Abstract
RET encodes the tyrosine kinase receptor of growth factors belonging to the glial-derived neurotrophic factor family. Recently, RET gene rearrangements with N-terminal of KIF5B gene were identified in lung adenocarcinomas from large-scale sequencing. We investigated RET mRNA expression by real-time reverse-transcriptase polymerase chain reaction (RT-PCR) assay using LightCycler, and KIF5B/RET gene rearrangements using newly established fluorescence in situ hybridization (FISH) analysis in surgically treated nonsmall cell lung cancer (NSCLC) cases. RET protein expression was also investigated by immunohistochemistry (IHC). This study included 157 surgically removed NSCLC cases for mRNA level analyses. The RET/β actin mRNA levels were not significantly different between lung cancer (6.359 ± 15.268) and adjacent normal lung tissues (8.205 ± 28.931, P = 0.6332). Tumor/normal (T/N) ratio of RET/β actin mRNA levels was not different within gender, stage, smoking status, and pathological subtypes. T/N ratio of RET/β actin mRNA levels was significantly higher in KIF5B/RET rearrangement samples (161.763 ± 123.488) than in wild-type samples (5.9013 ± 17.148, P = 0.044). Although RET IHC positivity was not perfectly correlated with KIF5B/RET arrangement, we have detected the KIF5B/RET rearrangements using FISH analysis. Thus, we have successfully introduced FISH for diagnosing KIF5B/RET positive lung adenocarcinoma. This method facilitates the molecular evaluation for RET fusions and could be applicable in clinical practice to detect lung cancer that may be responsive to RET inhibitors.
Keywords: FISH, KIF5B/RET, lung cancer, RET expression
Introduction
Lung cancer is a major cause of death from malignant diseases due to its high incidence, malignant behavior, and lack of major advancements in treatment strategy [1]. A great deal of progress has been made in the target therapy for nonsmall cell lung cancer (NSCLC), largely owing to the development of small-molecular inhibitors, such as epidermal growth factor receptor (EGFR) [2–6] and ALK [7]. The recent discovery of a fusion gene that joins the KIF5B and RET oncogene from large-scale sequencing [8, 9] in a subset of NSCLCs have added a novel molecular subtype to the classification scheme for adenocarcinomas. The importance of recognizing this molecular subtype was highlighted by an inhibition using RET inhibitor for KIF5B/RET overexpression cells [9]. Therefore, an accurate and practical assay is urgently needed to detect this molecular subset of lung cancer [9]. Currently, the methods available for detecting EML4/ALK rearrangement are reverse-transcriptase polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) [7, 10]. RT-PCR is a single detect test to detect the gene rearrangements; however, it generally requires good quality RNA and a multiplex system [11]. Thus, EML4/ALK status, at least that determined using FISH, does seem to be extremely important, and gold standard as a specific treatment sensitivity marker with respect to small-molecule inhibitors of ALK [12–14].
Our group has already screened the KIF5B/RET gene status using RT-PCR assay (Yokota et al., unpubl. ms.). In this study, we have investigated RET mRNA expression by real-time PCR using LightCycler (Roche Molecular Biochemicals, Mannheim, Germany), protein expression by immunohistochemistry (IHC) and KIF5B/RET gene rearrangement status using newly established FISH analysis in surgically treated NSCLC cases. The findings were compared with the clinicopathologic features and KIF5B/RET gene status.
Material and Methods
Patient samples
The study group included NSCLC patients who had undergone surgery at the Department of Surgery, Nagoya City University Hospital. All tumor samples were immediately frozen and stored at –80°C until assayed. Because Lipson et al. [9] demonstrated that the KIF5B/RET rearrangements were found within adenocarcinoma histology of NSCLC, we mainly focused on adenocarcinomas without EGFR mutations. The clinical and pathological characteristics of the 157 NSCLC patients for RET mRNA gene analyses were as follows: 104 (66.2%) were male and 53 were female; 127 (80.9%) were diagnosed as adenocarcinomas and 25 were diagnosed as squamous cell carcinoma; 105 (66.9%) were smoker and 52 were nonsmoker; and 105 (66.9%) were pathological stage I. KIF5B/RET rearrangements statuses were already investigated.
