Introduction
The seventh annual ARVO/Pfizer Ophthalmic Research Institute conference was held Friday and Saturday, April 29 and 30, 2011, at the Fort Lauderdale Hyatt Regency Pier 66, Fort Lauderdale, Florida. The conference, funded by The ARVO Foundation for Eye Research through a grant from Pfizer Ophthalmics, provided an opportunity to gather experts from within and outside ophthalmology to determine the state of knowledge pertaining to molecular biomarkers associated with glaucoma, as well as the methods to identify and validate them to predict (a) those who would be susceptible to development of glaucoma; (b) markers that will enable prediction of glaucoma progression; and (c) markers that will predict efficacy of treatment of glaucoma. Identification of such biomarkers will aid in prevention of glaucoma-related vision loss and blindness. The conference focused on an evaluation of glaucoma molecular biomarkers and progress needed for future validation of glaucoma biomarkers.
A working group of 21 glaucoma researchers, 7 scientists focused on diseases other than glaucoma and with expertise in areas such as proteomic biomarkers or molecular mechanisms for neurodegeneration, and 60 observers from ARVO, Pfizer, and clinical and basic ophthalmic research convened to evaluate current understanding of the molecular biomarkers of glaucoma. The meeting format emphasized discussion and concentrated on questions within areas of glaucoma molecular biomarker research:
Session I: How to define a biomarker in medicine? Current knowledge about biomarkers in human health and in glaucoma
Session II: Genetic biomarkers in glaucoma
Session III: Proteomic biomarkers in glaucoma
Session IV: Pre-immune and immune events: Immunoproteomics and its possible applications in glaucoma
Session V: From bench to bedside: How can a translational approach be successful?
Each session began with a 10-minute overview by a glaucoma researcher followed by a 30-minute presentation by an outside expert, with parallels between their fields of expertise and the eye included. Invited outside experts covered several areas of research, including proteomic biomarker discovery in cancer (Emanuel Petricoin, PhD, George Mason University, Maryland; and Akhilesh Pandey, MD, PhD, Johns Hopkins University, Maryland) and astroglial cells in neurodegeneration (Stephen D. Miller, PhD, Northwestern University, Illinois).
How to Define a Biomarker in Medicine? Current Knowledge about Biomarkers in Human Health and in Glaucoma
The increased sensitivity and accuracy of genomic, proteomic, and metabolomic techniques (see Figure) have brought about the potential to identify molecular entities that may serve as potentially useful markers, including (1) markers for early detection of a disease; (2) markers that will predict severity of a disease; (3) markers that will predict the rate of disease progression, and (4) markers that will serve as predictors of response to treatment. The severity of a disease may be very dissimilar in different individuals even if they are at an equivalent stage of the disease, owing to shortcomings in staging the disease process. On the other hand, the progression of the disease in different individuals, or even in different organs of the same individual, may occur at different rates. Glaucoma is an example of such asymmetric presentation. A patient with pseudoexfoliation glaucoma, often also referred to as exfoliation syndrome (ES), usually has asymmetry of involvement between the two eyes. Two-thirds of patients present unilaterally, and 50% of these develop the disease in the fellow eye within 15 years; rates of progression differ among individuals. The response to treatment also differs among individuals, and prediction of treatment outcome markers will be helpful to personalize treatment. The identification of quantitative biomarkers that reveal aspects of the disease process could especially help the clinician understand and monitor a patient's response to treatments.
Figure. .
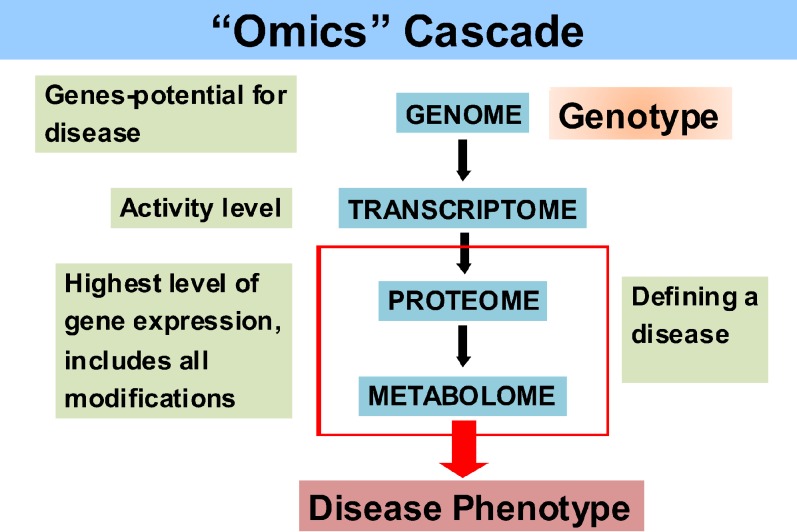
Flow diagram explaining the various “omics” relationships (courtesy of R. Beuerman).
Seventh ARVO/Pfizer Ophthalmics Research Institute Conference Working Group
Seventh ARVO/Pfizer Ophthalmics Research Institute Conference Working Group
Course Directors
Sanjoy Bhattacharya, Bascom Palmer Eye Institute, University of Miami Miller School of Medicine, Miami, FL
Franz Grus, University Medical Center Mainz, Mainz, Germany
Richard Lee, Bascom Palmer Eye Institute, University of Miami Miller School of Medicine, Miami, FL
Participants
Roger Beuerman, Singapore Eye Research Institute, Singapore
Alma Burlingame, University of California, San Francisco, CA
Antonio Coutinho, Instituto Gulbenkian de Ciencia, Oeiras, Portugal
John W. Crabb, Cleveland Clinic, Cleveland, OH
Jonathan Crowston, Center for Eye Research, University of Melbourne, Melbourne, Victoria, Australia
Richard Dodel, Philipps University of Marburg, Germany
John Fingert, University of Iowa, Iowa City, IA
Michael A. Hauser, Duke University Medical Center, Durham, NC
Simon John, Jackson Laboratory, Bar Harbor, ME
Inderjeet Kaur, L V Prasad Eye Institute, Hyderabad, India
Keith Martin, Cambridge University, Cambridge, UK
Stephen Miller, Northwestern University Medical School, Chicago, IL
Akhilesh Pandey, Johns Hopkins University, Baltimore, MD
Louis R. Pasquale, Massachusetts Eye and Ear Infirmary, Boston, MA
Margaret Pericak-Vance, University of Miami Miller School of Medicine, Miami, FL
Emanuel Petricoin, George Mason University, Manassas, VA
Norbert Pfeiffer, University Medical Center Mainz, Mainz, Germany
Robert Ritch, New York Eye and Ear Infirmary, New York, NY
Leopold Schmetterer, Medical University of Vienna, Vienna, Austria
Gülgün Tezel, University of Louisville, Louisville, KY
Fotis Topouzis, Aristotle University of Thessaloniki, Thessaloniki, Greece
Ananth Viswanathan, Moorfields Eye Hospital, London, UK
Robert Weinreb, University of California-San Diego, San Diego, CA
Janey L. Wiggs, Massachusetts Eye and Ear Infirmary, Boston, MA
Donald Zack, Wilmer Eye Institute, Johns Hopkins University, Baltimore, MD
Observers
Yong Zhou, Institute for Systems Biology, Seattle, WA
Nobuharu Asai, Nidek Co., Ltd., Gamagori, Aichi, Japan
Katharina Bell, University Medical Center Mainz, Mainz, Germany
Pierre Bitoun, Jean Verdier Hospital, CHU Paris-Nord, Paris, France
Jeffrey Boatright, Emory University School of Medicine, Atlanta, GA
Nils Boehm, University Medical Center Mainz, Mainz, Germany
Mojtaba Bonakdar, Alcon Research Ltd., Fort Worth, TX
Terete Borras, University of North Carolina, Chapel Hill, NC
Jingtai Cao, Regeneron Pharmaceuticals, Inc., Tarrytown, NY
Deborah Carper, National Eye Institute/National Institutes of Health, Bethesda, MD
Hemin Chin, National Eye Institute/National Institutes of Health, Bethesda, MD
Miguel Coca-Prados, Yale University School of Medicine, New Haven, CT
Cun-Jian Dong, Allergan, Inc., Irvine, CA
Douglas Faunce, Pfizer, Inc., Irvine, CA
Michael Fautsch, Mayo Clinic, Rochester, MN
John Flanagan, University of Toronto, Toronto Western Hospital, Toronto, Canada
Haiyan Gong, Boston University School of Medicine, Boston, MA
Hector Gonzalez, Fundacion de Investigacion Oftalmologica, Oviedo, Spain
Ruslan Grishanin, Rinat Laboratories, Pfizer, Inc., San Francisco, CA
Cynthia Grosskreutz, Novartis Institutes for Biomedical Research, Cambridge, MA
John Grunden, Pfizer, Inc., Pittstown, NJ
Samin Hong, Yonsei University College of Medicine, Seoul, Republic of Korea
Donald Hood, Columbia University, New York, NY
Won-Kyu Ju, University of California-San Diego, CA
Anna Junk, University of Miami, Miami, FL
Kui Dong Kang, Catholic University of Korea, Republic of Korea
Mike Karl, Center for Regenerative Therapies Dresden, Germany
Herbert Kaufman, Louisiana State University Eye Center, New Orleans, LA
Paul Kaufman, University of Wisconsin School of Medicine and Public Health, Madison, WI
Peng Khaw, Moorfields Eye Hospital and UCL Institute of Ophthalmology, London, Great Britain
In-Beom Kim, Catholic University of Korea, Republic of Korea
Tomoko Kirihara, Santen Pharmaceutical Co., Ltd., Ikoma, Nara, Japan
Andras Komaromy, University of Pennsylvania, School of Veterinary Medicine, Philadelphia, PA
Christina Kramann, University Medical Center Mainz, Mainz, Germany
Jeffrey Liebmann, New York University School of Medicine, New York, NY
