Abstract
It is reasonable to propose that gene expression profiles of purified stem cells could give clues for the molecular mechanisms of stem cell behavior. We took advantage of cDNA subtraction to identify a set of genes selectively expressed in mouse adult hematopoietic stem cells (HSC) as opposed to bone marrow (BM). Analysis of HSC-enriched genes revealed several key regulatory gene candidates, including two novel seven transmembrane (7TM) receptors. Furthermore, by using cDNA microarray techniques we found a large set of HSC-enriched genes that are expressed in mouse neurospheres (a population greatly enriched for neural progenitor cells), but not present in terminally differentiated neural cells. In situ hybridization demonstrated that many of them, including one HSC-enriched 7TM receptor, were selectively expressed in the germinal zones of fetal and adult brain, the regions harboring mouse neural stem cells. We propose that at least some of the transcripts that are selectively and commonly expressed in two or more types of stem cells define a functionally conserved group of genes evolved to participate in basic stem cell functions, including stem cell self-renewal.
Lymphohematopoietic cells are constantly replenished by common clonogenic precursors called hematopoietic stem cells (HSC; refs. 1 and 2). HSCs are the principal cells responsible for developing and maintaining lympho- and hematopoiesis during ontogeny and after BM transplantation (3, 4). One of the most intriguing properties of adult HSCs is a robust maintenance of the dynamic equilibrium between self-renewal and differentiation (5). Despite extensive phenotypic and functional characterization of HSCs (6, 7), little is known about the molecular mechanisms that regulate their self-renewal and differentiation. The likelihood that HSC function might require selective gene expression was recently supported by the identification of transcripts enriched in mouse fetal liver hematopoietic stem cells (8).
Central nervous system neural stem cells (NSC) have been identified in different regions of fetal and adult brain, and have come into focus as a new platform for research in neurodevelopment and neurobiology, as well as candidates for the potential treatment of neurodegenerative diseases and central nervous system injury (9–12). Similar to HSC, the NSC can self-renew and differentiate into all types of neural cells throughout life (13, 14). In both rodent and man, long-term neural cultures growing as adherent layers or as nonadherent neurospheres (NS) have been shown to contain clonal progenitors of neurons, astrocytes, and oligodendrocytes (14, 15).
Intriguingly, in the past few years several lines of evidence indicate that both hematopoietic and neural stem cells might demonstrate surprising plasticity. Highly purified HSC were shown to give rise to liver tissues (16) and bone marrow (BM)-derived cells were shown to give rise to muscle cells (17); in addition, there are reports claiming BM origin of a different type of neural cells, including mature neurons in brain (18, 19). In a similar fashion, NS-derived cells were shown in one case to give rise to hematopoietic cells (20) and in another case to participate in the formation of many other embryonic and adult tissues, but not blood (21). These results suggest that different stem cells, but not any other cells in the adult organism, may retain a general self-renewing and differentiating capacity or pluripotency (22) and thus it seems reasonable to propose that common basic molecular mechanisms, in addition to environmental clues, could be responsible for self-renewal properties of stem cells.
Here we approached the isolation of genes commonly and preferentially expressed in stem cells by identifying the genes that differentially expressed in adult HSC compared with its direct environment, BM. Furthermore, using cDNA microarray technology, we have identified a set of genes expressed in HSC (but not BM) that are also expressed in mouse neurospheres (but not in terminally differentiated neural cells). In vivo, many of these genes also selectively expressed in the regions of embryonic and adult brain that contain central nervous system stem cells and are the sites of continuous neurogenesis.
Materials and Methods
HSC Sorting and PCR Techniques.
About 3 × 105 double-sorted HSCs (23) from AKR/J mice and 6 × 105 BM cells were used to isolate RNA and make first-strand cDNA, using CLONTECH's SMART first-strand cDNA synthesis. The cDNA subtraction was performed with the PCR-Select protocol (CLONTECH) after 16 cycles of PCR amplification, according to manufacturer's instructions.
Sequence Analysis.
Analyses were performed on-line, except clustering/vector removal made with PHRAPP (Stanford Sequencing Center, Stanford, CA). The batch homology search was made with BLAST; sequence analysis and secondary structure prediction was preformed by using on-line packages from the ExPASy Molecular Biology Server (http://ExPASy.CH), European Molecular Biology Laboratory (EMBL), Institut Suisse de Recherches Experimentales sur le Cancer (Lausanne, Switzerland) (ISREC), and National Center for Biotechnology Information (NCBI) web sites, including CLUSTALW (EMBL) and BOXSHADE (ISREC).
Microarray Analysis and in Situ Hybridization.
Microarray analysis of HSC and WBM and in situ hybridization was performed exactly as described (24).
