Abstract
The plant pathogenic bacterium Dickeya dadantii has recently been shown to be able to kill the aphid Acyrthosiphon pisum. While the factors required to cause plant disease are now well characterized, those required for insect pathogeny remain mostly unknown. To identify these factors, we analyzed the transcriptome of the bacteria isolated from infected aphids. More than 150 genes were upregulated and 300 downregulated more than 5-fold at 3 days post infection. No homologue to known toxin genes could be identified in the upregulated genes. The upregulated genes reflect the response of the bacteria to the conditions encountered inside aphids. While only a few genes involved in the response to oxidative stress were induced, a strong defense against antimicrobial peptides (AMP) was induced. Expression of a great number of efflux proteins and transporters was increased. Besides the genes involved in LPS modification by addition of 4-aminoarabinose (the arnBCADTEF operon) and phosphoethanolamine (pmrC, eptB) usually induced in Gram negative bacteria in response to AMPs, dltBAC and pbpG genes, which confer Gram positive bacteria resistance to AMPs by adding alanine to teichoic acids, were also induced. Both types of modification confer D. dadantii resistance to the AMP polymyxin. A. pisum harbors symbiotic bacteria and it is thought that it has a very limited immune system to maintain these populations and do not synthesize AMPs. The arnB mutant was less pathogenic to A. pisum, which suggests that, in contrast to what has been supposed, aphids do synthesize AMP.
Introduction
Dickeya dadantii are plant pathogenic enterobacteria that provoke the soft rot disease in a wide range of plants. The factors required for its pathogeny are numerous, involving principally the production and the secretion of enzymes responsible for the degradation of plant cell wall components [1], [2]. Regulation of their expression has been studied in detail. The regulators of the virulence factors identified are either specific to D. dadantii and other plant pathogenic enterobacteria, such as KdgR, PecS and PecT, or are identical to those that can be found in animal and human pathogenic enterobacteria such as Escherichia coli and Salmonella enterica [3]. These regulators include H-NS, CRP, Fur, GacS-GacA and PhoP-PhoQ [4], [5], [6], [7]. The PhoP-PhoQ two component regulatory system controls virulence and Mg2+ homeostasis in many bacterial species. Its role has been well studied in S. enterica serovar typhimurium. In these bacteria, this system is involved in the regulation of Mg2+ uptake systems, survival in macrophages and resistance to antimicrobial peptides (AMP) [8]. This resistance occurs through the modification of LPS. Several enzymes, encoded by pagP, pagO, pmrC, pmrG, lpxO, pmrHFIJKLM, modify LPS, mostly by adding or modifying palmitate, phosphoethanolamine or 4-aminoarabinose to mask negative charges that allow interaction with cationic AMP [9]. Activation of these genes occurs either directly by PhoP-PhoQ or by PmrA-PmrB, a two component system that can be activated by PhoP through PmrD, a protein stabilizing PmrA in a phosphorylated state. Thus, the genes controlled by PmrA-B are induced both by the signals activating PmrB (macrophage phagosome, high Fe3+, low pH) and those activating PhoQ (low Mg2+, antimicrobial peptides, macrophage phagosome, low pH) [9]. However, the genes regulated by PhoP and the signal sensed by PhoQ may vary from one bacterium to the other [10]. For exemple, Edwarsiella tarda PhoQ is also able to sense temperature, Pseudomonas aeruginosa PhoQ does not respond to AMP, and Sodalis glossinidius PhoQ responds neither to Mg2+ nor to AMP [11], [12], [13]. An analysis of the transcriptome of a D. dadantii phoQ mutant showed that many genes are controlled by this regulator. The increased expression of ferric uptake systems and the decreased expression of pectate lyase genes let suppose that phoQ could control the bacterial virulence [14]. Llama-Palacios et al. [7] showed that the D. dadantii phoP and phoQ mutants have a reduced virulence and an increased sensitivity to the plant AMP thionin. A transcriptomic analysis of the response of D. dadantii to this AMP found 36 overexpressed genes. These induced genes are involved in regulation, transport and modification of the bacterial membrane [15].
D. dadantii is not only a plant pathogen but it can also infect animals. Presence of four insecticidal toxin-like genes in its genome, cytABCD [16], led to check for a possible pathogenicity towards different insects. It was found able to kill the pea aphid Acyrtosiphon pisum but not other insects tested [17]. Ingestion of as few as 100 bacteria can kill the aphid in 3–4 days. Bacteria cross the gut and after one day they are present in the gut in high number but also in the fat bodies and in the embryos. Their number increases of about one log each day and when it exceedes 107 bacteria/insect, the insect dies [18]. A mutant devoid of Cyt toxin genes is still pathogenic but kills the aphid more slowly, in 5 to 6 days. cyt genes are controlled by regulators of plant pathogenicity: they are activated by PecS and repressed by H-NS and VfmE. Mutants in other regulators, GacA, OmpR and PhoP, that do not control the cyt genes, have a reduced insect virulence. This let suppose that other factors are required for a full virulence to aphids [19]. However, no other known insect toxin gene was detected in the genome of D. dadantii.
Very few data are available on the expression pattern of genes of bacteria infecting insects. A transcriptome analysis listed the Yersinia pestis genes induced in flea, but Y. pestis colonizes only the flea gut [20]. Thus, this study gives a limited pattern of the bacterial response to conditions encountered in insects. A SCOTS analysis of Photorhabdus temperata and Xenorhabdus koppenhoeferi genes expressed in Rhizotrogus majalis identified genes mostly involved in virulence, stress response and metabolism [21]. A Transposon Site Hybridization assay identified Francisella novicida genes required to infect Drosophila melanogaster [22]. Moreover, in all these studies, infection was done by injection, which is not the most likely path to infection. To identify new factors involved in the virulence of D. dadantii towards A. pisum, we analyzed the transcriptome of D. dadantii infecting the insect. We show here that genes involved in resistance to AMP, efflux and motility are among the most induced genes while genes involved in plant pathogeny are repressed. Mutation of AMP resistance genes decreased bacterial virulence in insects which is a strong indication that, in contrast with current knowledge, aphids are producing this kind of molecules.
Results
Identification of D. Dadantii Genes with Modified Expression in A. Pisum
In order to identify genes that are important for growth inside the pea aphid and that may be required for virulence, we used CDS pangenomic microarrays to compare the transcriptome of D. dadantii grown in AP3 medium with that of the bacteria isolated from A. pisum three days after infection. AP3 medium, which contains a high concentration of sucrose, may mimic the phloemic sap on which aphids feed. Day three post infection was chosen as a stage where infection is well established. At this stage, aphids may contain up to 106 bacteria which are present in all the insect organs. For each condition, three independent cultures or infections were performed and the total bacterial RNA was extracted, treated and subsequently hybridized to separate arrays. Major modifications of the transcription pattern were observed since 164 genes were upregulated more than 5-fold and 328 genes were downregulated more than 5-fold with P values adjusted for multiple testing (FDR) of <0.002. The whole-genome GSEA (Gene Set Enrichment Analysis) analysis is reported in , and shows that the most affected bacterial gene classes by the insect infection process were membrane associated genes (GO cell compartment), both in up or down-regulated classes, RNA and ribosomal machinery genes, as well as many enzyme and sugar transporters (GO molecular function) and finally additional classes such as protein secretion, several biosynthetic processes, cell redox homeostasis and many central metabolic processes (GO biological processes). A list of selected induced or repressed genes that are discussed in the text is given in Table 1 (for the full list, see Table S2). The gene expression pattern obtained by microarray data results was confirmed by qRT-PCR on nine genes. Six up regulated (arnB, sotA, sotB, pmrC, GenID 19611 and GenID 15786) and three down regulated genes (pelE, kdgM and kdgN) were tested and gave a strong correlation with microarray results (Fig. S1).
