Abstract
Extensive measurements of the parts list of human skeletal muscle through transcriptomics and other phenotypic assays offer the opportunity to reconstruct detailed functional models. Through integration of vast amounts of data present in databases and extant knowledge of muscle function combined with robust analyses that include a clustering approach, we present both a protein parts list and network models for skeletal muscle function. The model comprises the four key functional family networks that coexist within a functional space; namely, excitation-activation family (forward pathways that transmit a motoneuronal command signal into the spatial volume of the cell and then use Ca2+ fluxes to bind Ca2+ to troponin C sites on F-actin filaments, plus transmembrane pumps that maintain transmission capacity); mechanical transmission family (a sophisticated three-dimensional mechanical apparatus that bidirectionally couples the millions of actin-myosin nanomotors with external axial tensile forces at insertion sites); metabolic and bioenergetics family (pathways that supply energy for the skeletal muscle function under widely varying demands and provide for other cellular processes); and signaling-production family (which represents various sensing, signal transduction, and nuclear infrastructure that controls the turn over and structural integrity and regulates the maintenance, regeneration, and remodeling of the muscle). Within each family, we identify subfamilies that function as a unit through analysis of large-scale transcription profiles of muscle and other tissues. This comprehensive network model provides a framework for exploring functional mechanisms of the skeletal muscle in normal and pathophysiology, as well as for quantitative modeling.
Keywords: skeletal muscle transcriptome, functional protein classification, functional protein network
human skeletal muscle is the most abundant tissue in the human body. Its major functional role is mechanical actuation, but skeletal muscle also plays critical roles in heat production during cold stress and in the management of whole body metabolism. The skeletal muscle composition includes muscle cells (long multinucleated fibers), connective tissue, blood vessels, and nerves. Detailed knowledge of the structure and organization of the muscle fiber is easily found in literature (30). In recent decades, genetic and molecular measurements in skeletal muscle have begun to reveal the molecular components and networks that comprise the cell and dictate its function. For example, gene expression measurements on skeletal muscle tissue samples (48, 56) have provided a partial parts list of genes that are expressed in the skeletal muscle under different conditions (22, 68, 72, 75), and proteomic assays have revealed the presence of many of the proteins involved in muscle function (85). The annotation of this gene and protein parts list has largely relied on legacy knowledge of protein function, as embedded in databases such as the protein and nucleic acid sequence databases or categorized by the “gene ontology (GO)” efforts (76) The GO classification, organized into three domain sets (list of molecular functions that are carried out, cellular components and locations where they act, biological processes in which they participate), encourages hyperconnectivity, and often there are memberships within sets that may not be synergistic (e.g., Z-disk with a fundamentally mechanical function and calcium-signaling proteins of a fundamental control function are both categorized under muscle contraction). The individual protein functions, however, provide little insight into the complex cellular pathways deployed by skeletal muscle. Notwithstanding this limitation, the assembly of the molecular “parts list” of skeletal muscle provides the scope for constructing functional models that can shed light on its normal and pathological function.
The formal basis for constructing functional models stems from the overall characterization of the function of the skeletal muscle as tissue engineered for specific functions. The large-scale functional demands of a skeletal muscle cell can be broadly divided into four major categories, each of which consists of a core set of proteins, and we refer to these as families. The excitation-activation (EA) family receives its excitation signal from the neuromuscular junction (NMJ) and activates the muscle (64). Specifically, the forward pathways depend on specialized protein complexes to transmit a transient motoneuronal signal into a spatial volume, which, in turn, transmits myriad calcium fluxes, resulting in binding of free calcium to troponin C sites on F-actin filaments. The energy-consuming reverse processes are used for pumping ions back across the potential gradients. This overall process of EA coupling triggers a cascade of activities, leading to the potential for force generation, as well as subtle remodeling signals related to calcium management over a wide temporal spectrum (15, 16). The mechanical transmission (MECH) family is a sophisticated three-dimensional force-producing (45) and transmitting (31, 50) apparatus that bidirectionally couples the millions of actin-myosin nanomotors within the cell volume with an external axial length and tensile force. This activation- and length-dependent force transmission is the major output of a skeletal muscle. The energy demands associated with EA and mechanical actuation are met by consuming ATP (8) that comes from the cellular metabolism, comprising largely mitochondrial processes of glycolytic and oxidative metabolism. The energy resource management is controlled by sensing the demands of the cell through signals that control metabolism, as well as gene transcription, as a first step toward protein production. The metabolic and bioenergetics (METB) family deals with this function. The signaling pathways also play a large role of remodeling the muscle fiber, which dynamically adjusts the function of the muscle based on mechanical need. The muscle fibers can be characterized as slow and fast (18), with its subtypes manifesting in the type of metabolism, size, contraction/relaxation time, and the capacity for force and power production (19, 53, 65); sensing and signaling pathways are critical to coordinated transition between fiber types, with changes in functional capacity reflected in all three of the other families. We describe the network of proteins involved in signaling and production to constitute the signaling and production (SIGP) family. Thus the four “skeletal muscle” functional families work synergistically to regulate the larger body functions of locomotion, body posture, metabolism, and heat production when warranted. This compartmentalization of function, albeit connected to each other via key signaling states, as in Fig. 4, provides an ideal basis for building pathway models that can be used to explore the muscle function in normal and diseased states. Importantly, these models can be formulated in mathematical terms.
Fig. 4.
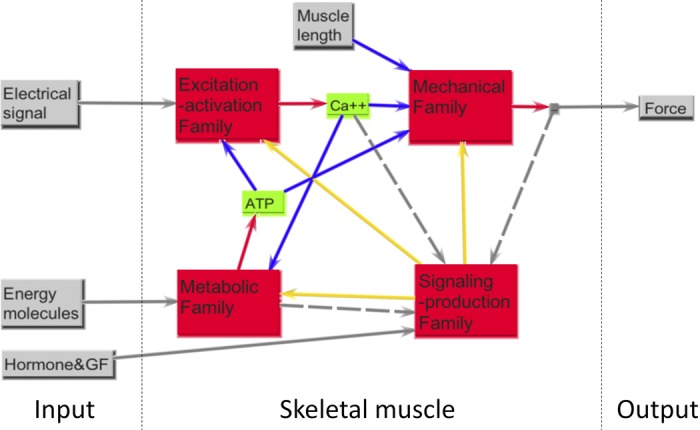
The functional protein families of skeletal muscle. EA model family addresses the challenge of near-synchronous temporal coactivation of spatially distributed nano-actuators. MECH model family manages transmission of forces between these nano-actuators and gross skeletal structures that can also generate forces (including force transmission across the mechanically vulnerable plasmalemma bilayer). METB model family manages production of energy to meet ongoing demands, including transport of energy fuels and materials such as oxygen. SIGP family “senses” key states and functional demands of the aforementioned families and up/downregulates transcriptional and proteomic levels so as to adapt materials and structure over timeframes of days to weeks. In the middle part (skeletal muscle), the blue (solid) lines are the output to input of a key highly regulated state variable operating between certain protein families, the gray (dashed) lines represent sensory signals to the SIGP family, and the yellow (solid) lines represent the slower remodeling signals continually sent back to the other three families.
In this work, we identify a parts list unique to human skeletal muscle by comparing the gene expression profiles of muscle tissue contrasted with other human tissues. From this large parts list, we build functional models based on the four functional families of EA, MECH, metabolism, and cell signaling. These models are then used to understand muscle function in molecular detail. We explain the functional and pathway differences between the healthy human skeletal muscle tissue and the tissue from Duchenne muscular dystrophy (DMD) and amyotrophic lateral sclerosis (ALS) patients in a companion paper (78a).
This paper is organized as follows. We outline the methods used in obtaining the skeletal muscle parts list and in the construction of functional pathway models. We present the results of model construction for each of the four families and discuss the functional role in the context of skeletal muscle contrasted with other tissues, including cardiac and smooth muscle tissue. We then discuss the consequence of the molecular description of each of the four functional families and elaborate on the functional roles of the proteins.
METHODS
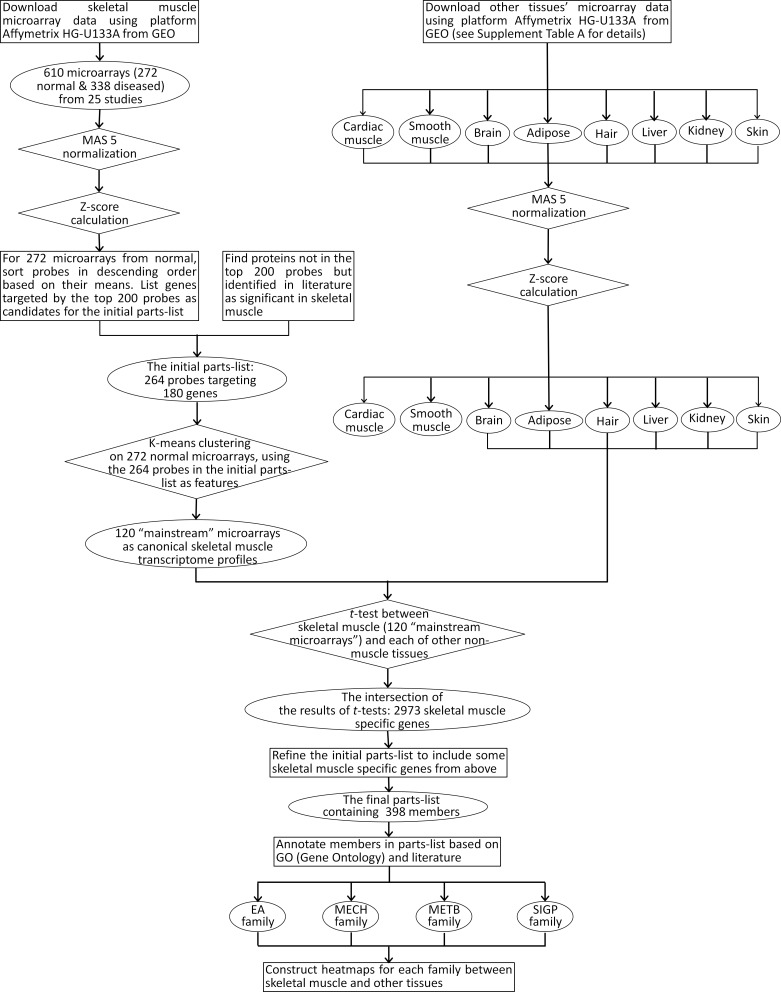
This section outlines the process involved in developing the skeletal muscle classification system from human-subject transcriptional profiles, annotations of skeletal muscle function, and incorporation of the legacy knowledge available in extant literature. The flowchart in Fig. 1 provides an overview of the procedures used in the construction of the parts list and the subsequent assembly into functional families.
Fig. 1.
The flow chart of the procedures used to construct the parts list and functional family. GEO, Gene Expression Omnibus; EA, excitation-activation; MECH, mechanical transmission; METB, metabolic and bioenergetics; SIGP, signaling and production.
Selection, Initial Filtering, and Clustering of Microarray Data for Skeletal Muscle and Other Tissues
The public repositories of gene expression data, namely Gene Expression Omnibus (GEO) from National Center for Biotechnology Information and ArrayExpress from the European Bioinformatics Institute were mined for tissue-specific expression data from human subjects. To avoid the noise of cross-platform analysis, only studies that used a single-array platform, namely, Affymetrix HG-U133A (22,283 probes), were chosen. We identified 68 tissue-specific expression data sets from adipose (3 data sets), brain (8 data sets), hair (1 data set), kidney (6 data sets), liver (5 data sets), skin (5 data sets), and muscle (33 data sets) tissues. The muscle data included smooth, cardiac, and skeletal muscles in normal and a host of skeletal muscle disease patients. For skeletal muscle, data from 272 microarrays of healthy and 338 microarrays of diseased subjects from 25 studies, with most studies carried out in triplicate, were chosen for analysis. Majority of the healthy tissues constituted the vastus lateralis (233 microarrays) with a few from quadriceps (14 microarrays), rectus abdominus (8 microarrays), and uncharacterized (28 microarrays). In the diseased cases, data from patients with DMD (33 microarrays), ALS (9 microarrays), facioscapulohumeral dystrophy (FSD) (53 microarrays), Emery-Dreifuss muscular dystrophy (8 microarrays), obesity (40 microarrays), and insulin resistance (72 microarrays) were chosen for analysis. The data were initially partitioned into “healthy” (part of 15 studies) and “diseased” (part of 14 studies), based on review of the information associated with each data set. The “healthy” data set, which represented a range of ages and both sexes (50.0% male, 37.9% female, 12.1% unknown), form the primary focus of this paper. A detailed analysis of the DMD and ALS patients in the context of skeletal muscle function is presented in the companion paper (78a). For cardiac muscle, a total of 64 arrays from patients with heart failure and for smooth muscle a total of 86 microarrays from studies carried out on smooth muscle from bladder (8 microarrays), coronary smooth muscle (9 microarrays), cerebral artery (9 microarrays), aorta (6 microarrays), and primary tracheal human airway smooth muscle (54 microarrays) were chosen in our study. A detailed listing of the data sets is presented in Supplemental Table SA. (The online version of this article contains supplemental data.)