Results
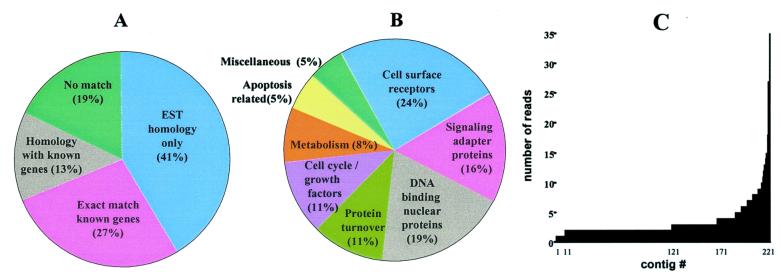
During mouse adulthood, virtually all hematopoietic stem cell activity is found within the c-kit+, Thy1.1low, Linneg/low, Sca-1+ population of the BM (7). BM cDNA was subtracted from HSCs cDNA to enrich for transcripts expressed in HSCs by using the PCR-Select procedure (CLONTECH). The subtracted library (named HB) was examined by high-throughput sequencing followed by the BLAST search of publicly available databases and sequence analysis. Clustering of 1,500 primary sequences resulted in a set of 223 nonredundant contigs—some of them (40%) representing known genes and their homologues, others (41%) matching novel sequences (expressed sequence tags, many of which originated from early embryonic tissues), and a substantial proportion (19%) producing no match in public databases (Fig. 1 A and B). Over 95% of the contigs were assembled from more than 1 read (up to 35 reads), suggesting an exhaustive sequencing of the HB library (Fig. 1C). Informative transcripts found in the HB library included a diverse set of novel and known genes (see Table 1, which is published as supplemental data on the PNAS web site, www.pnas.org). The well known mouse hematopoietic stem cell surface markers such as c-kit (7) and CD34 antigen (25) were found, confirming the veracity of subtraction. The most abundant transcripts, macro H2A1.2 and dnmt3b identified in the fetal liver stem cell screen (8) were not seen in the HB library, illustrating likely genetic differences between fetal and adult HSC. Among the common transcripts found in fetal liver and adult hematopoietic stem cells were Angiopoietin 1, a Tie2/TEK ligand (26) recently proposed to be responsible for HSC-promoted angiogenesis (27), CD27, a member of the TNF receptor family marking a subset enriched for the cells with short-term hematopoietic activities (28), and Evi-1, an oncogene involved in leukemogenesis (29). In general, a proportion of the analyzed transcripts from the HB library match the records in the Stem Cell Database (http://stemcell.princeton.edu), suggesting the overlapping but not identical expression profiles in fetal and adult HSC.
Figure 1.
Analysis of genes found in the HB library. (A) Categorization by sequence type. (B) Functional clustering of known (or highly homologous) genes. (C) Sequence composition of 223 nonredundant contigs.
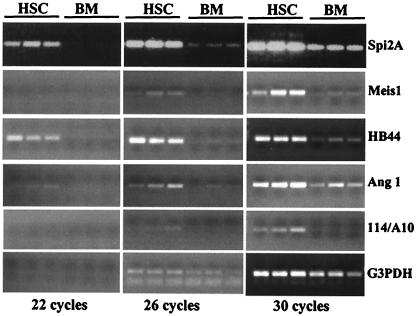
In parallel with sequencing efforts, dot blot hybridization with HSCs and BM cDNA probes was used to screen over 500 clones from the HB library. Clones with the most prominent differential expression were examined by semiquantitative reverse transcription–PCR analysis using gene specific primers and the RNA prepared from three independently double-sorted HSC and BM samples. Several transcripts were estimated to be over 100-fold (eight PCR cycles) enriched in HSC (Fig. 2), testifying to the efficiency of the subtraction procedure. PCR-analyzed clones showing robust differential expression were further scrutinized by Northern hybridization (see Fig. 5, which is published as supplemental data). In addition to strong differential expression in HSC, confirmed for all transcripts, most of the genes had a complex expression pattern, suggesting they play roles in other tissue and organs. In some cases (e.g., Meis1, HB44, and 114/A10), low to no expression was detected outside of the HSC compartment, suggesting that in adult mouse these genes could be functioning selectively in the HSC population.
Figure 2.
The reverse transcription–PCR analysis of selected genes found in the HB library. Each gene was tested by using three independently prepared samples of double-sorted HSC and BM.
Transcripts related to protease inhibitors were among the most abundant in the HB library. One of them, the Ctla-2α gene (incompletely spliced in the HB library), was found to be selectively expressed also in activated cytotoxic T cells and to share high homology to a cysteine protease proregion (30). Another serine protease inhibitor, Spi2A, was identified to be specifically expressed in multipotent hematopoietic progenitor FDCP-Mix cells (31). The Spi2A variant found in HSC has minor aa changes (G173S, N179K, E182Q, T191M, L344Q, T356K, C410S, M411E), possibly due to allelic variations. The abundant expression of Spi2A in HSC (at the level of β-actin) is highly differential (Fig. 2), with the lower level detected in spleen, thymus, and testis (31).
