Summary
The obligate intracellular bacterium Chlamydia has an unusual developmental cycle in which there is conversion between two forms that are specialized for either intracellular replication or propagation of the infection to a new host cell. Expression of late chlamydial genes is upregulated during conversion from the replicating to the infectious form, but the mechanism for this temporal regulation is unknown. We found that EUO, which is expressed from an early gene, binds to two sites upstream of the late operon omcAB, but only the downstream site was necessary for transcriptional repression. Using gel shift and in vitro transcription assays we showed that EUO specifically bound and repressed promoters of Chlamydia trachomatis late genes, but not early or mid genes. These findings support a role for EUO as a temporal repressor that negatively regulates late chlamydial genes and prevents their premature expression. The basis of this specificity is the ability of EUO to selectively bind promoter regions of late genes, which would prevent their transcription by RNA polymerase. Thus, we propose that EUO is a master regulator that prevents the terminal differentiation of the replicating form of chlamydiae into the infectious form until sufficient rounds of replication have occurred.
Introduction
One of the defining features of the genus Chlamydia is a biphasic developmental cycle in which the bacterium converts between an infectious elementary body (EB) and a non-infectious, replicating reticulate body (RB) within an infected host cell (Schachter, 1988). Members of this genus include Chlamydia trachomatis and Chlamydia pneumoniae, which are major human pathogens, and Chlamydia psittaci, which causes an uncommon zoonotic infection transmitted by birds (Stamm and Batteiger, 2009). The chlamydial developmental cycle has three distinct stages – an early stage where an EB that has just entered the cell converts into a RB, a mid stage where the metabolically active RB divides repeatedly by binary fission, and a late stage in which RBs convert back into EBs in an asynchronous manner (AbdelRahman and Belland, 2005). Chlamydial genes are transcribed as three main temporal groups that correspond to these three stages of the developmental cycle (Shaw et al., 2000; Belland et al., 2003a,b). Thus, the temporal expression of chlamydial genes is linked to the progression of the developmental cycle.
The expression of the relatively small number of late genes is a critical regulatory step in determining whether an individual RB continues to divide or terminally differentiates into an EB. Many of the 26 late genes that Belland and colleagues identified as being upregulated at the time of EB-to-RB conversion have EB-specific functions (Belland et al., 2003b). For example, omcAB encodes two cysteine-rich outer membrane proteins that are only found in EBs (Clarke et al., 1988; Liu et al., 2010). hctA and hctB encode histone-like proteins Hc1 and Hc2 respectively; these nucleoid-associated proteins bind DNA and cause chromosomal condensation, which is a characteristic feature of EBs (Barry et al., 1992; Brickman et al., 1993). Other late gene products include putative thioredoxin disulphide isomerases and membrane thiol proteases, which may have roles in the formation of the highly cross-linked outer membrane complex of an EB that provides protection against the extracellular environment (Newhall and Jones, 1983; Hackstadt et al., 1985; Wang et al., 2006; Liu et al., 2010). It is likely that the expression of these late genes is tightly regulated because premature expression prior to the end of the developmental cycle would induce conversion into EBs before the multiple rounds of RB division have been completed. If this were to happen, the infectious yield of the developmental cycle, which can be several hundred to a thousand EBs, would be greatly reduced.
Late chlamydial genes have been shown to be expressed as two separate subsets that are each regulated by a different transcriptional mechanism. One subset is transcribed by the major form of chlamydial RNA polymerase, which contains the major sigma factor, σ66 (Engel and Ganem, 1990; Koehler et al., 1990; Douglas et al., 1994). As σ66 RNA polymerase also transcribes early and mid genes, there must be regulatory mechanisms to control expression of each temporal class of σ66-dependent genes. A second subset of late genes is transcribed by a second form of RNA polymerase containing an alternative sigma factor σ28 instead of σ66 (Yu and Tan, 2003; Yu et al., 2006). σ28-dependent transcriptional activity has been proposed to be regulated by an anti-sigma factor RsbW that binds and sequesters σ28, although this model remains controversial and unproven (Hua et al., 2006).
It is not known how σ66-dependent late genes are regulated so that they are not prematurely expressed until late in the developmental cycle. Transcription of σ66-dependent late genes cannot be inhibited during early and mid times by an anti-sigma factor, as has been proposed for σ28, without also affecting transcription of early and mid genes. Mid genes have been proposed to be upregulated by increased DNA supercoiling in midcycle, but changes in DNA supercoiling are unlikely to regulate late genes because their promoters are not supercoiling-responsive (Niehus et al., 2008; Case et al., 2010). Instead, σ66-dependent late genes could be controlled by another major mechanism of prokaryotic gene regulation, which is the co-ordinate regulation of a group of genes by a transcription factor. For example, an activator could selectively upregulate the expression of σ66-dependent late genes at late times. Alternatively, a repressor could selectively prevent transcription of these late genes during early and mid stages, but not late in the development cycle.
Chlamydia encodes remarkably few transcription factors (Akers et al., 2011), but a chlamydial protein called EUO (early upstream ORF) has many of the features that would be predicted for a repressor of late genes. First, a candidate late repressor must be present starting at an early time in the intracellular infection in order to prevent premature expression of late genes. euo was one of the first early chlamydial genes to be identified, and euo transcripts and EUO protein are detectable as early as 1 h post infection (hpi) (Wichlan and Hatch, 1993; Zhang et al., 1998). Second, there is evidence that EUO is a DNA-binding protein that binds near the promoter of a known late gene. Specifically, EUO contains a helix–turn–helix motif and has been shown to bind the promoter region of the late operon omcAB, which encodes EB-specific cysteine-rich outer membrane proteins (Zhang et al., 2000). EUO shows a preference for binding A/T-rich sequences, and a 15 bp consensus DNA-binding site for EUO has been proposed based on DNA footprinting and a SELEX analysis (Zhang et al., 1998; 2000). Third, EUO caused a twofold decrease in transcription of the omcAB promoter in vitro (Zhang et al., 2000), suggesting that it may function as a transcription factor. However, EUO has not been shown to bind or modulate the promoter activity of other late genes, and it is not known if EUO has specificity for late and not early and mid chlamydial promoters.
To determine if EUO is a transcription factor that regulates the expression of late genes by σ66 RNA polymerase, we first examined EUO binding to its operator in the context of the omcAB promoter. We then used in vitro assays to test if EUO could bind and modulate transcription of representative promoters for early, mid and late chlamydial genes. We found that EUO selectively bound promoter regions of late genes and repressed transcription, providing support for this early expressed protein as a repressor of late genes in Chlamydia.
