Abstract
Microbes whose genomes are encoded by DNA and for which adequate information is available display similar genomic mutation rates (average 0.0034 mutations per chromosome replication, range 0.0025 to 0.0046). However, this value currently is based on only a few well characterized microbes reproducing within a narrow range of environmental conditions. In particular, no genomic mutation rate has been determined either for a microbe whose natural growth conditions may extensively damage DNA or for any member of the archaea, a prokaryotic lineage deeply diverged from both bacteria and eukaryotes. Both of these conditions are met by the extreme thermoacidophile Sulfolobus acidocaldarius. We determined the genomic mutation rate for this species when growing at pH 3.5 and 75°C based on the rate of forward mutation at the pyrE gene and the nucleotide changes identified in 101 independent mutants. The observed value of about 0.0018 extends the range of DNA-based microbes with rates close to the standard rate simultaneously to an archaeon and to an extremophile whose cytoplasmic pH and normal growth temperature greatly accelerate the spontaneous decomposition of DNA. The mutations include base pair substitutions (BPSs) and additions and deletions of various sizes, but the S. acidocaldarius spectrum differs from those of other DNA-based organisms in being relatively poor in BPSs. The paucity of BPSs cannot yet be explained by known properties of DNA replication or repair enzymes of Sulfolobus spp. It suggests, however, that molecular evolution per genome replication may proceed more slowly in S. acidocaldarius than in other DNA-based organisms examined to date.
Archaea isolated from geothermal environments grow optimally at temperatures that are lethal to all genetically well characterized microorganisms and damaging to DNA. The enzymes of these hyperthermophilic archaea are intrinsically thermostable due to a variety of structural features that discourage protein unfolding, which explains how metabolism can be maintained at extremely high temperatures (1). The strategy of intrinsic stabilization does not seem to apply to the chromosomes of these archaea, however, and does not address the problem of spontaneous DNA decomposition at physiological temperatures (2). Although information about the gene content, genome organization, and evolutionary divergence of hyperthermophilic archaea is expanding rapidly, their basic genetic processes remain largely unexplored. As a result, it is unclear how these organisms compare with well studied microbes with respect to genetic exchange, DNA repair, mutation, genetic exchange, and other fundamental processes important to their survival and evolution.
Rates of spontaneous mutation measured in microbial systems provide evidence of the biological importance of genetic fidelity. Accurate rates of spontaneous mutation per genome are available for only six DNA-based microbes: phage M13, phage λ, phage T2/T4, the bacterium Escherichia coli, the yeast Saccharomyces cerevisiae, and the filamentous fungus Neurospora crassa (3, 4). Despite their diverse life histories and genome sizes that differ by 104-fold, their individual genomic mutation rates deviate by less than 36% from the average value of 0.0034 mutations per genome replication. This average provides a benchmark value that has not, however, been tested among either archaea or organisms inhabiting intrinsically mutagenic environments. We addressed this deficiency by measuring the genomic mutation rate of the thermoacidophilic archaeon Sulfolobus acidocaldarius.
Estimating a genomic mutation rate requires a measure of the mutation rate in a representative target gene and a description of the kinds of mutations arising therein. The use of 5-fluoro-orotic acid (FOA) to select pyrimidine auxotrophs facilitated the measurement of an aggregate mutation rate for the pyrE and pyrF genes (about 3.3 × 10−7 mutations per cell division) in S. acidocaldarius (5), but no sequence information was available then for either gene. In the present study, we isolated numerous independent mutants resistant to FOA under similar conditions, identified their mutations by sequencing, and used this information to estimate the size of the mutational target and the fraction of base pair substitutions (BPSs) that were not detected. From this information, we estimate the genomic mutation rate to be about 0.0018 per chromosome replication, close to the standard rate for DNA-based microbes. We also found that the fraction of BPSs among spontaneous mutations is substantially lower than observed in other DNA-based organisms.
Materials and Methods
Organism and Growth Conditions.
S. acidocaldarius, obtained from the American Type Culture Collection (ATCC 33909), was clonally purified by streaking on solid medium and was confirmed to yield a pattern of genomic EcoRI restriction fragments identical to that of S. acidocaldarius strain DG6 (6). The laboratory designation for the resulting strain is DG185. Unless otherwise noted, growth media (pH 3.5) consisted of a mineral mixture supplemented with xylose, tryptone, and uracil; plates contained gellan gum for solidification (7). Incubation was at 75°C, which is near the midpoint of the temperature range for growth for this species (6).