Analysis of Transcriptional Profiles of Human Tissues and Identification of a Core Parts List of Skeletal Muscle Genes
The foundation for our parts lists analysis comes from the classification of microarrays that represent the healthy human skeletal muscle and contrasting these with those from other tissues.
Data normalization.
The raw data of those microarrays were downloaded from GEO database and normalized with MAS5 in Expression Console provided by Affymetrix. Then the data were further normalized by sample set to obtain the z-score data (67). For each microarray, assuming xi is the value of the ith probe of totally n probes (n = 22,283), then the z-score of the ith probe zi is
The z-score of any microarray data has the same mean (z̄ = 0) and standard deviation [stdev(z) = 1], and, therefore, all of the microarray data were normalized into the same comparable range.
In view of the interlaboratory variation of expression data, MAS5, which analyzes each microarray independently, was chosen instead of multichip analysis methods, such as RMA (Robust Multichip Average) and PLIER (Probe Logarithmic Intensity Error), with the caveat that MAS5 does not identify very small changes. Since most of the identified minimum number of significant proteins of skeletal muscle have above-average expression levels (see Candidate protein selection below), small differences, whether between skeletal muscle and other tissues, or between healthy skeletal muscle and disease conditions, are ignored.
Chip quality must be considered in any microarray data analysis. The intrinsic noise of the microarray may bring variance to the data, which affect mostly genes with low-expression level due to their high variances relative to their low values. Some statistical models, such as Vampire (43), were developed to remove these effects. In this study, because the low-expression genes were excluded from the parts list selection (see below), the intrinsic noise of the microarray had minimal effect on the subsequent data analysis.
Candidate protein selection.
The mean and SD of all probes from 272 microarrays of normal/control subjects were calculated, and all probes were sorted in descending order according to their means. One basic assumption of this study is that gene transcripts having high z-scores are more likely to be significant to skeletal muscle function. Generally, only transcripts that have z-scores above the median value (around −0.2 in most microarrays) in either healthy skeletal muscle or diseased conditions or both were considered, and those with negative mean z-scores (i.e., below average) in normal/control conditions were filtered out from the candidate protein selection process.
To ensure a number of gene transcripts that can be handled effectively by the K-means clustering algorithm, we arbitrarily set the maximum length of candidate list as the sample size (i.e., 272). As a further filtering process, we analyzed the sorted probe list for previously known skeletal muscle function. The initial candidate list contained 180 transcripts (264 probes, see Supplemental Table SB), most with very high z-scores. These then were used as the key features for clustering the microarray data in the next step.
Clustering.
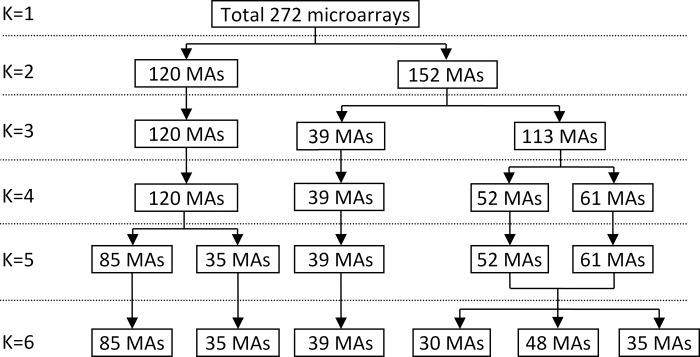
In the next step, the commonly used K-means clustering algorithm (54) was used to identify dominant clusters from expression profiles of tissues in healthy condition. As shown in Fig. 2, the K-means clustering yielded one main cluster encompassing 120 microarrays, with generally slightly higher z-scores and lower variance than the whole population for these 180 transcripts. This dominant cluster, derived from many studies, was also seen with a similar analysis of the “muscle pathology” data sets and was then used for all subsequent analysis, including statistical analysis for the full microarrays. The results of this K-means clustering are included in Supplemental Table SB.
Fig. 2.
The results of K-means classification of normal skeletal muscle microarrays (MAs). The same dominant cluster of 120 MAs, representing the gene expression pattern of normal skeletal muscle, were identified whether K = 2, 3, or 4; it was not until K = 5 or 6 that this cluster could be split into two (quite similar) clusters.
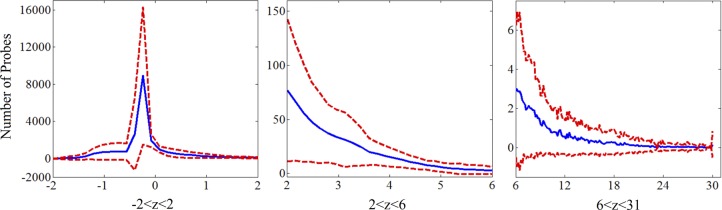
Figure 3 shows the z-score value distribution of the dominant 120 microarrays representing the normal skeletal muscle in subjects between ages 20 and 70 yr. A brief overview of the mRNA normal data is consistent with that expected from consideration of the skeletal muscle as a specialized tissue composed of highly differentiated cells; only ∼31% of the z-scores had positive values. Most of the high z-scores came from classic myo-structural proteins related to contractile filaments and energy-supporting structures, such as proteins in mitochondria (both a key focus of this study), and from various “production” and “bookkeeping” genes that are high for most tissues.
Fig. 3.
Z-score histogram of the 120 main normal MAs, with the y-axis (number of probes) being rescaled as the z-score increases and the number decreases. The blue line is the mean of the number of probes, and the red lines are the means ± SD. As expected, most of the probes (69%) have negative z-scores, indicating that many of these are essentially “off” or of very low expression for muscle tissue. Only 5.9% had a z-score value above 1.0, only 2.4% above 2.0, and only 0.6% above 5.0.
Comparison with other tissues.
The same procedure described above for skeletal muscle was applied to expression data from adipocyte, brain, hair, kidney, liver, and skin tissues. These tissues were selected because, for each of them, there are at least 15 microarrays of Affymetrix HG-U133A in GEO database. This procedure was also used for cardiac and smooth muscle tissues, although these were not used for our preliminary comparison (described below), owing to their similarity to skeletal muscle. For our subsequent more fine-tuned analysis, we compare all of the muscle types to provide isoform-specific classification of the parts lists.
This first stage was motivated by the desire to delineate skeletal muscle-specific z-scores, contrasting with those from other tissue types. This involved systematically comparing skeletal muscle z-score data to z-scores that we calculated for other tissues, with the aim of identifying statistically significant differences. The Student t-test was used to assess significance [with a sample size larger than 5, the t-test and other nonparametric methods are comparable (7)], and a threshold P value of 0.05 was chosen. If there were more than one probe targeting the same gene, a fuzzy integrative algorithm described in the appendix was applied to combine the results of t-tests of these probes based on the maximum mean intensity of each tissue, the percentage of significantly different probes, and the P values.
In each case, statistically significant z-scores were separated into those already on the initial list and those that were not. It was found that 2,973 genes (4,075 probes) of mRNA abundance levels were significantly higher in skeletal muscle than in nonmuscle tissues, with most of the proposed initial 180 candidate proteins (identified in the dominant cluster described in Candidate protein selection) on this list (Supplemental Table SC); transcripts for some essential proteins with z-scores that were not found to be significantly different from mRNA levels in certain other tissues (e.g., certain collagens and cytoskeletal linker proteins) were not eliminated, since there are many proteins that are functionally important for multiple types of tissues. Additionally, certain key muscle transcription factors can be expected to normally have low z-scores, but are known to upregulate with changes in functional demands. Each was subject to evaluation, and a superscript system is used to identify and delineate between members that are unique to skeletal muscle or to muscle in general.
Development of a Class Structure, and Process for Core Membership Assignment
The classification of these genes, identified in the skeletal muscle parts list above, was based on the functional physiology of the skeletal muscle and from the perspective of building mechanistic models amenable to computational analysis. While such functional classification has not been done previously, we used the parsimony principle (Occam's razor argument). The fundamental property of the skeletal muscle is contractility, where mechanical force is transduced in response to neuromotor signal process by the cell. The energetics for these processes is provided by mitochondrial function, which also acts in response to muscle fiber-type remodeling demands. The entire function of the muscle is orchestrated by intracellular signaling events that trigger changes in transcriptional and translational activity to produce the correct proteins at the appropriate time.
The four core functional models are summarized in Fig. 4, each of which has been the basis of mostly separate mathematical modeling foundations (26, 74). We propose that these can be used to form a conceptual framework for classifying families of key gene products associated with what amounts to nearly one-half of the protein content of the human body and ∼40% of the body mass. These families must structurally coexist within a compact physical space and yet are assumed to interact as models through several overriding dynamic signals that are highly regulated, as shown in Fig. 4. All but SIGP can be viewed as having a primary dynamic output of the model family that critical to real-time operation and performance, namely, Ca2+ activation, muscle force, and ATP supply. SIGP manages the “parameters” associated with these families that can gradually remodel (e.g., up- and downregulation), thus affecting future performance. Within each of these four families, we propose a small collection of three to five subfamilies, each of which contains transcripts associated with the key proteins or parts of proteins that code for a specified subfunction.
Importantly, the internal connectivity within each of these families is very different; this is a key strength of our dual model-family structure and why we also view these as functional models. For instance, connectivity between subfamilies and members within the MECH family can be viewed as (usually stiff) springs attached to bicausal (two-way) mechanical ports, reflecting the reality that, while there are biochemical (mostly covalent) connections between key structural proteins within the group, this is functionally a biomechanical lattice network model that connects between the extracellular matrix (ECM)/tendon structures and the collections of molecular myo-actuators that generate length- and velocity-dependent forces. Many members have embedded mechanochemical-sensing capabilities that affect networks within the SIGP family. In contrast, the EA family is primarily a unicausal information system composed of structures (often embedded within membranes) that enable a motoneuronal signal to be transmitted (ultimately via calcium) throughout the myotube volume; this family includes key subfamilies of proteins/transcripts that maintain quality control regarding how well calcium and other ions are managed and uses arrows to identify connectivity. Then, in contrast, the METB family is described using tools normally associated with biochemical systems modeling, including for material transport and energy utilization (e.g., storage, production, transport, consumption).
The key output signals from families include two relatively small diffusible molecules (calcium, ATP) that are highly regulated (often the aim of respective subfamilies described below) and of great interest to muscle physiologists. Such signals are assumed to be sensed and operated on by signaling cascades, often called signal-transduction networks, which then affect the transcription and translation processes that are captured by our signaling family (SIGP). The outputs of the SIGP family are the up/downregulation of protein products that, in turn, are the foundation for homeostatic adaption of strategic proteins of the other three families, thus completing the loops of connectivity among the four families shown in Fig. 4.
The core protein membership was further refined through case-by-case review (mainly through extensive legacy knowledge and GO functional association) of proteins associated with mRNAs with relatively high z-scores. We also took advantage of available data sets on certain classes of proteins, such as IUPHAR for receptors and ion channels (73), TransFac for transcription factors (81), and MitoProteome for mitochondrial proteins (39), within the context of our functional families (e.g., integrin subunit “receptors” are viewed as MECH structural proteins with some receptor sites).