Results
Examining how EUO regulates a target promoter
We first determined the location of EUO-binding sites relative to the promoter in the context of the C. trachomatis late operon omcAB. In an electrophoretic mobility shift assay (EMSA), recombinant C. trachomatis EUO bound to two 60 bp fragments containing different portions of the omcAB promoter region (Fig. 1A). EUO bound to sequences from –122 to –60 (upstream EUO-binding site) as well as from –58 to +5 (downstream EUO-binding site) relative to the transcription start site (Lambden et al., 1990). For each fragment, EUO bound in a concentration-dependent manner and produced a similar pattern of two shifted species consistent with EUO binding to DNA in different molar ratios. Using Western blot analysis of the EMSA gel, we showed that antibodies against recombinant His-tagged EUO recognized both shifted bands for the downstream omcAB fragment, indicating that both bands represent complexes containing EUO bound to DNA (Fig. 1B). These results demonstrate that there are at least two EUO-binding sites upstream of the omcAB promoter, and we propose that EUO can bind to each DNA site with either a 1:1 or 2:1 stiochiometry. These findings are consistent with published reports showing that recombinant and native EUO bound to the C. psittaci omcAB promoter in the region from –200 to +67, and that rEUO protected the region from –53 to –14 on the top strand and –62 to –9 on the bottom strand in DNase I footprinting analysis (Zhang et al., 1998).
Fig. 1.
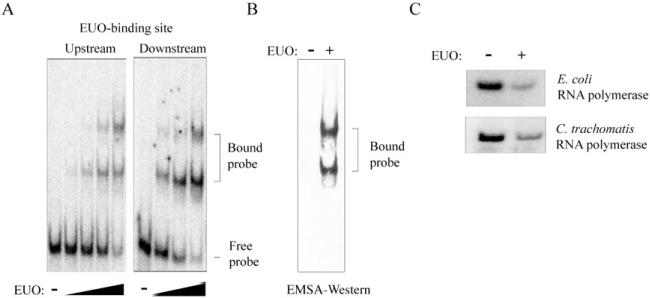
EUO binds and represses the C. trachomatis omcAB promoter.
A. EMSA experiments with upstream (nucleotides –122 to –60 relative to the start of transcription) or downstream regions (–58 to +5) of the omcAB promoter. The upstream probe was incubated with 0, 80, 160, 320 or 640 nM rEUO, while the downstream probe was incubated with 0, 80, 160 or 320 nM rEUO. Bands corresponding to the bound and free probes are indicated on the right.
B. Western blot verifying that the bound probe contains EUO. EMSA reaction for the downstream omcAB probe was performed in the absence or presence of 10 μM His-tagged rEUO and then transferred to nitrocellulose membrane and hybridized with anti-His antibodies. Bands corresponding to the bound DNA fragment are indicated to the right.
C. In vitro transcription of the omcAB promoter (pMT1150) by E. coli RNA polymerase or C. trachomatis RNA polymerase, as indicated, in the absence or presence of 2.5 μM EUO.
We performed in vitro transcription assays to determine if this binding by EUO leads to repression of the omcAB promoter. rEUO decreased transcription of the C. trachomatis omcAB promoter (contained on a template from –122 to +5) by 75% (Fig. 1C). The same level of repression was observed when the transcription reactions were performed with either C. trachomatis RNA polymerase (71% repression) or Escherichia coli RNA polymerase (75% repression). These results show that EUO binds to and represses transcription from the omcAB promoter and are in agreement with results from Hatch and colleagues that were performed with C. psittaci EUO (Zhang et al., 2000).
Using the proposed 15 bp EUO-binding site as a guide (Zhang et al., 2000), we predicted that the downstream EUO-binding site overlaps the –35 element of the C. trachomatis omcAB promoter (Fig. 2A). To test if EUO binds to this sequence, we designed mutant templates containing nucleotide substitutions and assayed if EUO binding was altered in EMSA experiments (Fig. 2A). Substitution of three nucleotides (mutants M1 and M2) in the putative EUO-binding site had minimal effects on EUO binding (Fig. 2B). However, substitution of nine of the 15 nucleotides in our predicted EUO-binding site (M3) completely disrupted binding of EUO (Fig. 2A and B). This result indicates that these sequences are necessary for EUO binding and provides support for the presence of an EUO operator overlapping the omcAB promoter.
Fig. 2.
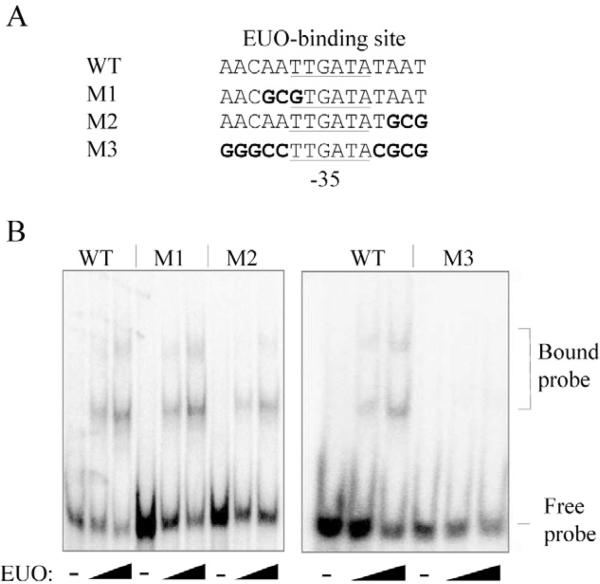
Mutational analysis to locate the downstream EUO-binding site of the omcAB promoter.
A. DNA sequence of the wild-type (WT) EUO-binding site and mutant templates containing nucleotide substitutions (highlighted in bold). The –35 promoter element is underlined – note that the 9 bp substitution in M3 did not alter the sequence of this –35 promoter element.
B. EMSA experiments with DNA probes containing these wild-type or mutant sequences. EMSA reactions were performed with 0, 80 or 160 nM rEUO. Bands corresponding to the bound and free probes are indicated on the right.
