Abstract
Background
Monosomic alien addition lines (MAALs) can easily induce structural variation of chromosomes and have been used in crop breeding; however, it is unclear whether MAALs will induce drastic genetic and epigenetic alterations.
Methodology/Principal Findings
In the present study, wheat-rye 2R and 5R MAALs together with their selfed progeny and parental common wheat were investigated through amplified fragment length polymorphism (AFLP) and methylation-sensitive amplification polymorphism (MSAP) analyses. The MAALs in different generations displayed different genetic variations. Some progeny that only contained 42 wheat chromosomes showed great genetic/epigenetic alterations. Cryptic rye chromatin has introgressed into the wheat genome. However, one of the progeny that contained cryptic rye chromatin did not display outstanding genetic/epigenetic variation. 78 and 49 sequences were cloned from changed AFLP and MSAP bands, respectively. Blastn search indicated that almost half of them showed no significant similarity to known sequences. Retrotransposons were mainly involved in genetic and epigenetic variations. Genetic variations basically affected Gypsy-like retrotransposons, whereas epigenetic alterations affected Copia-like and Gypsy-like retrotransposons equally. Genetic and epigenetic variations seldom affected low-copy coding DNA sequences.
Conclusions/Significance
The results in the present study provided direct evidence to illustrate that monosomic wheat-rye addition lines could induce different and drastic genetic/epigenetic variations and these variations might not be caused by introgression of rye chromatins into wheat. Therefore, MAALs may be directly used as an effective means to broaden the genetic diversity of common wheat.
Introduction
Modern cultivation procedures have narrowed the genetic base of common wheat (Triticum aestivum L.). A lot of wild relatives and related species were widely used to increase the genetic diversity available to wheat breeders. Rye (Secale cereale L.) has great potential for expanding the genetic variability of cultivated common wheat. Some newly synthesized triticale have been particularly revealing of rapid genomic and epigenomic changes [1]–[4]. The structural variations of rye chromosomes in triticale were also discovered [5]–[8]. Triticales are the beginning of introgression of rye chromatin into wheat genome and they were used to produce wheat-rye addition, substitution and translocation lines. Wheat-rye addition, substitution and translocation lines are the direct bridge materials for wheat improvement. The advantages of using addition lines are the possibility of locating alien gene(s) or species-specific characteristics to particular chromosomes, and the potential to transfer elite gene(s) between species. The alterations of rye telomeric/subtelomeric heterochromatin were observed in several sets of wheat-rye disomic addition lines [9]. Chromosome instability and genome rearrangements in wheat-rye disomic addition lines were also observed [10]–[11]. Almost all these previous studies focused on the wheat-rye disomic addition lines. Researchers should pay more attentions to monosomic alien addition lines (MAALs). It is an effective method to use MAALs to induce small-segment-translocation between species [12]. In rice (Oryza sativa L) breeding program, some useful genes were transferred into cultivated rice from its related wild species by MAALs [13]–[14]. MAALs were also used in breeding programs of sugar beet, oilseed rape, wheat and onion [12], [15]–[17]. Although many MAALs were used in crop breeding, it is unclear whether single alien chromosome added to plants can induce drastic genetic and epigenetic alterations. In present study, wheat-rye 2R and 5R monosomic addition lines and their selfed progeny were used to investigate the effects of single rye chromosome on the alterations of wheat genome.
Results
Identification of Wheat-rye Monosomic Addition Lines
From BC1F2 seeds, one wheat-rye 2R monosomic addition line and one wheat-rye 5R monosomic addition line were identified (Figure 1). The 2R and 5R monosomic addition lines were respectively named as MY+2RF2 and MY+5RF2. Then the 2R and 5R monosomic addition lines were bagged for self-fertilization. Twenty-two seeds (BC1F3) from the selfed progeny of MY+2RF2 and 17 seeds (BC1F3) from the selfed progeny of MY+5RF2 germinated. A 2R monosomic addition line (MY+2RF3) and a 5R monosomic addition line (MY+5RF3) were respectively identified from the selfed progeny of MY+2RF2 and MY+5RF2 (Figure 1). GISH analysis did not detect the translocations between wheat and rye chromosomes in MY+2RF2, MY+2RF3, MY+5RF2 and MY+5RF3 (Figure 1). The other 37 plants derived from MY+2RF2 and MY+5RF2 only contained 42 wheat chromosomes and they were named as 2RF3MY and 5RF3MY, respectively. ‘Mianyang11’, MY+2RF2, MY+2RF3, MY+5RF2 and MY+5RF3 were used for AFLP and MSAP analyses. In addition, 2RF3MY-4, 2RF3MY-8, 2RF3MY-10, 5RF3MY-1, 5RF3MY-5 and 5RF3MY-11 were randomly selected from the 37 plants. Although the parental rye ‘Kustro’ was kept by selfing, it was not used for AFLP and MSAP analyses because of its possible heterogeneity. GISH analysis did not detect small translocations between wheat and rye chromosomes in 2RF3MY-4, 2RF3MY-8, 2RF3MY-10, 5RF3MY-1, 5RF3MY-5 and 5RF3MY-11 (Figure 1). The result indicated that the six progeny only contained 42 wheat chromosomes. However, PCR reaction using rye-specific marker produced the target bands in 2RF3MY-4 and 5RF3MY-1 (Figure 2). These results indicated that rye chromatins have already introgressed into wheat genome and they can not be detected by GISH analysis. 2RF3MY-4 and 5RF3MY-1 were still considered as the progeny that only contained 42 wheat chromosomes because they just contained cryptic rye chromatins.