Table 1. A subset of D. dadantii genes whose expression varies in aphid.
| Gene product category and ID | Gene name | Fold changea | P-value | Description |
| Upregulated genes | ||||
| AMP response | ||||
| 15458 | pmrC | 90.8 | <0.002 | Phosphoethanolamine transferase |
| 16248 | arnB | 64.0 | <0.002 | UDP-4-amino-4-deoxy-L-arabinose alpha-ketoglutarate aminotransferase |
| 16247 | arnC | 36.1 | <0.002 | Undecaprenyl-phosphate 4-amino-4-deoxy-L-arabinose transferase |
| 18941 | pbpG | 12.7 | <0.002 | D-alanyl-D-alanine carboxypeptidase |
| 19383 | dltB | 32.0 | 0.02 | D-alanyl transfer protein |
| 19382 | dltA | 20.1 | 0.02 | D-alanine-activating enzyme |
| 19381 | dltC | 8.9 | 0.03 | D-alanine carrier protein |
| 16330 | eptB | 6.5 | 0.02 | Phosphoethanolamine transferase |
| Efflux system, transporter | ||||
| 15786 | 68.6 | <0.002 | Inner membrane component of multidrug resistance system | |
| 15787 | 48.2 | <0.002 | Membrane fusion component of multidrug resistance system | |
| 19611 | 37.7 | <0.002 | Transport protein (MFS family) | |
| 18887 | 24.4 | <0.002 | Transport protein (MFS family) | |
| 16087 | pecM | 24.1 | <0.002 | Transport protein |
| 20022 | sotB | 20.7 | <0.002 | Transport protein (MFS family) |
| 20031 | sotA | 14.4 | <0.002 | Transport protein (MFS family) |
| 15661 | 11.9 | <0.002 | Drug resistance efflux pump | |
| 15662 | 10.1 | <0.002 | Drug resistance efflux pump | |
| 18175 | 10.2 | 0.003 | ABC transport system | |
| 18174 | 5.6 | 0.01 | ABC transport system | |
| Motility | ||||
| 19858 | 43.4 | <0.002 | Methyl-accepting chemotaxis protein | |
| 19855 | 30.7 | <0.002 | Methyl-accepting chemotaxis protein | |
| 17672 | 8.3 | <0.002 | Methyl-accepting chemotaxis protein | |
| 15600 | 8.1 | <0.002 | Methyl-accepting chemotaxis protein | |
| 17668 | 7.5 | <0.002 | Methyl-accepting chemotaxis protein | |
| 17665 | 7.3 | <0.002 | Methyl-accepting chemotaxis protein | |
| 18761 | motA | 14.9 | <0.002 | Flagellar motor protein |
| 18760 | motB | 10.9 | <0.002 | Flagellar motor protein |
| Stress response | ||||
| 14750 | asr | 26.0 | <0.002 | Acid shock protein |
| 20273 | narI | 21.8 | <0.002 | Nitrate reductase. Anaerobiosis |
| 18171 | iscR | 11.9 | <0.002 | FeS cluster assembly, transcription factor |
| 15559 | rsxA | 10.1 | <0.002 | SoxR reducing complex |
| 15558 | rsxB | 9.2 | <0.002 | SoxR reducing complex |
| 17401 | nirB | 5,4 | <0.002 | Nitrite reductase. Anaerobiosis |
| Regulator | ||||
| 15788 | 61.2 | <0.002 | Transcription regulator | |
| 16073 | vfmE | 9.6 | 0.007 | Virulence regulator |
| Downregulated genes | ||||
| Pectin catabolism | ||||
| 19629 | kdgM | −92.2 | <0.002 | Oligogalacturonate porin |
| 15523 | kdgN | −55.0 | <0.002 | Oligogalacturonate porin |
| 19632 | paeX | −47.0 | <0.002 | Pectin acetylesterase |
| 19646 | pelE | −30.9 | <0.002 | Pectate lyase E |
| 18695 | exuT | −21.4 | <0.002 | Galacturonate transporter |
| 18698 | uxaB | −18.1 | <0.002 | Galacturonate catabolism |
| 20789 | pehN | −9.5 | <0.002 | Polygalacturonase N |
| 19699 | uxaA | −8.7 | <0.002 | Galacturonate catabolism |
| 19960 | kdgA | −6.7 | <0.002 | Galacturonate catabolism |
| 18229 | outC | −6.5 | <0.002 | Pectate lyase secretion |
| 47127 | kduD | −5.4 | 0.002 | Pectin catabolism |
| Galactan catabolism | ||||
| 18200 | ganL | −53.2 | <0.002 | Galactan porin |
| 18192 | ganE | −41.5 | <0.002 | Galactan transport |
| 18377 | mglB | −38.4 | <0.002 | Galactose transport |
| 18193 | ganF | −27.8 | <0.002 | Galactan transport |
| 18195 | ganG | −24.2 | <0.002 | Galactan transport |
| 18378 | mglA | −20.4 | <0.002 | Galactose transport |
| 18379 | mglC | −19.0 | <0.002 | Galactose transport |
| 18196 | ganA | −17.2 | <0.002 | Endogalactanase |
| 18198 | ganB | −7.3 | <0.002 | Exogalactanase |
| osmoregulation | ||||
| 19710 | betI | −72.5 | <0.002 | Betaine synthesis |
| 16548 | scrY | −53.2 | <0.002 | Sucrose porin |
| 19708 | betA | −52.2 | <0.002 | Betaine synthesis |
| 19709 | betB | −31.5 | <0.002 | Betaine synthesis |
| 16547 | scrA | −31.0 | <0.002 | Sucrose metabolism |
| 19635 | −22.0 | <0.002 | Osmotically induced lipoprotein | |
| Toxin | ||||
| 16662 | cytD | −3.5 | <0.002 | Insecticidal toxin |
| 16663 | cytC | −2.0 | <0.002 | Insecticidal toxin |
| 16664 | cytB | −2.8 | <0.002 | Insecticidal toxin |
| 16665 | cytA | −1.6 | <0.002 | Insecticidal toxin |
| stress | ||||
| 16995 | raiA | −92.6 | <0.002 | Cold shock protein |
| 16379 | uspA | −25.8 | <0.002 | Universal stress protein |
| 16377 | uspB | −9.5 | <0.002 | Universal stress protein |
| 19170 | cpxR | −12.4 | <0.002 | Transcriptional regulator |
| 19171 | cpxP | −12.3 | <0.002 | Stress resistance protein |
Positive values represent genes upregulated in aphids, whereas negative values represent genes downregulated.
Plant Virulence Genes are Repressed in Aphids
Our previous results showed that the regulators controlling cyt genes are identical to those regulating plant virulence factors, but that they act in an opposite way [19]. The microarray analysis confirms these observations on a wider basis since many genes involved in pectin degradation, which are induced in plant infection, are repressed. The genes of the two oligogalacturonate-specific porins [23], [24] are among the most repressed (kdgM, -92-fold change; kdgN, −55-fold change). Genes of some secreted enzymes involved in pectin degradation were also down regulated (pelE, paeX, pehN) (Table 1), as were genes involved in the transport and catabolism of pectin breakdown products (exuT, uxaB, uxaA, kduD, kdgA). The complete catabolic pathway of galactan, another component of plant cell walls, is also strongly repressed (−52-fold change for the gene of the porin GanL). Expression of the Out type II secretion system (T2SS), that secretes mostly pectinases is also reduced (outC, −6.5-fold change). In contrast, expression of the second T2SS, Stt, which secretes the pectin lyase PnlH is slightly increased (sttI, 5.3-fold change).
D. Dadantii Genes Induced in Aphids
The most induced genes in this transcriptome analysis probably correspond to those which are the most needed to infect and survive in aphids. It is remarkable to note that many of these genes encode exporters (Table 1). GenID 15786-7-8, with 68-, 48- and 64-fold change, respectively, encode a tripartite multidrug resistance system whose best homologues are found in plant pathogenic or plant-associated bacteria (Pseudomonas sp., Xanthomonas sp.) and also in insect pathogens (P. entomophila, Arsenophonus nasoniae) or some insect symbionts (Baumannia cicadellinicola). This transporter confers to D. dadantii resistance to phytoalexins and protamine [25]. Expression of four genes encoding transporters of the MFS family is also induced: GenID 19611 (37-fold change), GenID 18887 (24-fold change), sotA (14-fold change) and sotB (20-fold change). If the specificity of the two first exporters is not known, SotA and SotB have been shown to be able to export sugar and sugar derivatives that can be toxic to the bacteria [26]. The gene of the efflux protein PecM is induced 24-fold. pecM belongs to the pecS regulon and it has been proposed that it could efflux indigoidine, a blue pigment synthesized by the product of the indABC genes, which also belong to the pecS regulon. Surprisingly, the indABC genes are only weekly induced and other pecS-controlled genes are not induced, suggesting that PecM could be controlled by another regulator and efflux other types of molecules [27]. Induction of all these exporters shows that D. dadantii encounters hostile conditions in aphid and has to efflux many noxious compounds.