James Lindsey, University of California-San Diego, San Diego, CA
Katrin Lorenz, University Medical Center Mainz, Mainz, Germany
David Mackey, Lions Eye Institute, Tampa, FL
Sabri Markabi, Alcon Research Ltd., Fort Worth, TX
Mallory Mest, Scheie Eye Institute, Philadelphia, PA
Robert Miller, University of Minnesota, Duluth, MN
Sayoko Moroi, University of Michigan-Kellogg Eye Center, Ann Arbor, MI
Joan O'Brien, University of Pennsylvania, Philadelphia, PA
Colm O'Brien, Mater Misericordiae University Hospital, Dublin, Ireland
Toral Parikh, University of Miami, Miami, FL
Sung Chul Park, New York Eye and Ear Infirmary, New York, NY
Jacob Pe'er, Hadassah-Hebrew University Medical Center, Jerusalem, Israel
J. Mark Petrash, University of Colorado-Denver, Denver, CO
Juan Qu, Massachusetts Eye and Ear Infirmary, Boston, MA
Kay Rittenhouse, Pfizer, Inc., San Diego, CA
Steven Roberds, Pfizer Indications Discovery Unit, St. Louis, MO
Naj Sharif, Alcon Research Ltd., Fort Worth, TX
Allan Shepard, Alcon Research Ltd., Fort Worth, TX
Ruifang Sui, Peking Union Medical College Hospital, Peking, China
Carol Toris, University of Nebraska Medical Center, Omaha, NE
Carlo Traverso, University of Genoa, Genoa, Italy
Tracy Valorie, Pfizer, Inc., New York, NY
Roberto Weinmann, Pharmamar USA, New York, NY
Larry Wheeler, Allergan, Inc., Irvine, CA
Andy Whitlock, Lexicon Pharmaceuticals, The Woodlands, TX
Barbara Wirostko, University of Utah, Salt Lake City, UT
Use of molecular markers in the clinical setting to meet these goals will be exciting. However, there remain limitations to be overcome, including lack of common operating procedures for proper banking of biological tissues, analytical insensitivity of underpinning technologies, lack of standards, multiplexed complicated assays, and an ever-changing regulatory landscape.1,2 Proteomics is at the center of a number of these activities, since proteins are either the molecular therapeutic disease target or are the biomarkers used for early disease detection and monitoring.3 Another emergent group of potential biomarkers comprises lipids and other metabolites (Figure), which are important constituents of cell membranes and organelles and are also involved in cell signaling and myriad other biological processes.
However, identification of biomarkers from plasma or serum poses many challenges.2,4 (1) The samples are highly complex, with ranges of protein concentrations often spanning ∼11 orders of magnitude. (2) Biomarkers in biological samples (blood, other fluids, and tissue) often occur in extremely low concentrations, well below the detection limits of most sophisticated mass spectrometry instruments, which are the discovery engines for most proteomic efforts. Marker proteins often also undergo degradation during collection. (3) Modified proteins may be of greater interest than intact proteins, which are frequently lost in unmodified background protein. For example, for type 2 diabetes, it is the proteolytically processed proteins of HbA1c rather than the intact protein that may serve as biomarkers, but these modified proteins are often lost. (4) One of the biggest challenges in proteomic biomarker discovery, in contrast to transcriptomics, is the lack of general means to amplify proteins. The enrichment of peptides of interest prior to analysis using specific antibodies substantially reduces background interference and enables enrichment of low-abundance peptides from large initial volumes with dramatic enhancement of sensitivity. Enrichment has been shown to increase up to 104-fold sensitivity for L-plastin.5 Specific enhancement of N-terminal peptides is another very effective method that has been recently developed. Chemo-enzymatic labeling methods have high selectivity for N-termini and simplify each protein to one or a few peptides. Analogous strategies target other posttranslational modifications (PTMs) such as glycosylation and phosphorylation. Two such approaches have been considered: An additional positive charge (N,N-dimethyl lysine) may be placed at the N-terminus, enhancing detection by electron transfer detection (ETD) analysis; and for N-terminal–associated PTMs (e.g., O-glycosylation, lipid modifications), a modified subtiligase labeling scheme may be used.5 Reverse phase protein array (RPPA)6,7 and biomarker-harvesting nanoparticle technologies8,9 have also been developed to alleviate some of these problems. Nanoparticles composed of a hydrogel core shell, functionalized with internal affinity baits in solution phase in a single step, enable (a) molecular size sieving, (b) exclusion of abundant unwanted proteins, (c) target analyte affinity sequestration, and (d) complete protection of captured analytes from degradation, thereby alleviating the analytical challenges noted above, and facilitate identification by mass spectrometry or other analytical approaches. The nanoparticle-based approach also enables concentration of targeted classes of protein or other analytes sequestered by the particles and renders them stable during transportation and for the time duration prior to and during actual analyses. Technologies such as reverse phase protein array (RPPA), because of its tremendous analytical sensitivity (hundredths of cell equivalents per spot10), provide the opportunity to generate multiplexed pathway biomarker data from even laser capture microdissection (LCM)–procured cells from core needle biopsy samples and minute quantities of vitreous fluid.11 Indeed, a major attraction of the RPPA technology is its unique ability to quantitatively measure the activation state of hundreds of key signaling proteins at once from small samples. Since many of these signaling proteins (e.g., VEGFR) are the direct targets for many molecular targeted therapies, RPPA can be used to rapidly measure if the drug target is expressed and, most importantly, if it is activated and “in use.” Coupling RPPA with molecular profiling endeavors can provide the treating physician with key missing information that could help tailor treatment to a patient's specific molecular portrait.
Proteomics has emerged as a key technology to discover and validate candidate biomarkers in a number of disease areas, such as Alzheimer's disease,12 ovarian cancer,13 cardiovascular diseases,14 age-related macular degeneration,15–17 and ocular surface diseases.18 The discovery process often includes quantitative tandem mass spectrometric analysis of clinical samples that can range from body fluids (e.g., cerebrospinal fluid, serum, tears, aqueous humor, urine, synovial fluid) to tissue samples. For validation, mass spectrometry–based multiple reactions monitoring (MRM) assays are an attractive option to detect and quantify candidate biomarkers in a large number of samples. Eventually candidate biomarker validation platforms may employ different enrichment strategies and development of MRM–mass spectrometry in lieu of generating a suite of antibodies.19,20 Assessment of available evidence on associations between biomarkers and disease states, including data showing effects and interventions on both the biomarker and clinical outcomes, is needed for the further development of glaucoma biomarkers.3,21 Hence, a platform that integrates discoveries in genomics, proteomics, and metabolomics is necessary for integrative and meaningful discovery of biomarkers and their utilization.
Genetic Biomarkers in Glaucoma
Robust genetic biomarkers for open-angle glaucoma (OAG), a leading cause of optic nerve damage–related blindness, were largely lacking until recent times. Genetic linkage studies led to the identification of mutations in MYOC (myocilin), WDR36 (WD repeat domain 36), and OPTN (optineurin) associated with forms of OAG with familial inheritance; however, rare and common variants in these genes are associated with only a small percentage of primary open-angle glaucoma (POAG), an age-related optic neuropathy (which may actually consist of numerous disorders at the molecular and genetic levels) categorized by an excavated and atrophic optic nerve head without evident anterior segment abnormalities.22,23 Recent advances in human genomics transformed the field, yielding a bounty of new and robust genetic biomarkers that may reveal insight into the pathogenesis of OAG. This summary covers genetic biomarkers for OAG that have been confirmed by independent research efforts.