To determine if one or both of the two EUO-binding sites that we identified upstream of omcAB are necessary for repression, we repeated the transcription reactions with omcAB promoter templates containing only one EUO-binding site. To test the downstream site alone, we deleted sequences upstream of –55, which removed the upstream site (Fig. 3A). To test the upstream site alone, we used a template with the mutant downstream site (M3) from the EMSA experiment (Figs 2 and 3A). We had designed this 9 bp substitution so that it did not alter the sequence of the –35 promoter element, and we verified that promoter activity was not affected in an in vitro transcription assay compared to the wild-type promoter (data not shown). EUO produced a similar level of repression from a template containing both native binding sites or only the downstream site (73% vs. 67%, see Fig. 3B). However, there was less repression (22%) of an omcAB promoter template containing the upstream binding site alone (Fig. 3B). These results demonstrate that the downstream, and not the upstream, EUO-binding site of omcAB is important for repression. Furthermore, these studies provide additional experimental evidence that the operator for EUO overlaps the omcAB promoter. These findings are consistent with EUO functioning as a transcriptional repressor that binds its cognate operator in the vicinity of the promoter, blocking promoter recognition by RNA polymerase through steric hindrance.
Fig. 3.
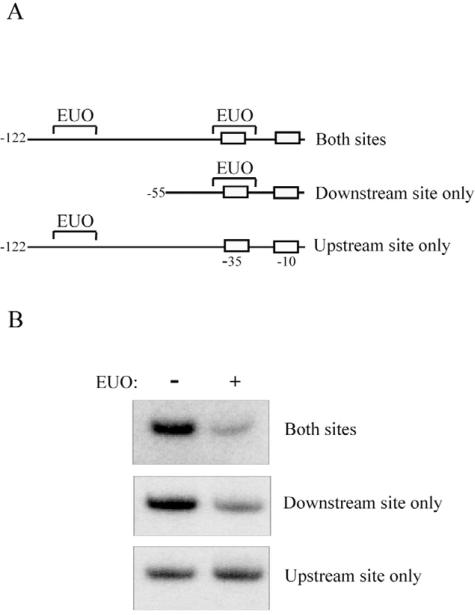
Only the downstream, and not the upstream, EUO-binding site in the omcAB promoter is important for repression by EUO.
A. Diagram showing transcription templates used to test the roles of the two EUO-binding sites in repression of the omcAB promoter. The template labelled ‘Both sites’ contains the two native EUO-binding sites upstream of omcAB in the region from +5 to –122. ‘Downstream site only’ is a truncation from –55 to +5 containing only the downstream EUO-binding site. ‘Upstream site only’ contains the omcAB promoter region from –122 to +5 in which the downstream EUO-binding site has been disrupted by a 9 bp substitution (M3 in Fig. 2). Positions of the EUO-binding sites, and the –10 and –35 promoter elements are shown.
B. In vitro transcription of these three transcription templates by E. coli RNA polymerase in the absence or presence of 2.5 μM C. trachomatis EUO.
EUO selectively binds and represses late promoters
To determine the role of EUO in temporal gene regulation in Chlamydia, we tested if it can bind and repress representative promoters for early, mid and late chlamydial genes. As an initial screen, we used competitive EMSA experiments to determine if a hundred-fold excess of unlabelled 60 bp DNA fragment containing promoter regions of representative early, mid or late genes could disrupt EUO binding to the omcAB promoter. Promoter fragments for the late genes ssc2 and ltuB, as well as omcAB itself, caused a loss of bound complexes, demonstrating that these sequences were able to compete for EUO binding (Fig. 4). In contrast, promoters for early (dnaK and groESL) and mid (cdsC) genes did not disrupt binding between the labelled probe and EUO (Fig. 4). We observed partial competition with the promoter for the mid gene fliF (Fig. 4).
Fig. 4.
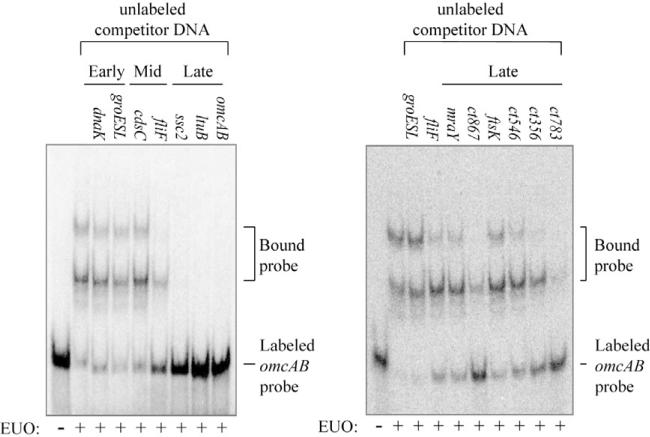
DNA fragments from late promoters, but not early or mid promoters, effectively compete for EUO binding. Competitive EMSAs were performed with the 60 bp labelled omcAB and 160 nM EUO in the presence of 50 nM unlabelled competitor DNA fragments (100-fold excess compared to the labelled omcAB probe). The promoter present on each DNA fragment is indicated for each reaction. Bands corresponding to the bound and free probes are indicated on the right.
To directly test the ability of EUO to bind to individual chlamydial promoters, we performed EMSA experiments with promoters representing the three temporal classes of chlamydial genes. EUO bound to DNA fragments containing the promoter region (–55 to +5 relative to the start of transcription) of the late genes ltuB, scc2 (Fig. 5) and cdsU (Table 1). For each of these late promoters, EUO produced two shifted species that were similar to those observed for each site in the omcAB promoter (Fig. 1A). In contrast, EUO did not significantly bind the promoters from two early genes and two mid genes (Fig. 5 and Table 1). In particular, there was no binding to the mid gene fliF, which was able to partially compete for EUO binding in the competitive EMSA. These results demonstrate that EUO binding is promoter-specific and suggest that EUO preferentially binds to late promoters.
Fig. 5.
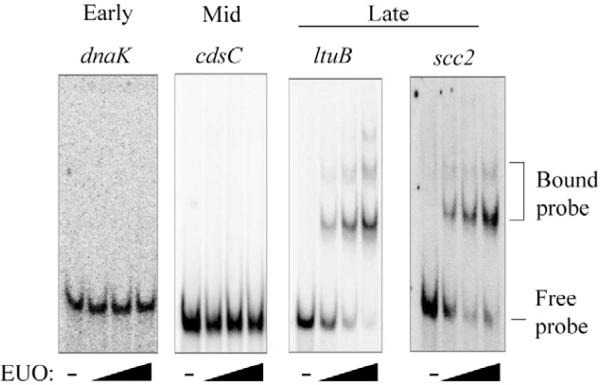
Differential binding of EUO to promoters of different temporal classes. For each promoter contained on a 60 bp DNA probe, EMSA reactions were performed in the absence of EUO or with 80, 160 or 320 nM EUO. Data for representative early (dnaK), mid (cdsC) and late promoters (ltuB and scc2) are shown. Bands corresponding to the bound and free probes are indicated on the right. Additional early, mid and late promoters were tested, and the results are shown in Table 1.
Table 1.
