedded cell line tissue arrays from SCLC and NSCLC samples. After incubation with fluorescent dye labeled aptamers, washing, and dehydration, stained array slides were mounted for array scanning and confocal imaging. Binding of aptamer probes was found to be specific to SCLC as only background level binding existed for NSCLC (Figure 3A). Notably, most aptamers bound to the periphery of target cells as shown in magnified confocal microscopy image (Figure 3B, Figure S4). This binding pattern is similar to that observed in tests of live cells, and further confirmed that aptamers indeed bind to their target molecules on fixed cells. These data indicate that specific recognition of cell line tissue array by aptamers is dependent on the presence of cell surface markers, which are still biochemically active after fixing cells.
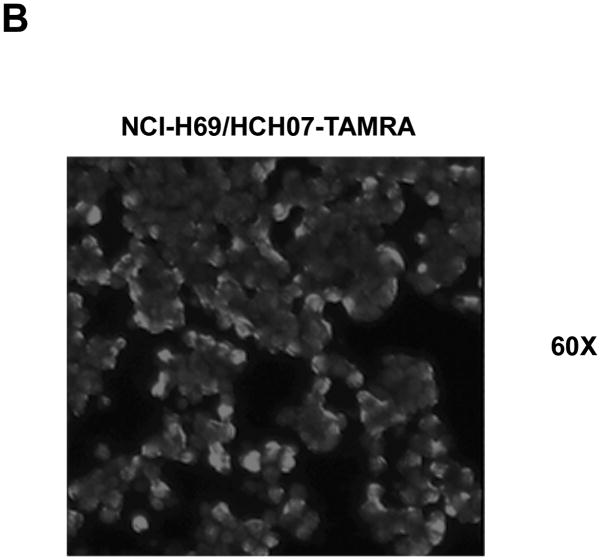
Figure 3. Specific Binding of Molecular Aptamers to Formalin-fixed, Paraffin-embedded SCLC Cell Line Tissue Array.
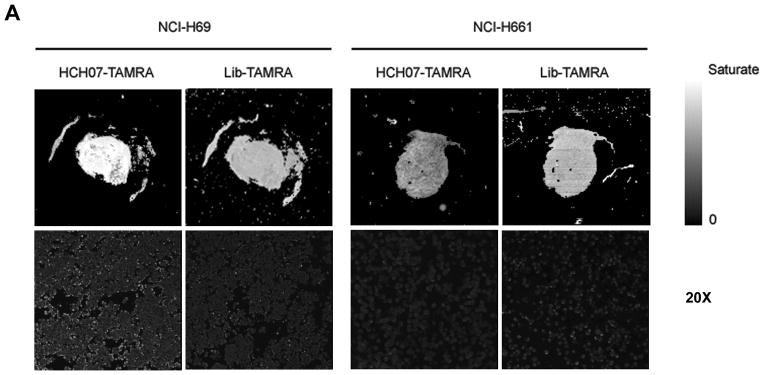
A) Recognition of SCLC and NSCLC cell line tissue arrays by fluorescent dye labeled aptamer. Sections of formalin-fixed SCLC and NSCLC cells embedded in paraffin were stained with TAMRA labeled aptamer and analyzed by array scanning (top panel) and confocal imaging at 20× magnification (bottom panel). TAMRA labeled ssDNA library was used as a control. Binding of aptamer is specific to SCLC cell line tissue array.
B) Fluorescent aptamer stained SCLC cell line tissue array shows similar binding pattern to live cells by magnified confocal imaging (60× magnification). Note the binding of fluorescent dye labeled aptamers to the periphery of fixed SCLC cells. Confocal image of fluorescent ssDNA library as control was shown in supporting information (Figure S4).
Clinical sample tests and detection of SCLC cells in whole blood samples
Next, we assessed the sensitivity and specificity of the aptamers for their ability to detect cancer cells in clinical sample from SCLC patient. Substantial change in fluorescence intensity was noted in the SCLC patient’s sample after incubation with dye labeled aptamers, indicating that aptamers developed for cultured cells are also able to recognize the cancer cells from SCLC patients (Figure 4A). This result clearly demonstrated the applicability of those aptamers to clinical samples, one important prerequisite for successful detection of SCLC patient cells in complex biological matrix by the aptamers.
Figure 4. Recognition of SCLC Cells in Patient Sample and Human Whole Blood with Molecular Aptamers.