The post-2007 revolution in OAG biomarker discovery is fueled by major advances in our understanding of genomic architecture, coupled with progress in bioinformatics and statistical methods. These advances have revolutionized genetic studies of complex diseases through the use of genome-wide association studies (GWAS). Availability of high-resolution genotyping arrays and new high throughput sequencing make high-resolution genomic analysis possible. Such large genomic studies have already been successfully carried out in myriad late-onset disorders, such as age-related macular degeneration24 and Parkinson's disease.25 The need for large sample sizes, a critical feature of this approach, has led to a number of highly collaborative projects, which systematically store DNA linked to clinical and other information. dbGaP (database of genotypes and phenotypes; http://www.ncbi.nlm.nih.gov/gap/) is one such database that includes phenotype and genotype data.
The first meaningful GWAS in glaucoma was performed for exfoliation syndrome,26 the leading cause of secondary OAG. In ES the intraocular pressure (IOP) tends to be higher than in POAG, and there are also associations with cataract, cataract surgery complications, retinal venous occlusive disease, hearing loss, and other systemic signs. The discovery by Icelandic researchers that common variants in LOXL1 (lysyl oxidase-like 1) are associated with ES has been reproduced in several other populations.27 While the prevalence of ES differs over an order of magnitude in populations studied thus far, variation in allele frequency for disease-associated LOXL1 single nucleotide polymorphisms (SNPs) cannot explain the disparity in the disease burden. Collectively these results suggest that these associated SNPs genetically tag the true functional variants, which have yet to be identified. Causative factors for ES could include altered LOXL1 regulatory mechanisms, other genetic loci, environmental influences, or gene–environment.
The application of methods to identify genetic biomarkers associated with quantitative traits related to the glaucoma phenotype has been a major advance for glaucoma genomics research. Quantitative trait-linked analyses correlating high throughput genotyping data to information on optic nerve features in a large population of European Caucasians showed that common variants in the ATOH7 (atonal homolog 7) genomic region are associated with disc size28 and that common variants in CDKN2B-AS1 (CDKN2B antisense RNA1) and SIX1/6 (SIX1 and SIX6 homeobox) are associated with cup/disc ratio.29 Furthermore, variants in TMCO1 (transmembrane and coiled-coil domains 1) and GAS7 (growth arrest-specific 7) are associated with variation in IOP,30 while polymorphisms in ZN469 (zinc finger protein 469) are associated with central corneal thickness.31 Mutations in the latter gene produce a rare form of corneal disease known as brittle cornea syndrome (BCS). Additional new genetic biomarkers for these quantitative traits are likely to be discovered in the near future.
Work by several groups worldwide has also led to the discovery of genetic biomarkers for POAG. As expected, some of the same biomarkers for glaucoma-related quantitative phenotypic traits are also associated with POAG. For example, Burdon et al. found a strong association between the CDKN2B-AS1 genomic region polymorphisms and POAG,32 an association that was also reported by Fan et al. using candidate genetic markers.33 A meta-analysis of two large studies from the United States—the Glaucoma Genes and Environment (GLAUGEN) study and the National Eye Institute Glaucoma Human Genetics Collaboration (NEIGHBOR) consortium—indicated that polymorphisms in the CDKN2B-AS1 region are also associated with normal pressure glaucoma (NPG), defined as POAG with no recorded IOP measurements above 21 mm Hg.34 The U.S. researchers also discovered a biomarker in an intergenic region on 8q22 that is also associated with NPG. This region contains a deoxyribonuclease I (DNase I) hypersensitivity site with probable regulatory effects that are active in nonpigmented and pigmented ciliary epithelium as well as choroid plexus epithelial cells. The latter cells are responsible for the formation of cerebrospinal fluid (CSF). Emerging evidence suggests that low CSF pressure may act similarly to elevated IOP to create deleterious trans-lamina pressure gradients that interfere with axoplasmic flow in retinal ganglion cells.35,36 The association between CDKN2B-AS1 gene variants and POAG has also been confirmed in Asian37,38 and African-derived populations.39 CDKN2B-AS1 is clearly an important biomarker for POAG, and case-only analyses suggest that it somehow alters optic nerve vulnerability to glaucomatous optic neuropathy.40 Other genetic biomarkers linked to glaucoma-related quantitative traits that are also associated with POAG by European investigators include TMCO1,32 SIX1, and ATOH7.41 Polymorphisms in an intergenic region between CAV1 (caveolin 1) and CAV2 (caveolin 2), initially associated with POAG in an Icelandic cohort,42 were confirmed in the GLAUGEN study.43 These genetic biomarkers showed a stronger association between CAV1/2 SNPs and POAG among women.43 The latter finding is interesting because caveolins, which are expressed in the trabecular meshwork,44 compartmentalize nitric oxide synthase activity in biological membranes.45 Work from the Nurses' Health Study shows interactions between NOS3 (nitric oxide synthase 3) SNPs and postmenopausal hormone use in high pressure glaucoma, defined as POAG with history of IOP >21 mm Hg.46
Overall, the new set of genetic biomarkers for POAG should be viewed as disease-associated risk factors and not causative loci. Future research efforts will include imputation and next-generation sequencing to provide more information about the genetic biomarkers that are tagged by this initial wave of genetic risk factors. Furthermore, as many of these biomarkers are reproduced in populations worldwide, there will be a push to discover their biochemical pathways and molecular significance in POAG pathogenesis. Finally, the recent formation of the International Glaucoma Genetics Consortium will capitalize on the opportunity for large-scale meta-analysis of GWAS results in glaucoma and its relevant quantitative traits, leading to the discovery of new genetic biomarkers.
Future successful clinical trials in OAG will identify patients with biomarkers associated with disease that are validated by virtue of recapitulating the ocular phenotype in animal models. The trial interventions would rescue or prevent further end organ damage in the animal system. For example, topical 4-phenylbutyrate reduced endoplasmic reticulum stress in the trabecular meshwork, lowered IOP, and protected the optic nerve in a murine model of OAG that involved a mutation in the third exon of MYOC.47 This research demonstrates the type of efforts that can be used to inform rational clinical trials in the treatment of OAG. Currently, the genetic landscape for most of the common OAGs is beginning to emerge. Hopefully, filling in the details regarding the complex web of biochemical events that collectively contribute to the OAGs will lead to a series of animal models relevant to the human OAG. The future is bright in terms of developing a whole new family of therapeutic interventions for the treatment of OAG.
Proteomic Biomarkers in Glaucoma
The term “normal-tension glaucoma” (NTG) or normal pressure glaucoma suggests the existence of risk factors other than or in addition to IOP.48 These risk factors can be present at any level of IOP; but when glaucoma develops with IOP in the normal range, they bear a proportionately greater etiologic responsibility for the resulting damage. Whether genetic, vascular, aging-oxidative, or immune-inflammatory, these risk factors are themselves, in turn, distinct disorders. It is also probable that multiple risk factors increase the chances of developing glaucoma and perhaps its severity and other phenotypic characteristics. Thus, our concept of biomarkers in this situation comprises information gained from blood (or other tissue) studies shedding light on measurable phenomena, whether genetic, immune, inflammatory, or otherwise biochemically distinct or present in abnormal amounts. So far, several genes and potential biomarkers have been discovered that are relevant to NTG.49–51
Biomarkers for either NTG or for the broad classifications of glaucoma, open- or closed-angle glaucoma, may originate from proteomic, imaging, genomics, or a combination of these studies. Proteomic or metabolomic biomarkers may be obtained from tissues or fluids, either from the eye or systemically. However, it is clear from the search for cancer biomarkers that the more locally obtained from the disease site, the greater the opportunity to discover biomarkers with high clinical significance. Recent work has shown the usefulness of the tears for discovering putative biomarkers for the inflammation that develops in patients on chronic glaucoma medication.52
Several genes and biomarkers have been discovered relevant to NTG,49–51 and discovery-based proteomics can yield molecular insights into disease mechanisms and identify multiple biomarker candidates. For glaucoma biomarkers, global quantitative proteomic analyses of ocular tissues involved in the early stages of the disease appear promising for identifying biomarker candidates. Subsequently, candidate biomarkers considered high priority can be targeted for validation in blood and ocular fluids from larger glaucoma/control study populations using SRM/MRM (select reaction monitoring/multiple reaction monitoring)–based technology. Promising candidate biomarkers can be chosen for clinical multi-site clinical trials using the MRM technique. This method would provide quantitative population-level values that can be used clinically for diagnosis or follow-up to treatment. However, these biomarkers should also represent some biological aspects for the disease so that each patient's condition can be understood on an individual basis. Recent proteomic studies of trabecular meshwork, retina, retinal ganglion cells, aqueous humor, and sera from glaucomatous and control group identified a number of proteins that show significant up- or downregulation.53–58 Although there are several methods for biomarker identification, such as protein arrays (both tissue and antibody based), these usually have limitations as to the number of entities that can be quantified, and the targets must be chosen beforehand. Thus, mass spectrometry (MS) is a state-of-the-art approach for proteomic biomarker discovery or metabolomic biomarkers; as with gene array technology, it is essentially unbiased and allows the greatest number of significant entities to be identified. Additionally, the high sensitivity and throughput of MS technology are important in trials with potentially hundreds of samples. Statistical validation of potential biomarkers is of course essential to establish effective prognostic tools. Study design is critical, as proteomics/lipidomics/metabolomics may often involve levels of measurements just as in urine, blood, local tissue, or local fluids (for example, aqueous humor) and thus the statistical power analysis.
