Abstract
Hd6 is a quantitative trait locus involved in rice photoperiod sensitivity. It was detected in backcross progeny derived from a cross between the japonica variety Nipponbare and the indica variety Kasalath. To isolate a gene at Hd6, we used a large segregating population for the high-resolution and fine-scale mapping of Hd6 and constructed genomic clone contigs around the Hd6 region. Linkage analysis with P1-derived artificial chromosome clone-derived DNA markers delimited Hd6 to a 26.4-kb genomic region. We identified a gene encoding the α subunit of protein kinase CK2 (CK2α) in this region. The Nipponbare allele of CK2α contains a premature stop codon, and the resulting truncated product is undoubtedly nonfunctional. Genetic complementation analysis revealed that the Kasalath allele of CK2α increases days-to-heading. Map-based cloning with advanced backcross progeny enabled us to identify a gene underlying a quantitative trait locus even though it exhibited a relatively small effect on the phenotype.
Many important traits in plant breeding are controlled by a series of genes and show continuous variation in progeny. Many efforts have been made to map such genes, known as quantitative trait loci (QTLs), because of their biological and agronomic importance. The development of a high-density linkage map based on DNA markers made it possible to map QTLs with high resolution (1). However, it has been difficult to identify genes underlying QTLs, because their individual effects on the phenotype are relatively small and are affected by environmental conditions.
Heading date is an important trait for the adaptation of crops to different cultivation areas. In rice, heading date is determined mainly by two factors: duration of the basic vegetative growth and photoperiod sensitivity (PS). Several research groups have identified many QTLs affecting heading date in rice (2–6). Five QTLs conferring heading date (Hd1 to Hd5) have been found in an F2 population derived from a cross between a japonica variety, Nipponbare, and an indica variety, Kasalath (7). Among them, two major QTLs, Hd1 and Hd2, exist in the middle of chromosome 6 and at the end of chromosome 7, respectively (7). Kasalath alleles on both loci greatly reduced days-to-heading. Hd3, Hd4, and Hd5 were detected on chromosome 6, 7 and 8, respectively, and Nipponbare alleles reduced days-to-heading (7). An additional QTL, Hd6, was detected on chromosome 3 in an analysis of advanced backcross progeny derived from the same cross combination, although it was not detected in the F2 population (8). Our colleagues also detected other minor QTLs by using several advanced backcross progeny or backcross inbred lines, and we have so far detected 13 chromosomal regions affecting rice heading date, including the QTLs mentioned above (ref. 9 and M.Y., unpublished data).
A series of chromosomal substitution lines or near-isogenic lines (NILs) produced by repetitive backcross and marker-assisted selection have been used for the analysis of QTLs. Using these lines allows the detailed function of each QTL to be clarified, excluding phenotypic variance because of other QTLs. Day-length treatment tests with an NIL of Hd6 [NIL(Hd6)] showed that Hd6 was involved in PS, and that the Kasalath allele increased days-to-heading under natural and long-day conditions but not under short-day conditions (8).
We can also use NILs for accurate linkage analysis, because the segregating populations obtained by recurrent crossing of NILs with their parents simplify genetic variations. In a previous study, linkage analysis of an advanced backcross progeny mapped Hd6 on the long arm of chromosome 3 as a single Mendelian factor (8). However, the resolution of the genetic map was too poor to allow map-based cloning of Hd6. In this study, to identify the gene corresponding to Hd6, we carried out a high-resolution fine-scale mapping of Hd6. Sequencing and complementation analysis led us to identify the Hd6 gene.
Materials and Methods
Plant Materials.
We crossed a japonica variety, Nipponbare, and an indica variety, Kasalath, and backcrossed a resultant F1 plant with Nipponbare. Repetitive backcrossing and marker-assisted selection produced several plants in which the region around Hd6 was heterozygous and almost all other regions were homozygous for Nipponbare. As an example, the graphical genotype of BC3F3-9-28 is shown in Fig. 1A. We used 2,807 self-pollinated progeny of these plants for the high-resolution mapping of Hd6. The plants were cultivated in a paddy field at the National Institute of Agrobiological Sciences, Tsukuba, Ibaraki, Japan in the natural growing season (April to August). Days-to-heading was scored as the duration from sowing to the appearance of the first panicle.