Expression of motility genes is strongly modified in the aphid. D. dadantii possesses 45 methyl-accepting chemotaxis proteins (MCP) which are often organized in clusters. Expression of some of these genes was increased. For example, in the four MCP gene cluster GenID 19858-GenID 19855-GenID 19852-Gen ID19851, expression of the two first increased 43-fold and 30-fold respectively, while that of the two last genes was unmodified (Table 1). Similarly, in the four MCP gene cluster GenID 17665-GenID 17668-GenID 17672-GenID 17674, expression of the three first genes increased 7-fold to 8-fold, while that of the last gene was unmodified. Expression of the genes of two components of the flagellar motor MotA and MotB increased 15-fold and 11-fold, respectively (Table 1). Colonization of the insect body occurs very quickly, within one day after infection [18]. Induction of these MCP and motility proteins could help the bacteria to quickly reach all the part of the insect body.
Osmotic Regulation of Cyt Toxin Genes
We have shown that the Cyt toxins are required only for infection by ingestion and that they can be detected in the gut but not in other organs, suggesting that they are not produced throughout all the infection process [18]. Moreover, their synthesis is induced in high osmolarity environment. These observations explain why cyt gene expression is surprisingly diminished in our results (Table 1). For the reference condition, bacteria were grown in the sucrose rich (20%) AP3 medium which strongly induces cyt gene expression. Bacteria collected in the aphids reside mainly outside of the gut, in the fat body or other organs of the insect in which osmolarity is low [18]. This difference of osmolarity between the two conditions could explain why cyt genes are globally less expressed in the aphids in our experiment. This is also the reason why some of the most repressed genes in this analysis are those involved in sucrose transport and metabolism such as the sucrose porin gene scrY (-53-fold change), or in osmoprotectant synthesis and transport (betA, -52-fold change and proX, -23-fold change) (Table 1). We thus checked whether the strong induction or repression observed for certain genes could result from an osmotic regulation. arnB and dltB expression (see below) were repressed only three-fold and 1.3-fold respectively by growth in 10% sucrose (data not shown). kdgM and kdgN expression is not regulated by osmolarity [23], [24]. Thus, osmoregulation seems to control only a limited subset of the genes induced or repressed in our transcriptomic analysis.
Response to Antimicrobial Peptides
Some of the most induced genes in this study are those homologous to genes conferring resistance to AMP in other enterobacteria by modifying LPS. A high induction (91-fold change) was observed for pmrC, a gene encoding the protein that adds phosphoethanolamine to lipid A (Table 1). pmrC is the first gene of an operon containing also pmrA and pmrB, which encode a two component regulation system that regulates genes in response to AMP in S. enterica. pmrA and B are induced about 3-fold. Proteins involved in the synthesis and addition to LPS of 4-aminoarabinose are encoded by the arnBCADTEF operon (also called pmrHFIJKLM or pbgP-E) which is strongly induced in D. dadantii infecting aphids (64-fold change for arnB). Modification of LPS by 4-aminoarabinose allows S. enterica to resist to AMP and it has been shown that this modification is required for full virulence of P. luminescens towards the greater wax moth, Galleria mellonella [28]. A last set of genes potentially involved in resistance to AMP can be noticed: dltD (or pbpG) (12-fold change) and the dltBAC operon (32-fold change) (Table 1). Homologues of these genes are found in Gram positive bacteria where their products modify teichoic acids by adding alanyl residues. Teichoic acids are cell wall glycopolymers which are not found in Gram negative bacteria. Their modification by alanylation masks their negative charges and confers to the bacteria resistance to AMPs [29]; [30]. In Gram negative bacteria dltABC and dltD genes are present in the phytopathogenic bacteria Dickeya sp, Pectobacterium sp. and in the entomopathogenic bacteria P. luminescens. However, the substrate potentially modified in these bacteria remains to be identified.
Induction of genes responsible for the resistance to AMPs in D. dadantii infecting aphids is surprising. The genes coding for AMPs usually produced by insects were not identified in the A. pisum genome and no AMP synthesized by the pea aphid has been detected by biochemical methods [31]. To determine if unknown AMP inducing this response could be synthesized by A. pisum, we tested whether the genes induced in aphids are indeed induced by known AMPs and whether PhoP and PmrA are involved in this regulation. We analyzed the resistance to AMPs of arnB and dltB mutants and examined their role during aphid infection.
Regulation of AMP Resistance Genes
Resistance to AMP has been extensively studied in S. enterica. We first looked for the presence and induction in D. dadantii of homologues of genes which have been shown to play a role in AMP resistance in S. enterica. The two component regulators PhoP-PhoQ and PmrA-PmrB are present in D. dadantii but PmrD, which links both systems and allows PmrA-regulated genes to be regulated by PhoP is absent. In addition to pmrC and to the arnB operon, the eptB gene, which codes for another phosphoethanolamine adding enzyme, is present and induced 6-fold (Table 1). The oxygenase encoding lpxO and the palmitoyl transferase encoding pagP are present in D. dadantii but are not induced in aphid and pagL is absent. Thus, most of the genes involved in LPS modification in response to AMP in S. enterica have a homologue in D. dadantii but all are not induced in an aphid infection.
Presence of AMP and growth in low Mg2+ medium are conditions inducing the PhoP-regulated genes in S. enterica and E. coli, including arnB-F. Regulation of D. dadantii arnB in these conditions was analyzed using a GUS reporter fusion. We tested two commonly used AMPs, polymyxin B and protamine. arnB was induced in the presence of both compounds (Fig. 1A and 1B). LB is a medium of low Mg2+ concentration [32], [33]. Addition of a high Mg2+ concentration (10 mM) in this medium repressed arnB expression five-fold (Fig. 1C). This repression is much lower than that described in S. enterica. To test whether the effect of polymyxin, protamine and Mg2+ on arnB occurs through PhoP or PmrA, the arnB-uidA fusion was assayed in these backgrounds. While its expression was unchanged in the pmrA mutant, it was reduced by one third in the phoP mutant, indicating that PhoP is an activator of arnB (Fig. 1D and 1E). In both backgrounds, repression by 10 mM Mg2+ was conserved, showing that neither PhoP nor PmrA are involved in this regulation (Fig. 1D). While induction by polymyxin was conserved in the phoP and pmrA mutant (data not shown), induction by protamine was abolished in the phoP background (Fig. 1E). This result indicates that regulation of arnB by AMPs involves two systems. One of them, the PhoP-PhoQ system, is sensing presence of protamine but is blind to polymyxin.
Figure 1. Regulation of arnB.
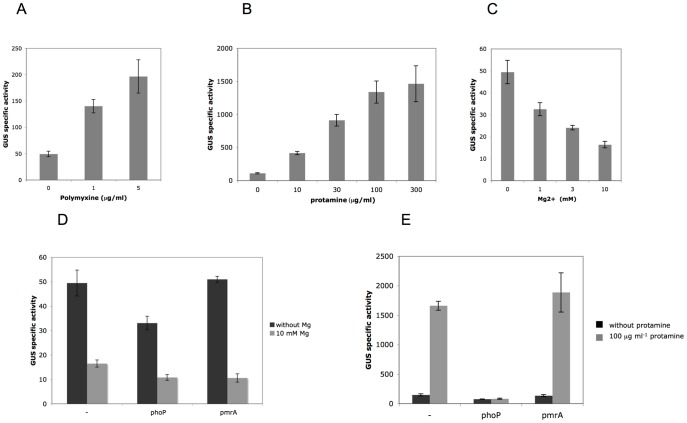
The arnB-uidA fusion of strain A5256 was assayed in the presence of increasing concentrations of polymyxin (A), protamine (B), and Mg2+ (C). Effect of phoP and pmrA mutations on arnB-uidA regulation by Mg2+ (D) and protamine (E) was assayed. Cultures were performed in LB medium for A, C and D and in M63 medium for B and E since protamine precipitates in LB medium. Activities are the mean value from at least four separate experiments and are expressed in µmoles of p-nitrophenol produced per minute and per milligram of bacterial dry weight ± standard deviation.
Some of the Genes Induced in Aphid are Induced by AMP
To know whether induction or repression of genes observed when D. dadantii is infecting A. pisum could be due to the presence of AMPs, we analyzed the effect of protamine and polymyxin on the expression of four upregulated genes (dltB, sotA, sotB, and sttE) and of three downregulated ones (outC, kdgM and kdgN). None of these genes was differencially expressed in the presence of 100 µg ml−1 protamine (data not shown). In contrast, while the expression of sotA, sotB, kdgM and kdgN was not significantly affected by polymyxin, that of outC was reduced by 40% and that of sttE was increased by 3-fold, respectively, in the presence of that AMP (Fig. 2A and 2B, Table S3). Thus, not all genes induced in aphids are induced by polymyxin and a putative AMP produced by aphids could be a signal that modifies the expression of some of them.