PCR assay for RET gene
Total RNA was extracted from NSCLC and adjacent normal lung tissues using Isogen kit (Nippon gene, Tokyo, Japan) according to the manufactures' instructions. RNA concentration was determined using Nano Drop ND-1000 Spectrophotometer (Nano Drop Technologies Inc., Rockland, DE). About 10 cases were excluded for each assay because tumor cells were too few to sufficiently extract tumor RNA. RNA (1 μg) was reverse transcribed using First strand cDNA synthesis kit with 0.5 μg oligo (dT)16 (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufactures' instructions. The reaction mixture was incubated at 25°C for 15 min, 42°C for 60 min, 99°C for 5 min, and then at 4°C for 5 min. The complementary DNA (cDNA) concentration was determined using Nano Drop ND-1000 Spectrophotometer. About 200 ng of each cDNA was used for PCR analysis. To ensure the fidelity of mRNA extraction and reverse transcription, all samples were subjected to PCR amplification with β actin primers kit (Nihon Gene Laboratory, Miyagi, Japan) using LightCycler FastStart DNA Master HybProbe Kit (Roche Diagnostics GmbH). The RET PCR assay reactions were performed using LightCycler FastStart DNA Master SYBR Green I kit (Roche Diagnostics GmbH) in a 20-μL reaction volume. The primer sequences for RET gene at kinase domain were as follows: the forward primer, 5′-ACAGGGGATGCAGTATCTGG-3′ (at exon 14) and the reverse primer, 5′-CCTGGCTCCTCTTCACGTAG-3′ (at exon 16). The cycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 10 sec, 61°C for 10 sec, and 72°C for 7 sec.
RET IHC
Seventy-two cases of NSCLC were immunostained by automated methods (Dako Japan Inc., Tokyo, Japan) for C-terminal of RET expression using the ready-to-use mouse monoclonal RET Oncoprotein clone 3F8 (Vector Laboratories, Burlingame, CA) and 78 for the rabbit monoclonal RET antibody (EPR2871, 1:250) (Epitomics, Inc., Burlingame, CA) with Dako linker kit using intercalated antibody-enhanced polymer (iAEP) method [15]. Slides were scored for cytoplasmic staining intensity and distribution. An IHC score was assigned to each case according to the following criteria showing the designated staining pattern: 3+, intense, granular cytoplasmic staining; 2+, moderately, smooth cytoplasmic staining; 1+, faint cytoplasmic staining; and no staining. The extent of the staining area was determined as 0–10%, 10–25%, 25–50%, 50–75%, 75–90%, and 90%. Cytoplasm was stained either granular (G1) or diffuse (G2).
RET FISH testing
Unstained 5-μm sections of formalin-fixed paraffin embedded tumor tissue were submitted to dual-color FISH analysis using four probe sets. The KIF5B/RET probe sets were developed at GSP Research Inc. (Kawasaki, Japan), labeled with TexRed and the FITC. Probe sets were as follows: set 1, 5′RET (490kb; around 43MB∼43.5MB)-TexRed and 3′RET (630kb; around 43.6MB∼44.2MB)-FITC at 10q11.2; set 2, 5′KIF5B (620kb)-TexRed and 3′RET (630kb)-FITC. FISH detection kit (K5599) was used (Dako Japan Inc.). Briefly, NSCLC slides were deparaffinized and then preincubated with K5599 vial 1 95∼99°C for 10 min. Five to eight drops of Protease digestion buffer at room temperature for 5∼15 min and then wash and dry up for the slides. Labeled probe sets (10 mL) were cohybridized at 37°C for 72 h after denaturing at 82°C for 5 min. Stringency wash was conducted at 65°C with saline-sodium citrate (SSC) for 10 min. Slides were counter stained with 4′6-diamidino-2-phenylindole (DAPI).
Statistical analysis
Statistical analyses were carried out using the Mann–Whitney U-test for unpaired samples and Wilcoxon's singed rank test for paired samples. Linear relationships between variables were determined by means of simple linear regression. Correlation coefficients were determined by rank correlation using Spearman's test and χ2 test. The overall survival of lung cancer patients was examined using the Kaplan–Meier methods, and differences were examined using the Log-rank test. All analysis was done using the Stat-View software package (Abacus Concepts Inc. Berkeley, CA), and was considered significant when the P-value was less than 0.05.