In addition to a number of DNA binding and nuclear proteins, well known transcription factors Meis1 and Evi-1 were found in the HB library. In the adult mouse, Meis1 mRNA was found at very low levels in several tissues, but strongly and differentially expressed in HSC compared with BM (see Fig. 5). Moreover, by using Meis1-specific antibodies (gift of Yakob Jakobs, Stanford University Medical School, Stanford, CA) high levels of Meis1 expression in HSC were confirmed on the protein level (A.V.T. and Y. Jakobs, unpublished observation).
Among the gene products implicated in signaling, several IFN-induced messages (GTP2/Tgtp, RNase-L, HB44) were identified in the HB library. The most abundant of them, GTP2/Tgtp, is induced in IFN-activated macrophages and T cells (32, 33). A new protein, HB44, is a member or a close homologue of an INF-inducible transmembrane receptor protein family (32, 34) and is closely related (76% aa identity) to the 9–27 gene, which was shown to form a complex relaying growth inhibitory and aggregation signals (35).
Messages coding for cell surface proteins represented the largest group in the HB library, including the well-known c-kit and CD34 HSC markers. Among the lesser known hematopoietic markers, the 114/A10 antigen (36) showed a very specific expression in HSC with almost no expression in other adult tissues (see Fig. 5). Indeed, the majority of hematopoietic colony-forming cells were found in the fraction (5%) of 114/A10 positive BM cells, consistent with its expression on hematopoietic progenitors (36).
Several novel transmembrane proteins were found in the HB library. We have focused on two proteins, Cyt28 and mETL1, both members of the secretin subfamily of seven transmembrane (7TM) receptors, based on hydropathy profile and multiple sequence alignments (see Fig. 6, which is published as supplemental data). The mouse homologue of rat ETL1 protein (gb|AAG33020.1|) mELT1 has high homology with rat latrotophilin receptor (CLA3B brain splice variant; ref. 37) and a human brain protein of unknown function (GenBank accession no. KIAA0768; ref. 38). Most of the homology is found within the 7TM region (56% identical, 74% positives) with no high score alignments for the putative extracellular part. Protein sequence analysis suggested that mETL1 has a leader peptide, one EGF-like domain and one or two tandem-organized, Ca-binding, EGF-like domains, a Latrophilin/CL-1-like GPS domain, and multiple sites for N- and O-linked glycosylation (see Fig. 6). The mETL1 protein has overall significant homology with CD97, a 7TM receptor for decay acceleration factor (DAF or CD55; refs. 39 and 40), and EMR1 (41), a 7TM protein with unknown function cloned from a neuroectodermal cDNA library. Northern blot analysis of adult mouse tissue detected moderate expression in heart and low levels in lung, kidney, brain, and liver (see Fig. 5).
Of particular interest was Cyt28 protein, a 7TM receptor also expressed in fetal liver stem cells (I. Lemischka, personal communication). The database search identified two human proteins, GPR56 (42) and TM7XN1 (43), that shared about 80% identity with Cyt28. GPR56 and TM7XN1 are 98% identical and differ mainly by the insertion in the first intracellular loop of GPR56 (see Fig. 6), most probably the result of allelic variations and/or alternative splicing. Thus, Cyt28 is most probably the mouse orthologue of GPR56/TM7XN1 human protein. Similar to mETL1, Cyt28 has a Latrophilin/CL-1-like GPS domain, and multiple N- and O-linked glycosylation sites reminiscent of mucin-like proteins. Northern blot analysis revealed high levels of Cyt28 expression in brain, kidney, and heart and moderate expression in lung and testis (see Fig. 5).