For mutant selection, 50 μg/ml FOA was added (7). In S. acidocaldarius, this concentration of FOA selects mutants deficient in either of two UMP biosynthetic enzymes: orotate phosphoribosyl transferase (OPRTase) and orotidine 5′-monophosphate decarboxylase, encoded by the pyrE and pyrF genes, respectively (5, 7). The selection reflects the fact that FOA is readily taken up and is not toxic per se, whereas the 5-F-UMP made from it by sequential action of the two enzymes kills cells. Loss of either enzyme thus spares the mutant in the presence of FOA, but also renders it dependent on exogenous uracil for growth. As an assay of spontaneous mutation, the FOA selection has the advantage of generality. In principle, any mutation that inactivates either gene product is selected, regardless of the nature of the sequence change.
Mutant Isolation and Mutation Rate Determination.
Fluctuation tests were performed as described (5) except that independent cultures were smaller and grown without mechanical agitation in microdilution plates. For each mutant-isolation experiment, one isolated colony was suspended in 3 ml of nonselective liquid medium and incubated until the culture reached a density of about 5 × 107 cells/ml. This culture then was diluted 104-fold in fresh nonselective medium and dispensed as 200-μl aliquots in the wells of a microdilution plate. The resulting cultures, each inoculated with about 103 cells, were incubated for 3 days, yielding visible turbidity. A 70-μl aliquot of each culture was plated on selective medium, and FOA-resistant colonies were analyzed after 8–10 days of incubation unless otherwise noted.
The parameters needed to calculate the rate of mutation under these conditions were derived from a set of 115 liquid cultures in individual wells. To calculate the average number of cells per culture, Nav, the volumes of four wells were measured at various positions within the plate and the viable titers in these wells were determined by serial dilution and plating on nonselective medium. The most probable number of mutational events per culture for the set, m, was determined by using the method of Lea and Coulson based on the tables of Koch (8). These tables yield one estimate of m for each of the three quartiles of the mutant frequency distribution. Three corresponding estimates of the mutation rate per cell generation, μ, were calculated by using the relationship μ = (ln2)(m/Nav). There was no apparent bias in the rate estimates as a function of the quartile used, reinforcing direct experimental evidence that possible distortions caused by phenotypic lag or differences in growth rate between mutant and parent strains in uracil-supplemented media are negligible (5).
Mutants were randomly chosen for sequence analysis by marking the back of the plate with a small dot when the plates were initially spread. The FOA-resistant colony that formed closest to this dot was picked from the plate and streaked for isolation on nonselective medium. A single, well isolated colony of each mutant then was grown in liquid medium. A portion of the resulting clonally pure culture was preserved at −70°C. Total genomic DNA was extracted from the remainder of the culture by the guanidinium thiocyanate procedure (9).
DNA Sequencing.
DNA samples were diluted with water to contain 33–100 ng of DNA per 50 μl, and the pyrE and pyrF regions were amplified by using the PCR and sequenced. The upstream PCR primer was 5′-ATGTTTCAATAACGCCCT-3′ and the downstream PCR primer was 5′-TCAGATATCCTAGCCAGT-3′. The resulting product from the wild-type template included 81 bp before the 594 bp encoding pyrE and 42 bp after the 660 bp encoding pyrF. The PCR consisted of 30 cycles of 1 min at 94°C, 1 min at 55°C, 1 min at 72°C, with a final extension time of 10 min at 72°C by using Taq large-fragment polymerase (Display Systems Biotech TAQ DNA polymerase from PGC Scientific, Gaithersburg, MD). The PCR products were purified with the QIAquick PCR Purification Kit (Qiagen, Valencia, CA). Sequencing was performed with an ABI Prism 377 DNA Sequencer using the dRhodamine Terminator Cycle Sequencing Kit (Perkin–Elmer). Each sample was sequenced with the two PCR primers above and with up to six internal primers. The latter consisted of the following pairs of forward and reverse primers: 5′-CAGTGGAGGAGGTAAGGA-3′ and 5′-TCCTTACCTCCTCCACTG-3′, 5′-TTGGACTGGGACGAGATA-3′ and 5′-TATCTCGTCCCAGTCCAA-3′, and 5′-GGTGTCCAAGGTGCCAA-3′ and 5′TTGGCACCTTGGACACC-3′. After noting that the first 64 samples to be sequenced contained almost exclusively pyrE mutations, sequencing of pyrF was discontinued except for rare instances in which no pyrE mutation was detected. Of the 49 leaky frameshift mutants at position 545–551, 14 were sequenced completely without finding an additional mutation in either gene. Sequencing detected no mutations in each of several preparations of DNA from the parental strain and detected no more than one mutation in any mutant.