Very promising, cutting-edge targeted SRM/MRM technology has the capacity to monitor hundreds of target peptides in a single 2-hour LC tandem mass spectrometry (MS/MS) run, and also the sensitivity to detect and quantify proteins at μg/mL level in blood. In addition, label-free58 and isobaric tags for relative and absolute quantitation (iTRAQ)-labeled56,57 quantitative proteomic analyses of trabecular meshwork, retina, and retinal ganglion cells from glaucomatous and control group result in identification of a number of proteins that exhibit up- or downregulation. Excitingly, the recently developed data-independent SWATH acquisition is believed to hold a capability “to systematically query sample sets for the presence and quantity of essentially any protein of interest.”58,59 On the other hand, front-end sample preparation strategies, such as immuno-affinity depletion, glycopeptide enrichment, and isoelectric focusing, can significantly reduce the proteomic complexity of specimens and enrich low-abundant targets,60 as recently demonstrated in biomarker discovery for brain diseases and hepatitis C virus–induced cirrhosis.61
Ion mobility instruments also offer high sensitivity and can reduce false positives. Prospects for automated shotgun lipidomics (identification and relative quantification) have emerged owing to software development.62–64 Dynamic time point quantitative data sets explored in a directed manner from static identified data sets will enable building protein–protein and protein–lipid dynamic interaction and their occupancy information.
Another discussion topic was whether the concept of predictive model building can be imported from other fields for biomarker-based predictions. A number of approaches have been used in recent years to predict failure in the field of finance.65 Some of these simulations have remarkable similarity with a dynamic interacting network of proteins and other biomolecules in biological systems. It remains open how recent developments in MS should be applied toward discovering glaucoma biomarkers that include both proteins and lipids. Identified proteins, lipids, and other metabolites can be tested for dynamic changes during the progression of glaucoma. Development of noninvasive methods will aid in these strategies. Significant developments are needed for the determination of dynamic interactions in a high throughput manner. The predictive models encompassing dynamic protein–protein and protein–lipid interaction data sets resulting in prediction of deleterious and advantageous interactions is currently far from reality. However, significant progress is likely to occur, and the predictive models used in other spheres such as finance will be helpful in building predictive models for dynamic biomolecular interaction data.
Finally, biomarkers have great value in the diagnosis of disease as well as in the understanding of an individual patient's response to treatment. The initial discovery stage for a putative biomarker can make use of a small but very carefully defined patient group. Larger studies and methods are now available to provide quantitative population parameters that can be used in conjunction with imaging or molecular genetics. Validated biomarkers that emerge from these studies can be used for drug development to lower costs and improve outcomes. It should be noted that the final clinical implementation could use various forms of MS, which in the case of a few samples would have a very rapid turnaround potentially while the patient is waiting.
Pre-Immune and Immune Events: Immunoproteomics and Its Possible Applications in Glaucoma
Several clinical and experimental studies point toward possible autoimmune involvement in the pathogenesis of glaucoma.66–69 Moreover, there is strong evidence that many patients undergo changes in their natural autoimmunity before and/or during the disease process.70–72 On account of this, one main focus in this session was the potential of natural autoantibodies. These are effectors of the innate immune system and can be considered regulatory factors,73 potentially able to modulate the activity of target molecules and influence their physiological functions. Further, natural autoantibodies may participate in a variety of physiological activities, from immune regulation, homeostasis, and repertoire selection to resistance to infections, transport, and functional modulation of biologically active molecules (reviewed in Coutinho et al.74). A relatively large fraction of serum immunoglobulins in healthy individuals are naturally occurring autoantibodies, so that complex profiles exist in healthy people.75,76 These specific profiles are consistent across populations and form the very stable so-called immunological homunculus.77 Hence, it can be important to investigate disease-specific changes in the complex profiles of naturally occurring autoantibodies as diagnostic markers, as elevated autoantibodies are found in several autoimmune diseases—for example, in patients suffering from systemic lupus erythematosus, primary antiphospholipid syndrome,78 primary Sjögren syndrome,79 and myasthenia gravis, which is an autoimmune response against the acetylcholine receptor.80,81
A controversial question that arose during discussions was whether autoantibodies are aberrant and contribute to disease pathogenesis or are beneficial, forming part of a protective mechanism. In the case of Alzheimer's disease (AD), data from animal experiments and clinical trials argue that autoantibodies may be part of a protective mechanism in innate immunity.82,83 Thus, naturally occurring antibodies could potentially be used for therapeutic purposes. Importantly, the application of intravenous immunoglobulin (IVIg), which has been proposed as a potential agent for AD immunotherapy,82–84 could improve cognitive function, and large-scale randomized trials are under way.85 However, the high coincidence of AD and glaucoma,86,87 and the fact that the targeting of different components of the Aβ formation and aggregation pathway can effectively reduce glaucomatous retinal ganglion cell (RGC) apoptosis in vivo,88 point to a possible IVIg treatment against glaucoma.
The enormous complexity of antibodies makes research in this field challenging, but studies on IgG antibody profiles have revealed specific antibody repertoires against ocular tissue in sera and aqueous humor of patients with NTG or POAG.68,89 These complex antibody profiles are very stable and exist among different study populations from Germany and the United States (Grus FH, et al. IOVS 2005;46:ARVO E-Abstract 1285).71 The antibody profiles could be validated with specific antigen microarrays,90 which are a promising approach. Importantly, not only could increased antibody reactivities in the glaucoma groups be compared to controls, but downregulated immunoreactivities could also be observed. Furthermore, the use of an artificial neural network in combination with a unique serum autoantibody pattern on prospective sera enabled the detection of glaucoma with a specificity and sensitivity of approximately 93%.90
To answer the question whether elevated antibody reactions can induce RGC loss in animal models, an experimental autoimmune glaucoma animal (EAG) model has been developed. For this purpose, Lewis rats were systemically immunized with biomarkers coming from clinical studies. The animals developed antibodies against the proteins, for example HSP60 (heat shock protein 60)91–93 or MBP (myelin basic protein),94 and elevated antibody reactions caused a distinct RGC loss in the retina. The immunization with MBP not only causes neuron loss in the RGC layer, but also triggers antibody reactivity against ocular tissue (Gramlich OW, et al. IOVS 2011;52:ARVO E-Abstract 292894). In addition, cell culture experiments showed that antibodies in sera from glaucoma patients can directly and specifically interact with RGC and influence their protein expression, leading to an additive effect when the RGCs are exposed to elevated pressure (Bell K, et al. IOVS 2011;52:ARVO E-Abstract 4598; Bell K, et al. IOVS 2010;51:ARVO E-Abstract 316995). Furthermore, studies suggest that autoantibodies can also be protective; antibodies that are downregulated in POAG patients were applied to RGC in vitro and increased their viability under stress conditions (Wilding C, et al. IOVS 2011;52:ARVO E-Abstract 3939). This suggests that the absence or the loss of some autoantibodies (e.g., anti-GFAP, anti-glial fibrillary acidic protein) possibly reflects a loss of natural protective autoimmunity, thus encouraging neurodegenerative processes.
In spite of these results, it is still unclear whether the changes found in autoantibody profiles are pathogenic or merely a consequence of glaucomatous optic neuropathy. Even if glaucoma is not considered a classic autoimmune disease, these changes in antibody profiles might be used as a screening test; the analysis of antibody profiles could be useful to detect glaucoma at an early stage, and they might be useful biomarkers in intervention trials to prevent manifestation before clinical signs appear. In many other diseases, antibody titers develop years before the clinical onset of the disease.96,97 This might also be the case in glaucoma, as the severity of changes in antibody patterns correlates with severity of the disease and with clinical Heidelberg retinal tomograph (HRT) parameters. Thus, these biomarkers could be used for diagnosis, prognosis, new treatment options, and personalized treatment options or for risk determination.
A final conclusion from this discussion was that there is a clear need for longitudinal studies that allow observation of the progression of glaucoma and permit the staging of the disease. Additionally, the effects of different treatments and the influence of IOP on antibody patterns over time are of great interest to gain deeper insights in the disease pathogenesis. Further open to question is the role of antigenic shifts, for example posttranslational modifications or oxidations, and their effects on cellular level.