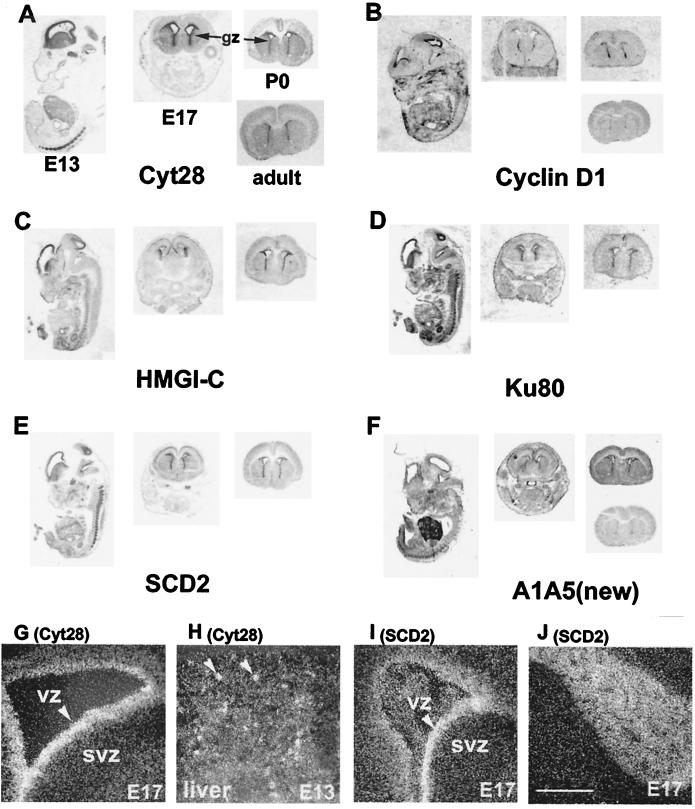
Homology searching and database mining revealed many transcripts found in the subtracted HB library, in addition to Cyt28, that were expressed in central and peripheral nervous tissues. These findings prompted us to look systematically for the genes that expressed specifically and in common in HSC and in neural stem/progenitor cells. Neurospheres derived from the subventricular zone (SVZ) of newborn mice can proliferate and retain pluripotency, differentiating into neurons, astrocytes, and oligodendrocytes [differentiated cells (DC)], under the proper growth factor conditions (44). Because this model system was recently used for genetic analysis of neural progenitor differentiation (24), it was ideal for testing the overlap between hematopoiesis and neurogenesis. To identify the HSC-specific genes expressed in NS, amplified cDNAs from HSC and BM were cohybridized with the array containing NS-specific transcripts. A large number of clones selectively hybridized with HSC cDNA. Those with 2-fold or greater enrichment in HSC over BM were chosen and differential expression of identified transcripts in HSC was reconfirmed by reverse transcription–PCR analysis using an independent set of double-sorted HSC and BM (data not shown). In addition to the genes already identified in the HB library (e.g., Cyt28, ml3-PSP, Nedd4), a number of transcripts with homology to known genes were identified (see Table 2, which is published as supplemental data). More than half of common-selective transcripts were novel; some showed homology to various expressed sequence tags from early embryonic and brain libraries and 25% of the sequences showed no matches in the public database. To determine whether common-selective genes expressed in zones of neurogenesis in vivo, we performed in situ hybridization onto tissue sections from developing mice ranging from embryonic day 13 to adult (Fig. 3). In all cases, labeling in the brain was generally restricted to the germinal zones at all stages examined, consistent with expression in neural progenitors. Predominant expression in the ventricular zone (VZ) versus the SVZ was observed for Cyt28, Cyclin D1, Ku80, and SCD2 genes; conversely, HMGI-C expression was greater in SVZ than in VZ (Fig. 3 G and I and data not shown). Distinct expression patterns were observed for some genes. Likewise for Cyt28 mRNA on E13, little hybridization was observed elsewhere in the embryo, except in the liver, where the distribution was patchy (Fig. 3H). For Cyclin D1 mRNA on E17 and P0, higher expression was seen in the dorsal and ventral parts of the forebrain germinal zone, and lower expression in the middle parts, possibly related to the production of different neuronal types. A very high expression level of HMGI-C was observed in developing meninges, structures proposed to play a role in neuron migration (ref. 45; Fig. 3C). In addition to the high expression level in VZ, heavy SDC2 labeling was seen in both cranial and spinal ganglia (Fig. 3 I and J). In addition to enriched expression in VZ, a strong expression in E13 liver, resembling ml3-PSP (24), was seen for a transcript A1A5 (Fig. 3F).
Figure 3.
In situ hybridization analysis of HSC-NS common-specific genes. (A–F) Bright field images: E13 whole embryo, E17 head, P1 brain, and adult brain. (G–J) Emulsion-dipped autoradiograms: G and I, brains; H, liver; J, cranial nerve 5. (Scale bar, 250 μm.)
Discussion
This study has identified a large number of genes enriched in adult HSC and demonstrated visible differences in gene expression profiles between adult and fetal liver HSC. Furthermore, we demonstrated an overlap between the genetic programs of hematopoietic and neural stem cells by identification and characterization of a set of transcripts that selectively and commonly expressed in adult HSC and neural stem/progenitor cells. We commence by describing the genes that differentially expressed in adult HSC as compared with BM. We took advantage of suppression subtractive hybridization (SSH; ref. 46), best optimized for limited amounts of starting materials (47, 48).
Mouse fetal liver HSC differ from adult BM HSC in a number of ways. Fetal liver HSC retain broader developmental potential than adult BM HSC—e.g., for producing fetal T cells with skin and female reproductive tract homing potential expressing γδ T-cell receptors (Vγ3Vδ1 for skin, Vγ4Vδ1 for vagina; refs. 49 and 50), B1 peripheral and marginal zone B cells (51), and CD4+, CD3−, LTB+, α4+, β7+ early lymph node cells (52). In addition, fetal liver HSC support the expansion of the HSC pool by the high rate of cell cycle entry (53), whereas steady-state adult BM HSC randomly enter cell cycle at the rate of 8% per day (54). Thus, we expected to find many ontogeny-specific changes in HSC, as well as many genes (such as Cyt28) in common.