For each family, we built a canonical protein network that included most or all of the proteins as described above. The connections in these model networks were set up based on KEGGS pathway database and public protein-protein interaction databases (e.g., Biomolecular Interaction Network Database, Information Hyperlinked over Proteins, and Human Protein Reference Database).
We present the detailed parts lists involved in the four families in Supplemental Table SD. To facilitate mechanistic description of the family models, we use functionally characterized proteins that are a subset of the total parts list. In a few instances, we use legacy knowledge of proteins or those that are differentially expressed in diseased muscle tissues, to incorporate them in the respective pathway diagrams. The expression heat maps that accompany the family pathway diagrams highlight the significance of these genes/proteins for the function of the skeletal muscle.
RESULTS
Based on comparative analysis between skeletal muscle tissue and other nonmuscle tissues, we identify 2,973 genes that have significant higher z-scores and 3,487 genes that have significant lower z-scores, respectively, in skeletal muscle compared with nonmuscle tissues (Supplemental Table SD), with results that appear mostly consistent with existing lists (4, 44, 72, 85, 87). Using our methodology, we use the principle of Occam's razor (selecting the hypothesis of fewest assumptions) to provide the following functional classification of model families and their subfamilies, presented graphically both as heat maps of transcripts and as functional protein networks.
EA Model Family
This family (see Fig. 5A), which normally occupies 3–5% of the physical muscle space, implements what is fundamentally a rapid unidirectional electrochemical information transmission system that transmits focally delivered motoneuronal action potential signals (the input) to millions of myofibrillar regulatory sites on thin filaments that are dispersed within the internal volume of the muscle cell. While often called excitation-contraction coupling (12, 17, 28), a more precise output state of this family is an “activation” signal defined by the degree of key troponin C sites on thin filaments that are bound by calcium ions (Ca2+). This is the definition of the “state” of “calcium activation” in dynamic models of muscle (40, 83); given the high binding affinity between troponin C and Ca2+ (e.g., fast equilibrium dynamics under 1 ms), this state maps closely to the concentration of Ca2+ within the local sarcoplasm.
Fig. 5.
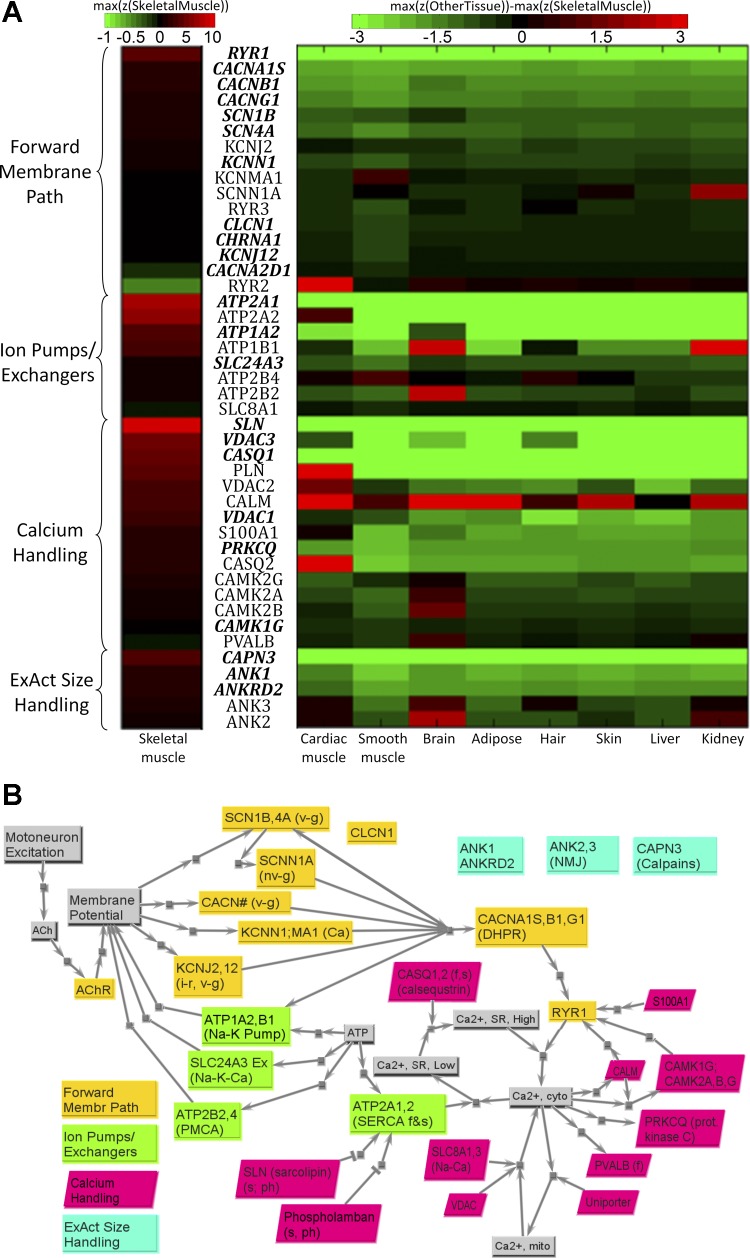
A: primary set of transcripts in EA model family, organized into four subfamilies. Transcripts with bold and italic font have higher z-scores in skeletal muscle than in other tissues (P < 0.05). For skeletal muscle, the color corresponds to the absolute z-score values, but with positive scaling from z = 0 to 10 (black to bright red), and negative only from 0 to −0.1 (black to bright green); for other tissues, the color corresponds to the difference between other respective tissue and skeletal muscle [i.e., z(other tissue) − z(skeletal muscle), with black indicating no saturation (z = 0), and color saturation at z = −3 (bright green) or z = 3 (bright red)]. B: key structural connectivity, as unicausal signals, of the four subfamilies of EA model. Forward membrane path subfamily includes proteins responsible for signal transmission, starting from the neuromuscular junction to the release of calcium from sarcoplasmic reticulum through ryanodine receptor (RYR1). Ion pumps/exchangers subfamily includes proteins within complexes that pump ions across key membranes against a concentration gradient. Calcium-handling subfamily includes proteins involved in the regulation of calcium homeostasis within cytoplasm. ExAct size-handling subfamily includes proteins that are regulators of the structural size and location of sarcoplasmic reticulum (SR). See text and supplemental tables for definitions of acronyms.
Forward membrane path subfamily.
This represents the transmembrane protein complexes responsible for spontaneous signal transmission from neural excitation at the NMJ through the release of a bolus of stored Ca2+. The first step after transduction across the NMJ involves high-fidelity transmission, both axially along the primary sarcolemma and also inwardly via t-tubules, and terminates at voltage sensor gauges within the dihydropyridine receptor (DHPR) protein complex. The DHPRs then trigger a signal amplification effect, which for skeletal muscle involves a mechanical coupling mechanism between the DHPR and ryanodine receptor (RYR) protein complexes of the sarcoplasmic reticulum (SR). This results in release of a bolus of Ca2+ from the high concentration reservoir stored within this region of the SR that accounts for over 99% of the released Ca2+ (58). The various transmembrane receptor gates each consist of a protein complex with multiple subparts defined by gene transcripts (16, 58). The highest z-scores are associated with the skeletal muscle-specific RYR1 and DHPR complex [CACNA1S (calcium channel, voltage-dependent, L type, α1S-subunit), CACNB1 (calcium channel, voltage-dependent, β1-subunit), and CACNG1 (calcium channel, voltage-dependent, γ-subunit 1)]; the heat map in Fig. 5A shows that these are also clearly specific to skeletal muscle, as are many members of this family. Interestingly, for the other ion protein complexes, there often appear to be certain subparts for a given type of channel that have a higher rate of turnover, as reflected by higher transcript z-scores (e.g., KCNS3 subunit for voltage-gated potassium channel, KCNJ2 subunit for inwardly-rectifying potassium channel, CLCN1 subunit for chloride channel); it is these “representative” gene transcripts that are included within our core list and in Fig. 5 (for a broader tabular list, see Ref. 10). In general, the highest transcript z-scores are only of moderate magnitude (e.g., often just below or above z = 1.0), which is consistent with a subfamily with proteins operating on membranes that occupy only a small portion of the volume. Many of these members are shared with cardiac muscle, brain, or hair. Two subunits of clear utilization are actually higher for another tissue (KCNMA1 for smooth muscle, SCNNA1 for kidney). The RYR for cardiac muscle, RYR2, is included, despite having a very low z-score, because we sometimes see it upregulated in the diseased muscle [discussed in the companion paper (78a)].
Ion pumps/exchangers subfamily.
This encompasses the energy-consuming pumps and exchangers for transporting ions (e.g., sodium, potassium, calcium) back across membranes against concentration gradients, so as to “prime” the electrochemical gradients for subsequent signal transmission by the first subfamily. These can be viewed as transmembrane machines, each composed of multiple subparts specified by genes (9). Each type of pump/exchanger has a peak “rate” of performance, and most require the direct consumption of energy via an ATPase site [e.g., for sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA)]. Our results show that there are often very different transcript z-scores for different subparts, presumably reflecting “use and high protein turnover” rates on these subparts that tend to have the highest transcript z-scores. We also find high z-scores for distinct “slow” and “fast” isoforms for the SERCA pumps [gene transcripts ATP2A1 for faster isoform, and ATP2A2 for the slower yet more energy-efficient isoform shared with cardiac muscle (52)]. While not the focus of this paper, the relative proportion of these transcript z-scores and the density of protein machines on SR can change (adapt) dramatically as a function of use history. Figure 5A also shows that ATP1A2 (Na+/K+ transporting) is shared with neural (brain) but not cardiac tissue, and ATP1B1 is actually higher for both brain and kidney. We note that most other z-scores are moderate, suggesting that the adaptive structural design for this subfamily is to maintain functional electrochemical gradients while occupying minimal spatial real estate and requiring energy resources from the METB family only to “reprime” the gradients.
Calcium-handling subfamily.
This represents aspects of the extensive calcium regulatory apparatus (15, 16, 58) that plays a direct role in managing the “real-time” state of free calcium within the sarcoplasm; calcium-related proteins that operate on slower time scales and primarily via transcription pathways are assumed to be part of the signaling family [e.g., peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), peroxisome proliferator-activated receptor (PPAR), histone deacetylase (HDAC), regulator of calcineurin 1 (RCAN1)/myocyte-enriched calcineurin interacting protein (MCIP), PKD, myocyte enhancer factor 2 (MEF2)]. Thus the calcium-signaling “toolkit” of Berridge's group (16) is functionally partitioned between these two families, based largely on temporal dynamics associated with “real-time” functional demands vs. “adaptive” remodeling functions. These subfamily members operate 1) as buffers that transiently bind free calcium in different ways and affect free calcium dynamics (e.g., calcineurin, calmodulin, protein kinase C); 2) as calcium-sensitive and/or phosphorylated dynamic regulators of the rates of calcium SERCA pumps [PLN (phospholamban), especially slow and cardiac fibers; SLN (sarcolipin), all types but especially fast fibers; and PVALB (parvalbumin), for transient buffering of Ca2+ pulses within fast fibers]; or 3) as specialized calcium flux managers within the SR storage space (e.g., calsequestrin, here including both CASQ1 for skeletal and CASQ2 for cardiac, since it is sometimes upregulated in disease) or mitochondrial storage space [e.g., Ca2+ uniporter, voltage-dependent anion channels (VDAC) 2 and 3]. We see in Fig. 5A that many of these are highly specific to skeletal muscle or even to a type of skeletal muscle. For instance, by slowing down SERCA pump rates for the slow SERCA isoform, dephosphorylated SLN and PLN can function (often in unison) to “smooth out” calcium transients during submaximal excitation rates, and yet SLN is shown to be associated with skeletal and has the highest z-score for this subfamily, and PLN is actually highest in cardiac (Fig. 5A). Also with a high z-score is CASQ1, reflecting the importance of effective flux of Ca2+ once within SR, so that it translocates to the proximity of the RYR gates.
EA size-handling subfamily.