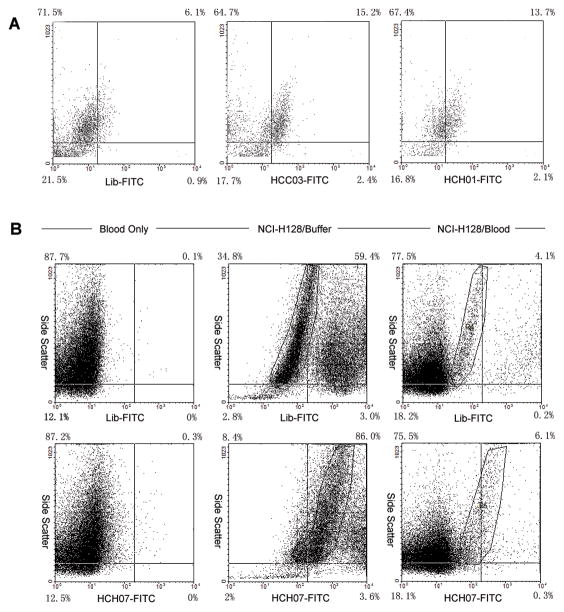
A) Clinical samples from SCLC patient were incubated with either FITC labeled ssDNA library or FITC labeled aptamers (HCC03 and HCH01). Samples were then analyzed by flow cytometry. SCLC cells were clearly identified in patient sample by aptamer probes.
B) Detection of SCLC cells in whole blood sample with aptamers. In human whole blood, blood cells were incubated with FITC labeled aptamer and ssDNA library as a control for specificity (left panel). SCLC cells in buffer were incubated with FITC labeled aptamer and ssDNA library to compare with SCLC cells in whole blood as a positive control (middle panel). Circled regions are live SCLC cells. The binding of aptamer to SCLC cells in human whole blood was also assessed (right panel). Circled regions are live SCLC cells. Aptamer retains the specificity to SCLC cells in human whole blood as determined by flow cytometry. No interference from blood cells was observed.
In addition, we sought to determine whether aptamers retain the ability to specifically recognize SCLC cells in the presence of human blood environment, another criterion for aptamers to be applied in clinical tests. The binding of fluorescently labeled aptamers to SCLC cells mixed with human whole blood was assessed by flow cytometry. As controls, we also tested aptamers with human whole blood and SCLC cells in buffer. Aptamers’ specificity in human whole blood is consistent with those obtained in buffer experiments. SCLC cells were recognized by aptamers specifically, and no interference from various blood cells was observed (Figure 4B). For the stability of aptamers in human blood environment, it has been found out that modification of aptamers with nonnatural nucleic acid can significantly improve the half-life while they still sustain the binding ability.[26] These results, together with the patient sample results, indicate the possibility for developed aptamer probes to be practically useful in clinical tests.
Extraction and detection of SCLC cells with aptamer conjugated nanoparticles
During the early stage of lung cancer, malignant lesions begin to shed circulating cells. Encoding valuable information for prognostic prediction, these exfoliated cells should be of great value for early lung cancer detection. Previously, the enrichment and detection of rare exfoliated cells were mainly performed by flow sorting and immunomagnetic cell sorting, which suffer from deficiency of sensitivity and specificity.[27,28] To evaluate the potential of the selected aptamers for early lung cancer detection, we prepared aptamer conjugated magnetic nanoparticles and aptamer conjugated fluorescent nanoparticles to isolate, enrich, and detect rare SCLC cells with a method previously established in our lab.[29] The spiked tumor cells were first incubated with aptamer conjugated magnetic and fluorescent nanoparticles. Magnetic nanoparticle bound cells were then isolated by magnetic separation. After recovery, we measured the fluorescence of the dye-doped nanoparticles, which also bound to the isolated cells through aptamer. Whereas two different types of SCLC cells were effectively isolated and detected, the extraction of NSCLC cells was very inefficient (Figure 5A). Additionally, very low background fluorescence signal was observed in the control experiment using DNA library conjugated nanoparticles, suggesting that nonspecific extraction of tumor cells is rare with this method. Effective enrichment and detection of SCLC cells were verified by confocal imaging results, which showed that the extracted tumor cells were indeed binding to aptamer conjugated nanoparticles (Figure 5B). Moreover, the dye-doped nanoparticles confer great sensitivity to the detection of extracted rare tumor cells. Therefore, this aptamer conjugated nanoparticle approach demonstrated its capability to enrich and detect rare lung cancer cells, which will be critical for early diagnosis of lung cancer.