Results
We performed three independent fluctuation tests from which a total of 108 FOA-resistant mutants were recovered, each arising in an independent culture. Of these 108, 101 eventually were found to contain intragenic pyrE mutations. (The 3′ end of pyrE overlaps the beginning of pyrF by 14 bases; mutations in the overlap region were classified as pyrE mutations.) Mutants from the first two tests (115 cultures each) were grouped together for subsequent analysis. Data from the second fluctuation test were used to confirm the genic mutation rate realized under our conditions. The value (3.37 ± 0.36) × 10−7, agrees well with an average value of (3.29 ± 1.35) × 10−7 determined previously for larger cultures under similar but not identical growth conditions (5). The 79 intragenic pyrE mutations from the first two tests were pooled and designated set I. The third fluctuation test (72 cultures) evaluated the effects of stronger selection stringency by limiting incubation of the selection plates to only 6 days. Under these conditions, only the resistant clones whose growth rate was not significantly retarded by the FOA yielded visible colonies. The resulting 22 intragenic pyrE mutations were designated set II.
Almost all of the FOA-resistant mutants were found to contain pyrE mutations; only 4/85 of the mutants from the first two fluctuation tests and 1/23 from the third fluctuation test contain pyrF mutations. Four of the five pyrF mutations are ±1 bp and one is a G⋅C → A⋅T mutation that generates an ochre (TAA) codon. Two additional FOA-resistant mutants contain large rearrangements; we recovered these by using the outermost PCR primers and found that the products were larger than that of the wild type. Using other PCR primers and diverse sequencing primers revealed that these two mutants contain sequence rearrangements (probably duplications), but their detailed structures were not further investigated. In sharp contrast to S. solfataricus, where most pyrE and pyrF mutations isolated by the same selection were caused by small transposable elements (IS elements) (10), we detected no IS insertions in any of our independent S. acidocaldarius mutants.
The intragenic pyrE mutations are summarized in Table 1 and Fig. 1. The BPSs comprise mostly transitions and, despite the fact that the pyrE gene contains only 36% G·C bp, the transitions are mostly G⋅C → A⋅T. The spectrum of mutants selected under standard conditions (set I) is dominated by one A7 → A6 frameshift hotspot at 545–551, which accounted for 49 of the 79 mutations. In addition to being the most abundant allele recovered under standard conditions, the A7 → A6 frameshift was the most sensitive to an increase in selection stringency, as shown by its absence from the 23 mutants of set II. When this mutation is ignored, comparison of sets I and II further suggests that elevated selection somewhat decreases the representation of BPS mutations (Table 1). This is the expected result of some missense mutations failing to completely inactivate the OPRTase.
Table 1.
Classes of spontaneous pyrE mutations
| Kind of mutation | No. in set I | No. in set II | Special aspects |
|---|---|---|---|
| Total | 79 | 22 | |
| BPSs | 10 | 2 | No CT mutations |
| Transitions | 7 | 2 | 8/9 = G⋅C → A⋅T |
| Transversions | 3 | 0 | |
| Intragenic additions and deletions | 69 | 20 | |
| +1 bp in runs | 8 | 4 | |
| −1 bp in runs | 53 | 7 | 49/53 and 0/7 A7 → A6 at 545–551 |
| +1 bp not in runs | 3 | 3 | |
| −1 bp not in runs | 2 | 0 | |
| +3 or +6 at fugue sequence | 1 | 2 | In CTACTACT |
| Intragenic deletions | 0 | 1 | |
| Tandem duplications | 2 | 3 |
This tabulation omits two large rearrangements and five pyrF mutations that are described in the text.