Binding and transcriptional repression of chlamydial promoters by EUO.
| Promoter | Bindinga | Repression (%)b | |
|---|---|---|---|
| Early genes | dnaK (ct396) | No | 6 ± 5 |
| groESL (ct110) | No | 21 ± 13 | |
| rRNA | n.t. | 7 ± 8 | |
| Mid genes | cdsC (ct674) | No | 12 ± 6 |
| fliF (ct719) | No | 11 ± 12 | |
| ct665 | n.t. | 21 ± 11 | |
| ompA (ct681) | n.t. | 12 ± 10 | |
| Late genes | omcB (ct443) | Yes | 60 ± 18 |
| ltuB (ct080) | Yes | 69 ± 9 | |
| ssc2 (ct576) | Yes | 75 ± 2 | |
| cdsU (ct091) | Yes | 46 ± 19 | |
| mraY (ct757) | Yes | n.t. | |
| ct783 | Yes | n.t. | |
| ct867 | Yes | n.t. | |
| ftsK (ct739) | Yes | n.t. | |
| ct356 | Yes | n.t. |
Binding was determined by EMSA using EUO protein over a range of 80–320 nM for each 60 bp labelled DNA probe containing the respective promoter.
Per cent repression was calculated as the ratio of transcript levels in the presence/absence of EUO. Results are from at least three independent experiments and are reported as the mean and standard deviation for each promoter.
n.t., not tested.
We next tested if the ability of EUO to selectively bind late promoters resulted in transcriptional repression of late but not early and mid chlamydial promoters. Using in vitro transcription assays, we found that EUO decreased transcription from the three late promoters ltuB, cdsU and scc2 by 46–75% compared to transcription in the absence of EUO (Fig. 6 and Table 1). In contrast, EUO did not significantly alter transcription from promoters of three early (dnaK, groESL and rRNA) and four mid genes (cdsC, fliF, ct665 and ompA) (Fig. 6 and Table 1). Together, these results demonstrate that EUO selectively binds to and represses promoters of late chlamydial genes but not early and mid genes. These findings support a role for EUO as a repressor of late genes in Chlamydia.
Fig. 6.
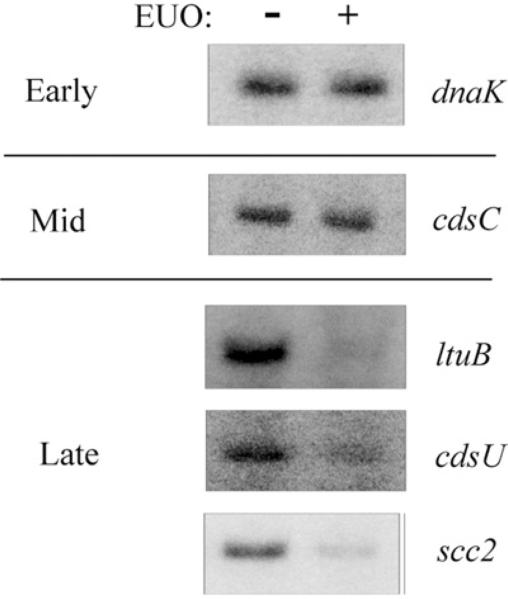
EUO represses late, but not early or mid promoters. Transcription templates containing the promoter regions of early (dnaK), mid (cdsC) and late (ltuB, cdsU and scc2) were transcribed with E. coli RNA polymerase in the absence or presence of 2.5 μM EUO. Additional early, mid and late promoters were tested, and the results are shown in Table 1.
We explored if EUO could regulate additional late genes by testing its ability to bind the promoter regions of five additional genes whose transcripts are upregulated at late times in the developmental cycle (Belland et al., 2003b). We based these predicted promoter regions on the transcription start sites identified in a deep sequencing study by Albrecht et al. (2010). In EMSA experiments, EUO bound to the predicted promoter regions of each of these five late genes, albeit with lower affinity than the omcAB promoter, while EUO did not bind the groESL promoter (Fig. 7). These five predicted late promoters and promoters of other late genes have not previously been studied. Experiments to determine if EUO also represses their transcription will first require that these predicted promoters be shown to be transcriptionally active. However, our studies provide experimental evidence that EUO regulates at least nine late chlamydial genes.
Fig. 7.
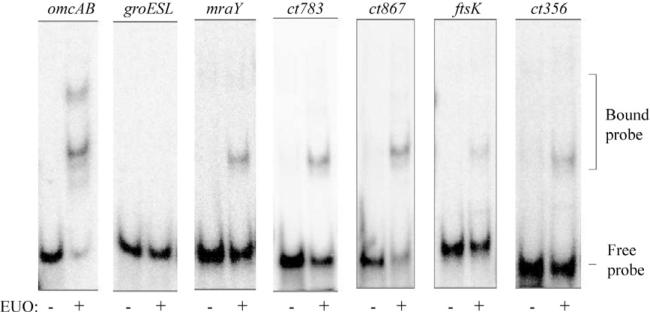
EUO binds to the predicted promoter regions of five additional late genes. For each promoter region contained on a 60 bp DNA probe, EMSA reactions were performed in the absence of EUO or with 320 nM EUO. Bands corresponding to the bound and free probes are indicated on the right.
Discussion
The developmental regulation of chlamydial genes as three main temporal classes is a hallmark of the intracellular Chlamydia infection. Prior to this study, it was not known how late genes transcribed by the major form of chlamydial RNA polymerase, σ66 RNA polymerase, are regulated so that they are only expressed at the time of conversion from RBs to infectious EBs. We now present data showing that the early chlamydial protein EUO specifically represses promoters of late genes, but not early and mid genes. We also demonstrate that the basis of this specificity is the ability of EUO to selectively bind the promoter regions of late genes. EUO bound to an operator that overlaps the –35 element of the omcAB promoter, and the presence of this operator was necessary for EUO-mediated repression. These findings are consistent with a mechanism of repression in which RNA polymerase is selectively blocked from transcribing target late genes by steric hindrance.
We thus propose that EUO specifically regulates σ66-dependent late genes because only these target genes, and not early and mid genes, contain an EUO operator in the vicinity of their promoters. EUO has been shown to preferentially bind A/T-rich sequences (Zhang et al., 1998), and promoters for σ66-dependent late chlamydial genes have been noted to be A/T-rich relative to mid promoters (Niehus et al., 2008). In all, we show that EUO binds upstream of nine late genes. Our strongest data are for four late genes, including omcAB, which encodes two EB-specific cysteine-rich outer membrane proteins, and cdsU and scc2, which encode proteins involved in the type III secretion system for secreting chlamydial effectors into the inclusion membrane and the host cytosol. For these late genes, we have identified candidate EUO-binding sequences overlapping –35 or –10 promoter elements, based on the proposed 15 bp DNA-binding site for EUO (Zhang et al., 2000) (Fig. 8). However, A/T-rich sequences resembling the proposed 15 bp EUO-binding site are also present in the promoter regions of early and mid promoters that were not bound by EUO in our studies. Thus with the available data, EUO target genes cannot be predicted by sequence alone, which underscores the importance of defining EUO operators with functional studies.