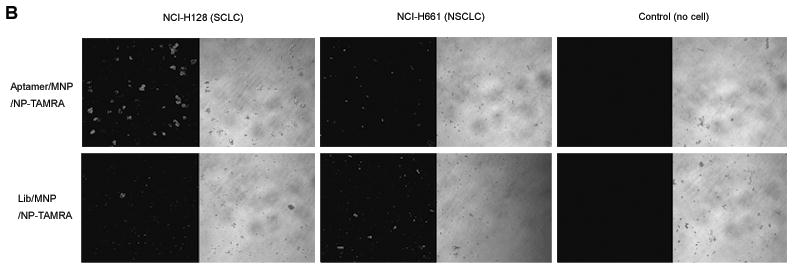
Figure 5. Extraction and Detection of SCLC Cells for Enrichment and Early Diagnosis with Aptamer Conjugated Magnetic/Fluorescent Nanoparticles.

A) Same amount of the spiked SCLC (NCI-H69 and NCI-H128) and NSCLC (NCI-H661) cells were incubated with aptamer conjugated magnetic and fluorescent nanoparticles. Aptamer bound cells were subsequently isolated by magnetic separation. Total extracted cell amount after magnetic separation was determined by measuring the fluorescence signal of dye-doped nanoparticles that also bind to cells. Aptamer conjugated nanoparticles effectively extracted SCLC cells but not NSCLC cells (cyan). ssDNA library conjugated nanoparticles were used in control experiments and showed only limited non-specific extraction of tumor cells (yellow).
B) Confocal imaging of cells extracted by aptamer conjugated nanoparticles (top panel) and ssDNA library conjugated nanoparticles (bottom panel). Extraction and detection of SCLC cells with this aptamer based approach were verified.
Conclusion
We have developed a panel of aptamer probes with specificity to SCLC cells, using cell-SELEX. SCLC was chosen as the model system for its worst prognosis among all lung cancer subtypes. The molecular differences among different types of cells resulted in the evolution of specific aptamer probes. Aptamers developed in this way specifically recognize SCLC cells, whereas NSCLC cells are not recognized by them. The target molecules of these aptamers are therefore reasoned to be preferentially expressed in SCLC. The interaction between aptamers and their target molecules were further characterized and confirmed. We found that aptamers are indeed binding to their target molecules on cell surface, and this interaction can be affected by treating cells with protease. Moreover, it was proved that cell-SELEX strategy can be used as an effective tool to select molecular probes for specific SCLC recognition. The method can be used for both cell suspensions as previously demonstrated and cell aggregates, the typical morphology of SCLC.
Our expanded screening results showed that the aptamers generated for certain SCLC cell line are also able to recognize other SCLC cell lines of the same type, but seldom bind to other subtypes of lung cancer as well as other types of cancer (e.g., leukemia and liver cancer). This suggests that the developed aptamer probes have the potential to be used practically with clinical samples. In addition, the aptamers developed from live cells can also recognize fixed cells, the main assay format for retrospective analysis of preserved specimens in early detection study, as well as histological examination in clinical diagnosis of lung cancer. Notably, these aptamers exhibited the same specificity for cancer cells from SCLC patients as they did with cultured cells. In the complex biological environment such as human whole blood, this specific binding ability of aptamers was not compromised. These results indicate that developed aptamer probes could be practically used in clinical tests.
In this study we also tested aptamers for possible application in lung cancer early detection, particularly enrichment and detection of exfoliated tumor cells, by using aptamer conjugated magnetic nanoparticles and fluorescent nanoparticles. The high affinity and great specificity of aptamers resulted in effective extraction of SCLC cells by magnetic separation, and the dye-doped nanoparticles gave rise to the sensitive detection after cell extraction. Thus, the aptamer conjugated nanoparticle strategy may substantially improve the efficiency of detecting circulating tumor cells; thereby potentially benefit the early detection of lung cancer.
Early detection and local therapy are widely believed to reduce the overall mortality related with lung cancer, especially SCLC by halting or reversing the progression of premalignant lesions at early stage. Whereas various imaging based methods have been described for early detection, most of these methods delivered very limited effect on the cancer mortality due to their relatively low sensitivity. To improve this situation, molecular approaches were exploited for early detection by detecting specific molecular markers. However, these molecular-marker based techniques also showed unsatisfactory results. For example, among more than 100 monoclonal antibodies for SCLC and NSCLC,[30] none of their antigens are exclusively expressed in SCLC samples. Therefore, the antibodies used for lung cancer early detection do not have the specificity, and may cross react with normal, mildly atypical, moderately atypical exfoliated epithelial cells, and even normal bronchial epithelium. Additionally, the availability of antibodies is limited to those characterized in previous studies.