Figure 1. Wheat-rye monosomic addition lines and their progeny were identified by FISH and GISH analyses using pSc119.2 and rye genomic DNA as probes.
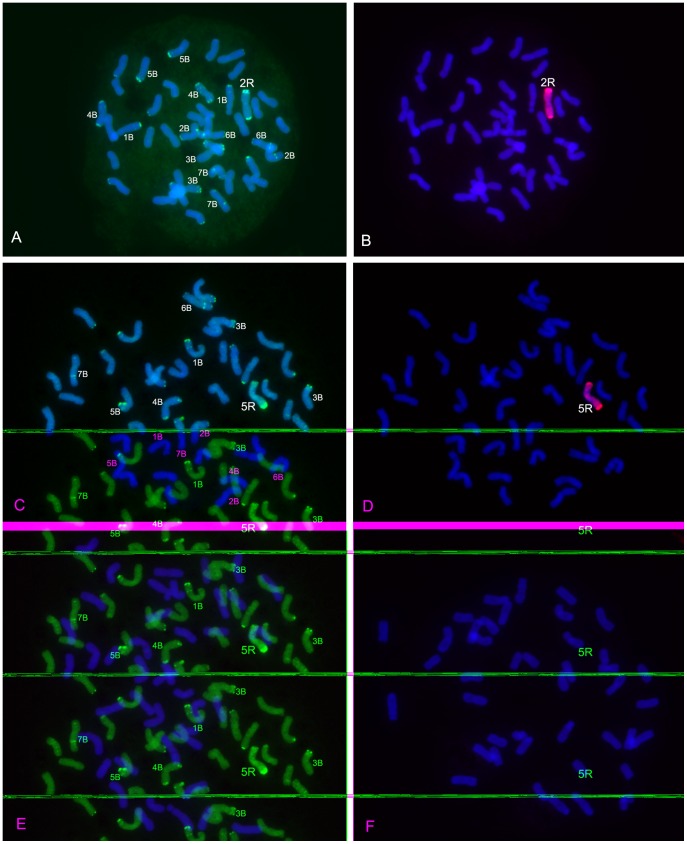
A, B Sequential FISH and GISH on the same spread of metaphase chromosomes of root tip of MY+2RF3 representing 2R monosomic addition line. 2R chromosome and B-genomic chromosomes were marked. C, D Sequential FISH and GISH on the same spread of metaphase chromosomes of root tip of MY+5RF3 representing 5R monosomic addition line. 5R chromosome and B-genomic chromosomes were marked. E GISH analysis on 2RF3MY-4. F GISH analysis on 5RF3MY-1. E and F represent the results of GISH analysis on the progeny that only contained 42 wheat chromosomes.
Figure 2. PCR amplification using rye-specific marker.
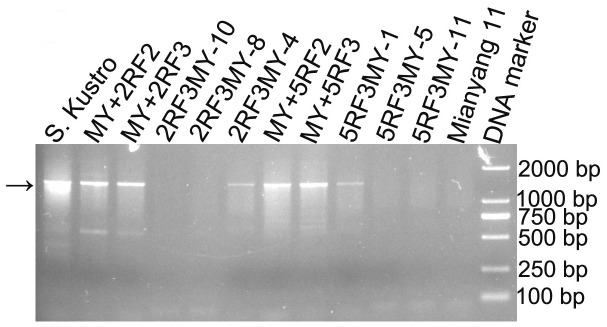
Rye-specific marker was used to investigate whether small rye chromatin has introgressed into the progeny that only contained 42 wheat chromosomes. Arrow indicates the target bands.
AFLP Analysis
Eight different AFLP selective primer combinations (Table 1) were first used to investigate the genetic variations of ten ‘Mianyang11’ plants. Almost no polymorphic amplified sites were found among the ten ‘Mianyang11’ plants (Figure 3). This result indicated that parental wheat plants were homozygous. Using the eight AFLP selective primer combinations, a total of 3065 bands were obtained from ‘Mianyang 11’ and its ten progeny used in this study. Polymorphic amplified sites were observed among the 11 materials (Figure 4). The numbers of absent parental bands and new bands in some progeny were listed in Table 2. Compared with parental wheat ‘Mianyang 11’, a total of 1135 changed bands including 531 lost and 604 new bands were detected in its six progeny, which only contained 42 wheat chromosomes (Table 2). Compared with MY+2RF2 (2R monosomic addition line, BC1F2), a total of 809 changed bands including 500 lost and 309 new bands were detected in its four selfed progeny (Table 2). When MY+5RF2 (5R monosomic addition line, BC1F2) was compared with its four selfed progeny, a total of 805 changed bands including 447 lost and 358 new bands were observed (Table 2). Great genetic variations were observed among the six progeny that only contained 42 wheat chromosomes (F = 9.10; F0.05 = 2.89). 2RF3MY-10 and 5RF3MY-1 exhibited great genetic variations when they were respectively compared with ‘Mianyang 11’ (t2RF3MY-10 = 3.41; t5RF3MY-1 = 3.88; t0.05 = 2.365). There were great genetic variations between MY+2RF2 and MY+2RF3 (t = 2.72; t0.05 = 2.365), and between MY+5RF2 and MY+5RF3 (t = 3.42; t0.05 = 2.365). Different genetic variations could be also observed when MY+2RF2 was compared with its selfed progeny and MY+5RF2 was compared with its selfed progeny (Table 2). These results indicated that wheat-rye monosomic addition lines could induce different and drastic genetic variation in their progeny.