Intriguingly, among the most abundant transcripts in the HB library were those coding for the protease inhibitors, CTLA-2, specifically found in activated T cells, and Spi2A, a potentially intracellular serpin (55) with close homology to secreted proteins. Curiously, Spi2A expression is not detected in resting T cells, but quickly induced upon Con A activation, mainly in CD8+ cytotoxic T cells (31). Serpins in HSC may inhibit the activity of regulatory proteases perhaps involved in the differentiation of HSC. Also, overexpression of serpins in HSC and activated T cells might serve to prevent apoptosis; for example, serpin 6 was likewise implicated in negative regulation of apoptosis in monocytes and granulocytes by inhibiting cathepsin G (56).
Among the transcription factors found in the HB library, both Meis1 and Evi-1 were identified as common loci of retroviral integration in myeloid leukemia in BXH-2 (57, 58) and AKXD (59) mice, respectively. Recent findings suggest that cooperating with Pbx and Hox family members, Meis1 may play a key role in the immortalization of myeloid progenitors (60). The Evi-1 oncoprotein was found to be involved in chromosomal translocation and connected with pathogenesis of human leukemias (61). Remarkably, many oncoproteins—e.g., MYC, BCL-2, C-ABL, EVI-1—play an important role both in normal leukemogenesis and oncogenesis. Among normal hematopoietic stem and progenitor cells, only HSC and especially long-term HSC have the capacity for long-term self-renewal (7). Specifically, prospectively isolated progenitors in the myeloerythroid pathway do not self-renew in vitro or in vivo (62), yet every leukemia in the myeloid-restricted pathway has acquired the property of self-renewal. It shall be important to determine whether genes regulated by Meis1 and Evi-1 play more general roles in self-renewal or prevention of differentiation, properties that functionally are similar in cancer cells and stem cells (63).
Abundant in HB were genes encoding two novel proteins, Cyt28 and mEML1, belonging to the 7TM family, which is one of the largest eukaryotic gene families (2–3% of all proteins) and mostly composed of G protein-coupled receptors (GPCR) involved in signal transduction (64). Both receptors possess the recently identified GPCR proteolytic site (GPC) domain (65) shown to encompass a cleavage site for the α-Latrotoxin receptor, whose N- and C-terminal parts remain strongly associated (66). We found mETL1 variants with one or two tandem-organized, Ca-binding, EGF-like domains, most probably the splice variants, which is a common phenomenon in 7TM receptors (67).
The Cyt28 receptor provided the first link to neurogenesis. The most prominent Cyt28 staining in vivo was seen within the proliferative layer(s) of cells immediately adjacent to the ventricles, previously demonstrated to harbor at least some types of neuronal stem cells (21), suggesting that Cyt28 is a potential marker for neural stem/progenitor cells. In addition, some clusters of cells in the E13 liver and spinal cord cells were found to be positively stained. It remains to be determined whether these are HSC clusters present in the liver at day 13 p.c. (53). Interestingly, a high expression level of TM7XN1, one of the Cyt28 human homologues, was found to positively correlate with the low metastatic capacity of melanomas (43). In light of the growing evidence of stem cell plasticity, it is tempting to speculate that commonly expressed receptors (like Cyt28) could account for the ability of stem cells to respond to different milieu, thus providing a molecular basis for the phenomena.
A systemic screen for genes selectively expressed in two different stem/progenitor cell populations, but not in differentiated cells (HSC vs. BM and NS vs. DC) yielded a number of candidates including known genes, their homologues, and 25% of novel genes. For the genes playing roles both in lympho- and neuropoiesis, a double phenotype would be expected in KO mice. Indeed, for Ku80, a DNA-binding subunit of DNA-PK found among common-specific genes, KO mice showed defects in both lymphogenesis and neurogenesis (68). Interestingly, by in situ hybridization, Ku80 expression was predominant in VZ compared with SVZ, whereas in Ku80 KO mice the massive cell death was found mainly in the SVZ (68). This finding raises the possibility that survival of migrating immature neurons depends on the function of Ku80, and possibly other components of the DNA repair machinery, in the neural stem/progenitor compartment. The finding of ERCC-1, a DNA repair gene, among common-selective genes and a critical role of DNA end-joining proteins XRCC-4 and DNA ligase in lymphogenesis and neurogenesis (69) support this idea.
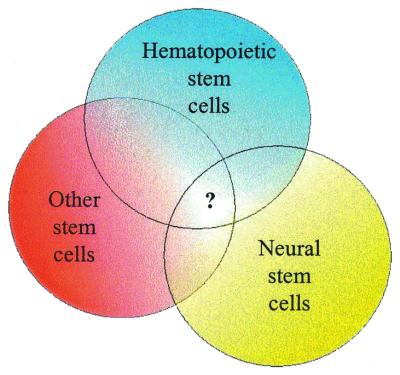
In addition to clearly observed correlates between hematopoiesis and neuropoiesis (70, 71), recent experiments suggested a much broader developmental capacity of HSC than previously thought (16, 20, 21). A close functional relationship was proposed between HSC, central nervous system stem cells (NSC), and possibly many other stem cell types; these unexpected progeny from tissue-specific multipotent could indicate either stem cell plasticity or pluripotency (21, 22, 63). Here, we have demonstrated a molecular similarity between the stem cells of hematopoiesis and of neural lineage cells. It shall be important to test whether these candidate molecules are involved in stem cell self-renewal, differentiation, or even plasticity.