In this study, developed aptamers showed great specificity for SCLC but not NSCLC. This is because these aptamers were generated based on the molecular differences between the two subtypes of lung cancer by cell-SELEX. Whether the specificity of these aptamers would eventually prevent cross-reactivity and generate fewer false positives in actual early detection tests needs to be studied more carefully. We have shown that aptamers developed in this work are suitable for multiple types of early detection studies. First, retrospective analysis of preserved specimens could be done with these aptamers by using assay formats including flow cytometry and confocal imaging as demonstrated in this work. Second, aptamer conjugated nanoparticles are able to isolate, enrich, and detect exfoliated tumor cells in peripheral blood. These aptamers could also be very useful for lung cancer subtyping during screening [31] and planning appropriate treatment, for example, avoiding excessive therapy in the case of resectable NSCLC.
With cell-SELEX, molecular aptamers can be readily developed for any cancer cells of interest without prior knowledge of cell surface marker proteins, and thus are more flexible and practical to use than other molecule marker based methods. It is noteworthy that a panel of aptamer probes for multiple cell surface differentiation markers can be developed by our strategy. The combination of multiple markers ultimately will be more accurate and predictive than single marker mainly used in previous studies. An additional, notable advantage of this aptamer based approach is that molecular markers are recognized at their native state on living cell surfaces. The molecular aptamers also may have important advantages over other methods for early lung cancer detection in terms of sensitivity, reproducibility, simplicity, robustness, production, and flexibility regarding modification. When coupled with appropriate assay formats, aptamers show great potential to be used in clinical tests.
Although the benefit of using this aptamer approach for lung cancer early detection remains to be determined in prospective trial, it might also be able to detect pre-invasive lesions even before the malignant cells exfoliated when local therapy has limited effect, or indicate the possible relapse in early stage [32] for proper therapy to prevent it if specific cell surface markers can be identified eventually. In addition, this approach would provide valuable information for the understanding of progressive neoplastic differentiation of lung cancer during early stage. However, the use of aptamer for lung cancer screening and whether it can eventually reduce the overall mortality must first be determined in a randomized clinical trial.
The evaluation of additional aptamers selected by cell-SELEX with affinity and specificity will also be continued. Meanwhile, identification of biologically relevant cell surface markers, which can be recognized by aptamers, is under investigation. In conjunction with the further selection of aptamers, biomarker identification, and assay development, the development of new drugs and novel targeted drug delivery methods may be accelerated in the future. The proteomics presented by aptamer approach, together with genomics, molecular imaging, and clinical factors, will in principle achieve the molecular profiling of lung cancer and provide tailored treatments, and therefore realize the personalized medicine.[33]
Experimental Section
Materials
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich and Fisher Scientific.
Buffers
Washing buffer was prepared by dissolving glucose (4.5 g/L), MgCl2 (5 mM), and BSA (1 mg/mL) in Dulbecco’s PBS (pH 7.3). Yeast tRNA (0.1 mg/ml) was added in washing buffer to prepare binding buffer with minimal nonspecific binding.
Cell culture
NCI-H69 (small cell carcinoma), NCI-H661 (large cell carcinoma), NCI-H146 (small cell carcinoma), NCI-H128 (small cell carcinoma), NCI-H23 (adenocarcinoma), NCI-H1385 (squamous cell carcinoma), CCRF-CEM (T cell acute lymphoblastic leukemia), and Ramos (B cell human Burkitt’s lymphoma) cells were purchased from American Type Culture Collection (ATCC), and maintained at 37°C and 5% CO2 in RPMI 1640 medium (ATCC) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (GIBCO) and 100 units/ml penicillin streptomycin (Cellgro). IMEA (liver cancer) and BNL (liver cancer) cells were obtained from the Department of Pathology at the University of Florida.
DNA synthesis and purification
An ABI 3400 DNA Synthesizer (Applied Biosystems) was used for synthesis of single stranded DNA library (71mer containing randomized 35 nucleotides and two primer binding sites, 5′-TACCAGTGCGATGCTCAG (N)35 CTGACGCATTCGGTTGAC-3′), PCR primers, and selected aptamers. The product was further purified by HPLC (ProStar, Varian) using a C18 column (Econosil, 5U, 250×4.6 mm, Alltech Associates) and a linear elution gradient. The HPLC purified product was then dried, detrityled, and re-suspended in buffer for use. UV-Vis measurements were performed with a Cary Bio-300 UV spectrometer (Varian) for DNA quantitation.