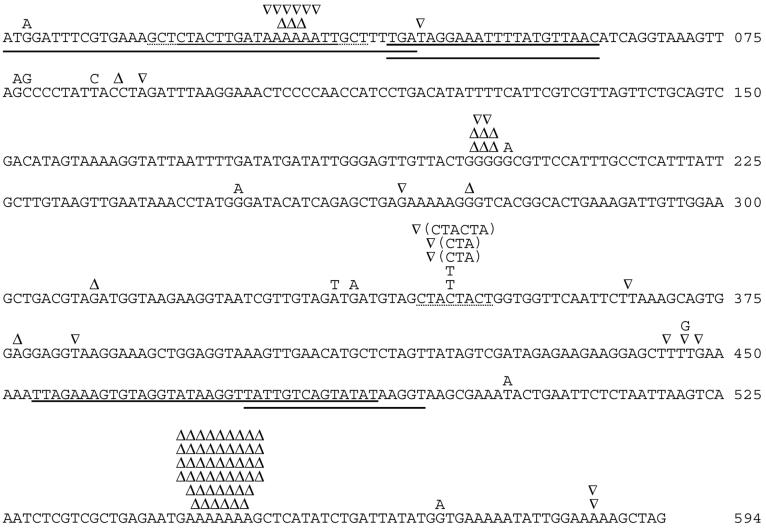
Figure 1.
The mutational spectrum. The sequence is that of the nontranscribed strand of the pyrE gene, with mutations of small extent shown above this sequence. A, T, G, and C denote BPSs. ▿ denotes a single-base insertion of a base identical to the base or bases below the symbol, except for the insertion of a G between T446 and T447 and the insertion of either three or six bases into the dotted-underlined sequence-fugue “CTACTACT” (for which we show only one of many possible insertions, as explained in the text). The five heavy underlines denote sequences that were tandemly duplicated. Δ denotes a single-base deletion of a base identical to the base or bases below the symbol. In the top line a deletion of 20 bases is shown comprising 17 bases (lightly underlined) between two triplet repeats (dotted underlined), one of which is also deleted.
The remaining mutants, which contain intragenic additions and deletions, exhibited the tight auxotrophic phenotype associated with complete loss of enzyme function (7). Those from sets I and II therefore can be pooled for analysis. Most are ±1 frameshift mutations that arose in runs of two or more identical bases. Of the six +1 mutations not arising in runs, five are tandem duplications of a base and one is an insertion of G within a TTTT run. Both −1 mutations not arising in runs have (by definition) lost a single base. The ±1 mutations arising in runs arose preferentially in longer runs. One ±1 arose among 19 runs of three bases, one ±1 arose among eight runs of four bases, 10 ±1 arose among five runs of five bases, nine ±1 arose in one run of six bases, and 49 ±1 arose in one run of seven bases. The pyrE target contains 379 A⋅T and 215 G⋅C bp, including 99 A⋅T in runs of ≥3 and 28 G⋅C in runs of ≥3, but the number of larger, more mutable runs is too small to detect a bias in frameshift mutability between A⋅T runs and G⋅C runs. In runs, ignoring the special case of the large hot spot, single-base additions and single-base deletions arise equally frequently (12 and 11 times, respectively).
Three additions of either three or six bp arose at 344-CTACTACT-351, generating either CTACTACTACT or CTACTACTACTACT. This eight-base sequence contains a large number of different repeat motifs: three overlapping triplet repeats (CTA CTA, TAC TAC, and ACT ACT), two overlapping tetrameric repeats (CTAC CTAC and TACT TACT), and an overlapping pentameric repeat (CTACT CTACT). The expansions detected in this remarkable fugue-like sequence could have arisen in many ways: slippage by three bases to insert CTA, TAC, or ACT, slippage by four bases (aligning the second member of a tetramer with the overlapping first member) to insert three bases, slippage by five bases (aligning the second CTACT with the overlapping first CAACT) to insert ACT, or slippage by six bases (aligning last CT with first CT) to insert ACTACT. A similarly complex fugue-like sequence of 14 bases occurs in the E. coli lacI gene (11).
The spectrum contains one larger deletion and five larger tandem duplications. A 20-base deletion runs between two GCTs and deletes one of these. This result fits the pattern that most deletions in most organisms arise between short repeats. Three of the tandemly duplicated sequences are not associated with repeated sequences—the sequence preceding the duplication starting at 001 is GGGTTAAA—whereas the tandem duplication of 476–494 is associated with the repeated sequence TATAAGGT. We observed no complex mutations, that is, mutations in which multiple close changes occur as a single event.