Fig. 8.
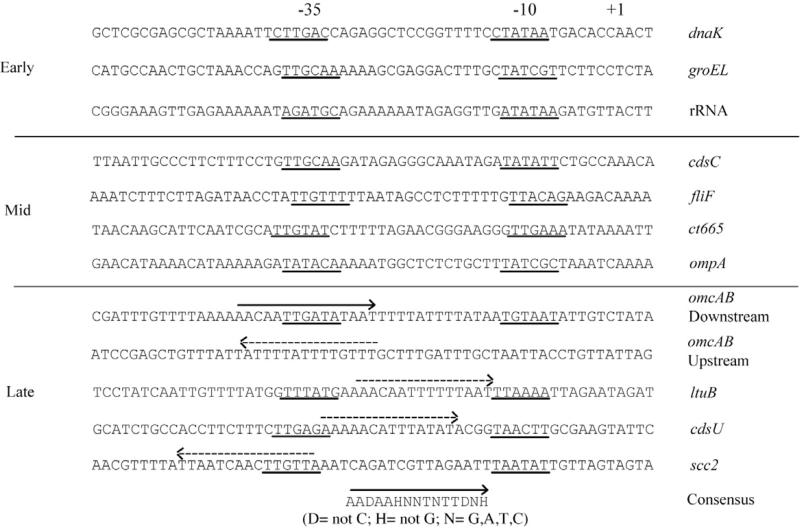
Identification of putative EUO operators in promoter regions of C. trachomatis genes tested in this study. Except for omcAB upstream, DNA sequences were aligned by the transcriptional start site (indicated by +1). The –35 and –10 promoter elements are underlined. The EUO-binding site for the downstream omcAB promoter region is indicated with a solid arrow above the sequence. The published consensus EUO-binding site is indicated at the bottom of the figure (Zhang et al., 2000). These two reference EUO-binding sites were used to identify putative binding sites (dashed arrows) for late, but not early or mid genes, on the basis of sequence similarity. Arrows pointing from right to left represent the EUO-binding site on the complementary DNA strand. DNA sequences for the promoter regions of the late genes tested in Fig. 7 are not listed, as the –35 and –10 elements have not yet been defined.
The operator sequence recognized by EUO appears to be a degenerate A/T-rich sequence rather than a specific DNA sequence. Hatch and colleagues performed in vitro binding studies and demonstrated that EUO has low DNA-binding specificity (Zhang et al., 2000). In agreement, we found that three-nucleotide substitutions in the omcAB operator had only slight effects on EUO binding and that a more extensive 9 bp substitution was necessary to disrupt binding (Fig. 2). Prokaryotic transcription factors that are global regulators have been noted to have a lower DNA-binding specificity (Zhang et al., 2000). Thus, the ability of EUO to bind to a degenerate binding site is consistent with a potential role as a global regulator whose regulon may include the 29 late genes identified in C. trachomatis (Belland et al., 2003b).
We identified a second EUO-binding site further upstream of the promoter elements and operator of omcAB. This upstream EUO-binding site, which was located between –122 and –60, was not necessary for EUO-mediated repression of the omcAB promoter, and its significance is not known. It is possible that this upstream binding site may compete with bona fide operators for EUO binding, or alternatively may increase the local concentration of EUO in the neighbourhood of the omcAB promoter. Two EUO-binding sites have also been identified in the omcAB promoter for C. psittaci (Zhang et al., 1998), but it is not known if this is a feature of other EUO-regulated genes.
There is evidence to support a role for EUO as a conserved regulator of late gene expression in Chlamydia. EUO is encoded by all eight sequenced Chlamydia spp. and the protein is well conserved with 62–86% identity and 77–92% similarity (based on a BLAST search for C. trachomatis EUO protein). An EUO homologue is also encoded by Chlamydia-like organisms, including Protochlamydia amoebophila, Simkania negevensis and Waddlia chondrophilia, which share a common ancestor with the Chlamydiaceae. Where it has been examined in C. pneumoniae and C. psittaci, EUO is also an early gene that is expressed during the early and mid stages of the developmental cycle (Zhang et al., 1998; Maurer et al., 2007). C. psittaci EUO binds and represses transcription of the omcAB promoter (Zhang et al., 2000), but its ability to selectively regulate promoters of other C. psittaci late genes has not been examined.
A major unresolved question is how EUO-mediated repression is relieved in order to allow transcription of its target genes at late times in the developmental cycle. The mechanism must account for asynchrony in the timing of RB-to-EB conversion, which occurs in a proportion of chlamydiae in a late inclusion while other RBs continue to divide. Thus, derepression of EUO-regulated late genes must be able to occur at the level of an individual chlamydia independently of other chlamydiae in the same inclusion. Two potential mechanisms for relieving EUO-mediated repression involve the regulation of EUO protein levels or the control of EUO activity levels by a cofactor.
Data from C. psittaci and C. pneumoniae indicate that EUO levels are downregulated at late times. In C. psittaci, euo transcript levels decreased after 4 hpi and EUO protein levels decreased by 20 hpi (Zhang et al., 1998). In a C. pneumoniae microarray analysis, euo mRNA levels were high at early times but decreased after 6 hpi (Maurer et al., 2007). In C. trachomatis, euo transcript levels from a microarray study did not decrease at late times (Belland et al., 2003b), although the temporal pattern of EUO protein levels is not known. If EUO levels fall below a certain threshold at late times in the developmental cycle, it could lead to derepression of target genes such as omcAB. Potential means of downregulating EUO levels include transcriptional, post-transcriptional and translational control mechanisms.