Results
KIF5B/RET gene alteration and RET mRNA status in Japanese lung cancer patients
We have sequenced for arrangement status of KIF5B/RET gene for 371 NSCLC samples (Yokota et al., unpubl. ms.). Briefly, of 371 patients, from direct sequencing using cDNA samples, we found three translocation cases. We have included these cases and investigated further studies. The RET/β actin mRNA levels were not significantly different between lung cancer (6.359 ± 15.268) and adjacent normal lung tissues (8.205 ± 28.931, P = 0.6332; n = 157). Tumor/normal (T/N) ratio of RET/β actin mRNA level was not correlated with gender (male vs. female, P = 0.7636), age (age ≤65 vs. >65, P = 0.3204), and smoking status (smoker vs. nonsmoker, P = 0.6693). The T/N ratio of RET/β actin mRNA level was not correlated with pathological stages, lymph node metastasis, tumor invasion statuses, and pathological subtypes (adeno-carcinoma vs. others, P = 0.2652). The T/N ratio of RET/ β actin mRNA level was more than 20 in seven cases including three translocation cases. The T/N ratio of RET/ β actin mRNA level was significantly higher in KIF5B/RET fusion cases (161.76 ± 123.488) when compared with the wild-type cases (5.901 ± 17.148, P = 0.044) (Table 1).
Table 1.
Clinicopathological data of 157 lung cancer patients
| RET | |||
|---|---|---|---|
| Factors | No. of patients (N = 157) | T/N ratio of RET/β-actin mRNA levels | P-value |
| Mean age (years) | 66.7 ± 9.0 | ||
| Stage | |||
| I | 105 (66.9%) | 8.534 ± 30.678 | NS |
| II | 23 (14.6%) | 6.981 ± 12.707 | |
| III–IV | 29 (18.5%) | 11.603 ± 40.194 | |
| Tumor status | |||
| T1 | 73 (46.5%) | 11.044 ± 39.731 | NS |
| T2 | 62 (39.5%) | 7.242 ± 21.568 | |
| T3 | 7 (4.5%) | 10.454 ± 16.874 | |
| T4 | 15 (9.6%) | 4.318 ± 13.079 | |
| Lymph node metastasis | |||
| N0 | 116 (73.9%) | 8.587 ± 29.636 | NS |
| N1 | 23 (14.6%) | 5.414 ± 10.982 | |
| N2 | 18 (11.5%) | 15.690 ± 49.890 | |
| Pathological subtype | |||
| Adenocarcinoma | 127 (80.9%) | 10.465 ± 33.907 | 0.2652 |
| Others | 30 (19.1%) | 2.137 ± 3.865 | |
| KIF5B/RET rearrangement | |||
| Positive | 3 (1.9%) | 161.76 ± 123.488 | 0.044 |
| Negative | 154 (91.8%) | 5.901 ± 17.148 | |
| Smoking | |||
| BI = 0 | 52 (33.1%) | 14.047 ± 45.428 | 0.6693 |
| BI > 0 | 105 (66.9%) | 6.312 ± 19.525 | |
| Age | |||
| 65≤ | 80 (51.0%) | 10.318 ± 3.579 | 0.9147 |
| >65 | 77 (49.0%) | 7.385 ± 3.350 | |
| Gender | |||
| Male | 104 (66.2%) | 6.473 ± 19.581 | 0.7636 |
| Female | 53 (33.8%) | 13.584 ± 45.087 | |
T/N, tumor/normal; NS, not significant; BI, Brinkman index.