Another focus of this session was on discussion of the role of mitochondrial dysfunction and aging in glaucoma. Epidemiological studies have shown that glaucoma prevalence increases quite markedly with age across all major populations,98 but the increase in prevalence of glaucoma is not entirely explained by increase in ocular hypertension. One interpretation is that in older people the non-IOP factors might become more and more important or that in older patients the optic nerve might become more vulnerable to any given insult that increases the susceptibility to glaucoma neurodegeneration with age. The mechanism of how aging relates to glaucoma pathogenesis may be multifactorial—for example, mechanical changes with age, such as changes in aqueous dynamics or lamina cribrosa and extracellular matrix compliance,99,100 or age-related changes in vascular autoregulation at the retina.101 In addition, accumulating evidence suggests that age-related mitochondrial dysfunction may play a central role in glaucoma, as aging mitochondria are associated with increased oxidative stress (reviewed in Kong et al.102).103 Any malfunction of the mitochondrial electron transport chain results in an excessive generation of free radicals, and this state of oxidative stress results in neural degeneration (reviewed in Tezel et al.104). While aging, the mitochondria acquire mutations within their genome, the mitochondrial DNA (mtDNA), and this might impact mitochondrial function; for example, reduction of oxidative phosphorylation (OXPHOS) complex function increases with age in muscle biopsies.105 Pilot data point to increased mtDNA mutations.106,107 Data from POAG lymphoblasts also show decreased complex I–linked adenosine triphosphate (ATP) production,108 and proteomics has revealed significant quantitative changes in mitochondrial proteins in normal and glaucomatous trabecular meshwork cells treated with TGFβ2 or dexamethasone.54,56 These results imply that mitochondrial dysfunction may represent risk factors for POAG.
How does mitochondrial dysfunction have an impact on optic nerve response to injury? This question was addressed through assessment of retinal neural vulnerability in a mouse model that accumulates high levels of mtDNA mutations owing to impairments in mitochondrial polymerase gamma (PolG) proofreading function in neuronal tissue.109,110 The increase in mtDNA mutations rendered retinal ganglion cells more vulnerable to IOP elevation,110 an insult known to produce mechanical, metabolic, and oxidative stress in the retina. Regarding the systemic deficiency in mitochondria, one would expect that glaucoma is associated with other diseases that involve oxidative stress and neurological deficits. Indeed, when we look at some aspects of hearing, a significant proportion of subjects with OAG have been found to have auditory dysfunction with mild defects in temporal processing of sound. This may point to the presence of a wider systemic neuronal vulnerability.111 This does not suggest that all glaucoma patients have mitochondrial dysfunction, but there may be some subsets of glaucoma patients who have more vulnerable nerves because of acquired mitochondrial DNA abnormalities. As a conclusion, biomarkers of oxidative stress and mitochondrial function could help to identify particular risk patients with low IOP, without the need to wait for diagnosis until the disease already has progressed.
Another relevant discussion topic was the role of oxidative stress in immune response pathways, including complement activation. Oxidative stress may be directly neurotoxic, but it may also induce secondary events with neurodestructive potential, such as stimulation of immune activity or glial dysfunction—for example, the stimulation of glial cytokine production and their antigen-presenting ability. Another oxidative stress–related event, the generation of advanced glycation end products (AGE), may also be immunostimulatory through a specific receptor (RAGE)–mediated signaling. Furthermore, oxidative protein modifications may alter antigenic features of proteins and induce autoantibody production. It is possible that oxidative stress may have an influence on several complement regulatory molecules, for example complement factor H (CFH).112 It may activate toll-like receptor (TLR) signaling113 and stimulate innate and adaptive immune responses (reviewed in Tezel et al.104, Tezel et al.114). Analyses of isolated astrocytes have resulted in the detection of prominent immunoregulator activation, which includes tumor necrosis factor-alpha (TNF-α), NF-κB–signaling, several TLR-related signaling molecules, Janus kinase (JAK) and signal transducer and activator of transcription (STAT)-signaling pathway, and also inflammasome formation. Although there are multiple lines of evidence supporting neurodegenerative potential of uncontrolled immune activity, the pathogenic importance of immune activation or dysregulation in glaucoma remains an open question.
What are the implications of oxidative stress for biomarker discovery? Serum samples from glaucomatous patients exhibited a significant increase in protein carbonyls (Tong MG, et al. IOVS 2011;52:ARVO E-Abstract 2431), which reflect oxidatively modified proteins, and the immunoreactivity of the glaucomatous patient sera against oxidatively stressed retinal cell culture proteins was augmented. Proteomic analyses validated that many of the immunoreactivities detected in glaucomatous serum samples against diseased retinal proteins corresponded to oxidized proteins identified in retina and serum samples (Luo C, et al. IOVS 2010;51:ARVO E-Abstract 2670). This suggests that by changing antigenic features of proteins, the oxidative modifications may stimulate autoantibody production and may serve as biomarkers. In this regard, evidence is growing that supports select oxidative modifications in plasma as biomarkers for age-related macular degeneration.15,16 In the case of glaucoma, oxidative modifications and oxidized serum proteins certainly hold promise as possible biomarkers but need further evaluation.
An interesting aspect of discussion was on the role of epitope spreading in T-cell–mediated autoimmune diseases, a mechanism that may contribute to RGC loss in glaucoma. A typical immune response against a self- or foreign protein is usually focused on one or two epitopes within that protein. Epitope spreading is defined as the diversification of epitope specificity and can occur intra- or intermolecularly, regardless of the initial antigenic stimulus. The specificity of the immune response can spread to include self-epitopes other than the one that initiated the inflammatory process. Specifically in the context of chronic tissue damage that releases endogenous antigens, epitope spreading can lead to autoimmunity (reviewed in Vanderlugt et al.115). If glaucoma is a chronic neurodegenerative disease, the release of self-antigens could lead to autoantibody production and epitope spreading. An interesting speculation was that posttranscriptional protein modifications of the target molecules could be a fundamental driver of the epitope spreading. This could certainly be the case, but in the context of epitope spreading in the experimental autoimmune encephalomyelitis (EAE) model, the T-cell epitopes are not influenced by posttranslational modifications. Thus, changing the folding and structure of a protein may also have an influence on antibody responses. In conclusion, autoimmune diseases are moving targets due to epitope spreading, and a tolerance-based immunotherapy can be used to treat ongoing autoimmune disease; but one needs to know what antigen to target, as tolerance is induced only by the spread epitope.
From Bench to Bedside: How Can a Translational Approach Be Successful?
In this session, the focus was on application of biomarkers to clinical trials, diagnosis, and treatment.
According to the FDA (U.S. Drug and Food Administration), a biomarker is defined as an anatomic, physiologic, biochemical, or molecular parameter associated with the presence and severity of specific disease states. A biomarker may be detectable and measurable by a variety of methods, including physical examination, laboratory assays, and medical imaging. Very different from a biomarker is a surrogate endpoint, which is defined as a laboratory measurement or physical sign that is used in therapeutic trials as a substitute for a clinically meaningful endpoint, which in turn is a direct measure of how a patient feels, functions, or survives and is expected to predict the effects of the therapy. Generally, a biomarker is a “candidate” surrogate marker, and for validation it has to be demonstrated that it fully captures net effects of treatment on clinical outcome, using an established scientific framework or body of evidence that elucidates the physiologic, toxicologic, pharmacologic, or clinical significance of the test results. But what is the present gold standard for glaucoma diagnosis with which new biomarkers should be compared, and what clinical study is needed to validate biomarkers for glaucoma? To facilitate the validation of biomarkers, statisticians must be included early in the process of development, as well as methods for pharmacokinetics, pharmacodynamics, and disease modeling. Furthermore, standardized schedules of efficacy and toxicity measurements, as well as multiple measurements of biomarkers at baseline, are required. In terms of treatment and outcome of clinical trials, a provocative stress test would be helpful—for example, raise IOP and quickly measure the changing parameters.
With regard to the variety of “glaucomas” with their numerous requirements and questions that have to be answered, different biomarkers are needed; for example, diagnostic biomarkers are demanded that indicate either a very high risk of onset or very early disease and have a very high specificity to avoid false-positive results. Additionally, a biomarker that indicates progression, the speed of RGC loss, or the number of remaining/dying RGC with a very high sensitivity would be very useful.