This represents regulators of the structural size and location of this family as a function of changing demands. This subfamily is inspired by the recent understanding of the ankyrin (ANK) collection of proteins, as ubiquitous linkers associated with a diverse set of binding partners, including many of the transmembrane pumps/exchangers and certain gates (e.g., RYR), but also the spectrin- and actin-based cytoskeleton (6). They appear to target primarily to EA performance capacity, ranging from myofibrillogenesis at the level of SR and the triads for skeletal muscle (e.g., ANK1, with a z-score in Fig. 5A that shows it is specific to skeletal muscle), which includes spliced isoforms associated with the SR (8) to neuromuscular synaptogenesis (e.g., ANK3, also important for the nervous system and cardiac). ANK appear to help provide an internal scaffolding for maintaining appropriate spatial configurations and alignment that are appropriate for “quality control” of EA signals (e.g., maintaining an effective NMJ and a uniform density of pumps, participating in splitting myofibrils that reach ∼2 μm in diameter, thus protecting calcium diffusion times). A representative transcript from the calpains with a high skeletal-specific z-score is also included; these are involved in breaking down Ca2+-dependent structural components (e.g., critical for SR size remodeling and myofibril splitting). The large macromolecule obscurin, with its various splice variations, appears to help regulate tethering of ANK and the SR with the contractile apparatus (e.g., via Z-disk and m-band connections with titin, discussed below) (5, 34) and suggests spatial regulation of components critical to EA function, such as Ca2+ diffusion distances to troponin; however, it is not yet placed here because 1) our evolving understanding of its roles may suggest the MECH or SIGP family; and 2) it is not included in Affymetrix HG-U133A platform.
MECH Model Family
The key attribute of members of this family is their functional role in bidirectional transmission of force, stiffness, and mechanical power. The contractile and force transmission apparatus, which normally occupies ∼70–85% of the muscle tissue space (depending on the type, role, and use history of a muscle), consists of structural proteins that include fundamental bond connectivity, which is mechanical with transmission of force through these bonds. This family includes the three most abundant proteins (by mass) in most mammals (collagen, actin, myosin) and their various “supporting cast members” that are critical to successful MECH under a wide range of mechanical conditions.
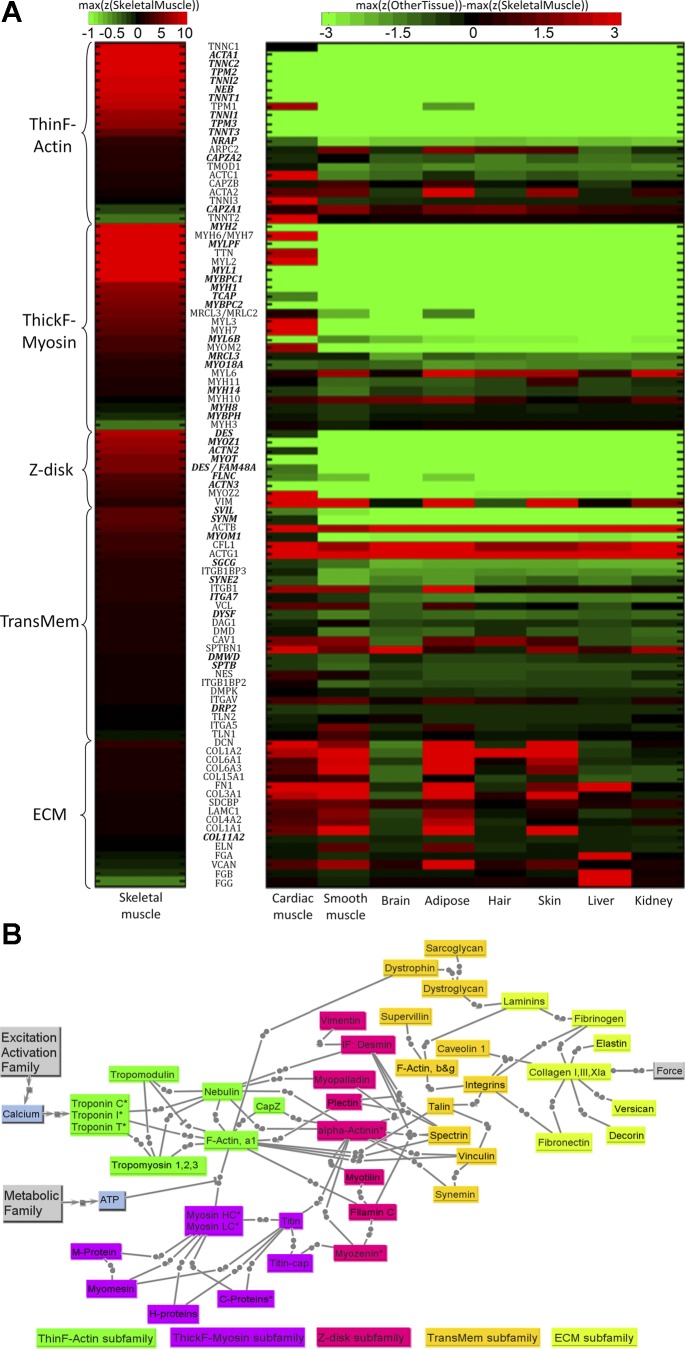
We present the MECH family gene product heat maps (Fig. 6A) and protein connectivity (Fig. 6B) obtained through methods described above. The heat map for the MECH family for skeletal muscle and other tissues (Fig. 6A) identifies a core set with consistently very high z-scores within the tissue sample. Five subfamilies are identified, each with a representative structural protein “pillar” (Fig. 6A) based on functional connectivity during actual operation, where forces are transmitted across this network over time scales on the order of milliseconds. The transient “state” and connectivity between these members is fundamentally mechanical rather than chemical or electrical (Fig. 6B); in mathematical models, force transmission and often strain and strain rate are of interest, especially since most of these building block parts contribute to the inherent viscoelasticity of muscle tissue (35), as reflected in muscle models (43, 82). Whenever viable, we lump parallel alternative isoforms together in Fig. 6B, thus embedding the biological tissue property of redundancy and overlap of function into our model; however, for the heat maps of Fig. 6A, all available isoforms are given individually. There was one case, actin, where the isoforms could not reduce below two: one set of isoforms that were members of the thin-filament subfamily, and one set that were functionally cytoskeletal and support transmembrane connectivity.
Fig. 6.
A: key transcripts in MECH model family. Transcripts with bold and italic font have higher z-scores in skeletal muscle than in other tissues (P < 0.05). See Fig. 5A legend for color-coding scheme. B: structural protein connectivity of the five subgroups composing the MECH model family. Connections ending with solid circle represent mechanical bonding. The “*” at the end of some proteins' names indicates that the protein has different isoforms in different types of fibers. Actin subfamily includes proteins associated mainly with the thin filament. Myosin subfamily includes proteins associated mainly with the thick filament. Z-disk subfamily includes proteins localized at the Z-disk and some intermediate filament proteins also associated with costameres. TransMem subfamily includes transmembrane “scaffolding” proteins just internal to or crossing the sarcolemma. ECM subfamily includes proteins composing the extracellular matrix for muscle tissue. See text and supplemental tables for definitions of acronyms.
Advantage of functional MECH approach.
Most of these members also have other connectivity (e.g., mechanosensing, assembly, other signaling), but what ties these together and delineates them from other families is their fundamental participation in structural mechanics. High z-scores for spanning core members of the F-actin/thin-filament and myosin/thick-filament subfamilies (e.g., often over z = 10 and usually over z = 5) with known interdependent connectivity reflects high rates of both gene products and, even more critically, protein production and coordinated assembly for subfamilies that span over one-half of the physical three-dimensional (3D) space. This is consistent with the reasonably high turnover rates for these key structural proteins [e.g., human skeletal muscle actin and myosin isoforms have a half-life of about 1 mo and can be as short as 1 wk (38, 79)]. Our z-score data also suggest isoforms for both slow and fast muscle, with results robustly consistent with the separate data set obtained for young normal [provided in companion paper (78a)].
F-actin/thin-filament subfamily.
It is useful to conceptually divide this subfamily into two functional subclasses: those participating in the repeat pattern where one component is needed for every seven actins (troponins C, T, and I, tropomyosin), and those where one to two components are needed per thin filament (nebulin, tropomodulin, capping protein z). There are consistently high z-scores for the functionally critical tropomyosin and the three troponin components, each with two to three isoforms that are associated with slow- and fast-muscle properties, and especially high for the troponin C isoforms that support the “state” of Ca2+ regulatory binding. Another remarkable finding is the high z-score for nebulin, since only two nebulin molecules span the length of the thin filament, which contains ∼200 times more actins and 30 times more tropomyosins and troponin subunits. This large molecule (∼600 kDa), representing 3–4% of the muscle protein content, is widely viewed as a sort of “ruler” that sets the thin-filament length through its ability to bind actin and tropomyosin via its repeat units along the length of the thin filament. The high z-scores for nebulin and the capping proteins, especially tropomodulin, suggest that detachment (e.g., for thin-filament parts adjustment) and replacement is frequent, most likely with a net addition of actins to the “barbed” end on the Z-disk (higher force) side that is regulated and capped by the cap z, and a net spillover at the dangling end on the “pointed” end that is capped by tropomodulin via actin and tropomyosin binding (24).
Myosin/thick-filament subfamily.
This includes the actual myosin components that are necessary for the roughly 300 units of myosin components per thick filament, each consisting of two wrapped myosin heavy chains (MHCs) with NH2-terminal “cross-bridge” heads and interconnected COOH-terminal tails that form much of the base of the thick filament, with two types of two customized myosin light chain (MLC) parts located near a “quasi-hinge” between these two functional MHC parts. All have reasonably high z-scores, including fast/slow isoforms. Also very high is the z-score for titin, a large (∼3 million kDa) multifunctional structural protein with very low overall stiffness that spans from the Z-disk to the thick filaments, and among other functions is involved in 3D stability of the thick filaments and in their assembly, and with possible strain-sensing sites. Two transcripts within these subfamilies with negative z-scores (representing isoforms that are essentially “off” for skeletal muscle), myosin, heavy chain 3, skeletal muscle, embryonic (MYH3) and troponin T type 2 (cardiac) (TNNT2), are included because we have seen pathological cases where their expression increases as skeletal muscle “reverts” toward use of more embryonic isoforms (e.g., MYH3) or toward use of cardiac isoforms (e.g., TNN2).
Z-disk subfamily.
These structural proteins, part of a localized 3D lattice that has a larger axial thickness in slow (vs. fast) muscle fibers, have mostly lower z-scores than for the aforementioned families (likely reflecting their smaller volumes within the muscle space). Four are relatively high: isoforms for α-actinin (ACTN2), clearly shared with cardiac in Fig. 6A, and to a less extent ACTN3, the critical “adhesive” proteins that help bind actin and other mechanical components; desmin (DES), an intermediate filament (IF) for both skeletal and cardiac muscle (Fig. 6A) that provides 3D structural support by connecting myofibrils at the Z-disks to each other and to transmembrane components and, as an IF, can switch between phosphorylated and unphosphorylated states with variable stiffness; myozenin (MYOZ1), the skeletal muscle isoform of this adhesive protein with binding that includes α-actinin (note in Fig. 6A that MYOZ2 is clearly more associated with cardiac, but still used for skeletal); and myotilin (MYOT), another adhesive protein for skeletal muscle with binding that includes α-actinin, f-actin, and titin.
Transmembrane subfamily.
Many of these cytoskeletal “scaffolding” and adhesive structures are also used by other types of cells (e.g., fibroblasts), such as for focal adhesions in nonmuscle cells (31); most accumulate at “costamere” structures that share many proteins with focal adhesions and tend to register laterally with Z-disks. Of special significance are two transmembrane mechanical linkage pathways composed of a complex of protein partners that mechanically protect the sarcolemma (33): the integrin complex (primary mode for focal adhesions) and the dystrophin-sarcoglycan (DSG) complex [a specialized apparatus for transmission forces across the membrane–the absence of dystrophin leads to DMD, as described in the companion paper (78a)]. Figure 6B illustrates a strategic subset of this large mechanical network of scaffolding proteins and of their connectivity. Z-scores for this subfamily tend to be much lower than that of the f-actin or myosin subfamilies and somewhat lower than that of the Z-disk subfamily, likely reflecting the lower volume of the scaffolding layer by the membrane shell, but, as diseases such as DMD indicate in our companion paper (78a), this is also a very important subfamily. The highest z-scores are for two cytoskeletal workhorse linker proteins, supervillin (SVIL; which binds f-actins to other scaffolding proteins including integrin complex) and synemin (SYNM; an IF with binding partners that include dystrophin). Also listed as representative are key muscle-specific subparts of the integrin complex [e.g., ITGA7 shared with only cardiac, the ITGB1BP3 subpart of the ITGB1 binding protein that is clearly also used for cardiac muscle (see Fig. 6A)], and some strategic proteins of the DSG complex including γ-sarcoglycan (SGCG), dystrophin (the largest known gene and involved in DMD), and dysferlin (DYSF). The dystrobrevin linkers, dystrobindin and synthrophin, of the DSG had moderate z-scores (highest probes of z-score = 0.6 to 1.6) were not included so that DSG is not overemphasized. Of note is that many of these structural elements not only transmit mechanical force, but are embedded with mechanosensor and other signaling sites (21, 31, 78).