Table 1. Sequences of AFLP and MSAP adapters and primers used in this study.
| Adaptors | |
| EcoR I-adaptors-F | 5′CTCGTAGACTGCGTACC3′ |
| EcoR I-adaptors-R | 5′AATTGGTACGCAGTC3′ |
| Mse I-adaptors-F | 5′GACGATGAGTCCTGAG3′ |
| Mse I-adaptors-R | 5′TACTCAGGACTCAT3′ |
| Hpa II/Msp I-adaptors-F | 5′GATCATGAGTCCTGCT3′ |
| Hpa II/Msp I-adaptors-R | 5′CGAGCAGGACTCATGA3′ |
| Preselective primers | |
| EcoR I-A | 5′GACTGCGTACCAATTCA3′ |
| Mse I-C | 5′GACGATGAGTCCTGAGTAAC3′ |
| Hpa II/Msp I-T | 5′ATCATGAGTCCTGCTCGGT3′ |
| Selective primer combinations used in AFLP | |
| EcoR I-AAC+ Mse I-CTC | EcoR I-ACC+ Mse I-CAA |
| EcoR I-AAC+ Mse I-CTG | EcoR I-AGG+ Mse I-CAA |
| EcoR I-ACT+ Mse I-CTG | EcoR I-ACG+ Mse I-CAA |
| EcoR I-ACA+ Mse I-CAA | EcoR I-ACG+ Mse I-CAA |
| Selective primer combinations used in MSAP | |
| EcoR I-AAC+ Hpa II/Msp I-TTCT (E1HM7) | EcoR I-AAG+ Hpa II/Msp I-TTGC (E2HM2) |
| EcoR I-AAC+ Hpa II/Msp I-TCGA (E1HM1) | EcoR I-AAG+ Hpa II/Msp I-TCGA (E2HM1) |
| EcoR I-AAC+ Hpa II/Msp I-TCCA (E1HM3) | |
Figure 3. Method of AFLP was used to analyze the ten parental wheat plants.
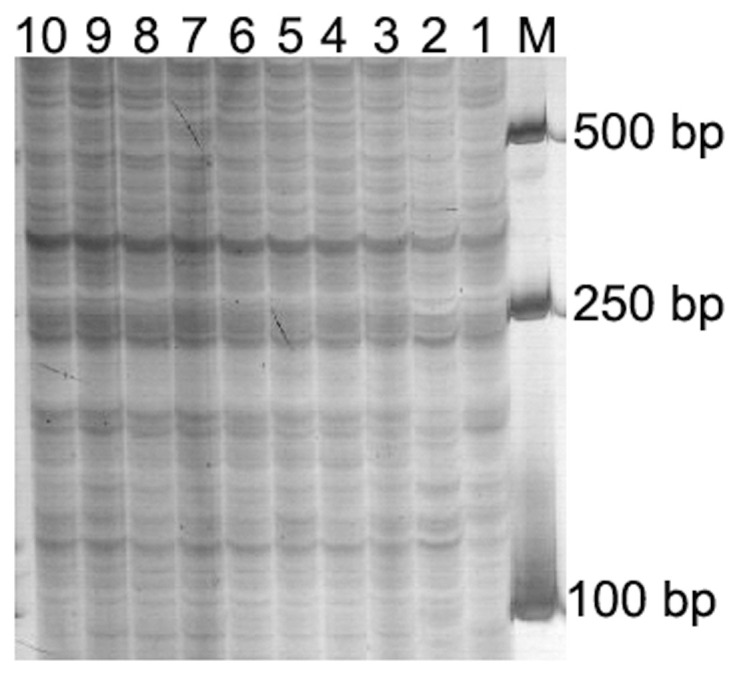
AFLP fingerprints of genomic DNA of ten ‘Mianyang 11’ plants displayed by selective primer combination EcoR I-ACG+ Mse I-CAA. M, DNA marker. 1–10, ten single ‘Mianyang 11’ plants.
Figure 4. Method of AFLP was used to analyze the 11 materials in present study.

AFLP fingerprints of genomic DNA of the 11 materials displayed by selective primer combination EcoR I-AAC+ Mse I-CTG. MY11, ‘Mianyang11’.
Table 2. Numbers of lost and new bands in progeny compared with their parental plants.
| Materials | Number of lost and new bands compared with parents | |||||
| Compared with ‘Mianyang 11’* | Compared with MY+2RF2 * | Compared with MY+5RF2 * | ||||
| Number of lost bands | Number of new bands | Number of lost bands | Number of new bands | Number of lost bands | Number of new bands | |
| ‘Mianyang11’ | – | – | – | – | – | – |
| MY+2RF2 | – | – | – | – | – | – |
| MY+2RF3 | – | – | 136 | 62 | – | – |
| 2RF3MY-4 | 67 | 79 | 86 | 41 | – | – |
| 2RF3MY-8 | 93 | 75 | 118 | 75 | – | – |
| 2RF3MY-10 | 132 | 171 | 160 | 131 | – | – |
| MY+5RF2 | – | – | – | – | – | – |
| MY+5RF3 | – | – | – | – | 154 | 120 |
| 5RF3MY-1 | 132 | 113 | – | – | 150 | 109 |
| 5RF3MY-5 | 57 | 51 | – | – | 86 | 38 |
| 5RF3MY-11 | 50 | 115 | – | – | 57 | 91 |
| Total | 531 | 604 | 500 | 309 | 447 | 358 |
‘–’indicates no comparison was done.