Once established, biomarkers could aid clinical trials of potential neuroprotective therapies with patient selection—for example, to identify who is most likely to benefit from a treatment or which patients progress most rapidly, to enrich the study population and have better chances to see any treatment effect. Such enrichment may be criticized as selective, and while it could make clinical trials faster and easier, it may also remove important results. However, the most relevant issue that a biomarker could help with would be in determining treatment effect and detecting changes earlier. Though there is no ideal measurement instrument to evaluate glaucoma progression, recently there has been a clear improvement in detection of structural and functional changes. In vivo live imaging with biomarkers permits identification of living, dying, or injured RGC; examples include in vivo imaging of RGC loss via counts of fluorescently labeled RGC116 and imaging axonal transport defects that are characterized by axonal swellings in the living eye (Keith Martin, unpublished data, 2011). Moreover, there is the potential to label injured cells. For example, Annexin 5, an early marker of apoptosis, has been combined with a noninvasive real-time imaging technique, using confocal laser-scanning ophthalmoscopy to visualize single nerve cell apoptosis in vivo, which allows longitudinal study of disease processes.117 Notably, an adaptive clinical trial design that is based on biomarkers could improve clinical research, as demonstrated for the BATTLE (biomarker-integrated approaches of targeted therapy for lung cancer elimination) study, which is a phase 2 trial that has demonstrated the feasibility of identifying groups of patients who are more likely to benefit from a specific agent.118,119 At the same time, patients who should not receive particular drugs could also be identified. The adaptive designs, which incorporated information gathered earlier in the trial to guide treatment for newer patients, could represent an innovative approach for neuroprotection in clinical trials. They allow predefined changes in the study designs affecting sample size, treatment arms, or stopping-early rules, based on interim analysis or by accommodating data from other studies.
Finally, there is probably no single “ideal” glaucoma biomarker that is going to cover all aspects of clinical disease including early detection, severity prediction, progression, and response to treatment. In consequence, we will have to decide which are the most useful and how they can be combined. With regard to new treatments, efforts should be put toward those biomarkers that predict responses after treatment, making clinical trials shorter and cheaper and necessitating fewer patients. Perhaps biomarkers that can be imaged directly in the eye are the best hope. The question is how to integrate biomarkers in the overall design of clinical studies. And how can they be used together with the structural and functional tests that we already have? These investigations will require translational research involving collaborations between basic scientists and clinicians.
Acknowledgments
We thank Roger Beuerman for the contribution of the figure (flow diagram explaining the various “omics” relationships) and Nadine von Thun und Hohenstein-Blaul for assistance in writing this manuscript.
Footnotes
Supported by the ARVO Foundation for Eye Research through a grant from Pfizer Ophthalmics, National Eye Institute (RKL), American Glaucoma Society (RKL), and American Health Assistance Foundation (RKL).
Disclosure: S.K. Bhattacharya, None; R.K. Lee, None; F.H. Grus, None
Participants in the Seventh ARVO/Pfizer Ophthalmics Research Institute Conference Working Group are listed on page 122.
References
- 1. Liotta LA, Ferrari M, Petricoin E. Clinical proteomics: written in blood. Nature. 2003; 425: 905 [DOI] [PubMed] [Google Scholar]
- 2. Liotta LA, Petricoin E. Cancer biomarkers: closer to delivering on their promise. Cancer Cell. 2011; 20: 279–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liotta LA, Kohn EC, Petricoin EF. Clinical proteomics: personalized molecular medicine. JAMA. 2001; 286: 2211–2214 [DOI] [PubMed] [Google Scholar]
- 4. Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002; 1: 845–867 [DOI] [PubMed] [Google Scholar]
- 5. Wildes D, Wells JA. Sampling the N-terminal proteome of human blood. Proc Natl Acad Sci U S A. 2010; 107: 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wulfkuhle JD, Speer R, Pierobon M, et al. Multiplexed cell signaling analysis of human breast cancer applications for personalized therapy. J Proteome Res. 2008; 7: 1508–1517 [DOI] [PubMed] [Google Scholar]
- 7. Paweletz CP, Charboneau L, Bichsel VE, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001; 20: 1981–1989 [DOI] [PubMed] [Google Scholar]
- 8. Tamburro D, Fredolini C, Espina V, et al. Multifunctional core-shell nanoparticles: discovery of previously invisible biomarkers. J Am Chem Soc. 2011; 133: 19178–19188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luchini A, Fredolini C, Espina BH, et al. Nanoparticle technology: addressing the fundamental roadblocks to protein biomarker discovery. Curr Mol Med. 2010; 10: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rapkiewicz A, Espina V, Zujewski JA, et al. The needle in the haystack: application of breast fine-needle aspirate samples to quantitative protein microarray technology. Cancer. 2007; 111: 173–184 [DOI] [PubMed] [Google Scholar]
- 11. Davuluri G, Espina V, Petricoin EF III, et al. Activated VEGF receptor shed into the vitreous in eyes with wet AMD: a new class of biomarkers in the vitreous with potential for predicting the treatment timing and monitoring response. Arch Ophthalmol. 2009; 127: 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer's disease. Lancet. 2011; 377: 1019–1031 [DOI] [PubMed] [Google Scholar]
- 13. Callesen AK, Mogensen O, Jensen AK, et al. Reproducibility of mass spectrometry based protein profiles for diagnosis of ovarian cancer across clinical studies: a systematic review. J Proteomics. 2012; 75: 2758–2772 [DOI] [PubMed] [Google Scholar]
- 14. Stastna M, Van Eyk JE. Secreted proteins as a fundamental source for biomarker discovery. Proteomics. 2012; 12: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ni J, Yuan X, Gu J, et al. Plasma protein pentosidine and carboxymethyllysine, biomarkers for age-related macular degeneration. Mol Cell Proteomics. 2009; 8: 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gu J, Pauer GJ, Yue X, et al. Assessing susceptibility to age-related macular degeneration with proteomic and genomic biomarkers. Mol Cell Proteomics. 2009; 8: 1338–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuan X, Gu X, Crabb JS, et al. Quantitative proteomics: comparison of the macular Bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol Cell Proteomics. 2010; 9: 1031–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou L, Beuerman RW. Tear analysis in ocular surface diseases. Prog Retin Eye Res. 2012; 31: 527–550 [DOI] [PubMed] [Google Scholar]
- 19. Prakash A, Rezai T, Krastins B, et al. Interlaboratory reproducibility of selective reaction monitoring assays using multiple upfront analyte enrichment strategies. J Proteome Res. 2012; 11: 3986–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whiteaker JR, Zhao L, Anderson L, Paulovich AG. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol Cell Proteomics. 2010; 9: 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knepper PA, Samples JR, Yue BYJT. Biomarkers of primary open-angle glaucoma. Expert Review of Ophthalmology. 2010; 5: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allingham RR, Wiggs JL, Hauser ER, et al. Early adult-onset POAG linked to 15q11-13 using ordered subset analysis. Invest Ophthalmol Vis Sci. 2005; 46: 2002–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramdas WD, van Koolwijk LM, Cree AJ, et al. Clinical implications of old and new genes for open-angle glaucoma. Ophthalmology. 2011; 118: 2389–2397 [DOI] [PubMed] [Google Scholar]
- 24. Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005; 308: 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009; 41: 1308–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thorleifsson G, Magnusson KP, Sulem P, et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007; 317: 1397–1400 [DOI] [PubMed] [Google Scholar]
- 27. Challa P. Genetics of adult glaucoma. International Ophthalmology Clinics. 2011; 51: 37–51 [DOI] [PubMed] [Google Scholar]