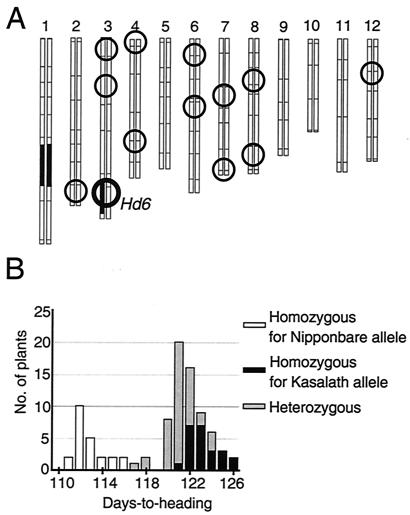
Figure 1.
Plant materials used in this study. (A) Graphical genotype of BC3F3-9-28. Black-and-white regions represent segments of the 12 chromosomes derived from Kasalath and Nipponbare, respectively. The circles indicate statistical positions of each QTL identified in QTL analyses of heading date (refs. 7–9; M.Y., unpublished data). (B) Frequency distribution of days-to-heading in selfed progeny derived from BC3F3-9-28. We used CAPS markers derived from R2404 and C1329 for genotyping and discovered plants in which recombination occurred between them. Ninety nonrecombinants were used in this analysis.
Progeny Test.
By progeny test, we determined the Hd6 genotypes of F2 plants in which recombination occurred around the Hd6 locus. Twenty or more selfed F3 progeny of each recombinant were grown in a paddy field or in a growth cabinet under long-day conditions, and the appearance of their first panicle was monitored. If the heading dates of the F3 population were the same as those of Nipponbare (early-fixed) or of NIL(Hd6) (late-fixed), the parent F2 plant was homozygous for the Nipponbare allele or the Kasalath allele at Hd6, respectively. If the F3 population showed a segregation of heading dates, the Hd6 genotype of the parent F2 plant was heterozygous.
Restriction Fragment-Length Polymorphism (RFLP) Analysis.
Total DNA used in the RFLP analysis was extracted from green leaves by the cetyltrimethylammonium bromide method (10). All experimental procedures for Southern blotting, hybridization, and linkage map construction have been described by Kurata et al. (10).
Cleaved Amplified Polymorphic Sequence (CAPS) Analysis.
DNA used in CAPS analysis was extracted as follows. A piece of leaf 2–3 cm long was placed into a microtube and broken mechanically in appropriate buffer. After incubation at 65°C, DNA was precipitated with isopropanol from the supernatant. The precipitated DNA was resuspended in 50 μl of 1/10 TE buffer (1 mM Tris⋅HCl, pH 8.0/0.1 mM EDTA), and 1 μl of solution was used for PCR amplification.
We used CAPS markers to select plants in which recombination had occurred near the Hd6 locus. We designed PCR primers on the basis of sequences of each cDNA clone assigned to the region near the Hd6 locus as RFLP marker (8). Digestion of the PCR products from Nipponbare and Kasalath genomic DNA with several restriction enzymes allowed us to identify sequence polymorphisms. We used CAPS markers derived from R2404 and G1015 as markers flanking to Hd6.
We also used CAPS markers to delimit the region in which recombination had occurred in each recombinant and to detect the introduced DNA fragment in transgenic plants. We made these CAPS markers on the basis of P1-derived artificial chromosome (PAC) sequences as described below.
Isolation of Yeast Artificial Chromosome (YAC) and PAC End-DNA Fragments.
YAC and PAC end-DNA fragments were cloned on the basis of the cassette-ligation-mediated PCR method (11). For isolation of PAC end-fragment clones, we used oligonucleotide primers AKI-2 (5′-GGCCGTCGACATTTAGGTGAC-3′, for the first PCR of the SP6 promoter side), AKI-4 (5′-CCGTCGACATTTAGGTGACACT-3′, for the second PCR of the SP6 promoter side), MICHI-1 (5′-AAGGAGCTGACTGGGTTGA-3′, for the first PCR of the T7 promoter side), and MICHI-4 (5′-CGGTCGAGCTTGACATTGTAGG-3′, for the second PCR of the T7 promoter side), as specific primers for the PAC vector.
Screening of PAC Library and CAPS Marker Development.
Using sequence-tagged site primers around the Hd6 region, we screened 8,352 PAC clones made from Nipponbare genomic DNA (12) and identified target clones.