Cell-SELEX
Target cell (NCI-H69) and control cell (NCI-H661) were counted and tested for viability before experiments. The ssDNA library (10 nmol in 1 mL binding buffer) was first denatured at 95°C for 5 minutes and kept on ice for 10 minutes. 2×106 target cells were washed, dissociated (0.53 mM EDTA/PBS), and then incubated with ssDNA library at 4°C for 30 minutes. After washing, the cell bound DNAs were eluted to 300 μL binding buffer by heating at 95°C for 5 minutes. The eluted DNAs were further incubated with excess control cells at 4°C for 30 minutes for counter selection (eliminated in first round of selection). After counter selection, the DNAs that don’t bind to control cells were collected, desalted, and PCR amplified with FITC and biotin labeled primers. The PCR product of first round of selection was then processed to generate single stranded DNAs for next round of selection. For the second round of selection, all product of first round was dissolved in 200 μL binding buffer as starting ssDNA pool. To increase the stringency of selection, the washing strength was enhanced by gradually increasing washing time (from 1 to 10 minutes), washing volume (from 1 to 3 mL), and washing round (from 3 to 5 times). The SELEX progress was monitored by flow cytometry.
Real-time PCR
At the end of every round of selection, target cell specific DNA molecules were PCR amplified to form the starting pool for next round of selection. Real-time PCR was first performed to determine the amount of DNA molecules to be amplified, using iTaq DNA polymerase (Bio-Rad) and a MyiQ real-time PCR system (Bio-Rad). SYBR green (Molecular Probes) was used for the detection of PCR products. PCR cycles were then optimized according to the template amount. The bulk of target cell specific DNA molecules was finally PCR amplified with the optimized PCR conditions. Primers for PCR amplification are:
Forward primer 5′-TACCAGTGCGATGCTCAG-3′,
Reverse primer 5′-GTCAACCGAATGCGTCAG-3′.
Unlabeled forward and reverse primers are used for real-time PCR detection with SYBR green. FITC labeled forward primer and triple-biotinylated (trB) reverse primer are used to generate PCR product for flow cytometry assay. TAMRA labeled forward primer and triple-biotinylated (trB) reverse primer are used to generate PCR product for confocal imaging.
PCR parameters consisted of 3 minutes of Taq activation at 95°C, and 15 cycles of PCR at 94°C for 30 s, 52°C for 30 s, 72°C for 15 s, followed by 5 minutes of extension at 72°C. Standard curves were generated for real-time PCR. Specificity of PCR amplification was verified by melt curve analysis. Amplification products were also resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
Single-stranded DNA generation
To generate single stranded DNA from PCR product for next round of selection, the sense ssDNA was separated from the biotinylated anti-sense ssDNA by streptavidin-coated sepharose beads (Amersham Pharmacia Biosciences). After elution with alkaline solution (0.2 M NaOH), the sense ssDNA was desalted with a Sephadex G-25 column (NAP-5, Amersham Pharmacia Biosciences), quantified by UV measurement, and dried in a SpeedVac. The product was then re-suspended in buffer to be used for next round of selection.
Molecular cloning
To isolate individual aptamers from selected pool, cloning was performed after 25 rounds of selection. The most selected ssDNA pool was PCR amplified with unlabeled primers, and inserted into the pCR 2.1-TOPO TA Cloning vector (Invitrogen). The vector was then transformed into Escherichia coli. Cultured monocolonies were picked up to extract the plasmids for sequencing.
Sequencing
Cloned sequences were determined with 454 Life Sciences DNA sequencing unit, GS20, at Interdisciplinary Center for Biotechnology Research (ICBR) of the University of Florida.
Multiple sequence alignment analysis
The sequencing results were subjected to the multiple sequence alignment analysis with the MEME/MAST SYSTEM, version 3.5.3 (developed by Timothy Bailey, Charles Elkan, and Bill Noble at the UCSD Computer Science and Engineering department with input from Michael Gribskov at Purdue University, http://meme.nbcr.net) to discover highly conserved motifs in groups of selected DNA sequences. The discovered consensus sequences with high repeats among selected pool were then synthesized and tested for specificity and affinity.
Flow cytometry
To monitor the enrichment of aptamers along with the progress of SELEX, FITC labeled ssDNA pools were incubated with 1×106 NCI-H69 or NCI-H661 cells in 400 μL binding buffer at 4°C for 30 minutes. Cells were washed twice after incubation and analyzed by flow cytometry. The binding of selected aptamers to SCLC cells, NSCLC cells, leukemia cells, and liver cancer cells were similarly analyzed. Flow cytometry was performed on a FACScan cytometer with CellQuest software (Becton Dickinson).