Despite very different genetic programs in vivo, under normal circumstances these two groups of cells share a fundamental common property: life-long self-renewal. Together with the ability to give rise to terminally differentiated cells, self-renewal is a specific and common feature of stem cells (by definition) that distinguishes them from the nearest environment. Thus, we would speculate that genes with common and differential expression patterns were evolved to participate in basic stem cell functions, including self-renewal (Fig. 4).
Figure 4.
Schematic representation of gene expression in different stem cell populations.
Supplementary Material
Acknowledgments
We thank Dr. Sergey Lukyanov for stimulating discussions, Dr. Slava Glukhov for help with primary sequence analysis, and Jing Ong and Dr. Robert Jackson for help with microarray analysis. A.V.T. is a fellow of the Irvington Institute for Immunological Research. This work was supported by the National Institute of Mental Health Grants MH60233 (to D.H.G.) and MH062800 (to H.I.K. and D.H.G.).
Abbreviations
- HSC
hematopoietic stem cells
- BM
bone marrow
- NS
neurospheres
- 7TM
seven transmembrane
- SVZ
subventricular zone
- VZ
ventricular zone
Footnotes
References
- 1.Till J, McCulloch E. Proc Natl Acad Sci USA. 1963;51:29–36. doi: 10.1073/pnas.51.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu A M, Till J E, Siminovitch L, McCulloch E A. J Exp Med. 1968;127:455–464. doi: 10.1084/jem.127.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissman I L. Immunity. 1994;1:529–531. doi: 10.1016/1074-7613(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 4.Aguila H L, Akashi K, Domen J, Gandy K L, Lagasse E, Mebius R E, Morrison S J, Shizuru J, Strober S, Uchida N, et al. Immunol Rev. 1997;157:13–40. doi: 10.1111/j.1600-065x.1997.tb00971.x. [DOI] [PubMed] [Google Scholar]
- 5.Morrison S J, Shah N M, Anderson D J. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- 6.Spangrude G J, Heimfeld S, Weissman I L. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 7.Morrison S J, Weissman I L. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 8.Phillips R L, Ernst R E, Brunk B, Ivanova N, Mahan M A, Deanehan J K, Moore K A, Overton G C, Lemischka I R. Science. 2000;288:1635–1640. doi: 10.1126/science.288.5471.1635. [DOI] [PubMed] [Google Scholar]
- 9.Gage F H, Ray J, Fisher L J. Annu Rev Neurosci. 1995;18:159–192. doi: 10.1146/annurev.ne.18.030195.001111. [DOI] [PubMed] [Google Scholar]
- 10.Suhonen J O, Peterson D A, Ray J, Gage F H. Nature (London) 1996;383:624–627. doi: 10.1038/383624a0. [DOI] [PubMed] [Google Scholar]
- 11.Morshead C M, Reynolds B A, Craig C G, McBurney M W, Staines W A, Morassutti D, Weiss S, van Der K D. Neuron. 1994;13:1071–1082. doi: 10.1016/0896-6273(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 12.Snyder E Y, Macklis J D. Clin Neurosci. 1995;3:310–316. [PubMed] [Google Scholar]
- 13.Johansson C B, Svensson M, Wallstedt L, Janson A M, Frisen J. Exp Cell Res. 1999;253:733–736. doi: 10.1006/excr.1999.4678. [DOI] [PubMed] [Google Scholar]
- 14.Gage F H. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 15.Uchida N, Buck D W, He D, Reitsma M J, Masek M, Phan T V, Tsukamoto A S, Gage F H, Weissman I L. Proc Natl Acad Sci USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagasse E, Connors H, Al Dhalimy M, Reitsma M, Dohse M, Osborne L, Wang X, Finegold M, Weissman I L, Grompe M. Nat Med. 2000;6:1229–1234. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari G, Cusella-De Angelis G, Coletta M, Paolucci E, Stornaiuolo A, Cossu G, Mavilio F. Science. 1998;279:1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 18.Brazelton T R, Rossi F M, Keshet G I, Blau H M. Science. 2000;290:1775–1779. doi: 10.1126/science.290.5497.1775. [DOI] [PubMed] [Google Scholar]
- 19.Mezey E, Chandross K J, Harta G, Maki R A, McKercher S R. Science. 2000;290:1779–1782. doi: 10.1126/science.290.5497.1779. [DOI] [PubMed] [Google Scholar]