ECM subfamily.
These are mostly expressed and produced by fibroblasts within muscle tissue, and various collagen isoforms are shared with certain other tissues (70). Decorin (DCN; a proteoglycan) and fibronectin 1 (FN1) have reasonably high z-scores, and some other representative ECM proteins, such as fibrinogens, are also included. While z-scores for this family are generally only low to moderate (reflecting their low relative volume in the tissue), of note is that, in disease conditions, both their overall and their relative contributions (e.g., of collagens) can change (most often increasing) as “hybrid” muscle tissue forms; endurance exercise training also causes upregulation of many ECM transcripts (77), with tight regulation between muscle and tendon (49). This subfamily includes several members with z-scores that are actually higher in certain other tissues, such as smooth muscle and adipose tissue for elastin and versican (Fig. 6A) and liver for fibrinogen, but that we find to be selectively upregulated for certain types of diseased muscle.
METB Model Family
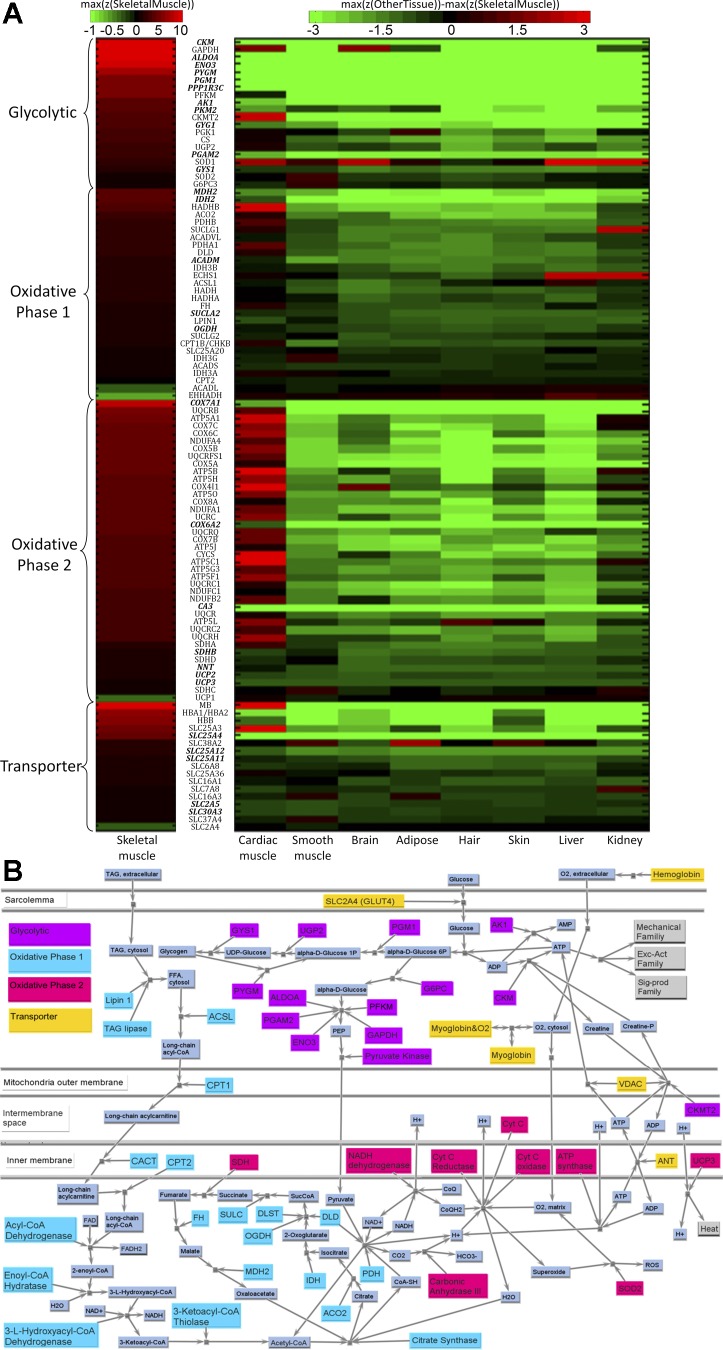
This family (see Fig. 7A) mainly deals with energy supply and consumption, designed to meet the fluctuating demands of the EA and MECH families. Compared with the resting state, when skeletal muscle is activated, the rate of ATP consumption can increase more than 100-fold, and most of the energy is consumed by Na+-K+-ATPase and Ca2+-ATPase from the EA family and actomyosin ATPase from the MECH family. However, the ATP concentration in sarcoplasm is known to be rather stable, even in fatigued muscle (2, 3). This nearly constant ATP supply is maintained by several complicated ATP replenishing systems dealing with immediate, short-term, and long-term energy requirements. Proteins involved in the immediate and short-term energy supply system were classified into the glycolytic/short-term subfamily, whereas proteins in long-term energy supply system were placed into the oxidative subfamily (e.g., enzymes involved in β-oxidation and TCA cycle) and the electron transport subfamily on the mitochondria inner membrane, which are responsible for high-yield ATP production. The transporter subfamily consists of carrier proteins that transport metabolic substrates, such as glucose and oxygen, from blood into muscle cells.
Fig. 7.
A: key transcripts in METB family. Transcripts with bold and italic font have higher z-scores in skeletal muscle than in other tissues (P < 0.05). See Fig. 5A legend for description of color-coded scheme. B: the protein network of METB model family. This model is partitioned into four subfamilies: oxidative, including members helping transfer lipid into mitochondria, enzymes involved in β-oxidation and TCA cycle; glycolytic, including members involved in supporting immediate energy, glycolysis, and glycogenesis; electron transport, including members generating the mitochondrial membrane potential and producing ATP; and transport, involved in transporting materials such as glucose and oxygen. See text and supplemental tables for definitions of acronyms. See text and supplemental tables for definitions of acronyms.
Glycolytic/short-term subfamily.
This is assumed to be composed of four pathways: two ATP replenishment mechanisms to meet immediate ATP demands, the consumption of glucose to generate ATP and pyruvate (glycolysis), and the transformation between glucose and glycogen polymers (i.e., gluconeogenesis and glycogenolysis). The first main source of immediate replacement of ATP, creatine phosphate, involves the enzyme CKM (creatine kinase, muscle), which had the highest z-score within this subfamily and is notably higher than for any other tissue, including cardiac muscle. CKM is normally stored in a bound form within the M-region of the myosin/thick-filament subfamily (42). In skeletal muscle, the second immediate energy replenishment mechanism is to generate ATP and AMP from ADP (see Fig. 7B), a reaction catalyzed by the skeletal muscle specific isoform adenylate kinase 1 (AK1), which also had higher z-score in skeletal muscle than in any other tissue. Proteins (and associated transcripts) included as part of the glycolytic subfamily include a number with very high z-scores (Fig. 7A): aldolase A fructose-bisphosphate (ALDOA), muscle phosphofructokinase (PFKM), phosphoglycerate mutase 2 (muscle) (PGAM2), enolase 3 (β, muscle) (ENO3), GAPDH, and muscle pyruvate kinase (PKM2), with ALDOA and ENO3 standing out as being remarkably higher for skeletal muscle compared with those in other tissues. Insulin receptors are included as part of the SIGP family. Of the protein involved in glycogenolysis (conversion of glycogen to glucose monomers) that are included in the parts list, one [phosphoglucomutase 1 (PGM1)] had significantly higher z-score than that for any other tissue including cardiac, as was one for gluconeogenesis [muscle glycogen phosphorylase (PYGM)].
Oxidative-phase 1 subfamily.
This process uses both lipids and the products of glycolysis within the fuel mix for satisfying longer term ATP requirement through aerobic oxidation. As Fig. 7B shows, enzymes supporting lipid oxidation implement three steps: the transportation of lipid into mitochondria, β-oxidation, and citric acid cycle (also called Krebs cycle). The first step of breaking triacylglycerol (TAG) into free fatty acid for transport into mitochondria (as long-chain acyl-CoA) involves TAG lipase and lipin, and then acyl-CoA synthetase. The translocation of long-chain acyl-CoA from cytosol to mitochondria is assisted by carnitine palmitoyltransferases and the carnitine/acylcarnitine translocase. The highest z-score for this process is for HADHB [hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase (trifunctional protein), β-subunit] (see Fig. 7A), which is even higher in the cardiac muscle, likely because a skeletal muscle tissue sample also includes fast muscle fibers, which make less use of HADHB. After long-chain acyl-CoA enters mitochondria, it goes through β-oxidation and transfers to acetyl-CoA, which starts the citric acid cycle. Acetyl-CoA is also generated from pyruvate, the end product of glycolysis. The z-scores for members of this ubiquitous subfamily are of moderate value, although higher than for most tissues, likely owing to the higher mitochondrial density. Enzymes with the highest z-scores in Fig. 7B are MDH2 (malate dehydrogenase) and IDH2 (isocitrate dehydrogenase). The citric acid cycle generates NADH and H+ used by the electron transport subfamily.
Oxidative-phase 2 subfamily.
This includes a chain complex composed of four parts, often called complexes I-IV, that align on the inner membrane, as seen in Fig. 7A, plus the ATP synthase machinery and proteins that are involved in proton/electron transfer, such as carbonic anhydrase. While complex II (SDH, succinate dehydrogenase) is involved in TCA, others transport H+ generated in TCA from the mitochondria matrix into the intermembrane space, which is later used by ATP synthase (represented by ATP5 subparts) to generate ATP. In general, the z-scores are high for this subfamily, with the most notable being COX7A1 [cytochrome c oxidase subunit VIIa, polypeptide 1 (muscle)] [and to a lesser extent COX6A2 (cytochrome c oxidase, subunit VIa, polypeptide 2)], which are seen to be specific to muscle and especially skeletal muscle (Fig. 7A). In contrast, many of the other subparts are even higher in cardiac muscle, reflecting the fact that the mixed-fiber content of skeletal muscle tissue samples, such as the vastus lateralis, include fast-glycolytic as well as slow-oxidative fibers within the tissue sample.
Transporter subfamily.
These members, critical to meeting functional demands, especially during high muscle use, include several oxygen transporters (hemoglobin within blood and myoglobin within muscle cells) with high z-scores, some proteins with high z-scores that are mitochondrial solute carriers (SLCs) across its inner membrane (SLC25), the VDACs that are also part of the EA calcium handler subfamily (see Fig. 5), and other key members of the SLC family, such as SCL2A/glucose transporter (GLUT) for transporting sugars, such as glucose (see Figs. 7A and 6B). Myoglobin has very high z-score (9.8) in skeletal muscle, approaching that in cardiac muscle (z-score: 13.9) and much higher than in other tissues (z-scores are all negative). The z-score of hemoglobin is rather high in skeletal muscle, but also similarly high in cardiac muscle and skin. A number of the SLC transporters of various metabolic substrates are significantly higher, with SLC25A4 (mitochondrial SLC; adenine nucleotide translocator, member 4), a gated pore at the mitochondrial inner membrane that translocates ATP from the mitochondrial matrix into the intermembrane space, standing out. VDACs, the porin ion channels at the mitochondrial outer membrane, not only help regulate Ca2+ flux, but are also the main channel for the exchange of metabolites between mitochondria and cytoplasm, with moderately high z-scores, especially for VDAC3 (Fig. 5A).