Figure 2. Regulation of sstE and outC.
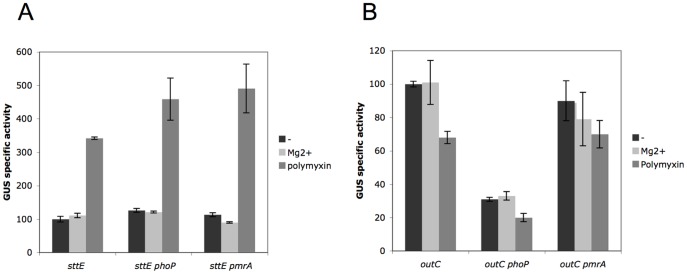
The sttE-uidA and outC-uidA fusions of strain A4206 and A1919, respectively, were assayed in the presence of 5 µg/ml of polymyxin or 10 mg/ml Mg2+ in the wt, phoP and pmrA backgrounds. Activities are the mean value from at least four separate experiments and are expressed in µmoles of p-nitrophenol produced per minute and per milligram of bacterial dry weight ± standard deviation.
The regulation of sttE and outC was studied in more details. Expression of none of these genes was controlled by Mg2+ concentration (Fig. 2A and 2B). Expression of sttE was not modified in a phoP or a pmrA background and in both cases polymyxin induction was conserved (Fig. 2A). The level of outC was identical in the pmrA background to that in the wt strain but was reduced three-fold in the phoP background, indicating that PhoP is an activator of outC. In both phoP and pmrA mutants, repression of outC by polymyxin was conserved (Fig. 2B). This confirms that PhoP and PmrA do not respond to polymyxin and that a gene regulated by PhoP is still sensitive to polymyxin in the absence of PhoP. In summary, regulation of genes involved in response to AMP is very different in D. dadantii from that described in S. enterica since neither polymyxin nor Mg2+ sensing occur through PhoP and PmrA and at least two regulators respond to different AMPs.
arnB and dltB are Involved in Resistance to AMP
To investigate whether arnB, dltB, phoP and pmrA are involved in resistance to AMP, a survival test to exposure to 1 µg ml−1 polymyxin was performed. The pmrA mutant was almost as resistant as the wt strain (Fig. 3). Thus PmrA-PmrB seems to play a limited role in resistance to AMP in D. dadantii. The phoP mutant showed more than 99% mortality after one hour exposure to polymyxin, showing that although PhoQ does not respond to that AMP, the PhoP-PhoQ system is involved in the regulation of genes required to resist to it. The dltB mutation had a strong effect on survival of the bacteria, leading to a 99% mortality. The most dramatic effect was observed with the arnB mutant with less than 0.1% of surviving bacteria after one hour (Fig. 3). This shows that LPS modification by the products of the arnBCADTEF operon is a main element of the resistance of D. dadantii to polymyxin and that absence of this modification leads to an increased bacterial susceptibility to this type of compounds. However, the dltB mutant survival was decreased, showing that, although its effect is limited when modification of LPS by 4-aminoarabinose can occur, alanylation of an unknown substrate by the dlt gene products is an important factor of resistance to AMP. No significant variation in the survival rate in the presence of polymyxin was observed when bacteria were grown in a LB medium containing 10 mM Mg2+ (data not shown).
Figure 3. Survival of various mutants to polymyxin.
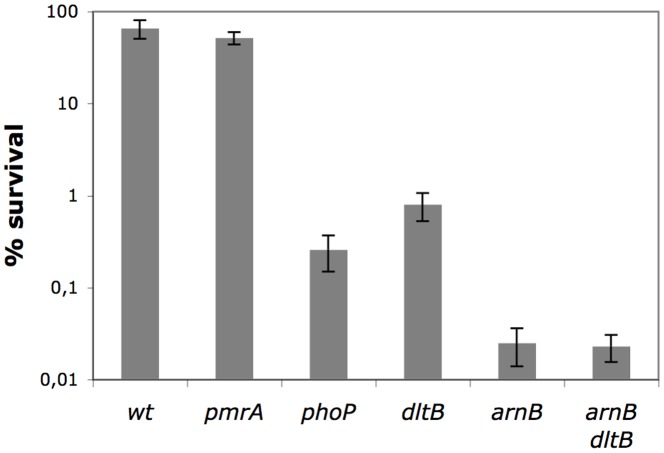
Wild type and various mutants in genes involved in resistance to AMP (phoP, pmrA, dltB, arnB) were incubated in the presence of 1 µg ml−1 polymyxin for 1 h. Samples were diluted and plated on LB agar plates to assess bacterial viability. Survival values are relative to the original inoculum. Data correspond to mean values of three independent experiments.
The arnB, the dltB and the arnB dltB mutants were then tested for their virulence in aphids. A comparison of the LT50s showed a slower mortality of aphids was observed with the arnB mutants but not with the dltB mutants (P = 0.042 for the wt versus arnB and P = 0.0671 for the wt versus arnB dltB double mutant comparisons, Log-rank non-parametric survival test), indicating a decreased ability of the arnB mutants to develop into the insect (Fig. 4). Thus, modification of LPS by the arn gene products is required for full virulence of the bacteria probably because it is involved in the resistance to AMPs produced by aphids.
Figure 4. Survival of pea aphids after oral infection by wt and mutants of D. dadantii 3937.
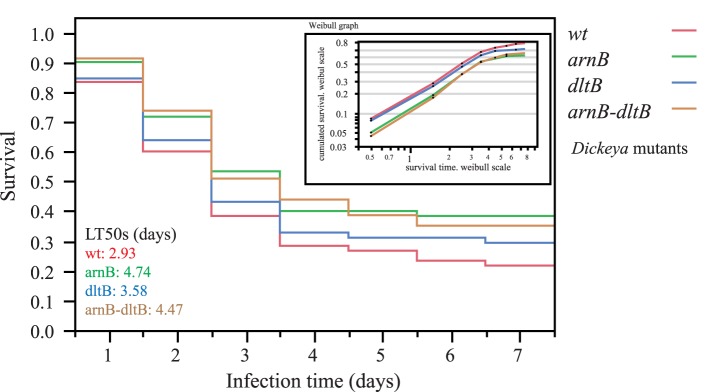
Survival is shown for aphids treated with wt bacteria (red), arnB (green), dltB (blue), and dltB arnB (brown) mutants. Results were obtained with 2×30 third instar aphid nymphs per treatment, including a diet-treated control (no mortality, not shown). The experiment was repeated twice with very similar results (p<0.06 in all comparisons of wt with arnB mutants). Median survival times (LT50s) were calculated with a Weibull fit (inlet), and give the following series [95% confidence intervals]: wt, 2.92 [2.18–3.92]; arnB, 4.74 [3.42–6.55]; dltB 3.58 [2.62–4.88] and arnB-dltB double mutant, 4.46 [3.23–6.18].
Inducible Peptides in Infected Pea Aphid
To try to identify the aphid AMPs, extracts from whole aphids, digestive tracts and hemolymph from control and Dickeya-challenged insects were separated by chromatography. All extracts gave very similar chromatographic traces at 280 nm (not shown) or 214 nm (Table S4 and Fig. S2). MALDI-ToF analysis of gut and haemolymph extract fractions did not show peptide peaks above 4500 Da, and few of them did differentiate the Dickeya-challenged modality (Table S4). Therefore, we focused our LC-MS analysis on the only samples that did show reproducible induced peaks in the bacteria-challenged samples (Figure S2), the digestive tract HPLC fractions (Table S4). Only one small peptide differentially induced in the Dickeya-challenged gut was found, namely the 13 amino acid C-terminal peptide from the ACYPI003154 gene product, a 14-3-3 family protein with conserved protein-protein interacting domains. This peptide, QDNDEPQEATGDN, has a net negative charge of -5 and could be an anionic AMP. This peptide was synthesized and its activity was tested against various bacteria. It had no antimicrobial activity against D. dadantii, E. coli and Bacillus subtilis. It was neither able to induce the expression of a D. dadantii arnB-uidA or dltB-uidA fusion (data not shown).