We determined the sequence of PCR products derived from Kasalath genomic DNA by using primers designed from the PAC sequence. We made CAPS markers according to differences in sequences between the PAC clone (Nipponbare) and the PCR products (Kasalath).
Rice Transformation.
We constructed about 80,000 cosmid clones made from Kasalath genomic DNA, screened them by hybridization of several DNA fragments around Hd6, and identified target clones. We digested a cosmid clone carrying the CK2α gene at Hd6 locus, completely with NheI and partially with BglII. After ligation of the digested DNA fragments to the XbaI and BamHI cloning sites of the binary vector pPZP2H-lac (13), PCR screening identified pPZPCKII-10A as containing the whole CK2α gene.
Transformation with the Agrobacterium system (14) was used for complementation testing. pPZPCKII-10A was introduced into Nipponbare callus in darkness at 25°C for 3 days. The callus was regenerated in a growth chamber, and the regenerated plants (T0) and their self-pollinated progeny (T1) were grown in an isolated greenhouse at the National Institute of Agrobiological Sciences. Days-to-heading of the T1 plants was scored under natural day-length condition (cultivated from May to September).
Results
Characterization of Hd6.
In a previous study, Hd6 was assigned to cosegregate with several RFLP markers (8). For more accurate genetic analysis, we selected an advanced backcross progeny (BC3F3-9-28) in which the region around Hd6 was heterozygous and almost all other regions were homozygous for Nipponbare (Fig. 1A). Using this line, we could delete the effects of genomic regions other than the Hd6 locus, which we could handle as a single Mendelian factor. In a population derived by self-pollination of the advanced progeny, we found clear segregation of early- and late-heading phenotypes (Fig. 1B). CAPS analysis revealed that the phenotypes corresponded clearly to the Hd6 genotype. The heterozygous plants headed slightly earlier than the plants homozygous for the Kasalath allele and clearly later than the plants homozygous for Nipponbare, which indicates that the Kasalath allele of Hd6 is semidominant against the Nipponbare allele.
High-Resolution Mapping of Hd6.
To determine the precise location of Hd6, we made a fine-scale high-resolution linkage map with 2,807 segregating plants derived from BC3F3-9-28 or from others with almost the same genotype (Fig. 2A). Hd6 was located on the map by progeny testing.
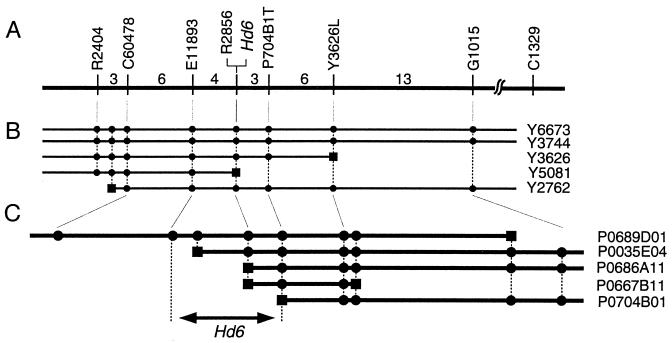
Figure 2.
Fine-scale linkage and physical maps around the Hd6 locus. (A) A linkage map made with 2,807 segregating plants. The vertical bars represent the positions of RFLP markers (8, 10). Numerals indicate the numbers of plants in which recombination occurred between each pair of adjacent markers. YAC (B) and PAC (C) contigs are also depicted in combination with the linkage map. After physical map construction, new RFLP markers from end-fragments of the YACs or PACs (markers preceded with Y or P, respectively) or from ESTs mapped on the YACs (C60478 and E11893) were used for further linkage analysis. Squares on the clones indicate the end-fragments of the clones. Circles indicate that RFLP markers or the end-fragment is contained in each clone. Sequencing analysis revealed that all of the end-fragments derived from one end of clones Y5081, P0686A11, and P0667B11 lay within the Hd6 gene.
We also constructed a YAC contig around Hd6 on the basis of information from a physical map (15) and from expressed sequence tag (EST) mapping by the Rice Genome Research Program (Fig. 2B). New RFLP markers from YAC end-fragments or from ESTs mapped on the YACs were used for further linkage analysis. We next screened a rice PAC library and found that a PAC clone P0689D01 contained the whole Hd6 candidate region (Fig. 2C). PAC end-fragments were also used for linkage analysis, and Hd6 was narrowed down to the region between RFLP markers E11893 and P704B1T.