Many of the mutants of set I were able to grow reasonably well in the absence of added uracil. The A7 → A6 frameshift at 545–551 subsequently was found in 42/44 of these mutants. This mutation was frequent in both fluctuation tests represented in set I and was found in two additional, independently derived mutants, one of which is strain DG96 (T.-L. Thia-Toong and D. Charlier, personal communication). Strain DG96 has been characterized biochemically. It has 13% residual OPRTase activity and a low level of FOA resistance and can grow to a limited extent in the absence of uracil supplementation (7). The fact that the A7 → A6 frameshift is both leakier and also much more frequent than most pyrE alleles explains prior observations that low-stringency FOA selection yields two phenotypically distinct classes of pyr mutants in approximately equal numbers (7). As deduced from the nucleotide sequence, this −1 frameshift modifies the carboxyl terminus of OPRTase by replacing the last 14 residues with SSYLIIW. The corresponding expansion (A7 → A8 at 545–551) should result in an OPRTase in which the last 13 residues are replaced by AHI. The expansion mutation was not recovered in either set I or set II, despite the fact that +1 and −1 mutations arise equally frequently in the other homopolymeric runs (Table 1). This finding suggests that the expansion at 545–551 yields little or no decrease in enzymatic activity under physiological conditions.
Discussion
The mutation rate (at 75°C at pH 3.5) of S. acidocaldarius to resistance to FOA was previously determined to be (3.29 ± 1.35) × 10−7 per cell division (5). In that study, phenotypic scoring (based on levels of FOA resistance indicated by colony size) was used to estimate separate mutation rates for pyrE and pyrF, but molecular analysis now shows that this phenotypic criterion does not accurately predict the site of mutation. For example, pyrE frameshift mutations depress levels of orotidine 5′-monophosphate decarboxylase (the product of pyrF), whereas other pyrE alleles confer high-level FOA resistance without affecting pyrF expression (12). Here we obtained a closely similar mutation rate of (3.37 ± 0.36) × 10−7 per cell division for mutations conferring FOA resistance. Almost all (101/108) of such mutations that we examined arose within pyrE. The spontaneous forward mutation rate of this gene is therefore about 3.37(101/108) = 3.15 × 10−7.
Before this rate can be converted into a genomic rate, it must first be adjusted for undetected mutations. One way to do this is to estimate total BPSs from numbers of chain-termination (CT) mutations, for instance by multiplying by (64 codons)/(3 CT codons) (3). However, we detected only 12 pyrE BPSs altogether and none of these created CT codons. We therefore collected published information providing values for the ratio (predicted total BPSs)/(observed BPSs). The average value over several reporter genes and organisms was 4.73 (see Table 3, which is published as supplemental material on the PNAS web site, www.pnas.org). Set II contained 69 addition and deletion mutations and 10 BPSs. The pyrE reading frame (Fig. 1) comprises 594 bases. Therefore, the estimated average spontaneous mutation rate per base is μb = (3.15 × 10−7)(69 + 4.73 × 10)/(79)(594) = 7.81 × 10−10.
To our knowledge, the size of the S. acidocaldarius genome has not yet been determined; a previously characterized genome (13) represents another Sulfolobus species. However, flow cytometry analyses of exponential-phase populations using three different DNA-specific fluorescent stains provide consistent estimates of DNA contents for S. acidocaldarius and S. solfataricus cells (14). The average ratio of DNA content for cells containing two complete chromosomes in figures 3 and 4 of ref. 14 is 0.75 ± 0.09. Therefore, because the genome of S. solfataricus has 2,990,993 bp (http://niji.imb.nrc.ca/sulfolobus/), the S. acidocaldarius genome has about 2,243,000 bp. Thus, the rate for S. acidocaldarius is about 0.0018 mutations per genome per replication. This value is close to or even slightly lower than the average rate for DNA-based microbes of 0.0034 (range 0.0025 to 0.0046, E. coli value = 0.0025) (4). Although the evolutionary forces that drive this rate remain unknown, they are likely to depend mainly on a balance between the deleterious effects of most mutations, which will tend to select for mutations that reduce mutation rates, and the costs of achieving this reduction (15).