- 20.Bjornson C R, Rietze R L, Reynolds B A, Magli M C, Vescovi A L. Science. 1999;283:534–537. doi: 10.1126/science.283.5401.534. [DOI] [PubMed] [Google Scholar]
- 21.Clarke D L, Johansson C B, Wilbertz J, Veress B, Nilsson E, Karlstrom H, Lendahl U, Frisen J. Science. 2000;288:1660–1663. doi: 10.1126/science.288.5471.1660. [DOI] [PubMed] [Google Scholar]
- 22.Weissman I L. Science. 2000;287:1442–1446. doi: 10.1126/science.287.5457.1442. [DOI] [PubMed] [Google Scholar]
- 23.Kondo M, Weissman I L, Akashi K. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 24.Geschwind D H, Qu J, Easterday M, Jackson R, Chen Z, Antoine H, Terskikh A V, Weissman I L, Nelson S F, Kornblum H. Neuron. 2001;29:325–339. doi: 10.1016/s0896-6273(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 25.Osawa M, Hanada K, Hamada H, Nakauchi H. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 26.Davis S, Aldrich T H, Jones P F, Acheson A, Compton D L, Jain V, Ryan T E, Bruno J, Radziejewski C, Maisonpierre P C, et al. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 27.Takakura N, Watanabe T, Suenobu S, Yamada Y, Noda T, Ito Y, Satake M, Suda T. Cell. 2000;102:199–209. doi: 10.1016/s0092-8674(00)00025-8. [DOI] [PubMed] [Google Scholar]
- 28.Wiesmann A, Phillips R L, Mojica M, Pierce L J, Searles A E, Spangrude G J, Lemischka I. Immunity. 2000;12:193–199. doi: 10.1016/s1074-7613(00)80172-7. [DOI] [PubMed] [Google Scholar]
- 29.Morishita K, Parganas E, William C L, Whittaker M H, Drabkin H, Oval J, Taetle R, Valentine M B, Ihle J N. Proc Natl Acad Sci USA. 1992;89:3937–3941. doi: 10.1073/pnas.89.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denizot F, Brunet J F, Roustan P, Harper K, Suzan M, Luciani M F, Mattei M G, Golstein P. Eur J Immunol. 1989;19:631–635. doi: 10.1002/eji.1830190409. [DOI] [PubMed] [Google Scholar]
- 31.Hampson I N, Hampson L, Pinkoski M, Cross M, Heyworth C M, Bleackley R C, Atkinson E, Dexter T M. Blood. 1997;89:108–118. [PubMed] [Google Scholar]
- 32.Lafuse W P, Brown D, Castle L, Zwilling B S. J Leukocyte Biol. 1995;57:477–483. doi: 10.1002/jlb.57.3.477. [DOI] [PubMed] [Google Scholar]
- 33.Carlow D A, Marth J, Clark-Lewis I, Teh H S. J Immunol. 1995;154:1724–1734. [PubMed] [Google Scholar]
- 34.Lewin A R, Reid L E, McMahon M, Stark G R, Kerr I M. Eur J Biochem. 1991;199:417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 35.Deblandre G A, Marinx O P, Evans S S, Majjaj S, Leo O, Caput D, Huez G A, Wathelet M G. J Biol Chem. 1995;270:23860–23866. doi: 10.1074/jbc.270.40.23860. [DOI] [PubMed] [Google Scholar]
- 36.Dougherty G J, Dougherty S T, Kay R J, Landsdorp P, Humphries R K. Exp Hematol. 1989;17:877–882. [PubMed] [Google Scholar]
- 37.Lelianova V G, Davletov B A, Sterling A, Rahman M A, Grishin E V, Totty N F, Ushkaryov Y A. J Biol Chem. 1997;272:21504–21508. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- 38.Nagase T, Ishikawa K, Suyama M, Kikuno R, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1998;5:277–286. doi: 10.1093/dnares/5.5.277. [DOI] [PubMed] [Google Scholar]
- 39.Hamann J, Eichler W, Hamann D, Kerstens H M, Poddighe P J, Hoovers J M, Hartmann E, Strauss M, van Lier R A. J Immunol. 1995;155:1942–1950. [PubMed] [Google Scholar]
- 40.Hamann J, Vogel B, van Schijndel G M, van Lier R A. J Exp Med. 1996;184:1185–1189. doi: 10.1084/jem.184.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baud V, Chissoe S L, Viegas-Pequignot E, Diriong S, N′Guyen V C, Roe B A, Lipinski M. Genomics. 1995;26:334–344. doi: 10.1016/0888-7543(95)80218-b. [DOI] [PubMed] [Google Scholar]
- 42.Liu M, Parker R M, Darby K, Eyre H J, Copeland N G, Crawford J, Gilbert D J, Sutherland G R, Jenkins N A, Herzog H. Genomics. 1999;55:296–305. doi: 10.1006/geno.1998.5644. [DOI] [PubMed] [Google Scholar]
- 43.Zendman A J, Cornelissen I M, Weidle U H, Ruiter D J, van Muijen G N. FEBS Lett. 1999;446:292–298. doi: 10.1016/s0014-5793(99)00230-6. [DOI] [PubMed] [Google Scholar]