To determine where recombinations nearest to Hd6 occurred, we made CAPS markers on the basis of the PAC clone sequence. Linkage analysis with these markers indicated that the Hd6 locus lay between 13B/BglII and 13G/NheI (Fig. 3). This region is about 26.4 kb long in Nipponbare.
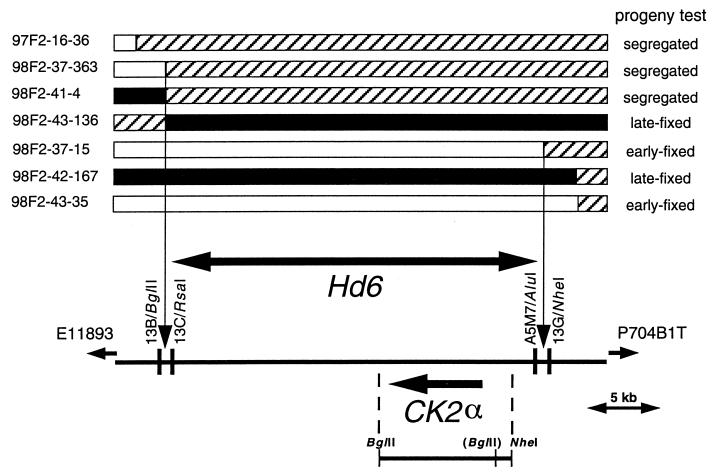
Figure 3.
Delimitation of Hd6. The upper rows show the genotype of seven plants in which recombination occurred between E11893 and P704B1T. Black, white, and hatched regions represent chromosomal segments that are homozygous for the Kasalath allele, homozygous for the Nipponbare allele, and heterozygous, respectively. The lower row depicts a part of P0689D01. CAPS markers derived from the PAC sequence revealed that three recombinations occurred between 13B/BglII and 13C/RsaI and one between A5 M7/AluI and 13G/NheI. Progeny testing of selfed progeny of each recombinant narrowed down the Hd6 locus into the region between 13B/BglII and 13G/NheI. An arrow indicates a CK2α gene found in this region [from transcription initiation site to poly(A) tail addition site]. The bar below the CK2α gene represents the DNA fragments used in complementation testing. The BglII site in parentheses is present only in the Kasalath genome.
Identification of Hd6 Gene.
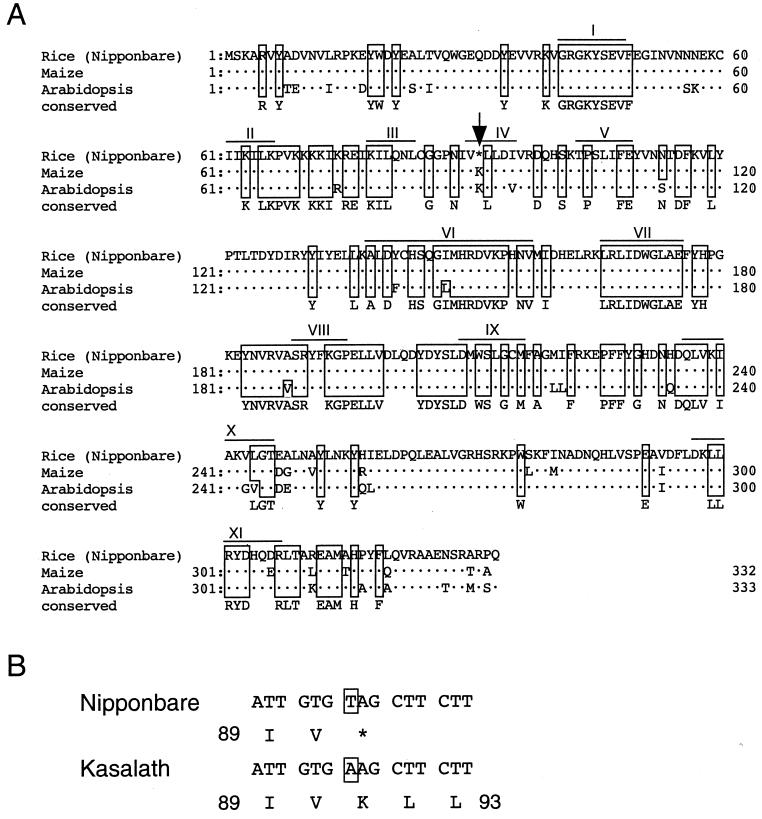
A blast homology search with this region against the Rice Genome Research Program ESTs (16) found only one gene encoding a cDNA clone, C10214 (data not shown). We determined the sequence of C10214 and found that its deduced amino acid sequence has extremely high homology (>90%) with the sequences of the α subunit of protein kinase CK2 (CK2α) in maize and Arabidopsis (Fig. 4A). We also found that C10214 has a premature stop codon (TAG), which truncated a predicted product. We determined the genomic sequence and the cDNA sequence of the Kasalath allele of CK2α. Within the coding region, only one nucleotide substitution occurred between the two alleles, which changed the premature stop codon (TAG) in Nipponbare to a lysine codon (AAG) in Kasalath (Fig. 4B).