EUO activity could instead be regulated by a cofactor such as a small molecule or a regulatory protein. A number of small molecules have been shown to serve as cofactors for chlamydial transcription factors. For example, amino acids such as tryptophan and arginine are cofactors for the repressors TrpR and ArgR respectively (Akers and Tan, 2006; Schaumburg and Tan, 2006). Similarly, Zn2+ is a co-repressor for the dual function YtgCYtgR repressor (CT069) (Akers et al., 2011), and nucleotides have been proposed to be a cofactor for NrdR, which is a regulator of DNA synthesis (Case et al., 2011). The molecular chaperone GroEL functions in an ATP-independent manner as a cofactor for the stress response regulator HrcA (Wilson et al., 2005; Chen et al., 2011). EUO repressor function could be dependent on a co-repressor that is present in chlamydiae during early and mid stages of the developmental cycle but depleted by late time points. Alternatively, repression by EUO could be antagonized by an inducer that prevents binding of EUO to its DNA targets when it is present at late times. However, our ability to demonstrate binding and repression by EUO in in vitro studies with defined components argues against a requirement for a co-repressor.
This proposed model for the negative regulation of σ66-dependent late genes stands in contrast to the positive regulation of midcycle chlamydial genes. Promoters for representative mid genes, but not late genes, have been shown to be upregulated by increased DNA super-coiling (Niehus et al., 2008; Case et al., 2010). As chlamydial supercoiling levels are highest in midcycle, mid genes have been proposed to be positively regulated by global DNA supercoiling (Niehus et al., 2008). In contrast, EUO provides a mechanism for σ66 RNA polymerase to transcribe early and mid genes while selectively repressing late genes. Intriguingly, supercoiling-dependent mid promoters are disproportionally G/C-rich (Niehus et al., 2008; Case et al., 2010), while EUO shows a strong preference for binding to A/T-rich DNA sequences, which are typical of late promoters (Fig. 8). These differences suggest that the temporal classes of chlamydial genes are hardwired via the sequence of their promoter regions to be differentially regulated.
The proposed role of EUO in repressing late genes may be relevant to chlamydial cell culture persistence, which is an altered growth state in which RB-to-EB conversion is blocked. When Chlamydia-infected cells are treated with factors such as IFN-γ or depleted of specific nutrients such as tryptophan, they enter this persistent state in which large aberrant RBs continue to replicate their DNA but do not divide (Beatty et al., 1993; 1994). Another feature of chlamydial persistence is that late gene expression, which is associated with RB-to-EB conversion, is downregulated (Belland et al., 2003a). Interestingly, euo is one of the few genes that is dramatically upregulated during persistence (Belland et al., 2003a). Our results predict that these high levels of EUO are the direct cause for the repression of late genes during cell culture persistence. The mechanism by which EUO expression is upregulated during chlamydial persistence remains to be determined. However, our findings suggest that by controlling late gene expression and the conversion of RBs into EBs, EUO is an important regulator of both the normal developmental cycle and chlamydial persistence.
In summary, we propose that EUO is a transcriptional repressor that prevents premature expression of σ66-dependent late genes until late in the developmental cycle. As EUO-regulated target genes are involved in RB-to-EB conversion, control of their expression is likely to play an important role in determining whether a RB continues to replicate or if it differentiates into an EB. Thus, we propose that EUO is a transcriptional regulator that determines the balance between the production of chlamydial progeny and the proportion of these progeny that are infectious.
Experimental procedures
Construction of in vitro transcription plasmids
Promoter sequences were amplified by PCR from C. trachomatis serovar D UW-3/Cx genomic DNA and cloned upstream of a promoterless G-less cassette transcription template in pMT1125, as previously described (Wilson and Tan, 2002). Plasmids used in this study are listed in Table 2.
Table 2.
List of transcription templates.
| Plasmid | Promoter (nucleotides relative to transcription start site) | Reference |
|---|---|---|
| pMT1013 | ltuB promoter region from –132 to +5; rRNA promoter region from –54 to +5 | Schaumburg and Tan (2000) |
| pMT1149 | euo promoter region from –90 to +5 | Adam Wilson (unpublished) |
| pMT1150 | omcAB promoter region from –122 to +5 | Wilson and Tan (2002) |
| pMT1178 | groESL promoter region from –137 to +5 | Wilson and Tan (2004) |
| pMT1225 | dnaK promoter region from –65 to +5 | Adam Wilson (unpublished) |
| pMT1441 | cdsU promoter region from –153 to +5 | Case et al. (2010) |
| pMT1442 | scc2 promoter region from –144 to +5 | Case et al. (2010) |
| pMT1513 | cdsC promoter region from –146 to +5 | Case et al. (2010) |
| pMT1515 | ct665 promoter region from –170 to +5 | Case et al. (2010) |
| pMT1516 | fliF promoter region from –154 to +5 | Case et al. (2010) |
| pMT1636 | omcAB promoter region from –55 to +5 | This work |
| pMT1637 | omcAB promoter region from –122 to +5 containing a 9 bp nucleotide substitution in the downstream EUO-binding site | This work |
For construction of pMT1636, primers T1710 (5′AGCGAATTCTAGACGATTTGTT; EcoRI site underlined) and SP6 (5′ CGCCAAGCTATTTAGGTGACACTATAG) were used to amplify the omcAB promoter region from –55 to +245 by PCR using pMT1150 as a template. The DNA fragment was cloned upstream of the promoterless G-less cassette template of pMT1125 at EcoRI and BamHI sites. For construction of pMT1637, primers T180 (5′ AGCGAATTCTTTGAATCCGAGCTGTTTATTATTT; EcoRI site underlined) and T1709 (5′ TTATAGAACAATATTACATTATAAAATAAAAGCCGTATCAAGGCCCTTTAAAACAATCGT; nucleotide substitutions indicated in bold) were used to amplify a mutant omcAB promoter by PCR from pMT1150. The DNA fragment was cloned upstream of the promoterless G-less cassette template of pMT1125 at EcoRI and EcoRV sites. All constructs were verified by sequencing (Genewiz).
Purification of recombinant EUO
Recombinant EUO containing a 6×His tag at the C-terminus was expressed from pMT1181 (Elizabeth DiRusso Case, unpublished), which contains the C. trachomatis euo gene cloned into the IPTG-inducible expression plasmid pRSETC (Invitrogen). E. coli strains containing pMT1181 were grown in 1 l LB to an OD600 of 0.4. Cultures were induced with 1 mM IPTG for 2 h at 37°C and cells were harvested by centrifugation. The cell pellet was resuspended in buffer N [10 mM Tris-HCl (pH 8.0), 0.3 M NaCl, 10 mM 2-mercaptoethanol] containing 20 mM imidazole. The resuspended cells were sonicated twice with a Branson digital sonifier 250D for 30 s at 22% output. The material was centrifuged and the supernatant was added to a 1 ml slurry of Qiagen Ni-NTA beads. The slurry was incubated for 1 h at 4°C and then applied to a 25 ml Poly-Prep chromatography column (Bio-Rad). The beads were washed with 500 ml of buffer N containing 20 mM imidazole. Recombinant EUO protein was eluted from the column with 2 ml of buffer N containing 250 mM imidazole. Eluted protein was then dialysed overnight against 1 l of storage buffer [10 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 0.1 mM EDTA, 100 mM NaCl, 10 mM 2-mercaptoethanol, 30% (v/v) glycerol]. Dialysed protein was aliquoted and stored at –70°C.