IHC for C-terminal of RET
Using the 3F8 antibody, three cases in the KIF5B/RET positive group were weakly positively stained (1+) for RET protein (Fig. 1a). The staining intensity was weak for each case. The staining pattern was less than 10% and cytoplasmic. We have additionally performed IHC for EGFR-Kras wild-type and KIF5B/RET wild-type patients. The staining intensity was strong (3+) in six cases, moderate (2+) in 12 cases, and weak (1+) in 28 cases. Other cases showed almost no stained for RET. Using the rabbit monoclonal RET antibody, the staining intensity was 3+ (90%, G2), 2+ (90%, G2), and 1+ (50–75%, G2) (Fig. 1b–d) for each KIF5B/RET fusion cases, respectively. On the other hand, in 75 wild-type cases, four were 3+ (two were 50–75%, G1, one was 50–75%, G2, and one was 90%, G1), 13 were 2+ (two were 75–90%, five were 50–75%, two were 25–50%, three were 10–25%, and one was 0–10%), 43 were 1+ (14 were 0–10%, 16 were 10–25%, seven were 25–50%, three were 50–75%, and three were 75–90%), and 14 cases did not stained at all. Sixteen were G1 and 44 were G2. As for increased RET mRNA cases without translocations, one had only nuclear staining (1+, 75–90%), one had predominantly membranous staining (3+, 50–75%), one had 2+ (50–75%, G2), and one had 1+ (0–10%, G1) cytoplasmic staining.
Figure 1.
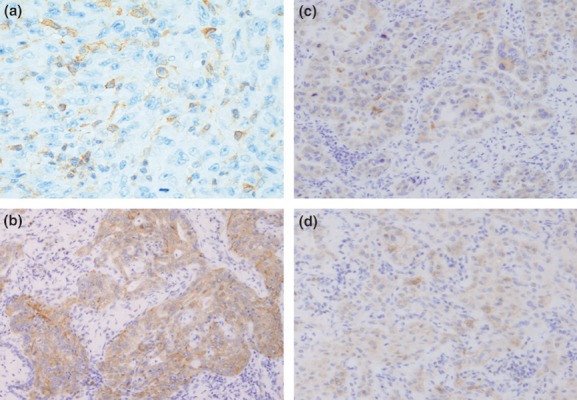
Using the 3F8 antibody, KIF5B/RET rearrangement case was weakly positively stained (1+) for RET protein (a). Using the rabbit monoclonal RET antibody, the staining intensity was 3+ (b), 2+ (c), and 1+ (d) for each KIF5B/RET fusion cases.
Cytogenic patterns of KIF5B/RET in FISH
Figure 2 showed the typical signals for set 1 probes classified as positive for RET rearrangement, showing the normal signal (fused 3′ 5′ RET), the split/single 3′ RET, and the split/single 5′ RET signal. Normal signals are frequently seen in lung cancer cells that also display RET gene rearrangements. The typical KIF5B/RET fusion case should show distinct red and green signals separated within the same cell. To compile the frequency of positive cells for RET rearrangements, the 15% of cells carrying split red and green and single red (RET 5′ region) signals was considered as positive. Figure 3 showed the typical signals for set 2 probes classified as positive for KIF5B/RET rearrangement, showing the normal signal (split red 5′-KIF5B and green 3′-RET signals), whereas if it was fusion, the signals showed only yellow signal per cells. Two of three KIF5B/RET cases showed positive signals for FISH analyses using two probe sets, whereas wild case showed negative signals.
Figure 2.
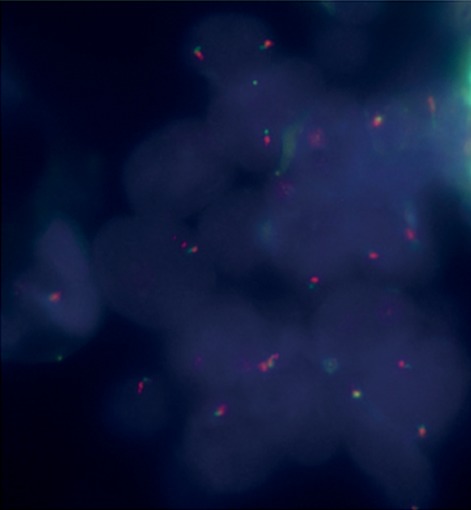
The typical signals for set 1 probes classified as positive for RET rearrangement, showing the normal signal (fused 3′ 5′ RET), the split/single 3′ RET, and the split/single 5′ RET signal.
Figure 3.