Figure 4.
The deduced amino acid sequence of CK2α. (A) Sequence alignment of CK2α. Dots represent amino acids identical to the rice sequence. We identified amino acids conserved among 13 CK2α genes from 10 organisms other than plants; these are listed on the bottom row (marked “conserved”). Amino acids identical to the conserved one are boxed. Roman numerals indicate the 11 conserved subdomains of protein kinases (16). The arrow indicates the position of the premature stop codon in the Nipponbare allele. The following accession numbers reference the sequences used for the alignments: maize, Y11526; Arabidopsis, D10246. (B) The nucleotide sequence and the deduced amino acid sequence around the Nipponbare premature stop codon. The stop codon is changed to a lysine codon in the Kasalath allele.
Complementation Test.
We introduced an 8.9-kb Kasalath genomic fragment carrying the CK2α gene (Fig. 3) into Nipponbare. In the T0 generation, because the plants were regenerated from callus, we could not synchronize the plants' growth enough to count the variance of their heading date. Using Southern analysis, we selected T0 plants that have as few copies of the transgene as possible and found two plants with only a few copies for progeny test. We monitored days-to-heading of self-pollinated progenies (T1) of each of selected plants under natural-day condition (Table 1). The T1 plants showed clear segregation of early- and late-heading phenotypes. Days-to-heading of plants headed early and plants headed late were almost the same as those of Nipponbare and NIL(Hd6), respectively. CAPS analysis revealed that plants with the transgene headed late, whereas those without headed early.
Table 1.
Frequency distribution of days-to-heading of selfed progeny of transgenic plants
| Days-to-heading
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 93 | 95 | 97 | 99 | 101 | 103 | 105 | 107 | 109 | 111 | 113 | 115 | 117 | |
| Nipponbare | 3 | 6 | 1 | ||||||||||
| NIL(Hd6) | 1 | 3 | 4 | ||||||||||
| 2Hlac-12 | 3 | 6 | 2 | ||||||||||
| CK2A-210 | 1* | 1* | 8 | 3 | |||||||||
| CK2A-55 | 1* | 1 | 2 | 1 | 1 | 1 | |||||||
The number of plants heading at first panicle was monitored every 2 days. 2Hlac-12, a line derived from Nipponbare transformed with a vector. CK2A-210 and CK2A-55, lines derived from Nipponbare transformed with the CK2α gene.
Plants that did not inherit the introduced DNA fragment carrying the CK2α gene.
In this test, we could not observe the semidominancy of the transgene. It might be observed, if we would test many more plants. Alternatively, growth conditions in the greenhouse prevented us from observing the semidominancy.
Discussion
We concluded that Hd6 encodes the α subunit of protein kinase CK2. First, high-resolution linkage analysis with 2,807 segregating plants narrowed the Hd6 locus to a 26.4-kb genomic region. In this region, we found only one EST (C10214), which showed high homology with the CK2α gene of Arabidopsis and maize. Second, the CK2α allele of Nipponbare has a premature stop codon that is not found in the same position in the Kasalath allele (Fig. 4B). The Kasalath mRNA has an ORF (333 aa) whose deduced amino acid sequence is highly homologous with those of the maize and Arabidopsis CK2α genes. In addition, Kasalath CK2α contains all of the amino acids conserved among various organisms from yeast to mammals. These facts strongly suggest that the Kasalath allele encodes a functional CK2α. The Nipponbare mRNA encodes only a truncated protein (90 aa) that lacks subdomains IV–XI (Fig. 4A). These missing subdomains are found in all protein kinases; some are implicated in ATP binding, catalytic reaction, amino acid specificity, or substrate specificity (17). Therefore, the truncated protein encoded by the Nipponbare allele must be nonfunctional. These facts coincide well with the dominance of the Kasalath allele against the Nipponbare allele.