MSAP Analysis
Because HpaII will not cut if either the outer or the inner cytosine of the ‘CCGG/GGCC’ site is fully (double-strand) methylated, whereas, MspI will not cut if the external cytosine is fully or hemi- (single-strand) methylated, the methylation states of the cytosine at ‘CCGG/GGCC’ sites would lead to a differential cleavage by the two isoschizomers. Thus, the band pattern from PCR amplification can reflect the methylation status at a certain site (Figure 5). The five pairs of EcoRI+HpaII/MspI selective primer combinations produced reproducible fragments at 134 sites (Table 3). For each of the materials used in this study, the genomic DNA methylation extent including full-methylation and hemi-methylation is list in Table 3. From Table 3, different methylation extent and pattern of the genomic DNA among the 11 materials can be observed. Among the six progeny that contained 42 wheat chromosomes, only 2RF3MY-10 and 5RF3MY-1 displayed great differences from ‘Mianyang 11’ in the extent of full methylation (t2RF3MY-10 = 3.10; t5RF3MY-1 = 3.27; t0.05 = 2.776), and only 5RF3MY-1 displayed great differences from ‘Mianyang 11’ in the extent of hemi-methylation (t = 2.90; t0.05 = 2.776). There was no great epigenetic variation between MY+2RF2 and MY+2RF3. The great epigenetic variation was not observed between MY+5RF2 and MY+5RF3, too. These results indicated that some of the progeny of monosomic addition lines displayed great epigenetic variations.
Figure 5. Method of MSAP was used to analyze the 11 materials in present study.
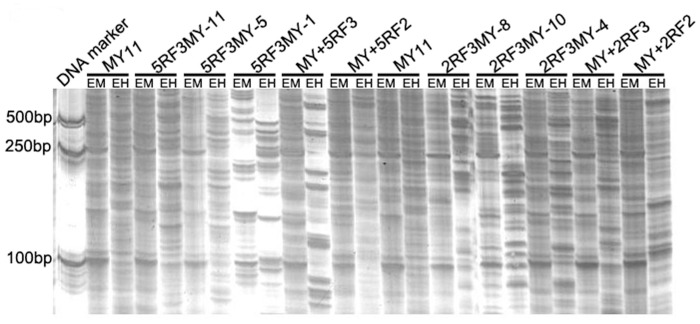
MSAP fingerprints of genomic DNA of the 11 materials and variation of methylation pattern displayed by selective primer combination EcoR I-AAC+ Hpa II/Msp I-TCGA. EM, EcoR I+Msp I; EH, EcoR I+Hpa II; MY11, ‘Mianyang11’.
Table 3. Number of bands amplified using five MSAP selective primer combinations in the 11 materials used in this study.
| Primer combinations* | E1HM7 | E1HM1 | E1HM3 | E2HM2 | E2HM1 | Total | Rate of methylated site (%) | ||||
| Number of amplification site | 31 | 19 | 26 | 27 | 31 | 134 | |||||
| Mianyang11 | Methylated site | Fully** | 2 | 1 | 1 | 2 | 4 | 10 | 54 | 7.46 | 40.30 |
| Hemi*** | 11 | 4 | 12 | 9 | 8 | 44 | 32.84 | ||||
| MY+2RF2 | Methylated site | Fully | 4 | 2 | 5 | 6 | 4 | 21 | 51 | 15.67 | 38.06 |
| Hemi | 8 | 1 | 6 | 5 | 10 | 30 | 22.39 | ||||
| MY+2RF3 | Methylated site | Fully | 2 | 3 | 7 | 5 | 5 | 22 | 56 | 16.42 | 41.79 |
| Hemi | 7 | 2 | 6 | 6 | 13 | 34 | 25.37 | ||||
| 2RF3MY-4 | Methylated site | Fully | 3 | 4 | 5 | 8 | 5 | 25 | 61 | 18.66 | 45.52 |
| Hemi | 10 | 6 | 3 | 4 | 13 | 36 | 26.87 | ||||
| 2RF3MY-8 | Methylated site | Fully | 1 | 1 | 3 | 8 | 8 | 21 | 65 | 15.67 | 48.51 |
| Hemi | 13 | 4 | 12 | 7 | 8 | 44 | 32.84 | ||||
| 2RF3MY-10 | Methylated site | Fully | 7 | 3 | 7 | 5 | 7 | 27 | 63 | 20.15 | 47.01 |
| Hemi | 8 | 6 | 8 | 7 | 7 | 36 | 26.87 | ||||
| MY+5RF2 | Methylated site | Fully | 4 | 2 | 5 | 5 | 7 | 23 | 46 | 17.16 | 34.33 |
| Hemi | 11 | 2 | 7 | 2 | 1 | 23 | 17.16 | ||||
| MY+5RF3 | Methylated site | Fully | 8 | 4 | 5 | 9 | 8 | 34 | 63 | 25.37 | 47.01 |
| Hemi | 10 | 1 | 4 | 4 | 10 | 29 | 21.64 | ||||
| 5RF3MY-1 | Methylated site | Fully | 8 | 1 | 3 | 10 | 8 | 30 | 52 | 22.39 | 38.81 |
| Hemi | 6 | 2 | 4 | 5 | 5 | 22 | 16.42 | ||||
| 5RF3MY-5 | Methylated site | Fully | 4 | 1 | 3 | 4 | 2 | 14 | 52 | 10.45 | 38.81 |
| Hemi | 6 | 0 | 11 | 6 | 15 | 38 | 28.36 | ||||
| 5RF3MY-11 | Methylated site | Fully | 4 | 0 | 4 | 3 | 8 | 19 | 65 | 14.18 | 48.51 |
| Hemi | 6 | 12 | 9 | 8 | 11 | 46 | 34.33 | ||||
The names of primer combinations are abbreviations and their corresponding full name are listed in Table 1.