Confocal imaging
The binding of selected ssDNA pools and individual aptamers to SCLC cells was evaluated by fluorescence confocal imaging. Cells were incubated with 250 nM TAMRA labeled aptamers in 100 μL binding buffer at 4°C for 30 minutes. After washing, 20 μL cell suspension was dropped on a covered glass slide for examination with confocal microscope. Fluorescence confocal imaging was performed on a Fluoview 500/IX81 inverted confocal scanning microscope system (Olympus). A 5-mW, 543-nm He-Ne laser was used as excitation source for TAMRA dye. The objective used for imaging was a 60× oil-immersion objective (PLAPO60XO3PH) with a numerical aperture of 1.40 (Olympus). A 20× objective with a numerical aperture of 0.7 (Olympus) was also used for imaging of large field. Staining of cell line tissue array by fluorescent aptamers and extraction of SCLC cells by aptamer conjugated nanoparticles were evaluated by confocal imaging as described above.
Saturation analysis
Saturation analysis was performed to measure the relative cell surface binding affinities of developed aptamers. Cells were incubated with FITC labeled aptamers at 4°C for 30 minutes, washed three times with 400 μL washing buffer, and finally re-suspended in 400 μL binding buffer containing 20% FBS. Cells were then assayed using flow cytometry. Concentrations of FITC labeled aptamers for the relative affinity measurements varied from 0 to 1 μM. The FITC labeled ssDNA library was used to determine nonspecific binding. The mean fluorescence intensity of aptamer bound cells (nonspecific binding of DNA library subtracted) was used to calculate bound aptamer fraction at different concentrations. All affinity measurements were performed in triplicate. The results are described as mean ± s.e.m. The equilibrium dissociation constants (Kd) were obtained by fitting the cell surface binding data of aptamers to a one-site saturation model with SigmaPlot 9.0 (Jandel Scientific).
Enzymatic treatment
To verify the binding of aptamers to SCLC cell surface markers, cells were examined by enzymatic treatment. 1×106 Cells were washed with 1 ml of PBS, and treated with 200 μL of 0.05% trypsin/0.53 mM EDTA in HBSS (Fisher Biotech) or 0.1 mg/mL proteinase K (Fisher Biotech) in PBS at 37°C for 2 minutes. FBS was then added to quench the enzyme activity. After washing with binding buffer, the cells were analyzed for aptamer binding with flow cytometry and confocal imaging as described above.
Cell line tissue array
Cultured SCLC and NSCLC cell lines were processed into homogeneous tissue arrays to evaluate the binding of aptamers to fixed cells. All cell line tissue arrays were prepared in the University of Florida Diagnostic Reference Laboratories. 10×106 cells grown in culture were first prepared as a cell suspension in minimal amount of medium (adherent cells were detached by trypsin-EDTA treatment). Cells were then fixed with 4% formaldehyde, and mixed with 1% agarose in isoosmotic PBS. The solidified cell blocks were cut into serial sections and processed on paraffin-embedded slides. Prepared cell line tissue arrays were stained with hematoxylin and eosin (H&E) for quality control.
Cell line tissue array staining
Cell line tissue arrays were first treated with xylene and ethanol (100%, 95%, and 70%) for deparaffinization. For antigen retrieval, the dried tissue arrays were rinsed with PBS and kept in 1 mM EDTA Tris buffer (pH 8.0) at 95°C for 15 minutes. Tissue arrays were then incubated with 200 μL of 0.25 μM TAMRA labeled aptamers in binding buffer at 4°C for 30 minutes. After washing and dehydration, the stained array slides were mounted for evaluation. Aptamer staining of cell line tissue arrays were analyzed by array scanning and by confocal imaging. For the array scanning, the stained array slides were scanned into a computer with a microarray scanner (2100 BioAnalyzer, Agilent) at 10 μm scan resolution, and analyzed using Agilent G2567AA Feature Extraction software (v.9.1). To confirm the array scanning results and show the binding details, the same stained array slides were imaged using an FV500-IX81 confocal microscope (Olympus) with a 543-nm excitation source. Images were collected with both 60× and 20× objectives as described above.
Clinical sample test
SCLC patient samples were obtained from the Department of Pathology at the University of Florida. Cells were washed and counted for incubation with aptamers. Cell surface binding of FITC labeled aptamers was analyzed by flow cytometry as detailed above.