- 28. Macgregor S, Hewitt AW, Hysi PG, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010; 19: 2716–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramdas WD, van Koolwijk LM, Ikram MK, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010; 6: e1000978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Koolwijk LM, Ramdas WD, Ikram MK, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012; 8: e1002611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu Y, Dimasi DP, Hysi PG, et al. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010; 6: e1000947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011; 43: 574–578 [DOI] [PubMed] [Google Scholar]
- 33. Fan BJ, Wang DY, Pasquale LR, Haines JL, Wiggs JL. Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest Ophthalmol Vis Sci. 2011; 52: 1788–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wiggs J, Yaspan B, Hauser M, et al. Common variants in the CDKN2BAS region on 9p21 and a novel region on 8q23 are associated with the normal tension form of open-angle glaucoma with increased susceptibility to optic nerve degeneration. PLoS Genet. 2012. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berdahl JP, Allingham R, Johnson DH. Cerebrospinal fluid pressure in primary open angle glaucoma. Ophthalmology. 2008; 115: 763–768 [DOI] [PubMed] [Google Scholar]
- 36. Ren R, Wang N, Zhang X, Cui T, Jonas JB. Trans-lamina cribrosa pressure difference correlated with neuroretinal rim area in glaucoma. Graefes Arch Clin Exp Ophthalmol. 2011; 249: 1057–1063 [DOI] [PubMed] [Google Scholar]
- 37. Nakano M, Ikeda Y, Tokuda Y, et al. Common variants in CDKN2B-AS1 associated with optic nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS One. 2012; 7: e33389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Osman W, Low SK, Takahashi A, Kubo M, Nakamura Y. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum Mol Genet. 2012; 21: 2836–2842 [DOI] [PubMed] [Google Scholar]
- 39. Cao D, Jiao X, Liu X, et al. CDKN2B polymorphism is associated with primary open-angle glaucoma (POAG) in the Afro-Caribbean population of Barbados, West Indies. PLoS One. 2012; 7: e39278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burdon KP, Crawford A, Casson RJ, et al. Glaucoma risk alleles at CDKN2B-AS1 are associated with lower intraocular pressure, normal-tension glaucoma, and advanced glaucoma. Ophthalmology. 2012; 119: 1539–1545 [DOI] [PubMed] [Google Scholar]
- 41. Ramdas WD, van Koolwijk LM, Lemij HG, et al. Common genetic variants associated with open-angle glaucoma. Hum Mol Genet. 2011; 20: 2464–2471 [DOI] [PubMed] [Google Scholar]
- 42. Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010; 42: 906–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wiggs JL, Hee Kang J, Yaspan BL, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011; 20: 4707–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Surgucheva I, Surguchov A. Expression of caveolin in trabecular meshwork cells and its possible implication in pathogenesis of primary open angle glaucoma. Mol Vis. 2011; 17: 2878–2888 [PMC free article] [PubMed] [Google Scholar]
- 45. Williams TM, Lisanti MP. The Caveolin genes: from cell biology to medicine. Ann Med. 2004; 36: 584–595 [DOI] [PubMed] [Google Scholar]
- 46. Kang JH, Wiggs JL, Rosner BA, et al. Endothelial nitric oxide synthase gene variants and primary open-angle glaucoma: interactions with gender and postmenopausal hormone use. Invest Ophthalmol Vis Sci. 2010; 51: 971–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zode GS, Bugge KE, Mohan K, et al. Topical ocular sodium 4-phenylbutyrate rescues glaucoma in a myocilin mouse model of primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2012; 53: 1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iwase A, Suzuki Y, Araie M, et al. The prevalence of primary open-angle glaucoma in Japanese: the Tajimi Study. Ophthalmology. 2004; 111: 1641–1648 [DOI] [PubMed] [Google Scholar]
- 49. Leibovitch I, Kurtz S, Kesler A, Feithliher N, Shemesh G, Sela BA. C-reactive protein levels in normal tension glaucoma. J Glaucoma. 2005; 14: 384–386 [DOI] [PubMed] [Google Scholar]
- 50. Sugiyama T, Moriya S, Oku H, Azuma I. Association of endothelin-1 with normal tension glaucoma: clinical and fundamental studies. Surv Ophthalmol. 1995; 39 (suppl 1): S49–56 [DOI] [PubMed] [Google Scholar]
- 51. Tatton W, Chen D, Chalmers-Redman R, Wheeler L, Nixon R, Tatton N. Hypothesis for a common basis for neuroprotection in glaucoma and Alzheimer's disease: anti-apoptosis by alpha-2-adrenergic receptor activation. Surv Ophthalmol. 2003; 48 (suppl 1): S25–37 [DOI] [PubMed] [Google Scholar]
- 52. Wong TT, Zhou L, Li J, et al. Proteomic profiling of inflammatory signaling molecules in the tears of patients on chronic glaucoma medication. Invest Ophthalmol Vis Sci. 2011; 52: 7385–7391 [DOI] [PubMed] [Google Scholar]
- 53. Boehm N, Wolters D, Thiel U, et al. New insights into autoantibody profiles from immune privileged sites in the eye: a glaucoma study. Brain Behav Immun. 2012; 26: 96–102 [DOI] [PubMed] [Google Scholar]
- 54. Bollinger KE, Crabb JS, Yuan X, Putliwala T, Clark AF, Crabb JW. Quantitative proteomics: TGFβ2 signaling in trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2011; 52: 8287–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Crabb JW, Yuan X, Dvoriantchikova G, Ivanov D, Crabb JS, Shestopalov VI. Preliminary quantitative proteomic characterization of glaucomatous rat retinal ganglion cells. Exp Eye Res. 2010; 91: 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bollinger K, Crabb J, Yuan X, Putliwala T, Clark A, Crabb J. Proteomic similarities in steroid responsiveness in normal and glaucomatous trabecular meshwork cells. Mol Vision. 2012. In press [PMC free article] [PubMed] [Google Scholar]
- 57. Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004; 3: 1154–1169 [DOI] [PubMed] [Google Scholar]
- 58. Ishihama Y, Oda Y, Tabata T, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005; 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- 59. Gillet LC, Navarro P, Tate S, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012; 11:O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007; 6: 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qin S, Zhou Y, Lok AS, et al. SRM targeted proteomics in search for biomarkers of HCV-induced progression of fibrosis to cirrhosis in HALT-C patients. Proteomics. 2012; 12: 1244–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009; 81: 4356–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schuhmann K, Herzog R, Schwudke D, Metelmann-Strupat W, Bornstein SR, Shevchenko A. Bottom-up shotgun lipidomics by higher energy collisional dissociation on LTQ Orbitrap mass spectrometers. Anal Chem. 2011; 83: 5480–5487 [DOI] [PubMed] [Google Scholar]
- 64. Herzog R, Schwudke D, Schuhmann K, et al. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011; 12: R8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haldane AG, May RM. Systemic risk in banking ecosystems. Nature. 2011; 469: 351–355 [DOI] [PubMed] [Google Scholar]
- 66. Wax MB. Is there a role for the immune system in glaucomatous optic neuropathy? Curr Opin Ophthalmol. 2000; 11: 145–150 [DOI] [PubMed] [Google Scholar]
- 67. Tezel G, Seigel GM, Wax MB. Autoantibodies to small heat shock proteins in glaucoma. Invest Ophthalmol Vis Sci. 1998; 39: 2277–2287 [PubMed] [Google Scholar]
- 68. Grus FH, Joachim SC, Hoffmann EM, Pfeiffer N. Complex autoantibody repertoires in patients with glaucoma. Mol Vis. 2004; 10: 132–137 [PubMed] [Google Scholar]
- 69. Tezel G, Wax MB. Glaucoma. Chem Immunol Allergy. 2007; 92: 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grus FH, Joachim SC, Wuenschig D, Rieck J. Autoimmunity and glaucoma. J Glaucoma. 2008; 17: 79–84 [DOI] [PubMed] [Google Scholar]
- 71. Grus FH, Joachim SC, Bruns K, Lackner KJ, Pfeiffer N, Wax MB. Serum autoantibodies to alpha-fodrin are present in glaucoma patients from Germany and the United States. Invest Ophthalmol Vis Sci. 2006; 47: 968–976 [DOI] [PubMed] [Google Scholar]
- 72. Joachim SC, Pfeiffer N, Grus FH. Autoantibodies in patients with glaucoma: a comparison of IgG serum antibodies against retinal, optic nerve, and optic nerve head antigens. Graefes Arch Clin Exp Ophthalmol. 2005; 243: 817–823 [DOI] [PubMed] [Google Scholar]
- 73. Poletaev A, Osipenko L. General network of natural autoantibodies as immunological homunculus (Immunculus). Autoimmun Rev. 2003; 2: 264–271 [DOI] [PubMed] [Google Scholar]
- 74. Coutinho A, Kazatchkine MD, Avrameas S. Natural autoantibodies. Curr Opin Immunol. 1995; 7: 812–818 [DOI] [PubMed] [Google Scholar]
- 75. Li WH, Zhao J, Li HY, et al. Proteomics-based identification of autoantibodies in the sera of healthy Chinese individuals from Beijing. Proteomics. 2006; 6: 4781–4789 [DOI] [PubMed] [Google Scholar]
- 76. Avrameas S. Natural autoantibodies: from “horror autotoxicus” to “gnothi seauton.” Immunol Today. 1991; 12: 154–159 [DOI] [PubMed] [Google Scholar]