The band pattern that absent for Hpa II but present for Msp I was regarded as full-methylation and the numbers of this kind of band pattern amplified by each primer pairs were scored in each material.
The band pattern that for present Hpa II but absent for Msp I was regarded as hemi-methylation and the numbers of this kind of band pattern amplified by each primer pairs were scored in each material.
Among the methylated sites, no monomorphic sites were observed, that is, all the 134 methylated sites showed polymorphism among the 11 materials. According to the extent of methylation variation in ‘CCGG/GGCC’ site, the methylated site which showed polymorphism patterns could be further divided into three types: (i) hypermethylation polymorphism site (HPS), the extent of cytosine methylation in this site was stronger than parental plants; (ii) demethylation polymorphism site (DPS), the extent of cytosine methylation in this site was weaker than parental plants; (iii) uncertain polymorphism site (UPS), the extent of cytosine methylation in this site could not be accurately qualitative compared with parental plants. The types of polymorphic methylated sites were determined by using ‘Mianyang11’ as reference. The criteria that were used to judge the patterns of polymorphic methylated sites were listed in Table 4. The polymorphism patterns of methylated sites in the progeny were listed in Table 5. From the Table 5, it could be noted that the numbers of HPS and DPS sites were different among the ten progeny. The six progeny that only contained 42 wheat chromosomes displayed great differences in both HPS and DPS sites (FHPS = 9.34; FDPS = 4.58; F0.05 = 3.15), however, no significant differences were observed between MY+2RF2 and MY+2RF3 in HPS and DPS sites. Similarly, there were no great differences between MY+5RF2 and MY+5RF3 in HPS and DPS sites. All these results mentioned above indicated that wheat-rye monosomic addition lines could induce different and drastic epigenetic variation in their progeny that only contained 42 wheat chromosomes.
Table 4. Criteria by which patterns of polymorphic methylated sites were judged.
| Methylation patterns | hypermethylationpolymorphism site (HPS) | demethylationpolymorphism site (DPS) | uncertainpolymorphism site (UPS) | ||||||||||
| Parental plants* | EM | − | − | − | + | + | + | + | + | − | − | − | + |
| EH | + | − | − | + | + | − | − | − | + | + | − | + | |
| Progeny* | EM | + | − | + | − | + | − | − | + | − | + | + | − |
| EH | − | + | − | + | − | + | − | + | − | + | + | − | |
EM, EcoR I+Msp I; EH, EcoR I+Hpa II; -, band absent; +, band present.
Table 5. Types of polymorphic methylated sites in the ten progeny of ‘Mianyang11’.
| Progeny | Polymorphicpattern* | Number of polymorphic site** |
| MY+2RF2 | HPS | 35 |
| DPS | 34 | |
| UPS | 18 | |
| MY+2RF3 | HPS | 35 |
| DPS | 30 | |
| UPS | 11 | |
| 2RF3MY-4 | HPS | 45 |
| DPS | 35 | |
| UPS | 13 | |
| 2RF3MY-8 | HPS | 32 |
| DPS | 22 | |
| UPS | 5 | |
| 2RF3MY-10 | HPS | 44 |
| DPS | 26 | |
| UPS | 15 | |
| MY+5RF2 | HPS | 31 |
| DPS | 37 | |
| UPS | 15 | |
| MY+5RF3 | HPS | 46 |
| DPS | 32 | |
| UPS | 8 | |
| 5RF3MY-1 | HPS | 37 |
| DPS | 38 | |
| UPS | 13 | |
| 5RF3MY-5 | HPS | 31 |
| DPS | 36 | |
| UPS | 6 | |
| 5RF3MY-11 | HPS | 34 |
| DPS | 23 | |
| UPS | 9 |
HPS, hypermethylation polymorphism site; DPS, demethylation polymorphism site; UPS, unpone.0054057.g001.tifcertain polymorphism site.
The numbers were obtained by comparing ‘Mianyang11’ with each of its ten progeny.
Sequencing of Polymorphic AFLP and MSAP Fragments
Seventy-eight lost and new AFLP fragments were gel isolated, reamplified and sequenced (JX518989-JX519066). Blastn search in NCBI revealed the characterization of these sequences. For 39 of the 78 AFLP sequences, no significant similarities were found. Among the rest 39 sequences, one, seven, six and 19 sequences were respectively involved in tandem repeat, DNA transposon, Copia-like retrotransposon and Gypsy-like retrotransposon. In addition, three sequences had over 90% similarity to cDNA, one sequence, which was cloned from one of new bands in 5RF3MY-1, only matched (81% similarity) to Secale cereale clone R4-6 genomic sequence (DQ414511.1) and two sequences had over 90% similarity to known wheat genomic DNA sequences that were anonymous.