The correction factor for undetected BPSs can be a major uncertainty in organisms in which BPSs considerably outnumber addition and deletion mutations. However, the correction factor has less weight for S. acidocaldarius because of the smaller fraction of BPSs: if no correction were made, the mutation rate would decrease by only 32%. In addition, the spectrum suggests that BPS detection within the S. acidocaldarius pyrE gene is fairly efficient. In most bacterial systems, CT mutations are far more frequent than 3/64 and may approach or exceed 50% when missense mutations are exceptionally poorly detected. Here, however, all 12 BPSs (at 11 different sites) are missense mutations, and the detection of missense mutations thus appears to be of better-than-average efficiency. One caution that must be applied to this conclusion, however, arises from the relative rarity of transversion mutations, which comprised only 3/12 of the BPSs in pyrE. All 81 of the pyrE codons that can convert to a CT codon by a single BPS must do so by a transversion, but the two kinds of transversions (G⋅C → C⋅G and A⋅T → T⋅A) recovered in pyrE have, between them, 46 codons as targets for conversion to CT codons.
Calculating a genomic mutation rate requires extrapolating from a mutation-reporter gene, which therefore should be representative of the genome as a whole. The S. acidocaldarius pyrE gene has a G⋅C content of 36.2%, which is close to the 37% for total DNA (16). The observed pyrE mutations and the presence of a frameshift-mutation hotspot are typical of most genes examined to date (whose spectra can be accessed through the references in Tables 3 and 4, which are published as supplemental material). The pyrE mutation spectrum is atypical only in its paucity of BPSs (see below). BPSs are unlikely to be poorly detected in our screen because it does detect mutations retaining sufficient residual OPRTase activity to sustain moderate growth in the absence of uracil. Even if the BPS frequency were 3-fold higher than estimated above, the genomic rate would be little affected, increasing to 0.0025 and even more closely approaching the standard rate. Nevertheless, the formal possibility remains that the S. acidocaldarius genomic mutation rate, and those of several other organisms, are either underestimated or overestimated because of some unrecognized property of the mutation-reporter gene.
Although the kinds of mutations described in Table 1 and Fig. 1 are commonly observed in the great majority of mutational spectra in diverse organisms, there is one striking quantitative difference between the S. acidocaldarius spectrum and that of most other DNA-based organisms: BPSs are less frequent in S. acidocaldarius. To assess this difference, we first tabulated %BPSs from a number of spectra (Table 2). In doing this, we wanted to exclude two frequent sources of bias. First, and particularly in bacteria, the insertion of transposable elements is quite variable, so that insertion sequence mutations may comprise from <2% to about 60% of all spontaneous mutations, depending on the species, strain and reporter gene (17). Second, the most intense mutational hotspots are additions and deletions at repeated sequences, and these, too, can dominate a spectrum in an unrepresentative manner. For instance, frameshift mutations at a particular site in the E. coli lacI gene can comprise as many as 88% of all mutations (see Table 4). Thus, to compare BPS frequencies among organisms, we decided first to eliminate high-frequency insertions and high-frequency addition-deletion hotspots. When this is done (Table 2), slightly more than 2/3 of detected mutations are BPSs among the organisms for which good spectra are available. In contrast, only 1/3 of the detected mutations in S. acidocaldarius are BPSs. In a χ2 test, this lower frequency is markedly different from the reference collection (P < 0.001).
Table 2.
Relative BPS frequencies
| Organism (reporter genes) | BPS | Total | % BPS |
|---|---|---|---|
| Bacteriophage M13 (lacZα) | 67 | 117 | 57.3 |
| Bacteriophage λ (cII) | 55 | 92 | 59.8 |
| Bacteriophage T4 (rI, ac) | 80 | 130 | 61.5 |
| Herpes simplex virus (supF) | 68 | 121 | 56.2 |
| E. coli (λcI, lactd, crp, supF, rpsL, tonB, lacI) | 855 | 1,299 | 65.8 |
| S. cerevisiae (SUP4, URA3, CAN1) | 333 | 385 | 86.5 |
| Mouse, rat, hamster, monkey, human (lacI, gpt, hprt, aprt, supF, tk, cI, cII) | 1,330 | 1,891 | 70.3 |
| Total | 2,788 | 4,035 | 69.1 |
| S. acidocaldarius (pyrE) | 10 | 30 | 33.3 |
For references and the underlying data, see Table 4. All entries are based on DNA sequencing. Insertions of transposable elements and strong frameshift hotspots were excluded from the values in the Total column (see text).