Purification of C. trachomatis RNA polymerase
RNA polymerase was partially purified from C. trachomatis LGV serovar L2 at 18 hpi by heparin-agarose chromatography as previously described (Tan and Engel, 1996). Essentially L929 mouse fibroblast cells were infected with C. trachomatis LGV serovar L2 EBs at an MOI (multiplicity of infection) of 3. Infected cells were harvested and resus-pended in PBS (pH 7.5). Cells were lysed by dounce homogenization and cell debris was removed by low-speed centrifugation. Chlamydiae (in the supernatant) were pelleted by high-speed centrifugation. The pellet was resuspended in lysis buffer [10 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM EDTA, 7.5% (v/v) glycerol, 0.6 M NaCl, 10 mM β-mercaptoethanol, 0.1% (v/v) Nonidet-40] followed by sonication three times in a Branson digital sonifier 250D for 10 s at 22% output. Cellular debris was removed by centrifugation and the supernatant was diluted with buffer I [10 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM EDTA, 7.5% (v/v) glycerol, 10 mM β-mercaptoethanol] resulting in a final NaCl concentration of 0.25 M and incubated with heparin agarose equilibrated in buffer I containing 0.25 M NaCl for 1 h. The slurry was applied to 25 ml Poly-Prep chromatography column (Bio-Rad) and the beads were washed with 50 ml of buffer I containing 0.25 M NaCl. RNA polymerase was eluted from the column with buffer I containing 0.6 M NaCl, followed by dialysis in 1 l of storage buffer overnight. Dialysed samples were aliquoted and stored at –80°C.
Electrophoretic gel mobility shift assays (EMSAs)
Annealed 60 bp complementary primers were labelled by T4 polynucleotide kinase (New England Biolabs) with approximately 30 μCi [γ-32P]-ATP (10 mCi ml–1, 6000 Ci mmol–1; MP Biomedicals). Free nucleotides were removed with a mini Quick Spin DNA column (Roche). Approximately 0.5 nM labelled DNA was incubated with rEUO over a range of concentrations from 80 to 320 nM in binding buffer [40 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 70 mM KCl, 125 μM EDTA, 100 μM dithiothreitol, 7.5% glycerol, 10 ng of salmon sperm DNA] at room temperature for 20 min. For competitive EMSAs, 50 nM of an unlabelled 60 bp DNA fragment was added to the binding reaction. Samples were loaded onto a 6% polyacrylamide EMSA gel at 150 V in 0.5× Tris-borate-EDTA (TBE) buffer (Read, 1996). After electrophoresis, the gel was dried on Whatman paper and exposed to a phosphorimager screen. The screen was scanned with a Bio-Rad Personal FX scanner.
EMSA–Western blot analysis
For detection by Western blot analysis, the preceding EMSA experiments were performed using higher DNA and protein concentrations: 50 ng (125 nM) of unlabelled 60 bp DNA fragment containing the omcAB promoter (–55 to +5) and 10 μM EUO. Bound DNA fragments were separated on a 6% polyacrylamide EMSA gel and then transferred to a Protran nitrocellulose membrane (Whatman). The membrane was preincubated with 5% milk, followed by incubation with anti-His antibody (1:10 000; Qiagen) overnight at 4°C. The membrane was then washed and probed with goat anti-mouse antibody (1:10 000) for 1 h at room temperature. Bands were visualized by chemiluminescence with exposure to HyBlot CL autoradiography film (Denville).
In vitro transcription assays
Approximately 3 nM plasmid DNA containing the transcription template was incubated with 5 μM rEUO at room temperature for 15 min. Transcription assays were initiated as described previously (Tan and Engel, 1996) using 2 μl of chlamydial RNA polymerase or 0.4 U E. coli RNA polymerase holoenzyme (Epicentre). However, transcription with the ompA promoter was performed with C. trachomatis RNA polymerase as this promoter is not transcribed by E. coli RNA polymerase (Douglas and Hatch, 1996). The transcripts were resolved by electrophoresis on an 8 M urea–6% polyacrylamide gel. The gels were then fixed, dried and exposed to a phosphorimager plate. The plate was scanned with a Bio-Rad Personal FX scanner and the amount of transcripts was quantified using Quantity One software (Bio-Rad). Repression was calculated as the percentage of transcripts in the presence/absence of EUO. For each plasmid, the transcription assays were performed as a minimum of three independent experiments, and values are reported as the mean of the repression [H11006] standard deviation.
Acknowledgements
We would like to thank Allan Chen, Eric Cheng, Kirsten Johnson and Jennifer Lee for critical reading of the manuscript. This work was supported by a grant from the NIH (AI 44198).