Forty-nine MSAP fragments that displayed methylation alteration were also gel isolated, reamplified and sequenced (JX519067-JX519115). Blastn search in NCBI indicated that the significant similarities of 22 sequences were not found, one sequence was involved in tandem repeat, one sequence was involved in DNA transposon, seven sequences had over 90% similarity to Copia-like retrotransposon and seven sequences had over 90% similarity to Gypsy-like retrotransposon. Additionally, five sequences had over 90% similarity to cDNA and six sequences had over 80% similarity to known wheat genomic DNA sequences that were anonymous. It is interesting that three of the 78 AFLP sequences were coding sequences, however, there were five coding sequences among the 49 MSAP sequences, and their difference was statistically significant (χ2<0.05).
Discussion
MAALs Induce Genomic and Epigenomic Variations
MAALs of several plant species such as rice, wheat, oilseed rape, sugar beet, cucumber, onion, and soybean have already been created [12], [14]–[19]. These MAALs were mainly used to locate genes or markers on chromosomes, introduce alien genes into plant cultivars, study the cytological behaviours of chromosomes and investigate the effect of single alien chromosome on phenotype. It has been reported that wheat-rye monosomic addition lines can easily induce structural variation of chromosome and high frequency of chromosome translocation [12], [20]. The genomic and epigenetic variations occurred in MAALs and their selfed progeny were seldom reported. In present study, great and different genomic/epigenetic variations were observed among 2R and 5R monosomic addition lines and their selfed progeny without rye chromosome. Furthermore, progeny that only contained 42 wheat chromosomes displayed different genetic/epigenetic variations and it seems that 2RF3MY-10 and 5RF3MY-1 displayed the highest extent of variations. These results indicated that drastic genomic and epigenetic variations of wheat can be induced by adding single alien chromosome into wheat background. It was interesting that great genetic variations were observed between MY+2RF2 and MY+2RF3, and between MY+5RF2 and MY+5RF3, however, drastic epigenetic variations were not detected. The reason why this phenomenon occurred is not clear. It is presumed that there might be no relationship between genetic and epigenetic variations in MAALs. As for the progeny that only contained 42 wheat chromosomes, their drastic genetic/epigenetic variations might be caused by losing the single alien chromosomes. The target products amplified by rye-specific marker from 2RF3MY-4 and 5RF3MY-1 indicated that the cryptic introgression of rye chromatin into wheat genome could be induced by wheat-rye monosomic addition lines. This may be one of the reasons why MAALs could induce genetic/epigenetic variations. However, 2RF3MY-10 displayed outstanding genetic/epigenetic alterations although there were no rye chromatins in this material. This case indicated that introgression of rye chromatins into wheat is not necessary for the drastic genetic/epigenetic alterations. Bento et al. [11] have assessed the genome rearrangements in wheat-rye disomic addition lines and indicated that genomic rearrangement events were not a direct consequence of backcrossing, but resulted from further genome structural rearrangements in the BC plant progeny. In this study, genome reshuffling and epigenetic variation were observed in two continuous generations of wheat-rye monosomic addition lines. The results in present study provided direct evidence to illustrate that MAALs could induce drastic genomic and epigenomic variations. Therefore, monosomic wheat-rye addition lines may be directly used as an effective means to broaden genetic diversity of common wheat.
Sequences Involved in Genomic and Epigenomic Variations
Restriction fragment length polymorphism (RFLP) analysis has been used to investigate newly synthesized amphiploids and the changes of specific low-copy noncoding and coding DNA sequences have been observed during allopolyploid formation in wheat [21]–[22]. AFLP (EcoRΙ-MseΙ combination) and MSAP (EcoRΙ-HpaII/MspI combination) methods were used to investigate the allopolyploidy-associated genetic and epigenetic alterations in wheat [23]. Results indicated that changed AFLP sequences corresponded to low-copy DNA and new AFLP bands corresponding to retroelements or DNA transposons were not found, whereas alterations in methylation patterns affected both repetitive DNA and low-copy DNA sequences in almost equal proportions [23]. AFLP and RFLP analyses were used in triticale and results suggested that high degree of coding sequences and repetitive sequences were involved in genomic variation of triticale [1]. These previous results indicated that sequences, which were involved in genetic and epigenetic variations during allopolyploidy, were mainly low-copy noncoding, coding DNA and repetitive DNA sequences. MSAP analysis was used to study the epigenetic variation among wheat-rye 1BL/1RS translocation lines and the alterations of methylation pattern also affected both repetitive and low-copy DNA sequences [24]. In present study, 78 fragments from changed AFLP bands and 49 fragments from changed MSAP bands were cloned and sequenced. For both the AFLP and MSAP sequences, almost half of them showed no significant similarity to known sequences. Among the AFLP sequences whose significant similarities were found, there were few low-copy coding DNA sequences, most of them were retrotransposons and most of the retrotransposons were Gypsy-like. Similarly, among the MSAP sequences whose significant similarities were found, there were few low-copy coding DNA sequences, most of them were retrotransposons, however, the numbers of Copia-like and Gypsy-like retrotransposons were equal proportions. Perhaps, the sequences that were involved in genetic and epigenetic alterations induced by MAALs are different from the ones induced by allopolyploidization. Although few low-copy coding DNA sequences were found in this study, significant difference was observed between ALFP and MSAP coding sequences. In previous studies on allopolyploidization, no great difference was observed between ALFP and MSAP in the variations of low-copy coding DNA sequences [23]–[24]. More evidences are needed to confirm that wheat-rye monosomic addition lines are more likely to induce the epigenetic variation than the genetic variation of coding sequences. In addition, a sequence, which had high similarity to rye genomic DNA sequence, was cloned from a new band in 5RF3MY-1. Rye-specific marker amplified products from 5RF3MY-1 and GISH analysis indicated that this material only contained 42 wheat chromosomes, therefore, these results confirmed that cryptic rye chromatin has integrated into wheat genome.