Third, some studies have revealed the involvement of CK2α in the plant phototransduction pathway. CK2 is a messenger-independent serine/threonine kinase that is present in all eukaryotic cells examined to date. It has a heterotetrameric structure, α2β2, consisting of two catalytic (α) and two regulatory (β) subunits. Some experiments have shown that CK2 can phosphorylate several transcription factors that bind to the promoter region of light-regulated genes (18–20). The expression of CK2α antisense RNA in Arabidopsis affected the expression of some light-regulated genes, although it had little effect on the degree of inhibition of hypocotyl elongation by red light (21). Recently, it was suggested that CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein (CCA1) in vitro (22). CCA1 is a MYB-related transcription factor and is involved in the control of flowering time (23). Overexpression of the β-subunit of CK2 (CK2β) shortened periods of rhythmic expression of CCA1, of late elongated hypocotyl (LHY), and of four other circadian clock-controlled genes, and caused early flowering in both long- and short-day conditions (24). Finally, our complementation testing showed that the introduced Kasalath allele of CK2α delayed the heading date of Nipponbare.
The experiments in Arabidopsis described above are insufficient to elucidate the role(s) of CK2 in photoperiodic flowering response, because CK2α antisense expression failed to abolish all CK2 activity (21), and because CK2β overexpression might disturb its authentic expression control. We can regard Nipponbare as a naturally occurring CK2α null mutant against NIL(Hd6). Therefore, these lines are useful in elucidating the function of CK2α. Our results indicated that CK2α plays a role in controlling plant PS, at least in rice.
Many studies indicated that CK2 is involved in a wide range of cellular functions. However, we could not see any phenotypic difference other than heading date between Nipponbare and NIL(Hd6). Although an RFLP marker R2856 cosegregated with Hd6 even in our high-resolution map, PCR analysis against YACs and PACs revealed that the gene encoding the cDNA clone R2856 (OsCKA1) did not exist around the Hd6 region (data not shown). Because the sequence of R2856 shows high homology with the CK2α gene corresponding to Hd6 (OsCKA2), R2856 was mapped as an RFLP marker cosegregating with Hd6, indicating that OsCKA1 exists in a chromosomal region other than the Hd6 locus. We think that OsCKA2 may function specifically in PS and that OsCKA1 does so universally. Alternatively, component(s) involved in PS are more sensitive to increased activity of CK2 caused by expression of OsCKA2, if it is increased. Sugano et al. similarly discussed the function of Arabidopsis CKB3 in the regulation of circadian rhythms (24).
Many kinds of QTLs have been detected in various organisms, and multiple efforts have been made to map a QTL precisely. However, to our knowledge, a major fruit-weight QTL of tomato (fw2.2) was the only case in which a QTL was isolated on a cloned DNA segment (<150 kb) (25). Quite recently, some groups have succeeded in identifying genes corresponding to QTLs. Frary et al. identified a gene for fw2.2 (26). Fridman et al. delimited a QTL for tomato sugar content (Brix9-2-5) to a 484-bp region within an invertase gene (27). M. Yano et al. also identified a gene for Hd1, a QTL with a large effect on rice PS (28). We mapped a minor QTL, Hd6, and have now succeeded in identifying the gene for it. This study shows that map-based cloning with advanced backcross progeny is an effective way to clone a gene for a QTL.
Research on PS has been greatly promoted by mutant analysis in Arabidopsis. The isolation of genes for QTLs may identify novel factors that will not be identified by mutant analysis. As rice is a short-day plant, unlike Arabidopsis, research on rice may elucidate the difference in genetic control of PS between short-day and long-day plants.