A spectrum of spontaneous mutation reflects numerous determinants, including unforced errors of DNA replication, mismatch correction by proofreading, postreplication DNA mismatch repair, DNA damage leading to mutations during inaccurate repair, and unrepaired DNA damage causing replication errors. One could hypothesize that the S. acidocaldarius cytoplasmic pH of about 6.0 (18) might promote base protonation and thus promote ionization-dependent modes of base mispairing (19). However, several DNA polymerases all exhibited decreased rather than increased mutation rates in vitro when the pH was reduced from 8–9 to 5–6, and this effect was manifested for both BPSs and frameshift mutations (20–22). Similarly, a high temperature would be expected to favor melting of the nascent strand terminus during DNA replication, thus increasing the frequency of mutagenic misalignments relative to simple base mispairs. However, spectra obtained at 70°C in vitro with DNA polymerase I of the thermophile Thermus aquaticus contained about 80% BPSs (23, 24). The same enzyme exhibited modest increases in mutation rates as the temperature was increased from either 22°C or 55°C to 70°C, without changing in the relative abundance of BPSs (23, 25). However, this enzyme probably contributes little to chromosomal replication (see below), and the replicative polymerase(s) might display different propensities. The only relevant studies in vivo were performed by using bacteriophage T4, whose replicative DNA polymerase belongs to the same family as those of the archaea (26). In T4, increasing the temperature from 30°C to 43°C promoted BPSs roughly 10-fold at A:T sites and 4-fold at G:C sites, and promoted frameshift mutations roughly 2-fold at a single tested site (27, 28). The replicative DNA polymerases of archaea are all family-B enzymes lacking homology with bacterial enzymes but exhibiting homology with eukaryotic replicative DNA polymerases (26). However, the eukaryotes, which are all mesophiles, generally display high BPS frequencies in vivo (Table 2), as do their replicative DNA polymerases in vitro (29), and no in vitro spectrum is yet reported for the replicative DNA polymerase of an archaeon. On balance, therefore, we have yet to identify a mechanistic explanation for the relatively low frequency of BPSs in S. acidocaldarius.
To the extent that DNA sequence evolution is driven mainly by neutral mutations (30) and that the pyrE gene fairly represents the S. acidocaldarius genome, this low rate of substitution mutations in S. acidocaldarius should affect its rate of molecular evolution. Our data predict that S. acidocaldarius should accumulate neutral mutations (mainly synonymous substitutions) at about half the rate per generation as does E. coli. Furthermore, S. acidocaldarius reproduces only about one-eighth as quickly as E. coli under optimal conditions (doubling time of 2.9 h at about 80°C) (6), although the relative numbers of microbial generations per year in nature cannot be inferred solely from this fact.
The modest rate of spontaneous mutation of all types in S. acidocaldarius is also relevant to understanding the nature and impact of DNA repair in hyperthermophilic archaea. To our knowledge, no cellular organism has been shown to depend solely on the accuracy of DNA synthesis plus proofreading for its overall genetic fidelity. In addition, the optimal growth temperatures of hyperthermophilic archaea should result in higher rates of spontaneous DNA damage than occur in the genetically better-characterized organisms. It is thus perplexing that hyperthermophilic archaea are the only cellular organisms in which several important DNA repair enzymes are not evident, even though genes encoding homologues of these enzymes occur in other archaea and hyperthermophilic bacteria (31). Among the most conspicuous of these missing enzymes are homologues of the bacterial mutHSL gene products, which provide a major defense against spontaneous mutation in both bacteria and eukaryotes. A genomic mutation rate similar to, or even lower than, the average of diverse mesophiles with classical mismatch repair suggests that S. acidocaldarius may have an effective but perhaps evolutionarily distinct error-correction system. Identifying this system and other determinants of the spectrum of spontaneous mutation will require a combination of enzymological and genetic studies using S. acidocaldarius and other hyperthermophilic archaea. Although technically challenging, such studies promise to clarify mechanisms of molecular evolution in hyperthermophilic archaea and identify enzymes that enforce genetic fidelity under harsh environmental conditions.