Binding assay in human whole blood
To evaluate the binding capacity of aptamers in complex biological environment, 2×106 SCLC cells were prepared as detailed above and mixed with 3.5 μL human whole blood (IPLA-WB1, Innovative Research, Inc.) in 300 μL of buffer. Human whole blood was prepared by mixing with the anticoagulant, sodium heparin. 100 μL of 1 μM FITC labeled aptamers was added to SCLC cells previously spiked in human whole blood. After incubation at 4°C and thorough washing, we assessed the binding of aptamers to SCLC cells in blood with flow cytometry. For controls, human whole blood and cells in buffer were incubated with aptamers and analyzed by flow cytometry. Background binding of aptamers to blood cells was negligible.
Aptamer conjugated magnetic and fluorescent nanoparticles
For the synthesis of aptamer conjugated magnetic nanoparticles, the 65-nm iron oxide-doped magnetic nanoparticles were first prepared by precipitating iron oxide as previously described.[29] The magnetite core particles were then coated with silica by the hydrolysis of tetraethoxyorthosilicate, and treated with TEOS. After washing, avidin coating was performed by incubating 0.1 mg/mL silica-coated magnetic nanoparticle solution with 5 mg/mL avidin solution at 4°C for 12 hours. The avidin coated magnetic nanoparticles were then washed with PBS, and stabilized by crosslinking with 1% glutaraldehyde at 25°C for 1 hour. After washing with Tris-HCl buffer, the 0.2 mg/mL avidin coated magnetic nanoparticles were incubated with excess biotinylated DNA aptamers and ssDNA library at 4°C for 12 hours. The prepared aptamer conjugated magnetic nanoparticles were washed and stored at a final concentration of 0.2 mg/mL at 4°C for use.
For the synthesis of aptamer conjugated fluorescent nanoparticles, TAMRA dye-doped nanoparticles were first prepared by the reverse microemulsion method as previously described.[29] After silica polymerization and stabilization treatment with TEOS, the dye-doped nanoparticles were coated with avidin as detailed above. Avidin coated dye-doped nanoparticles were further conjugated with excess biotinylated DNA aptamers and ssDNA library. The prepared aptamer conjugated fluorescent nanoparticles were washed and stored at a final concentration of 10 mg/mL at room temperature for use.
Extraction and detection of SCLC cells
For every experiment, 1.0×105 cells were prepared as detailed above and dispersed in 200 μL of cell media buffer. The specified amount of aptamer conjugated magnetic and fluorescent nanoparticles was then simultaneously added to the cell suspension. After 30 minute incubation and washing, cells were isolated from cell media buffer by magnetic extraction, and recovered in 20 μL of buffer for confocal imaging and fluorescence measurement. A 2-μL aliquot of the extracted sample was assessed by confocal imaging as described above. The rest samples were then added to 96-well plate, and the fluorescence of dye-doped nanoparticles bound to extracted cells was measured by a plate reader (Packard). ssDNA library conjugated magnetic and fluorescent nanoparticles were used for control experiments.
Supplementary Material
Acknowledgments
The authors thank Dr. Ying Li and Dr. Zehui Charles Cao for stimulating discussions, Ms. Kim Ahrens for help with cell culture and flow cytometry, Ms. Regina Shaw and Dr. William Farmerie for help with DNA sequencing, and the support of NIH, NCI, and Florida Department of Health grants.