SIGP Model Family
Unlike in the three families discussed above, this family relates primarily to tissue remodeling through up/downregulation of the protein within the families described above, rather than directly participating in real-time muscle action. Some proteins that are known to be important for intracellular signaling, but not unique to the skeletal muscle are included, as are muscle-specific transcription factors and transcriptional and translational regulators. Here we functionally partition the signaling family into longitudinal branches involved in calcium signaling and fiber remodeling and those involved in muscle regeneration and muscle atrophy. The organization also follows the time scale of signaling processes from seconds to days, spanning early signaling events to transcription and muscle regeneration.
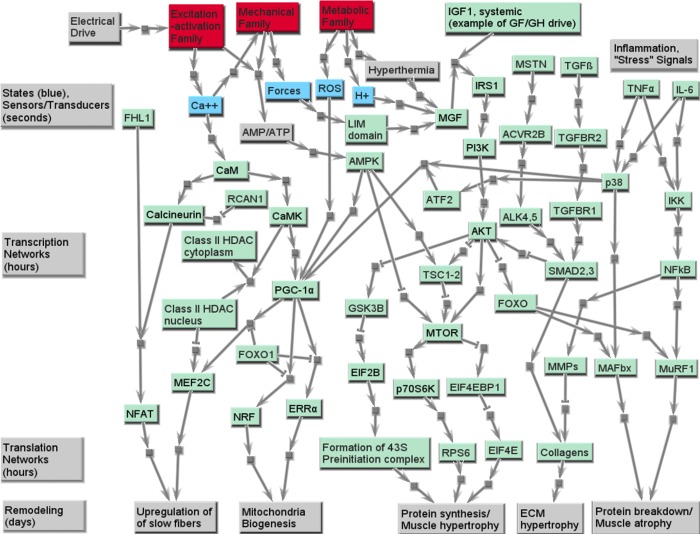
Several signaling molecules involved in skeletal muscle remodeling have been studied extensively that are either associated with calcium or mechanical variables, or are transcription factors that are downstream from calcium or mechanical signaling events (10, 80, 84, 87). Our analysis of the gene expression data from normal patient muscle tissues identifies the canonical list of players that are involved in remodeling of the muscle fiber. As an example, these include the calmodulin-calcineurin-nuclear factor of activated T-cells (NFAT) path proteins and calmodulin kinase-histone deacetylase-MEF2C (transcription factor) path proteins that are triggered by EA dysfunction. Calmodulin kinase also regulates a key mitochondrial master regulator, PGC-1α, that, in turn, through nuclear respiratory factors (NRF) and estrogen-related receptor-α (ERR-α) (transcription factors) regulates genes associated with mitochondrial biogenesis and thus affects the METB family (14, 32). Key pathways associated with the signaling events leading to fiber remodeling and mitochondrial biogenesis are presented in Fig. 8. As demonstrated in other systems, mitochondrial biogenesis is also regulated by reactive oxygen species, as well as mechanical forces within various MECH subfamilies. These paths are also represented in Fig. 8, with the skeletal muscle LIM family [e.g., FHL1 (four and one-half LIM domain protein 1), which has a very high z-score, and is shown separately] being representative of mechanotransduction signaling. Several pathologies are associated with inability of the muscle to switch fiber types, and we focus on aspects of cellular signaling defects in DMD and ALS in the companion paper (78a).
Fig. 8.
Signaling proteins involved in skeletal muscle remodeling processes. The green nodes are proteins in SIGP family, the red nodes are other protein families, and the blue nodes are key dynamic states in the final muscle model framework. Along the bottom are functional consequences related to phenotypic remodeling of skeletal muscle, including changes in the mass/size (atrophy and hypertrophy), fiber composition (percentage of slow type I and fast type II fibers), and fatigue resistance (proportion of oxidative vs. glycolytic fibers, and volume of mitochondria). See text and supplemental tables for definitions of acronyms.
The short-, medium-, and long-term function of the skeletal muscle is orchestrated by myriad signal transduction processes, which are described in Fig. 8. Our discussion below is centered around the signaling processes that span space and time in the cell and lead to changes in functional phenotypes, using our family structure as a framework. This will include fiber composition transformation, mitochondrial biogenesis, protein synthesis, leading to muscle hypertrophy, and protein breakdown, leading to muscle atrophy. In addition to phenotypes regulated by pathways such as calcium signaling, cellular signaling events also alter protein syntheses, and breakdown in skeletal muscle are also part of the SIGP family. Muscle hypertrophy can result from signaling pathways triggered by hormones and growth factors through the insulin receptor [insulin-like growth factor I receptor (IGF-IR)] stimulated phosphatidylinositol 3-kinase (PI3K) activity (36, 37) or by inhibition of myostatin through transforming growth factor-β (TGF-β) regulated SMAD pathways (51, 62). Also shown is triggering by use-related high forces within MECH family structural members that induce mechanosensor signaling events [e.g., FHL1, SMPX (small muscle protein, X-linked)], which stimulate the expression of mechanogrowth factor (MGF), an important local isoform (splice variant) of IGF-I (59). The IGF-I/MGF pathway involves AKT, mammalian target of rapamycin (mTOR) pathways that alter translation factor networks that regulate muscle hypertrophy. Alternately, inflammation and stress signals (e.g., due to damage of MECH family members) act through select cytokines resulting in NFκB driven changes to muscle atrophy factors MURF1 and MAFbx proteins. Key genes that code for the muscle hypertrophy and atrophy proteins are seen in our heat map and although common to several tissue types, these are significantly expressed also in the muscle. We note that normal patients whose muscle fibers are used in these studies have mixed use history, and we see expression of all the signaling proteins irrespective of specific fiber types. Fig. 8 also shows key signaling proteins involved in muscle hypertrophy and atrophy linked through canonical pathways (36, 37) associated PI3K-mTOR path (hypertrophy) and TNF-α-NF-κB path (atrophy). While a number of members of the signaling parts list are not unique to skeletal muscle, the specific paths and their dynamic activation are unique to the muscle and provide the type of minimal robust network model that can be used to help support emerging theories of evolutionarily well-conserved muscle signaling networks that are redundantly controlled by multiple higher level myogenic regulators, as well as local adaptive remodeling signals (11).
Some proteins that belong to the SIGP family are not explicitly included in the network in Fig. 8. These include the following: 1) proteins that serve as a biomarker of a disease or a remodeled status [e.g., SRPK3 (SFRS protein kinase 3), CHRNA1 (cholinergic receptor, nicotinic, α1, muscle) and RYR3]; and 2) proteins that are significantly differently expressed in skeletal muscle than in other tissues, but not included in other protein families [e.g., KTN1 (kinectin 1) and RXRA (retinoid X-receptor, α)]. The complete list of this protein family and related heat maps can be found in Supplemental Table SD. It would also be interesting to see the relationships between our families and the muscle-specific micro-RNAs, presumed to be macroregulators of mRNA abundance, that are gradually being identified (63).
DISCUSSION
There are several potential pitfalls along the steps of the process of collecting transcriptome data, from extracting a clean and representative muscle tissue sample that does not yet express a stress response to successfully navigating though the sample extraction process to avoid pitfalls of using so-called “bookkeeping” genes that may be varying. In this context, an important contribution of our approach is that we found that, through a careful normalization and clustering process, a remarkably robust cluster of genes is physiologically meaningful, with z-scores providing a greater range of mean values and yet lower standard deviations. Based on robust statistical analysis, we conclude that the 120 microarrays identified as the dominant cluster, coming from many studies, do in fact represent the “main-stream” of the normal gene expression pattern of skeletal muscle. First, z-scores associated with most of the known skeletal muscle-specific proteins, such as CKM, COX7A1, MYH2, ACTA1 (actin-α1, skeletal muscle), and so on, were among the top 1% highest z-scores. Second, the main structural proteins of thin and thick filaments (such as actin, troponin isoforms, tropomyosin isoforms, nebulin, titin, and myosin isoforms), SERCAs, the main proteins involved in glycolysis and oxidation (such as GAPDH, PYGM, ENO3, and COX7A1), and LIM domain proteins [such as FHL1 and PDLIM3 (PDZ and LIM domain 3)] had higher z-scores in the centers of this dominant cluster than of other clusters. Finally, the method also worked for integrating available young healthy samples [see companion paper (78a) and its supplemental data], where analysis of totally separate data sets yielded similar results (with occasional differences that made physiological sense in the context of developmental biology). Furthermore, other tissues (e.g., cardiac muscle tissue, certain disease muscle tissue samples) also benefited from clustering; some degree of abnormal data sets should be expected, but, with large enough samples, can be eliminated using appropriate statistical procedures. This provides validation to our method, increased confidence in the results of protein families, and further justification for muscle physiologists to utilize our approach and expand on our results.
The four functional families, with their subfamilies, and the associated model pathways presented above provide a detailed mechanistic and connected network view of the skeletal muscle. Our detailed networks also provide a spatiotemporal view of skeletal muscle function. Sampling of the normal skeletal muscle tissue shows significant expression of genes that code for proteins involved in muscle function, with most members of the MECH and EA families being unique to skeletal muscle, and with some shared with just a few other tissues; an exception is the ECM subfamily of the MECH family, as should be expected. Some subfamilies include isoforms representative of slow and fast fibers (many in MECH family, some in EA pumps/exchangers subfamily, and calcium-handling subfamily), with strong confirmation of cases where certain isoforms are shared with cardiac muscle. Also, metabolic processes that are associated with both glycolytic and oxidative machinery are confirmed and generally higher for skeletal and cardiac muscle, owing to their high demands that are over and above those of basal metabolic function (46); enzymatic levels decrease with disuse (23). Yet some explainable exceptions are illustrated in Fig. 7A (e.g., GAPDH is even higher for brain, SOD1 is higher for brain, kidney, liver). Furthermore, the results are robust, as will be seen in the companion paper (78a), for a parallel analysis using independent samples of young normal muscle. They are generally consistent with the microarray work of other groups (48, 55, 72) and provide normative data that can be helpful and complementary to differential microarray studies (4, 68).
Cellular signaling at multiple time scales dictate muscle function, remodeling, and regulation of metabolism, as reflected in the SIGP family. Skeletal muscle adapts to its use history. SIGP family includes proteins involved in the remodeling process of skeletal muscle, including receptors that detect and transmit mechanical and biochemical signals, intermediate proteins that pass these signals to transcription factors that alter gene production rates, and proteins that help with protein synthesis and/or breakdown. The electrical command signal communicated to the muscle triggers early signaling events involving transient receptors and channels of the EA family that lead to dynamic changes in intracellular calcium and, through activation of the MECH family, to forces that in turn lead [via mechanochemical transduction sites on many members of the MECH family, such as f focal adhesion kinase (FAK) signaling for mostly slow-oxidative fibers and TransMem subfamily members associated with integrins (27, 29)] to mechanical signaling events. While commonly there is synergy between these events, the bicausal nature of connectivity between MECH family members implies that forces are also a function of imposed loading and kinematics provided by skeletal structures and environment (e.g., for the same activation, there will be relatively higher forces with eccentric contractions and lower for low-loaded concentric movements). Furthermore, sustained EA (and thus MECH) demands for ATP energy supply, especially of causing energy-dependent difficulties in calcium handling (e.g., error signals associated with inadequate SERCA pumping), may also trigger stress-related signaling within the METB family members. Low use or disuse can also serve to trigger other (mostly competing) pathways. Figure 8 illustrates how all three families can provide sources for signaling pathways (top), and how the “demands” of a temporal use history, combined with secreted agents from other cells (here represented by IGF and TGF), cause integrative signaling that flows from sources at the top to phenotypic consequences (expressed in the three families) at the bottom. The key point is that the nature of the activation and force trajectories triggers muscle signaling events that can, over longer time periods, result in dynamic remodeling of the skeletal muscle between slow and fast fibers, as well as fiber size. Our SIGP family structure enables us to distinguish between the interplay between signaling and metabolism that dictates the remodeling processes. SIGP family proteins involved in the remodeling process of skeletal muscle include receptors that detect and transmit mechanical and biochemical signals, intermediate proteins that pass these signals to transcription level, and proteins that help protein synthesis and/or breakdown. Three aspects highlight this remodeling, as related to “capacity” criteria for each of our three families (see Fig. 4): the density of transmembrane proteins associated with our EA family, which affect transmission capacity; the muscle mass (or the cross-sectional area), which affects the peak contractile force capacity; and the oxidative capacity, which determines how long the skeletal muscle can work incessantly at submaximal intensity. The change of muscle mass normally refers to muscle atrophy/hypertrophy. And the oxidative capacity of a muscle fiber depends on the mitochondrial volume and the content of enzymes in it, which are also of the characteristics of different fiber types. The fiber composition of skeletal muscle is one criterion that determines its oxidative capacity.