Supplementary Material
Acknowledgments
We thank Joe Haseman for the statistical analysis of the data in Table 3, Katherine Schmidt for assistance in processing some of the mutants, and Youri Pavlov and Roel Schaaper for critical readings of the manuscript.
Abbreviations
- BPS
base pair substitution
- CT
chain-termination (mutation)
- FOA
5-fluoro-orotic acid
- OPRTase
orotate phosphoribosyl transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jaenicke R, Böhm G. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/s0959-440x(98)80094-8. [DOI] [PubMed] [Google Scholar]
- 2.Grogan D W. Mol Microbiol. 1998;28:1043–1049. doi: 10.1046/j.1365-2958.1998.00853.x. [DOI] [PubMed] [Google Scholar]
- 3.Drake J W. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drake J W, Charlesworth B, Charlesworth D, Crow J F. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs K L, Grogan D W. J Bacteriol. 1997;179:3298–3303. doi: 10.1128/jb.179.10.3298-3303.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grogan D W. J Bacteriol. 1989;171:6710–6719. doi: 10.1128/jb.171.12.6710-6719.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grogan D W, Gunsalus R P. J Bacteriol. 1993;175:1500–1507. doi: 10.1128/jb.175.5.1500-1507.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koch A L. Mutat Res. 1982;95:125–143. [Google Scholar]
- 9.Pitcher D G, Saunders N A, Owen R J. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 10.Martusewitsch E, Sensen C W, Schleper C. J Bacteriol. 2000;182:2574–2581. doi: 10.1128/jb.182.9.2574-2581.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaaper R M, Danforth B N, Glickman B W. J Mol Biol. 1986;189:273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- 12.Reilly M S, Grogan D W. J Bacteriol. 2001;183:2943–2946. doi: 10.1128/JB.183.9.2943-2946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo S, Yamagishi A, Oshima T. J Bacteriol. 1993;175:1532–1536. doi: 10.1128/jb.175.5.1532-1536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernander R, Poplawski A. J Bacteriol. 1997;179:4963–4969. doi: 10.1128/jb.179.16.4963-4969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura M. Genet Res. 1967;9:23–34. [Google Scholar]
- 16.Fuchs T, Huber H, Teiner K, Burggraf S, Stetter K O. Syst Appl Microbiol. 1995;18:560–566. [Google Scholar]
- 17.Galas D J, Chandler M. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 109–162. [Google Scholar]
- 18.Meyer W, Schäfer G. Eur J Biochem. 1992;207:741–746. doi: 10.1111/j.1432-1033.1992.tb17104.x. [DOI] [PubMed] [Google Scholar]
- 19.Goodman M R, Creighton S, Bloom L B, Petruska J. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 20.Eckert K A, Kunkel T A. Nucleic Acids Res. 1990;18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckert K A, Kunkel T A. J Biol Chem. 1993;268:13462–13471. [PubMed] [Google Scholar]
- 22.Eckert K A, Kunkel T A. Nucleic Acids Res. 1993;21:5212–5220. doi: 10.1093/nar/21.22.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tindall K R, Kunkel T A. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki M, Yoshida S, Adman E T, Blank A, Loeb L A. J Biol Chem. 2000;275:32728–32735. doi: 10.1074/jbc.M000097200. [DOI] [PubMed] [Google Scholar]
- 25.Eckert K A, Kunkel T A. In: Polymerase Chain Reaction I: A Practical Approach. McPherson M J, Quirke P, Taylor G R, editors. Oxford: IRL; 1991. pp. 227–246. [Google Scholar]
- 26.Edgell D R, Doolittle W F. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 27.Bessman M J, Reha-Krantz L J. J Mol Biol. 1977;116:115–123. doi: 10.1016/0022-2836(77)90122-x. [DOI] [PubMed] [Google Scholar]
- 28.Smith L A, Drake J W. Genetics. 1998;148:1611–1618. doi: 10.1093/genetics/148.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T A, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 30.Crow J E, Kimura M. An Introduction to Population Genetics Theory. New York: Harper & Row; 1970. [Google Scholar]
- 31.Grogan D W. Trends Microbiol. 2000;8:180–185. doi: 10.1016/s0966-842x(00)01729-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.