Acknowledgments
We thank T. Baba and K. Yamamoto for sequence analysis of the PAC clone P0689D01. This work was supported by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
Abbreviations
- QTL
quantitative trait locus
- PS
photoperiod sensitivity
- PAC
P1-derived artificial chromosome
- NIL
near-isogenic line
- RFLP
restriction fragment-length polymorphism
- CAPS
cleaved amplified polymorphic sequence
- YAC
yeast artificial chromosome
- EST
expressed sequence tag
Footnotes
References
- 1.Paterson A H, Lander E S, Hewitt J D, Peterson S, Lincoln S E, Tanksley S D. Nature (London) 1988;335:721–726. doi: 10.1038/335721a0. [DOI] [PubMed] [Google Scholar]
- 2.Xiao J, Li J, Yuan L, Tanksley S D. Genetics. 1995;140:745–754. doi: 10.1093/genetics/140.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, Pinson S R M, Stansel J W, Park W D. Theor Appl Genet. 1995;91:374–381. doi: 10.1007/BF00220902. [DOI] [PubMed] [Google Scholar]
- 4.Xiao J, Li J, Yuan L, Tanksley S D. Theor Appl Genet. 1996;92:230–244. doi: 10.1007/BF00223380. [DOI] [PubMed] [Google Scholar]
- 5.Lin H X, Qian H R, Zhuang J Y, Lu J, Xiong Z M, Min S K, Huang N, Zheng K L. Rice Genet Newsl. 1996;12:253–255. doi: 10.1007/BF00224031. [DOI] [PubMed] [Google Scholar]
- 6.Lu C, Shen L, Tan Z, Xu Y, He P, Chen Y, Zhu L. Theor Appl Genet. 1997;94:145–150. doi: 10.1007/s001220050393. [DOI] [PubMed] [Google Scholar]
- 7.Yano M, Harushima Y, Nagamura Y, Kurata N, Minobe Y, Sasaki T. Theor Appl Genet. 1997;95:1025–1032. doi: 10.1007/BF00223368. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto T, Lin H X, Sasaki T, Yano M. Genetics. 2000;154:885–891. doi: 10.1093/genetics/154.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S Y, Sasaki T, Yano M. Theor Appl Genet. 1998;96:997–1003. [Google Scholar]
- 10.Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio B A, Shomura A, Shimizu T, Lin S Y, et al. Nat Genet. 1994;8:365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- 11.Isegawa Y, Sheng J, Sokawa Y, Yamanishi K, Nakagomi O, Ueda S. Mol Cell Probes. 1992;6:467–475. doi: 10.1016/0890-8508(92)90043-w. [DOI] [PubMed] [Google Scholar]
- 12.Baba T, Katagiri S, Tanoue H, Tanaka R, Chiden Y, Saji S, Hamada M, Nakashima M, Okamoto M, Hayashi M, et al. Bull Natl Inst Agrobiol Resour. 2000;14:24–36. [Google Scholar]
- 13.Fuse, T., Sasaki, T. & Yano, M. (2001) Plant Biotechnol., in press.
- 14.Toki S. Plant Mol Biol Rep. 1997;15:16–21. [Google Scholar]
- 15.Tanoue H, Shimokawa T, Wu J, Sue N, Umehara Y, Ashikawa I, Kurata N, Sasaki T. DNA Res. 1997;4:133–140. doi: 10.1093/dnares/4.2.133. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto K, Sasaki T. Plant Mol Biol. 1997;35:135–144. [PubMed] [Google Scholar]
- 17.Hanks S K, Quinn A M, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 18.Klimczak L J, Schindler U, Cashmore A R. Plant Cell. 1992;4:87–98. doi: 10.1105/tpc.4.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klimczak L J, Collinge M A, Farini D, Giuliano G, Walker J C, Cashmore A R. Plant Cell. 1995;7:105–115. doi: 10.1105/tpc.7.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terzaghi W B, Cashmore A R. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- 21.Lee Y, Lloyd A M, Roux S J. Plant Physiol. 1999;119:989–1000. doi: 10.1104/pp.119.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugano S, Andronis C, Green R M, Wang Z Y, Tobin E M. Proc Natl Acad Sci USA. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z Y, Tobin E M. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 24.Sugano S, Andronis C, Ong M S, Green R M, Tobin M. Proc Natl Acad Sci USA. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alpert K B, Tanksley S D. Proc Natl Acad Sci USA. 1996;93:15503–15507. doi: 10.1073/pnas.93.26.15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frary A, Nesbitt T C, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert K B, Tanksley S D. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 27.Fridman E, Pleban T, Zamir D. Proc Natl Acad Sci USA. 2000;97:4718–4723. doi: 10.1073/pnas.97.9.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]