Discussion
D. dadantii has the abilities to be both a plant and an insect pathogen. While the mechanisms of virulence towards plants are now well understood, factors required for the development of bacteria in aphids and to kill them are unknown. Cyt toxins seem to play only an accessory role in this process since a mutant devoid of toxin genes can still kill aphids. An analysis of the transcriptome of D. dadantii infecting aphids was performed to better understand the mechanisms of pathogenicity of bacteria towards insects. This technique was preferred to IVET or TraSH techniques used in other studies since it allows a global analysis of the genes induced and repressed which is not restricted to virulence factors required for survival in insects and allows to detect redundant factors that could be missed by other methods [21], [22]. The results showed a very important modification of the pattern of gene expression. However, no characterized toxin gene was found among the induced genes. New putative toxins are perhaps to be identified among the induced proteins of unknown function. Other factors required for pathogenesis may not have been detected because they are expressed early in the infection. Another possibility is that no additional toxin is required for D. dadantii to kill aphids or that toxins are expressed only during the early stage of infection. The high number of bacteria found in all the insect organs could be sufficient to provoke the death by bacteremia, as observed for Pseudomonas aeruginosa PA14 infecting D. melanogaster [34]. Genes induced in aphid body would just favor multiplication of the bacteria.
Insects are used as alternative models to identify virulence factors required for infection of more complex organisms, plant or animals, since a factor is often active in different models [22], [35], [36]. D. dadantii pathogenicity towards insect and plant seems to require a totally different set of genes since all the known plant virulence factors are strongly repressed or at least not affected in A. pisum. This opposition had already been observed for cyt gene regulation [19]. This reinforces the hypothesis that this bacterium may be faced with these two types of ecosystems and it has adaptive mechanisms to cope with a rapidly changing environment (e.g. ingestion by an insect). Such a dramatic change in gene expression profile has been observed for Candidatus Phytoplasma asteris OY-M grown in planta or in its insect host [37].
The transcriptome also reflects the response of the bacteria to the insect innate immune system. It consists of encapsulation, phagocytosis, melanisation and antimicrobial peptides and lysozyme synthesis [38]. For the two first factors, no known specific bacterial response has been identified. Melanisation catalyzed by the polyphenoloxidases produces reactive oxygene species which damage DNA. Bacteria respond to this stress by destroying reactive oxygene and reparing DNA. In F. novicida infecting D. melanogaster a large set of DNA repair and detoxification genes are induced [22]. In our experiment a very limited number of genes potentially involved in these processes are overexpressed (mutM, iscR, rsxA, rsxB), which could indicate that oxidative stress felt by bacteria is low. The low level of polyphenoloxidase activity measured in aphids with systemic infection by bacteria confirms that this pathway is of limited importance to fight bacterial infections [38].
Among the most induced genes, a number are involved in efflux and in a typical response to AMP. This let suppose that synthesis of this type of molecules could be the main response of A. pisum to bacterial infection. Whether A. pisum does produce AMP was up to date unknown, in spite of explicit searches. Genes coding for classical insect AMP were not found in the A. pisum genome and biochemical tests to identify them were unsuccessful [31], [38], [39]. However, induction of genes involved in AMP resistance typical of Gram negative (arnB) and Gram positive (dltB) bacteria strongly suggests that aphids do produce AMP. The reduced virulence to aphids of the arnB mutant probably results from its increased sensitivity to AMPs. Efflux transporters expel from the bacteria all kind of toxic molecules. Interestingly, the mutant in one of the efflux system induced in this study, GenID 15786-7-8, is more sensitive to protamine. Given the broad spectrum of this type of exporters, involvement of some of them in AMP efflux is possible.
AMPs are very diverse in sequence and structure but most of them are positively charged, allowing their interaction with the bacterial envelope. Modifications of LPS to mask the negative charges that allow interaction with AMP are one of the main responses to these compounds in many Gram negative bacteria. This response is generally induced by growth in Mg2+-limiting conditions or in the presence of AMP. Sensing these conditions occurs most often through PhoP-PhoQ and PmrA-PmrB. The wiring between the signals (Mg2+ and AMP), the regulators (PhoP and PmrA) and their targets may vary from species to species [32]; [10]. Our data show that the sensing capacities of PhoP-PhoQ and the PhoP-regulated genes in D. dadantii are different from what has been previously described in other bacteria. Growth in low Mg2+-medium is an inducing condition for the LPS-modifying gene arnB. However induction is low and is independent of the regulation of arnB by PhoP (Fig. 1C). The induction observed suggests that another Mg2+ sensing regulator controls arnB expression (Fig. 1C). Presence of an acidic patch in the sensor PhoQ is required for sensing Mg2+ in S. enterica and E. tarda [11], [40]. This acidic patch is not conserved in D. dadantii (SSEDKPT versus EDDDDAE in S. enterica PhoQ) which explains the blindness of PhoQ to Mg2+. A similar blindness of PhoQ due to an absence of acidic patch has been shown in S. glossinidius, an endosymbiont of the tse-tse fly [13]. This has been correlated to the adaptation of the bacteria to a steady environment where Mg2+ sensing is no longer required. A low Mg2+ concentration is supposed to mimic the conditions encountered in phagosomes by intracellular pathogens. As a plant pathogen, it is always found outside cells where divalent cation concentration (Mg2+, Ca2+) is high. Thus, a link between low Mg2+ and response to AMP may not be required for this bacteria.
A recent transcriptome analysis of D. dadantii treated with thionin, a plant AMP, identified 36 induced genes, most of them being regulated by phoP [15]. Very few of them are common with those upregulated in our analysis: the arnB operon genes, the vfmE regulator and an ABC tranporter (GenID 18174 and 18175). Thus, D. dadantii PhoP-PhoQ is able to sense certain AMPs since induction of arnB expression by protamine and thionine is abolished in a phoP mutant (Fig. 1E and [15]). However, all AMPs are not sensed by PhoP-PhoQ since induction by polymyxin of arnB and outC (Fig. 2 A and 2B) is conserved in a phoP background. This suggests existence of another AMP sensing system. In P. aeruginosa, arnB, which is induced by polymyxin, is not regulated by PhoP-PhoQ but by an other two-component system ParR-ParS [41], that has no homologue in D. dadantii. Polymyxin is inducing or repressing several genes that are induced or repressed in our transcriptomic analysis (dltB, sttE, outC). They could all be regulated by a polymyxin-sensing regulator that could also sense a yet unknown AMP synthesized by aphids.
A. pisum AMPs could not be identified by homology-based methods, indicating that they are probably limited in number and under rapid diversifying evolution. In our attempts to identify such an AMP by a biological method, we unambiguously characterized only one small peptide that was differentially present in bacteria-infected tissue extracts. This peptide is the C-terminal end of a 14-3-3 protein, a member of a well-known family of protein-protein interactants, with recent reported involvement in insect immunity, including control of AMP secretion [42], [43]. Although this peptide has the characteristics of an anionic AMP, it has no activity on D. dadantii or other tested bacteria. The mode of action of this type of AMPs is less known [44] but it could be active on Buchnera aphidicola, the aphid endosymbiont, which has an atypical membrane that does not contain LPS [45]. Alternatively, this peptide might be part of a signal transduction pathway of infected aphid gut, and thus falls outside the scope of the present study.
Analysis of induced genes allowed to detect other signals sensed by the bacteria when they infect A. pisum. In contrast to E. coli or S. enterica, PmrA does not control the D. dadantii arnB operon. pmrA inactivation modifies only weakly the resistance of the bacteria to polymyxin. However, it activates some genes which are induced in aphids but not by polymyxin and it allows their induction by Fe3+ (data not shown). Thus, in D. dadantii, pmrA does not seem involved in the response to AMP and it is not known if Fe3+ is the only signal it can sense. Induction of the synthesis of the nitrate and nitrite dehydrogenases (NarI and NirB) probably results from anaerobic conditions in some aphid compartments. Our work allowed for the first time to decipher the global gene expression of a bacterium during an insect infection and showed that induction of genes in insects could be the result of many different signals, including several AMPs, the PmrA signal, anaerobiosis, sensed by a complex set of regulators.
Methods
Bacterial Strains and Growth Conditions
Bacterial strains, phages, plasmids and oligonucleotides used in this study are described in Table S5. D. dadantii and E. coli cells were grown at 28 and 37°C respectively unless otherwise stated in LB medium or M63 minimal medium supplemented with a carbon source (0.2%, w/v). When required antibiotics were added at the following concentration: ampicillin, 100 mg l−1, kanamycin and chloramphenicol, 25 mg l−1, tetracycline, 20 mg l−1. For aphid ingestion tests, bacteria were resuspended in AP3 medium [46]. Media were solidified with 1.5% (w/v) Difco agar. All mutations were in the wt background for insect tests and in the A350 background for enzymatic assays. Transduction with phage ΦEC2 was performed according to Résibois et al. [47].