References
- AbdelRahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Akers JC, Tan M. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J Bacteriol. 2006;188:4236–4243. doi: 10.1128/JB.01660-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers JC, Hodac H, Lathrop RH, Tan M. Identification and functional analysis of CT069 as a novel transcriptional regulator in Chlamydia. J Bacteriol. 2011;193:6123–6131. doi: 10.1128/JB.05976-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C, Hayes S, Hackstadt T. Nucleoid condensation in Escherichia coli that express a chlamydial histone homolog. Science. 1992;256:377–379. doi: 10.1126/science.256.5055.377. [DOI] [PubMed] [Google Scholar]
- Beatty WL, Byrne GI, Morrison RP. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci USA. 1993;90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect Immun. 1994;62:3705–3711. doi: 10.1128/iai.62.9.3705-3711.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Nelson DE, Virok D, Crane DD, Hogan D, Sturdevant D, et al. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci USA. 2003a;100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA. 2003b;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Barry CE, III, Hackstadt T. Molecular cloning and expression of hctB encoding a strain-variant chlamydial histone-like protein with DNA-binding activity. J Bacteriol. 1993;175:4274–4281. doi: 10.1128/jb.175.14.4274-4281.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case ED, Peterson EM, Tan M. Promoters for Chlamydia type III secretion genes show a differential response to DNA supercoiling that correlates with temporal expression pattern. J Bacteriol. 2010;192:2569–2574. doi: 10.1128/JB.00068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case ED, Akers JC, Tan M. CT406 encodes a chlamydial ortholog of NrdR, a repressor of ribonucleotide reductase. J Bacteriol. 2011;193:4396–4404. doi: 10.1128/JB.00294-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AL, Wilson AC, Tan M. A Chlamydia-specific C-terminal region of the stress response regulator HrcA modulates its repressor activity. J Bacteriol. 2011;193:6733–6741. doi: 10.1128/JB.05792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke IN, Ward ME, Lambden PR. Molecular cloning and sequence analysis of a developmentally regulated cysteine-rich outer membrane protein from Chlamydia trachomatis. Gene. 1988;71:307–314. doi: 10.1016/0378-1119(88)90047-9. [DOI] [PubMed] [Google Scholar]
- Douglas AL, Hatch TP. Mutagenesis of the P2 promoter of the major outer membrane protein gene of Chlamydia trachomatis. J Bacteriol. 1996;178:5573–5578. doi: 10.1128/jb.178.19.5573-5578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AL, Saxena NK, Hatch TP. Enhancement of in vitro transcription by addition of cloned, overexpressed major sigma factor of Chlamydia psittaci 6BC. J Bacteriol. 1994;176:3033–3039. doi: 10.1128/jb.176.10.3033-3039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J, Ganem D. A PCR-based approach to cloning sigma factors from eubacteria and its application to the isolation of a sigma70 homolog from Chlamydia trachomatis. J Bacteriol. 1990;172:2447–2455. doi: 10.1128/jb.172.5.2447-2455.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Todd WJ, Caldwell HD. Disulfide-mediated interactions of the chlamydial major outer membrane protein: role in the differentiation of chlamydiae? J Bacteriol. 1985;161:25–31. doi: 10.1128/jb.161.1.25-31.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua L, Hefty PS, Lee YJ, Lee YM, Stephens RS, Price CW. Core of the partner switching signalling mechanism is conserved in the obligate intracellular pathogen Chlamydia trachomatis. Mol Microbiol. 2006;59:623–636. doi: 10.1111/j.1365-2958.2005.04962.x. [DOI] [PubMed] [Google Scholar]
- Koehler JE, Burgess RR, Thompson NE, Stephens RS. Chlamydia trachomatis RNA polymerase major sigma subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli sigma 70 and Bacillus subtilis sigma 43. J Biol Chem. 1990;265:13206–13214. [PubMed] [Google Scholar]
- Lambden PR, Everson JS, Ward ME, Clarke IN. Sulfur-rich proteins of Chlamydia trachomatis: developmentally regulated transcription of polycistronic mRNA from tandem promoters. Gene. 1990;87:105–112. doi: 10.1016/0378-1119(90)90500-q. [DOI] [PubMed] [Google Scholar]
- Liu X, Afrane M, Clemmer DE, Zhong G, Nelson DE. Identification of Chlamydia trachomatis outer membrane complex proteins by differential proteomics. J Bacteriol. 2010;192:2852–2860. doi: 10.1128/JB.01628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer AP, Mehlitz A, Mollenkopf HJ, Meyer TF. Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 2007;3:e83. doi: 10.1371/journal.ppat.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhall WJ, Jones RB. Disulfide-linked oligomers of the major outer membrane protein of chlamydiae. J Bacteriol. 1983;154:998–1001. doi: 10.1128/jb.154.2.998-1001.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus E, Cheng E, Tan M. DNA supercoiling-dependent gene regulation in Chlamydia. J Bacteriol. 2008;190:6419–6427. doi: 10.1128/JB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read M. Electrophoretic mobility shift assay (EMSA). In: Docherty K, editor. Gene Transcription: DNA Binding Proteins. John Wiley & Sons; Chichester, West Sussex, UK: 1996. pp. 6–11. [Google Scholar]
- Schachter J. The intracellular life of Chlamydia. Curr Top Microbiol Immunol. 1988;138:109–139. [PubMed] [Google Scholar]
- Schaumburg CS, Tan M. A positive cis-acting DNA element is required for high level transcription in Chlamydia. J Bacteriol. 2000;182:5167–5171. doi: 10.1128/jb.182.18.5167-5171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg CS, Tan M. Arginine-dependent gene regulation via the ArgR repressor is species specific in Chlamydia. J Bacteriol. 2006;188:919–927. doi: 10.1128/JB.188.3.919-927.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AC, Christiansen G, Roepstorff P, Birkelund S. Genetic differences in the Chlamydia trachomatis tryptophan synthase alpha-subunit can explain variations in serovar pathogenesis. Microbes Infect. 2000;2:581–592. doi: 10.1016/s1286-4579(00)00368-3. [DOI] [PubMed] [Google Scholar]
- Stamm WE, Batteiger BE. Introduction to Chlamydia and Chlamydophila. In: Mandell GL, Bennett JE, Dolin R, editors. Mandell: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Churchill Livingstone/Elsevier; Philadelphia, PA: 2009. pp. 2439–2442. [Google Scholar]
- Tan M, Engel JN. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J Bacteriol. 1996;178:6975–6982. doi: 10.1128/jb.178.23.6975-6982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Zhang YX. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 2006;15:122–134. doi: 10.1110/ps.051616206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichlan DG, Hatch TP. Identification of an early-stage gene of Chlamydia psittaci 6BC. J Bacteriol. 1993;175:2936–2942. doi: 10.1128/jb.175.10.2936-2942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Tan M. Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J Bacteriol. 2002;184:6566–6571. doi: 10.1128/JB.184.23.6566-6571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Tan M. Stress response gene regulation in Chlamydia is dependent on HrcA-CIRCE interactions. J Bacteriol. 2004;186:3384–3391. doi: 10.1128/JB.186.11.3384-3391.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Wu CC, Yates JR, 3rd, Tan M. Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein. J Bacteriol. 2005;187:7535–7542. doi: 10.1128/JB.187.21.7535-7542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HHY, Tan M. Sigma 28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol Microbiol. 2003;50:577–584. doi: 10.1046/j.1365-2958.2003.03708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HHY, Kibler D, Tan M. In silico prediction and functional validation of sigma 28-regulated genes in Chlamydia and Escherichia coli. J Bacteriol. 2006;188:8206–8212. doi: 10.1128/JB.01082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Douglas AL, Hatch TP. Characterization of a Chlamydia psittaci DNA binding protein (EUO) synthesized during the early and middle phases of the developmental cycle. Infect Immun. 1998;66:1167–1173. doi: 10.1128/iai.66.3.1167-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Howe MM, Hatch TP. Characterization of in vitro DNA binding sites of the EUO protein of Chlamydia psittaci. Infect Immun. 2000;68:1337–1349. doi: 10.1128/iai.68.3.1337-1349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