In conclusion, single rye chromosome added into wheat background could induce drastic genetic/epigenetic variations and the introgression of cryptic rye chromatins into wheat genome. The introgression of rye chromatins into wheat is not necessary for the drastic genetic/epigenetic alterations. Retrotransposons were mainly involved in genetic and epigenetic variations. Genetic variations basically affected Gypsy-like retrotransposons, whereas alterations of methylation pattern affected Copia-like and Gypsy-like retrotransposons equally. Genetic and epigenetic variations seldom affected low-copy coding DNA sequences. The sequences that were involved in genetic and epigenetic alterations induced by MAALs might be different from the ones induced by allopolyploidization. Monosomic wheat-rye addition lines may be directly used as an effective means to broaden genetic diversity of common wheat.
Materials and Methods
Plant Materials
Octoploid triticale MK25 were obtained by crossing between common wheat T. aestivum L. ‘Mianyang11’ and rye S. cereale L. ‘Kustro’. The parental wheat and rye plants were maintained by strict selfing. Some BC1F2 and BC1F3 seeds were obtained by the controlled backcrossing of MK25 with ‘Mianyang11’ followed by self-fertilization.
Identification of Monosomic Addition Lines
The root mitotic metaphase cells of BC1F2 and BC1F3 plants were analyzed through fluorescence in situ hybridization (FISH) using repetitive DNA sequence pSc119.2 as probe and genomic in situ hybridization (GISH) using genomic DNA of ‘Kustro’ as probe. The repetitive DNA sequences pSc119.2 was labeled with Chroma Tide Alexa Fluor-488-5-dUTP (Invitrogen). The root tips and chromosome spreads were prepared according to the methods described by Han et al. [25]. Probe labeling and hybridization were also operated according to the protocols from Han et al. [25].
DNA Extraction
Mature seeds of ‘Mianyang11’, ‘Kustro’, wheat-rye 2R, 5R monosomic addition lines and the selfed progeny of the two addition lines were surfaced-sterilized with 70% ethanol and were placed in 15 cm Petri dishes on two layers of soaked filter paper. After germination, plant seedlings were grown in a chamber at 25±2°C under 16 h of artificial daylight and 8 h of darkness for two weeks. Then, leaves were collected from seedlings and they were divided equally. Genomic DNA was extracted from leaves according to the methods described by Zhang et al. [26].
Investigation of Progeny that Only Contained 42 Wheat Chromosomes Using Rye-specific Marker
To determine whether small rye chromatin has introgressed into the progeny, which only contained 42 wheat chromosomes, a pair of rye-specific DNA primers (5′-GATCG CCTCT TTTGC CAAGA-3′; 5′-TCACT GATCA CAAGA GCTTG-3′) [27] was used to detect the presence of small rye chromatin. The polymerase chain reaction (PCR) was performed according to the procedures described by Katto et al. [27].
Amplified Fragment Length Polymorphism (AFLP) and Methylation-sensitive Amplification Polymorphism (MSAP) Analyses
EcoR I and Mse I were used for AFLP analysis, and HpaII/MspI and EcoR I were used for MSAP analysis. The AFLP and MSAP procedures were performed according to Zhang et al. [24]. The EcoR I, Mse I and HpaII/MspI adaptor, the preselective primers, and the selective primer combinations were listed in Table 1.
In both AFLP and MSAP procedures, repeats were carried out and patterns resulting from two independent digestions were compared for each sample. In addition, for both AFLP and MSAP gels, the upper part and the lower part of the gel, where resolution was not satisfactory, were not used for band scoring. Only stable and repeatable patterns were retained for analysis.
Statistical Analysis
The degree of genomic/epigenetic variations was determined by the analyses of variance, t-test and Chi2 test.
Isolation and Sequencing of AFLP and MSAP Fragments
The polymorphic AFLP and MSAP fragments were isolated from polyacrylamide gels, reamplified by PCR, and sequenced. The procedures were performed according to Zhang et al. [24].
Sequences Analysis
AFLP and MSAP sequences were analyzed for similarity to known sequences in public databases using BLAST package 2.2 on National Center for Biotechnology Information sever (http://www.ncbi.nlm.nih.gov/Blast).
Acknowledgments
The authors are most grateful to Yong Zhang for his excellent technical assistance.