Glossary
- DNA
deoxyribonucleic acid
- NSCLC
non-small cell lung cancer
- PCR
polymerase chain reaction
- SCLC
small cell lung cancer
- SELEX
systematic evolution of ligands by exponential enrichment
Footnotes
Supporting information for this article is available on the WWW under http://www.chemmedchem.org or from the author.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu JQ, Thun MJ. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman PC, Mauer AM, Vokes EE. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 3.Mulshine JL, Sullivan DC. N Engl J Med. 2005;352:2714–2720. doi: 10.1056/NEJMcp042630. [DOI] [PubMed] [Google Scholar]
- 4.Jackman DM, Johnson BE. Lancet. 2005;366:1385–1396. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 5.Rom WN, Hay JG, Lee TC, Jiang YX, Tchou-Wong KM. Am J Respir Crit Care Med. 2000;161:1355–1367. doi: 10.1164/ajrccm.161.4.9908012. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch FR, Franklin WA, Gazdar AF, Bunn PA. Clin Cancer Res. 2001;7:5–22. [PubMed] [Google Scholar]
- 7.McWilliams A, MacAulay C, Gazdar AF, Lam S. Oncogene. 2002;21:6949–6959. doi: 10.1038/sj.onc.1205831. [DOI] [PubMed] [Google Scholar]
- 8.Mulshine JL. Nat Rev Cancer. 2003;3:65–73. doi: 10.1038/nrc972. [DOI] [PubMed] [Google Scholar]
- 9.Patz EF, Goodman PC, Bepler G. N Engl J Med. 2000;343:1627–1633. doi: 10.1056/NEJM200011303432208. [DOI] [PubMed] [Google Scholar]
- 10.Bernal SD, Elias AD. J Clin Oncol. 1988;6:1676–1678. doi: 10.1200/JCO.1988.6.11.1676. [DOI] [PubMed] [Google Scholar]
- 11.Wistuba II, Berry J, Behrens C, Maitra A, Shivapurkar N, Milchgrub S, Mackay B, Minna JD, Gazdar AF. Clin Cancer Res. 2000;6:2604–2610. [PMC free article] [PubMed] [Google Scholar]
- 12.Mao L. Oncogene. 2002;21:6960–6969. doi: 10.1038/sj.onc.1205564. [DOI] [PubMed] [Google Scholar]
- 13.Mao L, Hruban RH, Boyle JO, Tockman M, Sidransky D. Cancer Res. 1994;54:1634–1637. [PubMed] [Google Scholar]
- 14.Levin NA, Brzoska PM, Warnock ML, Gray JW, Christman MF. Genes Chromosomes Cancer. 1995;13:175–185. doi: 10.1002/gcc.2870130307. [DOI] [PubMed] [Google Scholar]
- 15.Tockman MS, Gupta PK, Myers JD, Frost JK, Baylin SB, Gold EB, Chase AM, Wilkinson PH, Mulshine JL. J Clin Oncol. 1988;6:1685–1693. doi: 10.1200/JCO.1988.6.11.1685. [DOI] [PubMed] [Google Scholar]
- 16.Yanagisawa K, Shyr Y, Xu BGJ, Massion PP, Larsen PH, White BC, Roberts JR, Edgerton M, Gonzalez A, Nadaf S, Moore JH, Caprioli RM, Carbone DP. Lancet. 2003;362:433–439. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 17.Chanin TD, Merrick DT, Franklin WA, Hirsch FR. Curr Opin Pulm Med. 2004;10:242–247. doi: 10.1097/01.mcp.0000130321.11513.13. [DOI] [PubMed] [Google Scholar]
- 18.Blank M, Weinschenk T, Priemer M, Schluesener H. J Biol Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 19.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. Proc Natl Acad Sci U S A. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shangguan D, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, Sefah K, Yang CYJ, Tan WH. Proc Natl Acad Sci U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carney DN. Lancet. 1992;339:843–846. doi: 10.1016/0140-6736(92)90286-c. [DOI] [PubMed] [Google Scholar]
- 22.Ettinger DS, Aisner J. J Clin Oncol. 2006;24:4526–4527. doi: 10.1200/JCO.2006.07.3841. [DOI] [PubMed] [Google Scholar]
- 23.Osborne SE, Ellington AD. Chem Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 24.Wilson DS, Szostak JW. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 25.Famulok M, Mayer G, Blind M. Acc Chem Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 26.Shangguan D, Tang ZW, Mallikaratchy P, Xiao ZY, Tan WH. ChemBio Chem. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- 27.Kraemer PS, Sanchez CA, Goodman GE, Jett J, Rabinovitch PS, Reid BJ. Cytometry A. 2004;60A:1–7. doi: 10.1002/cyto.a.20041. [DOI] [PubMed] [Google Scholar]
- 28.Iinuma H, Okinaga K, Adachi M, Suda K, Sekine T, Sakagawa K, Baba Y, Tamura J, Kumagai H, Ida A. Int J Cancer. 2000;89:337–344. doi: 10.1002/1097-0215(20000720)89:4<337::aid-ijc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 29.Herr JK, Smith JE, Medley CD, Shangguan DH, Tan WH. Anal Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 30.Souhami RL, Beverley PCL, Bobrow LG. Lancet. 1987;2:325–326. doi: 10.1016/s0140-6736(87)90904-4. [DOI] [PubMed] [Google Scholar]
- 31.Doyle LA, Giangiulo D, Hussain A, Park HJ, Yen RWC, Borges M. Cancer Res. 1989;49:6745–6751. [PubMed] [Google Scholar]
- 32.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, Chan WK, Li HN, Liu CC, Singh S, Chen WJ, Chen JJW, Yang PC. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 33.Herbst RS, Lippman SM. N Engl J Med. 2007;356:76–78. doi: 10.1056/NEJMe068218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