The skeletal muscle atrophy/hypertrophy phenotypic changes (19, 66) and signaling pathways (16, 20, 71, 86) have been widely studied and reviewed. As Fig. 8 shows, the hypertrophy pathway starts with the short period, but intensive tension that the muscle experienced, which is sensed by mechanical sensors (e.g., FHL1 and SMPX) and stimulates the expression of MGF. MGF is a local isoform of IGF-I. Like systemic IGF-I, it can stimulate the protein synthesis through PI3K/AKT pathway (69). The hypertrophy effect of PI3K/AKT is through two independent mediators, mTOR and glycogen synthase kinase-3β (GSK-3β), which then promote the ribosome proteins at translational level and stimulate the protein synthesis, resulting in hypertrophy. AKT also blocks the atrophy pathways by inhibiting FOXO (forkhead box O1), which is a promoter for two ubiquitin ligases [muscle atrophy F-box (MAFbx) and muscle RING finger 1 (MuRF1)], the key players in muscle atrophy process. Among these proteins, insulin receptor substrate 1 (IRS1) and eukaryotic translation initiation factor 2B, subunit-2β, 39 kDa (EIF2B) have higher, while ribosomal protein S6 kinase, 70 kDa, polypeptide 1 (RPS6) has lower, expression level in skeletal muscle than in any other tissue (see Supplemental Table SC).
As mentioned above, the type of a muscle fiber indicates its oxidative capacity. Generally, the type I (slow) fibers have higher oxidative capacity, while the type IIx fibers are more glycolytic. Glycolytic fibers may transform to oxidative fibers, along with the increasing mitochondrial volume. Endurance exercise promotes mitochondria biogenesis, a process that may happen in days (41) and is controlled by PGC-1α (1). On the other hand, calcineurin/NFAT and MEF2C can promote the expression of slow fibers (57), which utilizes a different isoform of MHC. In skeletal muscle, MEF2 and PGC-1α have higher expression level than in any other tissues, while FHL1, a promoter of the calcineurin/NFAT pathway, is significantly overexpressed (see Supplemental Table SD).
Our parts list and network framework for skeletal muscle function pave the way for discussing muscle pathologies, as illustrated in the companion paper (78a). Furthermore, these networks provide mechanistic insights into the nature of the dysfunction and thus provide pathways for therapeutic interventions. Most importantly, our model family structure serves as a highly valuable framework for more quantitative modeling that can help connect muscle mechanical models used for task analysis (40, 82) with calcium flux models (60, 61) and with metabolic models (13, 25, 47), which can be used to complement targeted experimental investigations of human skeletal muscle physiology that include use of muscle biopsy data.
GRANTS
This work is supported by the following grants: National Heart, Lung, and Blood Institute Grant 5 R33 HL087375-02 (S. Subramaniam), National Science Foundation (NSF) Grant DBI-0641037 (S. Subramaniam), NSF Collaborative Grant DBI-0835541 (S. Subramaniam), and NSF Collaborative Grant STC-0939370 (S. Subramaniam).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: Y.W., J.M.W., and S.S. conception and design of research; Y.W., J.M.W., and S.S. performed experiments; Y.W., J.M.W., and S.S. analyzed data; Y.W., J.M.W., and S.S. interpreted results of experiments; Y.W., J.M.W., and S.S. prepared figures; Y.W., J.M.W., and S.S. drafted manuscript; Y.W., J.M.W., and S.S. edited and revised manuscript; Y.W., J.M.W., and S.S. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Robert Byrnes for assistance in figure generation, Dr. Brian Saunders for assistance in data management, and Richard L. Lieber for valuable discussions.
APPENDIX
Fuzzy Algorithm to Integrate t-test Results
We present below the fuzzy algorithm used to integrate t-test results for comparing transcriptome of the “main stream” skeletal muscle to those of other tissues. For every type of other tissue:
Step 1: Get the z-scores of every microarray data, including both skeletal muscle and tissue T.
Step 2: Compare means of every probe of skeletal muscle to tissue T, the result r is 1 (skeletal muscle is no less than tissue T) or −1 (skeletal muscle is less than tissue T).
Step 3: Do t-test of skeletal muscle and tissue T for every probe.
Step 4: Divide result of step 2 into two groups: consistent, which means that the results of all probes pointing to the same gene are all 1 or all −1; and inconsistent, which means that the results of all probes pointing to the same gene have both 1 and −1.
Step 5: For consistent group, assuming n probes point to the same gene with P value pi (i = 1, …, n), calculate the degree of significance yi for each pi. The total significance y = Uyi, where U is a fuzzy OR operation (a U b = a + b − ab). If y is higher than 0.75 and r is 1, then the n probes go to ConsistHigh subgroup; if y is higher than 0.75 and r is −1, then the n probes go to ConsistLow subgroup; if y is not higher than 0.75, then the n probes go to ConsistNonsig subgroup. The value 0.75, which means P value is 0.0075 for n = 1, is arbitrarily picked.
- Step 6: While the significance is the most and only important parameter when processing consistent probes, both significance and number of probes must be considered when processing inconsistent ones. For inconsistent probes, assuming n probes with positive r (P value: xpi) and m probes with negative r (P value: xni) [total (n + m) probes], the pseudo-code is,
The threshold for pos/sig is 2/3, which means significant higher: significant lower = 2:1 for InconsistHigh subgroup (or vice versa for InconsistLow subgroup) is arbitrarily picked. A value of 3/4 for pos/sig ratio, produced only about 1% of difference in the result.
Step 7: Combine ConsistHigh and InconsistHigh subgroups into SK-High-Than-TissueT group, which represents the higher expressed genes in skeletal muscle compared with tissue T.
REFERENCES
- 1. Adhihetty PJ, Irrcher I, Joseph AM, Lyubicic V, Hood DA. Plasticity of skeletal muscle mitochondria in response to contractile activity. Exp Physiol 88: 99–107, 2003. [DOI] [PubMed] [Google Scholar]
- 2. Allen DG, Lamb GD, Westerblad H. Skeletal muscle fatigue: cellular mechanisms. Physiol Rev 88: 287–332, 2007. [DOI] [PubMed] [Google Scholar]
- 3. Allen DG, Lännergren J, Westerblad H. Intracellular ATP measured with luciferin/luciferase in isolated single mouse skeletal muscle fibers. Pflügers Arch 443: 836–842, 2002. [DOI] [PubMed] [Google Scholar]
- 4. Almon RR, DuBois DC, Yao Z, Hoffman EP, Ghimbovschi S, Jusko WJ. Microarray analysis of the temporal response of skeletal muscle to methylprednisolone: comparative analysis of two dosing regimens. Physiol Genomics 30: 282–299, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armani A, Galli S, Giacomello E, Bagnato P, Barone V, Rossi D, Sorrentino V. Molecular interactions with obscurin are involved in the localization of muscle-specific small ankyrin 1 isoforms to subcompartments of the sarcoplasmic reticulum. Exp Cell Res 312: 3546–3558, 2006. [DOI] [PubMed] [Google Scholar]
- 6. Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol 160: 245–253, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519, 2001. [DOI] [PubMed] [Google Scholar]
- 8. Barany M. ATPase activity of myosin correlated with speed of muscle shortening. J Gen Physiol 50: 197–218, 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barton K, MacLennan D. The proteins of sarcotubular system. In: Myology: Basic and Clinical (3rd Ed.), edited by Engel AG, Franzini-Armstrong C. New York: McGraw Hill, 2004, p. 307–323. [Google Scholar]
- 10. Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem 75: 19–37, 2006. [DOI] [PubMed] [Google Scholar]
- 11. Baugh LR, Hunter CP. MyoD, modularity, and myogenesis: conservation of regulators and redundancy in C. elegans. Genes Dev 20: 3342–3346, 2006. [DOI] [PubMed] [Google Scholar]
- 12. Beam KG, Horowitz P. Excitation-contraction coupling in skeletal muscle. In: Myology: Basic and Clinical (3rd Ed.), edited by Engel AG, Franzini-Armstrong C. New York: McGraw Hill, 2004, p. 257–280. [Google Scholar]
- 13. Beard DA. A biophysical model of the mitochondrial respiratory system and oxidative phosphorylation. PLoS Comput Biol 1: e36, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benton CR, Wright DC, Bonen A. PGC-1alpha-mediated regulation of gene expression and metabolism: implications for nutrition and exercise prescriptions. Appl Physiol Nutr Metab 33: 843–862, 2008. [DOI] [PubMed] [Google Scholar]
- 15. Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiol Rev 80: 1216–1265, 2000. [DOI] [PubMed] [Google Scholar]
- 16. Berridge MJ, Bootman MD, Roderick HL. Calcium signaling: dynamics, homeostasis and remodeling. Nat Rev Mol Cell Biol 4: 517–529, 2003. [DOI] [PubMed] [Google Scholar]
- 17. Bers DM. Excitation-Contraction Coupling and Cardiac Contractile Force (2nd Ed.). Dordrecht, the Netherlands: Klumer Academic, 2001. [Google Scholar]
- 18. Buller AJ, Eccles JC, Eccles RM. Interactions between motoneurones and muscles in respect of the characteristic speed of their responses. J Physiol 150: 417–439, 1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caiozzo VJ. Plasticity of skeletal muscle phenotype: mechanical consequences. Muscle Nerve 26: 740–768, 2002. [DOI] [PubMed] [Google Scholar]
- 20. Carlson CJ, Fan Z, Gordon SE, Booth FW. Time course of the MAPK and PI3-kinase response within 24 h of skeletal muscle overload. J Appl Physiol 91: 2079–2087, 2001. [DOI] [PubMed] [Google Scholar]
- 21. Chen CS. Mechanotransduction–a field pulling together? J Cell Sci 121: 3285–3292, 2008. [DOI] [PubMed] [Google Scholar]
- 22. Chen YW, Nader GA, Baar KR, Fedele MJ, Hoffman EP, Esser KA. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J Physiol 545: 27–41, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chi MM, Hintz CS, Coyle EF, Martin WH, Ivy JL, Nemeth PM, Holloszy JO, Lowry OH. Effects of detraining on enzymes of energy metabolism in individual human muscle fibers. Am J Physiol Cell Physiol 244: C276–C287, 1983. [DOI] [PubMed] [Google Scholar]
- 24. Cooper JA, Schafer DA. Control of actin assembly and disassembly at filament ends. Curr Opin Cell Biol 12: 97–103, 2000. [DOI] [PubMed] [Google Scholar]
- 25. Dash RK, Li Y, Kim J, Saidel GM, Cabrera ME. Modeling cellular metabolism and energetic in skeletal muscle: large-scale parameter estimation and sensitivity analysis. IEEE Trans Biomed Eng 55: 1298–1318, 2008. [DOI] [PubMed] [Google Scholar]
- 26. De Beaudrap P, Witten G, Biltz G, Perrier E. Mechanistic model of fuel selection in the muscle. J Theor Biol 242: 151–63, 2006. [DOI] [PubMed] [Google Scholar]
- 27. De Boer MD, Selby A, Atherton P, Smith K, Seynnes OR, Maganaris CN, Maffulli N, Movin T, Narici MV, Rennie MJ. The temporal responses of protein synthesis, gene expression and cell signaling in human quadriceps muscle and patellar tendon to disuse. J Physiol 585: 241–251, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dulhunty AF. Excitation-contraction coupling from the 1950s into the new millennium. Clin Exp Pharmacol Physiol 33: 763–772, 2006. [DOI] [PubMed] [Google Scholar]