Funding Statement
This study was supported by National Natural Science Foundation of China (No. 31000713) and by the National High Technology Research and Development Program (“863” Program) of China (No. 2011AA100101; 2011AA100103). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ma XF, Fang P, Gustafson JP (2004) Polyploidization-induced genome variation in triticale. Genome 47: 839–848. [DOI] [PubMed] [Google Scholar]
- 2. Ma XF, Gustafson JP (2006) Timing and rate of genome variation in triticale following allopolyploidization. Genome 49: 950–958. [DOI] [PubMed] [Google Scholar]
- 3.Bento M, Pereira HS, Rocheta M, Gustafson P, Viegas W, et al. (2008) Polyploidization as a rectraction force in plant genome evolution: sequence rearrangements in Triticale. PloS ONE 3: e1402. Available: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0001402. Accessed 2012 Dec 15. [DOI] [PMC free article] [PubMed]
- 4. Tang ZX, Wu M, Zhang HQ, Yan BJ, Tan FQ, et al. (2012) Loss of parental coding sequences in an early generation of wheat-rye allopolyploid. Int J Plant Sci 173: 1–6. [Google Scholar]
- 5. Appels R, Gustafson JP, May CE (1982) Structural variation in the heterochromatin of rye chromosomes in triticales. Theor Appl Genet 63: 235–244. [DOI] [PubMed] [Google Scholar]
- 6. Gustafson JP, Lukaszewski AJ, Bennett MD (1983) Somatic deletion and redistribution of telomeric heterochromatin in the genus Secale and in Triticale . Chromosoma 88: 293–298. [Google Scholar]
- 7. Lapitan NLV, Sears RG, Gill BS (1984) Translocations and other karyotypic structural changes in wheat × rye hybrids regenerated from tissue culture. Theor Appl Genet 68: 547–554. [DOI] [PubMed] [Google Scholar]
- 8. Tang Z X, Fu SL, Ren ZL, Zhou JP, Yan BJ, et al. (2008) Variation of tandem repeat,regulatory element, and promoter regions revealed by wheat-rye amphiploids. Genome 51: 399–408. [DOI] [PubMed] [Google Scholar]
- 9. Alkhimova AG, Heslop-Harrison JS, Shchapova AI, Vershinin AV (1999) Rye chromosome variability in wheat-rye addition and substitution lines. Chromosome Res 7: 205–212. [DOI] [PubMed] [Google Scholar]
- 10. Szakács É, Molnár-Láng M (2010) Molecular cytogenetic evaluation of chromosome instability in Triticum aestivum-Secale cereale disomic addition lines. J Appl Genet 51: 149–152. [DOI] [PubMed] [Google Scholar]
- 11. Bento M, Gustafson P, Viegas W, Silva M (2010) Genome merger: from sequence rearrangements in triticale to their elimination in wheat-rye addition lines. Theor Appl Genet 121: 489–497. [DOI] [PubMed] [Google Scholar]
- 12. Ren ZL, Zhang HQ (1997) Induction of small-segment-translocation between wheat and rye chromosomes. Sci China Ser C 40: 323–331. [DOI] [PubMed] [Google Scholar]
- 13. Brar DS, Khush GS (1997) Alien introgression in rice. Plant Mol Biol 35: 35–47. [PubMed] [Google Scholar]
- 14. Multani DS, Khush GS, delos Reyes BG, Brar DS (2003) Alien genes introgression and development of monosomic alien addition lines from Oryza latifolia Desv. To rice, Oryza sativa L. Theor Appl Genet 107: 395–405. [DOI] [PubMed] [Google Scholar]
- 15. Reamon-Ramos SM, Wricke G (1992) A full set of monosomic addition lines in Beta vulgaris from Beta webbiana: morphology and isozyme markers. Theor Appl Genet 84: 411–418. [DOI] [PubMed] [Google Scholar]
- 16. Chevre AM, Eber F, This P, Barret P, Tanguy X, et al. (1996) Characterization of Brassica nigra chromosomes and of blackleg resistance in B. napus-B.bigra addition lines. Plant Breed 115: 113–118. [Google Scholar]
- 17. van Heusden AW, Shigyo M, Tashiro Y, Ginkel RV, Kik C (2000) AFLP linkage group assignment to the chromosomes of Allium cepa L. via monosomic addition lines. Theor Appl Genet 100: 480–486. [Google Scholar]
- 18. Singh RJ, Krishna P, Hymowitz T (1998) monosomic alien addition lines derived from Glycine max (L.) merr. And G. tomentella hayata: production, characterization, and breeding behavior. Crop Sci 38: 1483–1489. [Google Scholar]
- 19. Chen JF, Luo XD, Qian CT, Jahn MM, Staub JE, et al. (2004) Cucumis monosomic alien addition lines: morphological, cytological, and genotypic analyses. Theor Appl Genet 108: 1343–1348. [DOI] [PubMed] [Google Scholar]
- 20. Ren ZL, Lelley T, Röbbelen G (1990) the use of monosomic rye addition lines for transferring rye chromatin into bread wheat. Plant Breed 105: 257–264. [Google Scholar]
- 21. Liu B, Vega JM, Segal G, Abbo S, Rodova M, et al. (1998) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. Ι. Changes in low-copy noncoding DNA sequences. Genome 41: 272–277. [DOI] [PubMed] [Google Scholar]
- 22. Liu B, Vega JM, Feldman M (1998) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. ΙΙ. Changes in low-copy coding DNA sequences. Genome 41: 535–542. [DOI] [PubMed] [Google Scholar]
- 23. Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y, Liu ZH, Liu C, Yang ZJ, Deng KJ, et al. (2008) Analysis of DNA methylation variation in wheat genetic background after alien chromatin introduction based on methylation-sensitive amplification polymorphism. Chinese Sci Bull 53: 58–69. [Google Scholar]
- 25. Han FP, Lamb JC, Birchler A (2006) High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Pro Natl Acad Sci USA 103: 3238–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang HB, Zhao XP, Ding X, Paterson AH, Wing RA (1995) Preparation of megabase-sized DNA from plant nuclei. Plant J 7: 175–184. [Google Scholar]
- 27. Katto MC, Endo TR, Nasuda S (2004) A PCR-based marker for targeting small rye segments in wheat background. Genes Genet Syst 79: 245–250. [DOI] [PubMed] [Google Scholar]