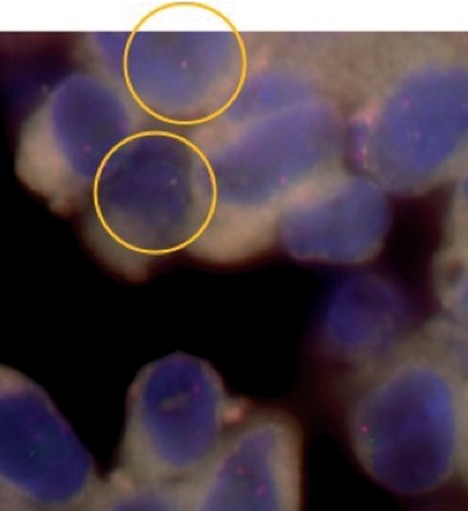
The typical signals for set 2 probes classified as positive for KIF5B/RET rearrangement, showing the normal signal (split red 5′-KIF5B and green 3′-RET signals), whereas if it was fusion, the signals showed only yellow signal per cells.
Discussion
In this study, we have introduced newly developed methods for the detection of KIF5B/RET rearrangement using FISH, and the results showed a concordance between RT-PCR assay and FISH. We also investigated RET expression by IHC using two antibodies for C-terminal of RET; however, those did not correspond well part with the gene arrangement results evaluated with FISH. Although RET protein overexpression was not perfectly matched with the RET rearrangements, FISH analysis might be one of the candidate method for detecting RET gene status in lung cancer.
The KIF5B/RET fusion gene contains the entire kinase domain of RET and resulted in increased RET kinase activity. Oncogenic PTC/RET translocations detected in thyroid cancer and cell lines are sensitive in vitro and in vivo to RET inhibitors [16–19], suggested the possibility that sunitinib, sorafinib, vandetanib, or other RET inhibitors might also be clinically effective in KIF5B/RET lung cancers. Ba/F3 cells that were introduced with the full length KIF5B-RET gene (variant 1) showed interleukin 3 (IL-3) independent growth consistent with oncogenic transformation [9]. As cells with KIF5B/RET fusion gene overexpress the chimeric RET receptor tyrosine kinase and show spontaneous cellular transformation [20], inhibiting RET receptor tyrosine kinase may suppress tumor progression of KIF5B-RET fusion-positive lung cancer [20, 21]. The KIF5B/RET Ba/F3 cells were sensitive to sunitinib, sorafenib, and vandetanib, multitargeted kinase inhibitors that inhibit RET, but not to gefitinib, an EGFR kinase inhibitor [9]. Sunitinib, but not gefitinib, inhibited RET phosphorylation in the KIF5B-RET Ba/F3 cells [9].
The recent study has been reporting the novel KIF5B/RET gene rearrangement found to occur in 2% (9/405) of Asian lung adenocarcinoma patients screened [9]. In our previous screen (Yokota et al., unpubl. ms.), we could only confirm the existence of 3/270 (1.1%) the KIF5B/RET rearrangements in lung adenocarcinomas. Several explanations exist for this discrepancy. Lung cancer encompasses a broad range of clinical subtypes, and the makeup of the two cohorts differed. Next, a racial difference between the studies about mutant KIF5B/RET may be existed, like EGFR gene mutations [2–6]. Because the arrangements were originally identified from massively parallel sequencing to generate comprehensive genomic profiles, we cannot conclude that differences were owing to our methodology as our PCR and sequencing methods. The technique used by Lipson et al. to identify RET arrangement was RT-PCR [8, 9]. This assay requires extraction of RNA from clinical samples, is not routinely performed in many diagnostic pathology laboratories, and suffers from some important limitations. Each PCR assay is RET fusion specific, and although assays may be multiplexed [11], they may not detect all possible KIF5B/RET gene arrangements, especially those involving novel fusion partners [15], a consequence of which may be false-negative results. Identification of a small fraction of lung cancers patients responsive to a molecular target therapy may have large clinical impact as highlighted by the recent FDA approval of crizotinib for ALK-rearrangement lung cancers [13]. This was based on the results of clinical trials for advanced lung cancers with ALK rearrangements, and early responses in ALK rearrangement assessed by FISH [13, 14]. Accurate and rapid screening of rearrangement is important in lung cancers, especially in determining their eligibility for molecular target therapy. Thus, we forward to step FISH analysis, which is gold standard for detection of EML4/ALK rearrangement. FISH enables the pathologist to perform a more reliable quantification of the genomic alteration. Takeuchi et al. [21] had undergone KIF5B/RET FISH analyses using BAC clones from National Center for Biotechnology Information. However, the length of each probe might be between 160 and 300 kb. In our analyses, we mixed 3–5 BAC clones from GSP Research Inc. (Figs. 4 and 5). Thus, the length of each probes for our KIF5B/RET analyses were between 490 and 630 kb. This may improve the specificity and sensitivity for KIF5B/RET rearrangements detection. In our preliminary experiments, the increase in the probe length increased the brightness of our FISH results.
