Abstract
Recent work in computational genomics has shown that a functional association between two genes can be derived from the existence of a fusion of the two as one continuous sequence in another genome. For each of 30 completely sequenced microbial genomes, we established all such fusion links among its genes and determined the distribution of links within and among 15 broad functional categories. We found that 72% of all fusion links related genes of the same functional category. A comparison of the distribution of links to simulations on the basis of a random model further confirmed the significance of intracategory fusion links. Where a gene of annotated function is linked to an unclassified gene, the fusion link suggests that the two genes belong to the same functional category. The predictions based on fusion links are shown here for Methanobacterium thermoautotrophicum, and another 661 predictions are available at http://fusion.bu.edu.
High-throughput sequencing projects are producing large numbers of genes of unknown function, creating the need for computational means of ascribing putative functions. Sequence similarity searches, traditionally the predominant computational tool used to investigate uncharacterized genes, are now complemented by such methods as phylogenetic profiling (1, 2), chromosomal proximity (3, 4), microarray expression profiles (5), fusion links (6, 7), protein–protein interactions (8), and combinations of these methods (9–11).
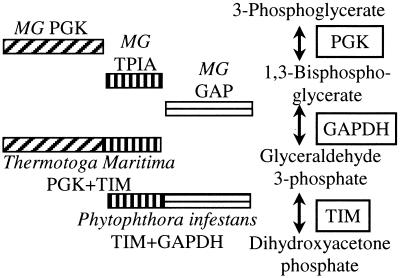
The fusion method was introduced by Marcotte et al. (6) and Enright et al. (7) to infer a functional relationship between two distinct genes in an organism by finding an instance in which the genes are fused as a continuous sequence in another organism. The composite gene suggests a link between the component genes, and because these genes are not necessarily similar to each other in sequence, this method can complement sequence analysis approaches to assigning functions. For example, Fig. 1 shows two fusion genes that link three nonhomologous Mycoplasma genitalium genes that are functionally related in that they encode sequential steps in glycolysis. This example is typical, because three of every four fusion links in Escherichia coli have been shown to relate two metabolic genes (12).
Figure 1.
Correspondence between functional associations and genes linked by the fusion method. Independent genes in one genome may be found as one continuous gene in other genomes. These fusion links can confirm known functional relationships between genes: M. genitalium genes phosphoglycerate kinase (PGK), triosephosphate isomerase (TPIA), and glyceraldehyde-3-phosphate dehydrogenase (GAP), all sequential agents in glycolysis, are linked by fusion events elsewhere. These links may be used to infer putative functions when one of the component genes is of an unknown function.
Marcotte et al. (6) discussed the applicability of the gene fusion method for the detection of protein–protein interactions in light of a model for the evolution of interacting surfaces. On the basis of the observation that the fusion of two proteins greatly increases their effective concentrations, they postulated that two noninteracting proteins could evolve a strong affinity after being fused, perhaps subsequently separating to become interacting proteins.
The availability of dozens of fully sequenced genomes allows a large-scale analysis of fusion links. Here, we identify the fusion links between genes in 30 complete microbial genomes and study the functional correlation among the genes on the basis of broad functional categories such as transcription and energy production. For each genome, we consider every possible pair of individual genes and search in a comprehensive sequence database (i.e., not only in the 30 microbial genomes) for a gene that is a fusion of both sequences. We then tabulate the distribution of fusion links within and among the broad functional categories, and to determine the significance of this distribution, compare it with a simulation based on a random model. We find that fusion links tend to relate genes of the same functional category with significant frequency. In cases in which an uncharacterized gene is fusion-linked to a gene of known function, the latter provides a strong hint regarding the function of the former (6, 7).
Methods
Clusters of Orthologous Groups (COGs) Database.
The COGs database (2, 13) is a phylogenetic classification of proteins or domains into sets of orthologs, i.e., each COG presumably contains the “same” gene or domain from multiple organisms. Of particular interest to this article is the classification of the COGs into 15 functional categories (see Fig. 3) in addition to general function prediction only (R) and function unknown (S).
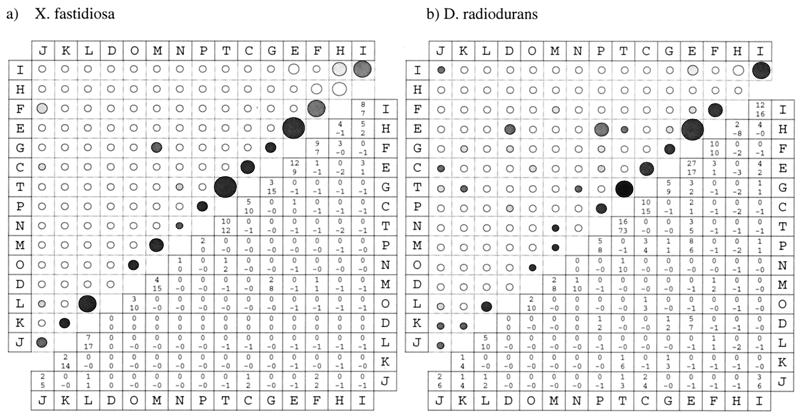
Figure 3.