Strain Construction
To construct strain A5256 that contains an arnB-uidA fusion, a 1.8 kb DNA fragment containing arnB was amplified with primers arnB+ (ggatggatgaatgtttgcggctg) and arnB- (cgcggccgaattgtcgctgctg). The resulting DNA fragment was inserted into the pGEM-T plasmid (Promega). A uidA-kanR cassette was prepared from plasmid pUIDK1 [48] by digestion with EcoRI and inserted into the unique MunI site of arnB. To construct strain A5248 that contains a pmrA-CmR insertion, a 1.9 kb DNA fragment containing pmrA was amplified with primers pmrA+ (ggaatatcaggcgccgcttg) and pmrA- (atatggtgtatgcccggcgg). The resulting DNA fragment was inserted into the pGEM-T plasmid (Promega). A Ω-CmR cassette was prepared from plasmid pHP45-ΩCm [49] by digestion with BamHI and inserted between the two BamHI sites of pmrA. To construct strain A5394 and A5399 that contain a dltB-uidA fusion and a dltB-CmR insertion respectively, a 1.7 kb DNA fragment containing dltB was amplified with primers dltB+ (aaagtgctgcgacattctgg) and dltB- (cgcgggcttgagtaatgccg). The resulting DNA fragment was inserted into the pGEM-T plasmid (Promega). A uidA-kanR cassette was prepared from plasmid pUIDK3 by digestion with XmaI and inserted into the unique BspEI site of dltB. A CmR cassette was prepared from plasmid CKC15 [4] by digestion with XmaI and inserted into the unique BspEI site of dltB. All the resulting constructs were inserted into the D. dadantii chromosome by recombination in low phosphate medium [50]. Recombinations were checked by PCR.
Aphid Strains and Infection Experiments
The aphid clone used was LL01, an alfalfa collected clone long-established in the lab and grown on broad beans (Vicia faba cv. Aquadulce). Inoculations by ingestion were performed as described in Grenier et al. [17]. Forty third instar aphid nymphs, fed on broad beans, were maintained for 24 h on an AP3 diet, containing bacteria at 106 bacteria ml−1 before being placed back onto bean leaves at 20°C ( = day 1). To evaluate the survival rate of infected pea aphids the number of survivors was counted every day for 7 days. For each strain, two independent biological replicates were tested.
RNA Preparation
To extract RNA from D. dadantii 3937 grown AP3 medium, bacteria grown for 24 h (OD600>2) were harvested by centrifugation and total RNA were extracted using TRIzol® reagent (Invitrogen) according to the manufacturer recommendations. To extract D. dadantii 3937 RNA from infected aphids, bacterial inoculation by ingestion was performed as described in [17]. For each assay, 40 third instar aphid nymphs, fed on broad beans, were maintained for 24 h on an AP3 diet containing 106 bacteria ml−1, before being placed back onto the beans at 20°C. After 3 days of infection aphids were crushed in two microcentrifuge tubes (2×20 aphids), with a sterilized pestle, in 250 µl of RNAprotect Bacteria Reagent (Qiagen). 250 µl of cold DEPC-treated water were then added. The suspension (≈ 1 ml) was centrifuged at 10000 g for 10 min at 4°C on a discontinuous 10% to 50% (w/w) sucrose gradient in Tris-HCl 10 mM pH 8.0, 1 mM EDTA in order to eliminate mitochondria, host nuclei and cellular fragments. Bacterial cells, localized in a green band, were collected with a glass pipette (V ≈ 1.5 ml) and pooled. Total RNA were then extracted using TRIzol® reagent (Invitrogen) following the manufacturer recommendations.
Bacterial RNA from all samples was treated with DNase I of the TURBO DNAfree™ kit (Ambion) following the manufacturer instructions. Absence of genomic DNA contamination was checked by PCR with two primer pairs cytA-up - cytB-down and cytC-up - cytD-down [19]. Purified RNA was quantified on the basis of its absorption at 260 nm using an ND 100 Nanodrop spectrophotometer and visualized on an agarose gel to check quality. Enrichment for mRNA was required to increase the signal intensity on the microarray and reach the NimbleGen® sample requirements. A step of RNA amplification was performed with the MicrobEnrich™ kit (Ambion) following the manufacturer recommendations. In order to ensure that this additional step had no influence on the relative gene expression pattern, a part of purified RNA from all samples was stored at −80°C for later confirmation by RT-qPCR. Amplified RNA was finally quantified using Nanodrop spectrophotometer and visualized on an agarose gel to check quality before being sent to NimbleGen for hybridization on D. dadantii microarrays.
Microarray Design
The microarrays used in this study were custom designed and produced by NimbleGen Systems, Inc. (Madison, WI), based on the annotated sequence (version number 6) of D. dadantii (available at https://asap.ahabs.wisc.edu/asap/logon.php), which comprised 4753 coding sequences (CDS). The microarrays consisted of 70-mer oligonucleotides with 5 perfect match probes per CDS, in three blocks on the array. For microarray analyses, cDNA was synthesized, labeled, and hybridized by NimbleGen Systems, Inc. For each condition, three independent biological replicates were tested. The experimental design of array, and all raw and processed data, were deposited as a GEO archive (accession number GSE42585).
Data Analysis and Statistical Procedures
All statistical tests, including aphid survival analyses (non parametric and parametric models, of which a Weibull adjustment was found to best describe the insect survival trend), were performed with JMP software (V 9.0.3, SAS Inc Cary 27513-2414 USA). Aphid EST analysis was performed with the MacVector software, including the assembler module (V 12.6, MacVector Cary NC 27519 USA). The two aphid gut libraries ID0AFF (control) and ID0AAG (Dickeya-treated) [51] were assembled altogether with MacVector, blasted against V2.1 A. pisum genome assembly (mRNAs), and all hits were discarded, in order to retrieve all small and non-conventional gene models not present in the V2.1 genome assembly. The 527 contigs retrieved were then filtered-off for contaminating Buchnera proteins, translated and the resulting partial ORFs used for additional target analysis in the LC MS-MS analyses. Micro-arrays were analyzed using the expression module from the CLC-Bio Main Workbench V6 package. Quality control was checked by data analysis and principal component and group cluster analysis, which easily grouped biological conditions, biological replicates and technical replicates in this order. Normalized data (scaling, median) were analyzed by K-means hierarchical clustering of genes (5 clusters) and by gene set enrichment analysis, GSEA [52] on GO classifications.
Quantitative Real-time PCR Control
After microarray data results, qRT-PCR were performed on several genes to confirm the relative gene expression pattern. Same RNA samples used for RNA amplification and next array hybridization were used for these experiments. RT was performed using SuperScript II reverse transcriptase (Invitrogen) with 500 ng of total RNA and 25 ng of random hexamer primers, according to the manufacturer’s protocol (first strand cDNA synthesis). One microliter of the RT reaction mixture was added as a template to the Qbiogen Sybr Green mix for PCR with gene-specific primers. Primers used in this work are listed in Table S6. The thermal cycling reactions were performed using a LightCycler (Roche) according to the following conditions: an initial step at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, 55°C for 15 s, and 72°C for 20 s. Two housekeeping genes, rpoA and ffh, were used as standards to obtain normalized target gene expression ratios as described in a previous study of D. dadantii transcriptome [53]. The qPCR efficiency was 1.80 and 1.96 for rpoA and ffh, respectively. The statistical program used to analyze the data was the Relative Expression Software Tool (REST 2009 V2.0.13) [54]. The specificity of the PCR primers was verified with a melting curve analysis using the LightCycler® 480 Software (Roche).
Peptide Extraction and Chromatographic Analysis
Aphids were extracted as young day 2–5 parthenogenetic females, using either a flash-hemolymph extraction technique [31], or a gut dissection following the same TFA 0.1% extraction procedure [51]. Phenyl-thiourea was used for SDS-PAGE hemolymph purity controls, but omitted when HPLC was used subsequently. Aphid acid extracts (whole-body WB, haemolymph HY or digestive tract DT) were then separated using a Dionex UPLC System Ultimate 3000 and a Waters XBridge BEH130 C18 column (2.1×150 mm, 5 µm particle size, 130-Å porosity) at 40°C, with a precolumn. Absorbance was monitored at 214 and 280 nm. The solvent system was 0.1% TFA in water (Solvent A) and 0.09% TFA in 70% acetonitrile/water (Solvent B) with a flow rate of 0.4 ml min−1 and the gradient is shown on figures. Fractions were collected every 30 sec in ELISA plates and dried using a Speed-Vac concentrator.