Figure 4.
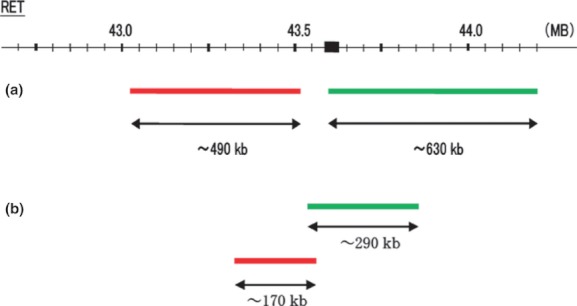
RET split probes. (a) 5′ (∼490kb) and 3′ (∼630kb) probes for RET gene (set 1). (b) 5′ (∼170kb) and 3′ (∼290kb) possible probes using BAC clones from National Center for Biotechnology Information [21].
Figure 5.
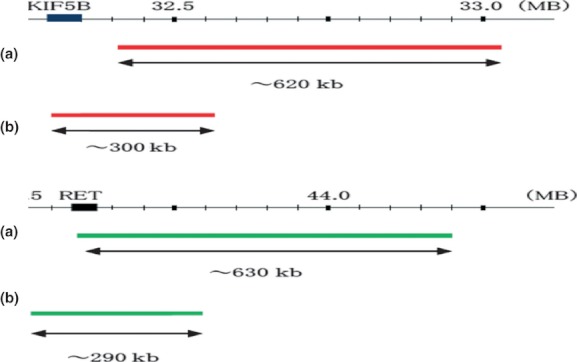
KIF5B/RET fusion probes. (a) 5′ KIF5B (∼620kb) and 3′ RET (∼630kb) probes for fusion gene (set 2). (b) 5′ KIF5B (∼300kb) and 3′ RET (∼290kb) possible probes using BAC clones from National Center for Biotechnology Information [21].
However, even if we would like to expand the study, because the availability of tumor samples in the cohort was limited, we were unable to perform FISH in most patients. Most patients with advanced stage of NSCLC had only small tissue samples and the samples were mostly used for clinical diagnosis, leaving limited residual samples for molecular diagnosis. FISH methods need specific sample handling, such as tissue processing and method of fixation, standardization of reagents and protocols, standardized reporting criteria, and implementation of quality assurance procedures, including the requirement for laboratory accreditation and external proficiency assessment [22]. Paik et al. [23] showed that EML4/ALK rearrangement detection using IHC staining correlated well with FISH. IHC staining could be applied broadly in routine biopsy tissue diagnosis in clinical practice and thus we performed IHC using commercially available antibody and iAEP methods. Yang et al. [24] expand on previous experience [25] by evaluating the approach of initial testing IHC with the commercially available monoclonal ALK antibody and an enhanced detection system paired with subsequent FISH testing. The report suggested that IHC test was sensitive but not specific [24]. Similar strategies have been proposed using another commercially available antibody [26]. Thus, development and validation of strategies to improve effective identification of the patient population with strategies incorporating IHC or other techniques are important and likely to assume a place in clinical practice. Diffuse cytoplasmic staining (G2), more than 50% using EPR2871 might be correlated with RET translocation. There was a case where there was an elevated RET mRNA and protein expression, but no detectable rearrangement. This would suggest the presence of a novel translocation. Prospective study is needed to compare the usage of RT-PCR, FISH, and iAEP IHC. We only validated the result of RT-PCR using FISH or iAEP IHC using commercially available antibody in a limited number of patients.
Acknowledgments
The authors thank Mrs. Miki Mochizuki for her excellent technical assistances. This work was supported by Grants-in-Aid for Scientific Research, Japan Society, for the Promotion of Science (JSPS) (Nos, 24592097, 23659674, 21390394, 21591820) and a grant for cancer research of Program for developing the supporting system for upgrading the education and research (2009) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of Interest
None declared.