- 44.Rao M S, Anderson D J. J Neurobiol. 1997;32:722–746. doi: 10.1002/(sici)1097-4695(19970620)32:7<722::aid-neu7>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 45.Hartmann D, Ziegenhagen M W, Sievers J. Neurosci Lett. 1998;244:129–132. doi: 10.1016/s0304-3940(98)00154-2. [DOI] [PubMed] [Google Scholar]
- 46.Gurskaya N G, Diatchenko L, Chenchik A, Siebert P D, Khaspekov G L, Lukyanov K A, Vagner L L, Ermolaeva O D, Lukyanov S A, Sverdlov E D. Anal Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- 47.Bogdanova E, Matz M, Tarabykin V, Usman N, Shagin D, Zaraisky A, Lukyanov S. Dev Biol. 1998;194:172–181. doi: 10.1006/dbio.1997.8828. [DOI] [PubMed] [Google Scholar]
- 48.Vasiliev O L, Lukyanov S A, Belyavsky A V, Kazanskaya O V, Zaraisky A G. Int J Dev Biol. 1997;41:877–882. [PubMed] [Google Scholar]
- 49.Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, Chien Y H, Weissman I L. Cell. 1990;62:863–874. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 50.Havran W L, Chien Y H, Allison J P. Science. 1991;252:1430–1432. doi: 10.1126/science.1828619. [DOI] [PubMed] [Google Scholar]
- 51.Hardy R R, Li Y S, Allman D, Asano M, Gui M, Hayakawa K. Immunol Rev. 2000;175:23–32. [PubMed] [Google Scholar]
- 52.Mebius R E, Rennert P, Weissman I L. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 53.Morrison S J, Hemmati H D, Wandycz A M, Weissman I L. Proc Natl Acad Sci USA. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheshier S H, Morrison S J, Liao X, Weissman I L. Proc Natl Acad Sci USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korpula-Mastalerz R, Dubin A. Acta Biochim Pol. 1996;43:419–429. [PubMed] [Google Scholar]
- 56.Scott F L, Hirst C E, Sun J, Bird C H, Bottomley S P, Bird P I. Blood. 1999;93:2089–2097. [PubMed] [Google Scholar]
- 57.Steelman S, Moskow J J, Muzynski K, North C, Druck T, Montgomery J C, Huebner K, Daar I O, Buchberg A M. Genome Res. 1997;7:142–156. doi: 10.1101/gr.7.2.142. [DOI] [PubMed] [Google Scholar]
- 58.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mucenski M L, Taylor B A, Ihle J N, Hartley J W, Morse H C, III, Jenkins N A, Copeland N G. Mol Cell Biol. 1988;8:301–308. doi: 10.1128/mcb.8.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang C P, Jacobs Y, Nakamura T, Jenkins N A, Copeland N G, Cleary M L. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jolkowska J, Witt M. Leuk Res. 2000;24:553–558. doi: 10.1016/s0145-2126(00)00031-x. [DOI] [PubMed] [Google Scholar]
- 62.Akashi K, Traver D, Miyamoto T, Weissman I L. Nature (London) 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 63.Weissman I L. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 64.Schoneberg T, Schultz G, Gudermann T. Mol Cell Endocrinol. 1999;151:181–193. doi: 10.1016/s0303-7207(99)00017-9. [DOI] [PubMed] [Google Scholar]
- 65.Ponting C P, Hofmann K, Bork P. Curr Biol. 1999;9:R585–R588. doi: 10.1016/s0960-9822(99)80379-0. [DOI] [PubMed] [Google Scholar]
- 66.Krasnoperov V G, Bittner M A, Beavis R, Kuang Y, Salnikow K V, Chepurny O G, Little A R, Plotnikov A N, Wu D, Holz R W, et al. Neuron. 1997;18:925–937. doi: 10.1016/s0896-6273(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 67.Kilpatrick G J, Dautzenberg F M, Martin G R, Eglen R M. Trends Pharmacol Sci. 1999;20:294–301. doi: 10.1016/s0165-6147(99)01355-3. [DOI] [PubMed] [Google Scholar]
- 68.Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt F W. Proc Natl Acad Sci USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 70.Quesenberry P J, Hulspas R, Joly M, Benoit B, Engstrom C, Rielly J, Savarese T, Pang L, Recht L, Ross A, et al. J Neurotrauma. 1999;16:661–666. doi: 10.1089/neu.1999.16.661. [DOI] [PubMed] [Google Scholar]
- 71.Shamblott M J, Axelman J, Littlefield J W, Blumenthal P D, Huggins G R, Cui Y, Cheng L, Gearhart J D. Proc Natl Acad Sci USA. 2001;98:113–118. doi: 10.1073/pnas.021537998. . (First Published December 26, 2000; 10.1073/pnas.021537998) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.