Peptide Identification by Mass Spectrometry
Plate-collected UPLC fractions were dissolved in 10 µl H2O/acetonitrile 99/1 acidified by 0.1% of TFA. The analysis was performed on a nanoACQUITY Ultra-Performance-LC (UPLC, Waters, Milford, MA). 4µL of each sample were loaded on a 20×0.18 mm, 5 µm Symmetry C18 precolumn (Waters Corp.), and the peptides were separated on a ACQUITY UPLC® BEH130 C18 column (Waters Corp.), 75 µm×200 mm, 1.7 µm particle size. The solvent system consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). Trapping was performed during 3 min at 5 µL min−1 with 99% of solvent A and 1% of solvent B. Elution was performed at a flow rate of 300 nL min−1, using 1–40% gradient (solvent B) over 35 min at 50°C followed by 65% (solvent B) over 5 min. The MS and MS/MS analyzes were performed on the SYNAPT™, an hybrid quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer (Waters, Milford, MA) equipped with a Z-spray ion source and a lock mass system. The capillary voltage was set at 3.5 kV and the cone voltage at 35 V. Mass calibration of the TOF was achieved using phosphoric acid (H3PO4) on the [50;1800] m/z range in positive mode. Online correction of this calibration was achieved using lock-mass on product ions derived from the [Glu1]-fibrinopeptide B (GFP). The ion (M+2H)2+ at m/z 785.8426 is used to calibrate MS data and the fragment ion (M+H)+ at m/z 684.3469 is used to calibrate MS/MS data during the analysis. For tandem MS experiments, the system was operated with automatic switching between MS and MS/MS modes (MS 0.5 s/scan on m/z range [250;1500] and MS/MS 0.7 s/scan on m/z range [50;1800]). The 3 most abundant peptides (intensity threshold 60 count s−1), preferably doubly and triply charged ions, were selected on each MS spectrum for further isolation and CID fragmentation. Fragmentation was performed using argon as the collision gas. The complete system was fully controlled by MassLynx 4.1 (SCN 712, Waters, Milford, MA). Raw data collected during nanoLC-MS/MS analyses were processed and converted with ProteinLynx Browser 2.4 (Waters, Milford, MA) into.pkl peak list format. Normal background substraction type was used for both MS and MS/MS with 5% threshold and polynomial correction of order 5, and deisotoping was performed. The MS/MS data were analyzed using the MASCOT 2.4 algorithm (Matrix Science) to search against the aphid proteome database (v 2.1), concatenated with additional translation of normal/infected digestive tract EST contigs [51], locally assembled with Vector 12.6 (Mac Vector Inc. Cary NC 27519 USA).
Enzymatic Assays
β-glucuronidase assays were performed on toluenized extracts of cells grown to exponential phase by the method of Bardonnet et al. [48] using p-nitrophenyl-β-D-glucuronate as substrate.
Antimicrobial Peptide Resistance Assays
Stationary phase grown cultures of D. dadantii strains were diluted 103-fold in LB medium. 1 ml of dilution was placed in an 1.5 ml Eppendorf tube and AMP was added. After 1 hour of incubation at room temperature, bacteria were diluted and plated on LB plates. Colonies were counted after 2 days of growth at 28°C.
Supporting Information
Validation of microarray results by qRT-PCR. (A) Expression ratios of D. dadantii genes during aphid infection versus growth in liquid medium measured by qRT-PCR. Expression of each gene was normalized to the expression of the two housekeeping genes rpoA and ffh. A positive expression ratio indicates upregulated genes during aphid infection, and a negative expression ratio indicates downregulated genes during aphid infection. Standard error ranges were calculated from the data from three independent biological replicates. (B) Comparison of gene expression measurements by microarray approach and real-time qRT PCR. The correlation coefficient (R2) is given.
(DOC)
HPLC traces at 214 nm of haemolymph and digestive tract acidic extracts from the pea aphid challenged or not by Dickeya dadantii . The horizontal bar shows the collected (differential) fractions.
(EPS)
GSEA analysis of Gene Ontology categories in insect-infecting bacteria vs control Dickeya dadantii cells.
(DOCX)
D. dadantii genes induced or repressed in aphid with a P value <0.002.
(XLSX)
Expression of selected genes in the presence of polymyxin.
(DOC)
Summary of peptide LC MS analysis.
(DOCX)
Bacterial strains used in this study.
(DOC)
Oligonucleotides used for RT-qPCR experiments.
(DOC)
Acknowledgments
We thank Marie-Hélène Metz-Boutigue, Geraldine Effantin, Gabrielle Duport and Alain Clavel for technical help, Sylvie Reverchon and Nicole Cotte-Pattat for reading the manuscript.
Funding Statement
This study was funded by an ANR-Genoplante grant to GC, including a post-doctoral fellowship to DC, coordinated by YR (ANR07GPLA002-Aphicibles. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S (1996) Regulation of pectinolysis in Erwinia chrysanthemi . Annu Rev Microbiol 50: 213–257. [DOI] [PubMed] [Google Scholar]
- 2. Charkowski A, Blanco C, Condemine G, Expert D, Franza T, et al. (2012) The role of secretion systems and small molecules in soft-rot enterobacteriaceae pathogenicity. Annu Rev Phytopathol 50: 425–449. [DOI] [PubMed] [Google Scholar]
- 3. Sepulchre JA, Reverchon S, Nasser W (2007) Modeling the onset of virulence in a pectinolytic bacterium. J Theor Biol 244: 239–257. [DOI] [PubMed] [Google Scholar]
- 4. Lebeau A, Reverchon S, Gaubert S, Kraepiel Y, Simond-Cote E, et al. (2008) The GacA global regulator is required for the appropriate expression of Erwinia chrysanthemi 3937 pathogenicity genes during plant infection. Environ Microbiol 10: 545–559. [DOI] [PubMed] [Google Scholar]
- 5. Reverchon S, Expert D, Robert-Baudouy J, Nasser W (1997) The cyclic AMP receptor protein is the main activator of pectinolysis genes in Erwinia chrysanthemi . J Bacteriol 179: 3500–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nasser W, Faelen M, Hugouvieux-Cotte-Pattat N, Reverchon S (2001) Role of the nucleoid-associated protein H-NS in the synthesis of virulence factors in the phytopathogenic bacterium Erwinia chrysanthemi . Mol Plant Microbe Interact 14: 10–20. [DOI] [PubMed] [Google Scholar]
- 7. Llama-Palacios A, Lopez-Solanilla E, Rodriguez-Palenzuela P (2005) Role of the PhoP-PhoQ system in the virulence of Erwinia chrysanthemi strain 3937: involvement in sensitivity to plant antimicrobial peptides, survival at acid pH, and regulation of pectolytic enzymes. J Bacteriol 187: 2157–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zwir I, Shin D, Kato A, Nishino K, Latifi T, et al. (2005) Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica . Proc Natl Acad Sci U S A 102: 2862–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gunn JS (2008) The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16: 284–290. [DOI] [PubMed] [Google Scholar]
- 10. Perez JC, Shin D, Zwir I, Latifi T, Hadley TJ, et al. (2009) Evolution of a bacterial regulon controlling virulence and Mg(2+) homeostasis. PLoS Genet 5: e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chakraborty S, Li M, Chatterjee C, Sivaraman J, Leung KY, et al. (2010) Temperature and Mg2+ sensing by a novel PhoP-PhoQ two-component system for regulation of virulence in Edwardsiella tarda . J Biol Chem 285: 38876–38888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prost LR, Daley ME, Bader MW, Klevit RE, Miller SI (2008) The PhoQ histidine kinases of Salmonella and Pseudomonas spp. are structurally and functionally different: evidence that pH and antimicrobial peptide sensing contribute to mammalian pathogenesis. Mol Microbiol 69: 503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pontes M, Smith K, de Vooght L, van den Abbeele J, Dale C (2011) Attenuation of the sensing capabilities of PhoQ in transition to obligate insect-bacterial association. PLoS Genet 7: e1002349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Venkatesh B, Babujee L, Liu H, Hedley P, Fujikawa T, et al. (2006) The Erwinia chrysanthemi 3937 PhoQ sensor kinase regulates several virulence determinants. J Bacteriol 188: 3088–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rio-Alvarez I, Rodriguez-Herva JJ, Cuartas-Lanza R, Toth I, Pritchard L, et al. (2012) Genome-wide analysis of the response of Dickeya dadantii 3937 to plant antimicrobial peptides. Mol Plant Microbe Interact 25: 523–533. [DOI] [PubMed] [Google Scholar]