- 29. Durieux AC, D'Antona G, Desplanches D, Freyssenet D, Klossner S, Bottinelli R, Fluck M. Focal adhesion kinase is a load-dependent governor of the slow contractile and oxidative muscle phenotype. J Physiol 587: 3703–3717, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Engel AG, Franzini-Armstrong C. Myology: Basic and Clinical (3rd Ed.). New York: McGraw Hill, 2004. [Google Scholar]
- 31. Ervasti JM. Costameres: the Achilles' heel of herculean muscle. J Biol Chem 278: 13591–13594, 2003. [DOI] [PubMed] [Google Scholar]
- 32. Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 116: 615–622, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Franzini-Armstrong C, Horwitz AR. The cytoskeleton: maintenance of muscle fiber integrity. In: Myology: Basic and Clinical (3rd Ed.), edited by Engel AG, Franzini-Armstrong C. New York: McGraw Hill, 2004, p. 443–454. [Google Scholar]
- 34. Fukuzawa A, Lange S, Holt M, Vihola A, Carmignac V, Fereiro A, Udd B, Guatel M. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band–implications for hereditary myopathies. J Cell Sci 121: 1842–1851, 2008. [DOI] [PubMed] [Google Scholar]
- 35. Fung YC. Biomechanics. Mechanical Properties of Living Tissues (2nd Ed.). New York: Springer-Verlag, 1993. [Google Scholar]
- 36. Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 37: 1974–1984, 2005. [DOI] [PubMed] [Google Scholar]
- 37. Glass DJ. Signaling pathways perturbing muscle mass. Curr Opin Clin Nutr Metab Care 13: 225–229, 2010. [DOI] [PubMed] [Google Scholar]
- 38. Goldspink DF. Exercise-related changes in protein turnover in mammalian striated muscle. J Exp Biol 160: 127–148, 1991. [DOI] [PubMed] [Google Scholar]
- 39. Guda P, Subramaniam S, Guda C. Mitoproteome: human heart mitochondrial protein sequence database. Methods Mol Biol 357: 375–383, 2007. [DOI] [PubMed] [Google Scholar]
- 40. Hatze H. A general myocybernetic control model of skeletal muscle. Biol Cybern 28: 143–157, 1978. [DOI] [PubMed] [Google Scholar]
- 41. Hood DA. Contractile activity-induced mitochondria biogenesis in skeletal muscle. J Appl Physiol 90: 1137–1157, 2001. [DOI] [PubMed] [Google Scholar]
- 42. Hornemann T, Kempa S, Himmel M, Hayess K, Fürst DO, Wallimann T. Muscle-type creatine kinase interacts with central domains of the M-band proteins myomesin and M-protein. J Mol Biol 332: 877–878, 2003. [DOI] [PubMed] [Google Scholar]
- 43. Hortobágyi L, Dempsey D, Fraser D, Zheng G, Hamilton J, Lambert Dohm L. Changes in muscle strength, muscle fiber size and myofibrillar gene expression after immobilization and retraining in humans. J Physiol 524: 293–304, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hsiao A, Worrall DS, Olefsky JM, Subramaniam S. Variance-modeled posterior inference of microarray data: detecting gene-expression changes in 3T3–L1 adipocytes. Bioinformatics 20: 3108–3127, 2004. [DOI] [PubMed] [Google Scholar]
- 45. Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature 233: 533–538, 1971. [DOI] [PubMed] [Google Scholar]
- 46. Ingjer F. Effects of endurance training on muscle fiber ATP-ase activity, capillary supply and mitochondrial content in man. J Physiol 294: 419–432, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jenson JAL, Westerhoff HV, Kuchmerick MJ. A metabolic control analysis of kinetic controls in ATP free energy metabolism in contracting skeletal muscle. Am J Physiol Cell Physiol 279: C813–C832, 2000. [DOI] [PubMed] [Google Scholar]
- 48. Kandarian SC. Large scale gene expression profiles as tools to study skeletal muscle adaptation. In: Skeletal Muscle Plasticity in Health and Disease, edited by Bottinelli KR, Reggiani C. New York: Springer, 2006, p. 29–54. [Google Scholar]
- 49. Kjaer M, Magnusson P, Krogsgaard M, Moller JB, Oleson J, Heinemeier K, Hansen M, Haraldsson B, Koskinen S, Esmarck B, Langberg H. Extracellular matrix adaptation of tendon and skeletal muscle to exercise. J Anat 208: 445–450, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kreis T, Vale R. Guidebook to the Cytoskeletal and Motor Proteins (2nd Ed.). London: Oxford University Press, 1999. [Google Scholar]
- 51. Lee SJ, McPherron AC. Myostatin and the control of skeletal muscle mass. Curr Opin Genet Dev 9: 604–607, 1999. [DOI] [PubMed] [Google Scholar]
- 52. Lytton J, Westlin M, Burk SE, Shull GE, MacLennan DH. Functional comparisons between isoforms of the sarcoplasmic or endoplasmic reticulum family of calcium pumps. J Biol Chem 267: 14483–14489, 1992. [PubMed] [Google Scholar]
- 53. MacIntosh BR, Gardiner PF, McComas AJ. Skeletal Muscle: Form and Function (2nd Ed). Champaign, IL: Human Kinetics, 2006. [Google Scholar]
- 54. MacQueen J. Some methods for classification and analysis of multivariate observations. In: Proc Fifth Berkeley Sympos Math Statist and Probability. I. Statistics Berkeley, CA: Univ. of California Press, 1967, p. 281–297. [Google Scholar]
- 55. Maher AC, Fu MH, Isfort RJ, Varbanov AR, Qu XA, Tarnopolsky MA. Sex differences in global mRNA content of human skeletal muscle. PLos One 4: e6335, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mahoney DJ, Tarnopolsky MA. Emerging molecular tends in muscle damage research. In: Skeletal Muscle Damage and Repair, edited by Tiidus PM. Champaign, IL: Human Kinetics, 2008, p. 89–102. [Google Scholar]
- 57. Mallinson J, Meissner J, Chang KC. Chapter 2. Calcineurin signaling and the slow oxidative skeletal muscle fiber type. Int Rev Cell Mol Biol 277: 67–101, 2009. [DOI] [PubMed] [Google Scholar]
- 58. Martonosi AN, Pikula S. The network of calcium regulation in muscle. Acta Biochim Pol 50: 1–30, 2003. [PubMed] [Google Scholar]
- 59. Matheny RW, Jr, Nindl BC, Adamo ML. Minireview: Mechano-growth factor: a putative product of IGF-I gene expression involved in tissue repair and regeneration. Endocrinology 151: 865–875, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maurya MR, Subramaniam S. A kinetic model for calcium dynamics in RAW 264.7 cells. 1. Mechanisms, parameters, and subpopulational variability. Biophys J 93: 709–728, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maurya MR, Subramaniam S. A kinetic model for calcium dynamics in RAW 264.7 cells. 2. Knockdown response and long-term response. Biophys J 93: 729–40, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murphy KT, Chee A, Gleeson BG, Naim T, Swiderski K, Koopman R, Lynch GS. Antibody-directed myostatin inhibition enhances muscle mass and function in tumor-bearing mice. Am J Physiol Regul Integr Comp Physiol 301: R716–R726, 2011. [DOI] [PubMed] [Google Scholar]
- 63. Nielsen S, Scheele C, Yfanti C, Akerström T, Nielsen AR, Pedersen BK, Laye MJ. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J Physiol 588: 4029–4037, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Perry SV. Activation of the contractile mechanism by calcium. In: Myology: Basic and Clinical (3rd Ed.), edited by Engel AG, Franzini-Armstrong C. New York: McGraw Hill, 2004, p. 281–306. [Google Scholar]
- 65. Pette D. Historical Perspectives: Plasticity of mammalian skeletal muscle. J Appl Physiol 90: 1119–1124, 2001. [DOI] [PubMed] [Google Scholar]
- 66. Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol Endocrinol Metab 273: E99–E107, 1997. [DOI] [PubMed] [Google Scholar]
- 67. Quackenbush J. Microarray data normalization and transformation. Nat Genet 32, Suppl: 496–501, 2002. [DOI] [PubMed] [Google Scholar]
- 68. Reverter A, Hudson NJ, Wang Y, Tan SH, Barris W, Byrne KA, McWilliam SM, Bottema CD, Kister A, Greenwood PL, Harper GS, Lehnert SA, Dalrymple BP. A gene coexpression network for bovine skeletal muscle inferred from microarray data. Physiol Genomics 28: 76–83, 2006. [DOI] [PubMed] [Google Scholar]
- 69. Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol 3: 1009–1013, 2001. [DOI] [PubMed] [Google Scholar]
- 70. Sanes JR. The extracellular matrix. In: Myology: Basic and Clinical (3rd Ed.), edited by Engel AG, Franzini-Armstrong C. New York: McGraw Hill, 2004, p. 471–487. [Google Scholar]
- 71. Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R, Sandri M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol 296: C1248–C1257, 2009. [DOI] [PubMed] [Google Scholar]
- 72. Schon EA. The tools of molecular genetics and their application to the study of muscle diseases. In: Myology: Basic and Clinical (3rd Ed.), edited by Engel AG, Franzini-Armstrong C. New York: McGraw Hill, 2004, p. 915–958. [Google Scholar]
- 73. Sharman JL, Mpamhanga CP, Spedding M, Germain P, Staels B, Dacquet C, Laudet V, Harmar AJ. NC-IUPHAR IUPHAR-DB: new receptors and tools for easy searching and visualization of pharmacological data. Nucleic Acids Res 39: D534–D538, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shorten PR, O'Callaghan P, Davidson JB, Soboleva TK. A mathematical model of fatigue in skeletal muscle force contraction. J Muscle Res Cell Motil 28: 293–313, 2007. [DOI] [PubMed] [Google Scholar]
- 75. Stevenson EJ, Giresi PG, Koncarevic A, Kandarian SC. Global analysis of gene expression patterns during disuse atrophy in rat skeletal muscle. J Physiol 551: 33–48, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Timmons JA, Jansson E, Fischer H, Gustafsson T, Greenhaff PL, Ridden J, Rachman J, Sundberg CJ. Modulation of extracellular matrix genes reflects the magnitude of physiological adaptation to aerobic exercise training in humans. BMC Biol 3: 19, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu Rev Biophys Biomol Struct 35: 459–488, 2006. [DOI] [PubMed] [Google Scholar]
- 78a. Wang Y, Winters J, Subramaniam S. Functional classification of skeletal muscle networks. II. Applications to pathophysiology. J Appl Physiol; doi: 10.1152/japplphysiol.01515.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Waterlow JC. Protein Turnover. Cambridge, MA: CABI, 2006. [Google Scholar]
- 80. West DW, Burd NA, Staples AW, Phillips SM. Human exercise-mediated skeletal muscle hypertrophy is an intrinsic process. Int J Biochem Cell Biol 42: 1371–1375, 2010. [DOI] [PubMed] [Google Scholar]
- 81. Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Prüss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 28: 316–319, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Winters JM, Stark L. Analysis of fundamental movement patterns through the use of in depth antagonistic muscle models. IEEE Trans Biomed Eng 32: 826–839, 1985. [DOI] [PubMed] [Google Scholar]
- 83. Winters JM. Hill based muscle models: a systems engineering perspective. In: Multiple Muscle Systems: Biomechanics and Movement Organization, edited by Winters JM, Woo SY. New York: Springer Verlag, 1990, p. 69–93. [Google Scholar]
- 84. Wright DC. Mechanisms of calcium-induced mitochondrial biogenesis and GLUT4 synthesis. Appl Physiol Nutr Metab 32: 840–845, 2007. [DOI] [PubMed] [Google Scholar]
- 85. Yi Z, Bowen BP, Hwang H, Jenkinson CP, Coletta DK, Lefort N, Bajaj M, Kashyap S, Berria R, De Filippis EA, Mandarino LJ. Global relationship between the proteome and transcriptome of human skeletal muscle. J Proteome Res 7: 3230–3241, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yoshida T. MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol 28: 8–17, 2008. [DOI] [PubMed] [Google Scholar]
- 87. Zhao P, Hoffmann E. Transcriptional cascades in muscle regulation. In: Skeletal Muscle Repair and Regeneration, edited by Schiafino S, Parthridge T. New York: Springer, 2008, p. 85–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.