- 77. Nobrega A, Haury M, Grandien A, Malanchere E, Sundblad A, Coutinho A. Global analysis of antibody repertoires. II. Evidence for specificity, self-selection and the immunological “homunculus” of antibodies in normal serum. Eur J Immunol. 1993; 23: 2851–2859 [DOI] [PubMed] [Google Scholar]
- 78. George J, Gilburd B, Shoenfeld Y. The emerging concept of pathogenic natural autoantibodies. Hum Antibodies. 1997; 8: 70–75 [PubMed] [Google Scholar]
- 79. Terzoglou AG, Routsias JG, Avrameas S, Moutsopoulos HM, Tzioufas AG. Preferential recognition of the phosphorylated major linear B-cell epitope of La/SSB 349-368 aa by anti-La/SSB autoantibodies from patients with systemic autoimmune diseases. Clin Exp Immunol. 2006; 144: 432–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Graus YM, De Baets MH. Myasthenia gravis: an autoimmune response against the acetylcholine receptor. Immunol Res. 1993; 12: 78–100 [DOI] [PubMed] [Google Scholar]
- 81. Hughes BW, Moro De Casillas ML, Kaminski HJ. Pathophysiology of myasthenia gravis. Semin Neurol. 2004; 24: 21–30 [DOI] [PubMed] [Google Scholar]
- 82. Dodel R, Neff F, Noelker C, et al. Intravenous immunoglobulins as a treatment for Alzheimer's disease: rationale and current evidence. Drugs. 2010; 70: 513–528 [DOI] [PubMed] [Google Scholar]
- 83. Dodel R, Balakrishnan K, Keyvani K, et al. Naturally occurring autoantibodies against beta-amyloid: investigating their role in transgenic animal and in vitro models of Alzheimer's disease. J Neurosci. 2011; 31: 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Relkin NR, Szabo P, Adamiak B, et al. 18-Month study of intravenous immunoglobulin for treatment of mild Alzheimer disease. Neurobiol Aging. 2009; 30: 1728–1736 [DOI] [PubMed] [Google Scholar]
- 85. Hughes RA, Dalakas MC, Cornblath DR, Latov N, Weksler ME, Relkin N. Clinical applications of intravenous immunoglobulins in neurology. Clin Exp Immunol. 2009; 158 (suppl 1): 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Janciauskiene S, Westin K, Grip O, Krakau T. Detection of Alzheimer peptides and chemokines in the aqueous humor. Eur J Ophthalmol. 2011; 21: 104–111 [DOI] [PubMed] [Google Scholar]
- 87. Yoneda S, Hara H, Hirata A, Fukushima M, Inomata Y, Tanihara H. Vitreous fluid levels of beta-amyloid((1-42)) and tau in patients with retinal diseases. Jpn J Ophthalmol. 2005; 49: 106–108 [DOI] [PubMed] [Google Scholar]
- 88. Guo L, Salt TE, Luong V, et al. Targeting amyloid-beta in glaucoma treatment. Proc Natl Acad Sci U S A. 2007; 104: 13444–13449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Grus FH, Joachim SC, Pfeiffer N. Analysis of complex autoantibody repertoires by surface-enhanced laser desorption/ionization-time of flight mass spectrometry. Proteomics. 2003; 3: 957–961 [DOI] [PubMed] [Google Scholar]
- 90. Boehm N, Wolters D, Thiel U, et al. New insights into autoantibody profiles from immune privileged sites in the eye: a glaucoma study. Brain Behav Immun. 2011; 26: 96–102 [DOI] [PubMed] [Google Scholar]
- 91. Wax MB, Tezel G, Yang J, et al. Induced autoimmunity to heat shock proteins elicits glaucomatous loss of retinal ganglion cell neurons via activated T-cell-derived fas-ligand. J Neurosci. 2008; 28: 12085–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Joachim SC, Grus FH, Kraft D, et al. Complex antibody profile changes in an experimental autoimmune glaucoma animal model. Invest Ophthalmol Vis Sci. 2009; 50: 4734–4742 [DOI] [PubMed] [Google Scholar]
- 93. Joachim SC, Wax MB, Seidel P, Pfeiffer N, Grus FH. Enhanced characterization of serum autoantibody reactivity following HSP 60 immunization in a rat model of experimental autoimmune glaucoma. Curr Eye Res. 2010; 35: 900–908 [DOI] [PubMed] [Google Scholar]
- 94. Gramlich OW, Joachim SC, Gottschling PF, et al. Ophthalmopathology in rats with MBP-induced experimental autoimmune encephalomyelitis. Graefes Arch Clin Exp Ophthalmol. 2011; 249: 1009–1020 [DOI] [PubMed] [Google Scholar]
- 95. Bell K, Funke S, Pfeiffer N, Grus FH. Serum and antibodies of glaucoma patients lead to changes in the proteome, especially cell regulatory proteins, in retinal cells. PLoS One. 2012; 7: e46910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shmerling RH. Autoantibodies in systemic lupus erythematosus--there before you know it. N Engl J Med. 2003; 349: 1499–1500 [DOI] [PubMed] [Google Scholar]
- 97. Arbuckle MR, McClain MT, Rubertone MV, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003; 349: 1526–1533 [DOI] [PubMed] [Google Scholar]
- 98. Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006; 90: 262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Albon J, Purslow PP, Karwatowski WS, Easty DL. Age related compliance of the lamina cribrosa in human eyes. Br J Ophthalmol. 2000; 84: 318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Albon J, Karwatowski WS, Easty DL, Sims TJ, Duance VC. Age related changes in the non-collagenous components of the extracellular matrix of the human lamina cribrosa. Br J Ophthalmol. 2000; 84: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Harris A, Ciulla TA, Chung HS, Martin B. Regulation of retinal and optic nerve blood flow. Arch Ophthalmol. 1998; 116: 1491–1495 [DOI] [PubMed] [Google Scholar]
- 102. Kong GY, Van Bergen NJ, Trounce IA, Crowston JG. Mitochondrial dysfunction and glaucoma. J Glaucoma. 2009; 18: 93–100 [DOI] [PubMed] [Google Scholar]
- 103. Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956; 11: 298–300 [DOI] [PubMed] [Google Scholar]
- 104. Tezel G. Oxidative stress in glaucomatous neurodegeneration: mechanisms and consequences. Prog Retin Eye Res. 2006; 25: 490–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Trounce I, Byrne E, Marzuki S. Decline in skeletal muscle mitochondrial respiratory chain function: possible factor in ageing. Lancet. 1989; 1: 637–639 [DOI] [PubMed] [Google Scholar]
- 106. Abu-Amero KK, Morales J, Bosley TM. Mitochondrial abnormalities in patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006; 47: 2533–2541 [DOI] [PubMed] [Google Scholar]
- 107. Abu-Amero KK, Morales J, Osman MN, Bosley TM. Nuclear and mitochondrial analysis of patients with primary angle-closure glaucoma. Invest Ophthalmol Vis Sci. 2007; 48: 5591–5596 [DOI] [PubMed] [Google Scholar]
- 108. Lee S, Sheck L, Crowston JG, et al. Impaired complex-I-linked respiration and ATP synthesis in primary open-angle glaucoma patient lymphoblasts. Invest Ophthalmol Vis Sci. 2012; 53: 2431–2437 [DOI] [PubMed] [Google Scholar]
- 109. Kasahara T, Kubota M, Miyauchi T, et al. Mice with neuron-specific accumulation of mitochondrial DNA mutations show mood disorder-like phenotypes. Mol Psychiatry. 2006; 11: 577–593, 523 [DOI] [PubMed] [Google Scholar]
- 110. Kong YX, Van Bergen N, Trounce IA, et al. Increase in mitochondrial DNA mutations impairs retinal function and renders the retina vulnerable to injury. Aging Cell. 2011; 10: 572–583 [DOI] [PubMed] [Google Scholar]
- 111. Rance G, O'Hare F, O'Leary S, et al. Auditory processing deficits in individuals with primary open-angle glaucoma. Int J Audiol. 2012; 51: 10–15 [DOI] [PubMed] [Google Scholar]
- 112. Tezel G, Yang X, Luo C, et al. Oxidative stress and the regulation of complement activation in human glaucoma. Invest Ophthalmol Vis Sci. 2010; 51: 5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Luo C, Yang X, Kain AD, Powell DW, Kuehn MH, Tezel G. Glaucomatous tissue stress and the regulation of immune response through glial Toll-like receptor signaling. Invest Ophthalmol Vis Sci. 2010; 51: 5697–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tezel G. The immune response in glaucoma: a perspective on the roles of oxidative stress. Exp Eye Res. 2011; 93: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Vanderlugt CL, Miller SD. Epitope spreading in immune-mediated diseases: implications for immunotherapy. Nat Rev Immunol. 2002; 2: 85–95 [DOI] [PubMed] [Google Scholar]
- 116. Higashide T, Kawaguchi I, Ohkubo S, Takeda H, Sugiyama K. In vivo imaging and counting of rat retinal ganglion cells using a scanning laser ophthalmoscope. Invest Ophthalmol Vis Sci. 2006; 47: 2943–2950 [DOI] [PubMed] [Google Scholar]
- 117. Cordeiro MF, Guo L, Luong V, et al. Real-time imaging of single nerve cell apoptosis in retinal neurodegeneration. Proc Natl Acad Sci U S A. 2004; 101: 13352–13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Printz C. BATTLE to personalize lung cancer treatment. Novel clinical trial design and tissue gathering procedures drive biomarker discovery. Cancer. 2010; 116: 3307–3308 [DOI] [PubMed] [Google Scholar]
- 119. Trial watch: Adaptive BATTLE trial uses biomarkers to guide lung cancer treatment. Nat Rev Drug Discov. 2010; 9: 423 [DOI] [PubMed] [Google Scholar]