- 16. Glasner JD, Yang CH, Reverchon S, Hugouvieux-Cotte-Pattat N, Condemine G, et al. (2011) Genome sequence of the plant pathogenic bacterium Dickeya dadantii 3937. J Bacteriol 193: 2076–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grenier AM, Duport G, Pages S, Condemine G, Rahbe Y (2006) The phytopathogen Dickeya dadantii (Erwinia chrysanthemi 3937) is a pathogen of the pea aphid. Appl Environ Microbiol 72: 1956–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Costechareyre D, Balmand S, Condemine G, Rahbe Y (2012) Dickeya dadantii, a plant pathogenic bacterium producing Cyt-Like entomotoxins causes septicemia in the pea aphid Acyrthosiphon pisum . PLoS One 7: e30702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Costechareyre D, Dridi B, Rahbe Y, Condemine G (2010) Cyt toxin expression reveals an inverse regulation of insect and plant virulence factors of Dickeya dadantii . Environ Microbiol 12: 3290–3301. [DOI] [PubMed] [Google Scholar]
- 20. Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ (2010) Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis . PLoS Pathog 6: e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. An R, Sreevatsan S, Grewal PS (2009) Comparative in vivo gene expression of the closely related bacteria Photorhabdus temperata and Xenorhabdus koppenhoeferi upon infection of the same insect host, Rhizotrogus majalis . BMC Genomics 10: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moule MG, Monack DM, Schneider DS (2010) Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog 6: e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blot N, Berrier C, Hugouvieux-Cotte-Pattat N, Ghazi A, Condemine G (2002) The oligogalacturonate-specific porin KdgM of Erwinia chrysanthemi belongs to a new porin family. J Biol Chem 277: 7936–7944. [DOI] [PubMed] [Google Scholar]
- 24. Condemine G, Ghazi A (2007) Differential regulation of two oligogalacturonate outer membrane channels, KdgN and KdgM, of Dickeya dadantii (Erwinia chrysanthemi). J Bacteriol 189: 5955–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maggiorani-Valecillos A, Rodriguez-Palenzuela P, Lopez-Solanilla E (2006) The role of several multidrug resistance systems in Erwinia chrysanthemi pathogenesis. Mol Plant Microbe Interact 19: 607–613. [DOI] [PubMed] [Google Scholar]
- 26. Condemine G (2000) Characterization of SotA and SotB, two Erwinia chrysanthemi proteins which modify isopropyl-beta-D-thiogalactopyranoside and lactose induction of the Escherichia coli lac promoter. J Bacteriol 182: 1340–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reverchon S, Rouanet C, Expert D, Nasser W (2002) Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J Bacteriol 184: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bennett H, Clarke D (2005) The pbgPE operon in Photorhabdus luminescens is required for pathogenicity and symbiosis. J Bacteriol 187: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weidenmaier C, Peschel A (2008) Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6: 276–287. [DOI] [PubMed] [Google Scholar]
- 30. Abi Khattar Z, Rejasse A, Destoumieux-Garzon D, Escoubas JM, Sanchis V, et al. (2009) The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J Bacteriol 191: 7063–7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, et al. (2010) Immunity and other defenses in pea aphids, Acyrthosiphon pisum . Genome Biol 11: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winfield MD, Groisman EA (2004) Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc Natl Acad Sci U S A 101: 17162–17167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia Vescovi E, Soncini FC, Groisman EA (1996) Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84: 165–174. [DOI] [PubMed] [Google Scholar]
- 34. Limmer S, Haller S, Drenkard E, Lee J, Yu S, et al. (2011) Pseudomonas aeruginosa RhlR is required to neutalize the cellular immune response in a Drosophila melanogaster oral infection model. Proc Natl Acad Sci U S A 108: 17378–17383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mahajan-Miklos S, Rahme LG, Ausubel FM (2000) Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol Microbiol 37: 981–988. [DOI] [PubMed] [Google Scholar]
- 36. Shirasu-Hira MM, Schneider DS (2007) Confronting physiology: how do infected flies die? Cell Microbiol 9: 2775–2783. [DOI] [PubMed] [Google Scholar]
- 37. Oshima K, Ishii Y, Kakizawa S, Sugawara K, Neriya Y, et al. (2011) Dramatic transcriptional changes in an intracellular parasite enable host switching between plant and insect. PLoS One 6: e23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laughton AM, Garcia JR, Altincicek B, Strand MR, Gerardo NM (2011) Characterisation of immune responses in the pea aphid, Acyrthosiphon pisum . J Insect Physiol 57: 830–839. [DOI] [PubMed] [Google Scholar]
- 39. Altincicek B, ter Braak B, Laughton AM, Udekwu KI, Gerardo NM (2011) Escherichia coli K-12 pathogenicity in the pea aphid, Acyrtosiphon pisum, reveals reduced antibacterial defense in aphids. Developmental and comparative immunology 35: 1089–1095. [DOI] [PubMed] [Google Scholar]
- 40. Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, et al. (2005) Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122: 461–472. [DOI] [PubMed] [Google Scholar]
- 41. Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, et al. (2010) Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54: 3372–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ulvila J, Vanha-aho LM, Kleino A, Vaha-Makila M, Vuoksio M, et al. (2011) Cofilin regulator 14-3-3zeta is an evolutionarily conserved protein required for phagocytosis and microbial resistance. J Leukoc Biol 89: 649–659. [DOI] [PubMed] [Google Scholar]
- 43. Shandala T, Woodcock JM, Ng Y, Biggs L, Skoulakis EM, et al. (2011) Drosophila 14-3-3epsilon has a crucial role in anti-microbial peptide secretion and innate immunity. J Cell Sci 124: 2165–2174. [DOI] [PubMed] [Google Scholar]
- 44. Harris F, Dennison SR, Phoenix DA (2009) Anionic antimicrobial peptides from eukaryotic organisms. Curr Protein Pept Sci 10: 585–606. [DOI] [PubMed] [Google Scholar]
- 45. Charles H, Balmand S, Lamela A, Cottret L, Pérez-Brocal V, et al. (2011) A genomic reappraisal of symbiotic function in the aphid/Buchnera symbiosis: reduced transporter sets and membrane organisations. PLoS One 6: e29296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rahbé Y, Febvay G (1993) Protein toxicity to aphids: an in vitro test on Acyrthosiphon pisum . Entomol Exp Appl 67: 149–160. [Google Scholar]
- 47. Résibois A, Colet M, Faelen M, Schoonejans E, Toussaint A (1984) phiEC2, a new generalized transducing phage of Erwinia chrysanthemi . Virology 137: 102–112. [DOI] [PubMed] [Google Scholar]
- 48. Bardonnet N, Blanco C (1991) Improved vectors for transcriptional signal screening in corynebacteria. FEMS Microbiol Lett 68: 97–102. [DOI] [PubMed] [Google Scholar]
- 49. Fellay R, Frey J, Krisch H (1987) Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52: 147–154. [DOI] [PubMed] [Google Scholar]
- 50. Roeder DL, Collmer A (1985) Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi . J Bacteriol 164: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sabater-Munoz B, Legeai F, Rispe C, Bonhomme J, Dearden P, et al. (2006) Large-scale gene discovery in the pea aphid Acyrthosiphon pisum (Hemiptera). Genome Biol 7: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hommais F, Oger-Desfeux C, Van Gijsegem F, Castang S, Ligori S, et al. (2008) PecS is a global regulator of the symptomatic phase in the phytopathogenic bacterium Erwinia chrysanthemi 3937. J Bacteriol 190: 7508–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of microarray results by qRT-PCR. (A) Expression ratios of D. dadantii genes during aphid infection versus growth in liquid medium measured by qRT-PCR. Expression of each gene was normalized to the expression of the two housekeeping genes rpoA and ffh. A positive expression ratio indicates upregulated genes during aphid infection, and a negative expression ratio indicates downregulated genes during aphid infection. Standard error ranges were calculated from the data from three independent biological replicates. (B) Comparison of gene expression measurements by microarray approach and real-time qRT PCR. The correlation coefficient (R2) is given.
(DOC)
HPLC traces at 214 nm of haemolymph and digestive tract acidic extracts from the pea aphid challenged or not by Dickeya dadantii . The horizontal bar shows the collected (differential) fractions.
(EPS)
GSEA analysis of Gene Ontology categories in insect-infecting bacteria vs control Dickeya dadantii cells.
(DOCX)
D. dadantii genes induced or repressed in aphid with a P value <0.002.
(XLSX)
Expression of selected genes in the presence of polymyxin.
(DOC)
Summary of peptide LC MS analysis.
(DOCX)
Bacterial strains used in this study.
(DOC)
Oligonucleotides used for RT-qPCR experiments.
(DOC)