References
- 1.Ginsberg RJ, Kris K, Armstrong G. Cancer of the lung. In: De Vita VT Jr, Hellman S, editors. Principles and practice of oncology. 4th ed. Philadelphia: Lippincott; 1993. pp. 673–682. [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki H, Endo K, Konishi A, Takada M, Kawahara M, Iuchi K, et al. EGFR mutation status in Japanese lung cancer patients: genotyping analysis using LightCycler. Clin. Cancer Res. 2005;11:2924–2929. doi: 10.1158/1078-0432.CCR-04-1904. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki H, Shimizu S, Endo K, Takada M, Kawahara M, Tanaka H, et al. EGFR and erbB2 mutation status in Japanese lung cancer patients. Int. J. Cancer. 2006;118:180–184. doi: 10.1002/ijc.21301. [DOI] [PubMed] [Google Scholar]
- 6.Fukuoka M, Wu Y-L, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, et al. Biomarker analyses and first overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J. Clin. Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 7.Soda H, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 8.Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Massively parallel sequencing assay identifies novel ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat. Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak EL, Bang YL, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancers. N. Engl. J. Med. 2010;363:1692–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin. Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 12.Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly-defined subset of non-small cell lung cancer. J. Thorac. Oncol. 2009;4:1450–1454. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- 13.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy BR, Costa BR, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J. Clin. Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou SH, Bezhenova L, Camidge DR, Solomon BJ, Herman J, Kain T, et al. Rapid and dramatic radiographic and clinical response to an ALK inhibitor (Crizotinib, PF02341066) in an ALK translocation-positive patient with non-small cell lung cancer. J. Thorac. Oncol. 2010;5:2044–2046. doi: 10.1097/JTO.0b013e318200f9ff. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin. Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 16.Henderson YC, Ahn SH, Kang Y, Clayman GL. Sorafenib potently inhibits papillary thyroid carcinomas haboring RET/PTC1 rearrangement. Clin. Cancer Res. 2008;14:4908–4914. doi: 10.1158/1078-0432.CCR-07-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlomagno F, Anaganti S, Guida T, Salvatore G, Trancone G, Wilhelm SM, et al. BAY 43–9006 inhibition of oncogenic RET mutants. J. Natl. Cancer Inst. 2006;98:326–334. doi: 10.1093/jnci/djj069. [DOI] [PubMed] [Google Scholar]
- 18.Kim DW, Jo YS, Jung HS, Chung HK, Song JH, Park KC, et al. An orally administered multitarget tyrosine kinase inhibitor, SU11248, is a novel potent inhibitor of thyroid oncogenic RET/papillary thyroid cancer kinases. J. Clin. Endocrinol. Metab. 2006;91:4070–4076. doi: 10.1210/jc.2005-2845. [DOI] [PubMed] [Google Scholar]
- 19.Dawson S, Conus NM, Toner GC, Raleigh JM, Hicks RJ, McArthur G, et al. Sustained clinical responses to tyrosine kinase inhibitor sunitinib in thyroid carcinoma. Anticancer Drugs. 2008;19:547–552. doi: 10.1097/CAD.0b013e3282fc6cf7. [DOI] [PubMed] [Google Scholar]
- 20.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 22.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 23.Paik JH, Choe G, Kim H, Choe JY, Lee HJ, Lee CT, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer correlation with fluorescence in situ hybridization. J. Thorac. Oncol. 2011;6:466–472. doi: 10.1097/JTO.0b013e31820b82e8. [DOI] [PubMed] [Google Scholar]
- 24.Yang P, Kulig K, Boland JM, Erickson-Johnson MR, Oliveira AM, Wampfler J, et al. Worse disease-free survival in never-smokers with ALK+ lung adenocarcinoma. J. Thorac. Oncol. 2012;7:90–97. doi: 10.1097/JTO.0b013e31823c5c32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi ES, Boland JM, Maleszewski JJ, Roden AC, Oliveira AM, Aubry MC, et al. Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J. Thorac. Oncol. 2011;6:459–465. doi: 10.1097/JTO.0b013e318209edb9. [DOI] [PubMed] [Google Scholar]
- 26.Mino-Kenudson M, Chirieac LR, Law K, Hornick JL, Lindeman N, Mark EJ, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin. Cancer Res. 2010;16:1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]