Global views of fusion links in X. fastidiosa and D. radiodurans. The figure shows the number of pairwise links between functional categories resulting from fusion events in X. fastidiosa (a) and D. radiodurans (b) in both graphic and tabular form. Each element in the lower triangle indicates the number of fusion links followed by the significance expressed as the number of standard deviations above or below (−) the number expected; the latter is omitted if unavailable (see Methods). In the upper triangle, the size of the circle indicates the number of fusion links, and the shading indicates the significance of this number. The categories are: J, translation, ribosomal structure, and biogenesis; K, transcription; L, DNA replication, recombination, and repair; D, cell division and chromosome partitioning; O, posttranslational modification, protein turnover, and chaperones; M, cell envelope biogenesis and outer membrane; N, cell motility and secretion; P, inorganic ion transport and metabolism; T, signal transduction mechanisms; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme metabolism; and I, lipid metabolism. The figure was made by using spotfire (http://www.spotfire.com).
Genome Sequences and Functional Annotations.
Protein sequences from 30 microbial genomes and their annotations were retrieved from the COGs database. In this well curated database, orthologous proteins are clustered and assigned to a functional category (see Fig. 3). We did not consider the COG functional categories general function prediction (R) only and function unknown (S) in the analysis of correlated functional categories between the fusion-link partners, because they do not contain a categorized function. Different domains of the same protein may belong to different COGs; therefore the set of sequences from a COG typically represents a single domain. Consequently, we investigated fusion links between domains rather than whole genes and thus avoided potential errors of transitivity in the functional annotation of multidomain genes. We ignored the relatively few COGs that are annotated with more than one functional category.
Identification of Fusion Links.
Fusion links between genes were found by a BLASTP (14) search of the translated sequences against nrdb90 (15), a representative composite of major protein sequence databases in which no two sequences have sequence identity of 90% or greater. We used the translated sequences because they yield a significant improvement in sequence similarity searches. We deemed two proteins to be fusion-linked if each had an alignment of at least 80 residues (16) to the same nrdb90 protein with a maximum expectation value of 10−10 and with a maximum overlap of 20 residues between the two alignments. We detected fusion links between homologous domains, including self-fusions, by using the permissive E value threshold of 10−3 and excluded these from consideration. Because we searched for fusion links between domains rather than whole genes, we also ignored links between homologous multidomain genes, which were identified by using an E value of 10−3. For example, given two homologs X and Y, with domains AB and A′B′, respectively, A fuses with B′ and B with A′, but because X and Y are similar in sequence overall, they were not counted here as fusion links. We counted at most one link between any pair of COGs, so that paralogous genes did not artificially inflate the number of fusion links. In instances in which a gene was fusion-linked to multiple nonhomologous genes, all possible links were counted.
Model for Determining the Significance of the Number of Fusion Links Observed Among Functional Categories.
To estimate the significance of the number of fusion links observed among functional categories, we developed a random model that served as a negative control. We generated random fusion links that were subject to two constraints: the total number of links and the number of links for each gene were preserved. We repeated this entire process 1,000 times and then calculated the means and standard deviations of the numbers of fusion links among functional categories. The significance of the number of fusion links among functional categories was estimated as the number of standard deviations by which the observed number differed from the random mean (i.e., positive if observed exceeded random and negative otherwise). In some cases, the significance cannot be calculated, either because genes in a category are not involved in any fusion links or are so infrequent that no fusion links occur in the random simulations. All fusion links that included genes of poorly characterized function, namely those in categories general function prediction only (R) and function unknown (S), were excluded when the significance was estimated. The significance concerns the number of fusions links within and among functional categories, not individual fusion links.
Model for Determining the Significance of the Intracategory Links in a Given Genome.
To estimate the significance of the number of intracategory fusion links observed, we developed a random model that served as a negative control. As in the method discussed above, we generated random fusion links such that the number of fusion links for each gene was preserved and determined the fraction of intracategory links among all fusion links. We repeated this entire process 1,000 times and then calculated the means and standard deviations of the fraction of intracategory links. The significance of the fraction of intracategory fusion links was estimated as the number of standard deviations by which the observed fraction differed from the random mean (again, positive if observed exceeded random and negative otherwise).
Results
Fusion Links Among M. genitalium Genes.
The parasitic bacterium M. genitalium, the genome of which contains only 468 genes, makes for an appropriate starting point for the analysis of the correspondence of function between genes related by fusion links, because there are expectedly few fusion links among its genes. We found a strong correspondence of function between the partners in all 20 fusion links (16 of which are pairs in which both members are present in COGs), consistent with the 28 detected by Huynen et al. (10) with more permissive search criteria and without the collapsing of degenerate links (see Methods). There are five fusion links between pairs of genes, the products of which act in adjacent steps in metabolic pathways. Fig. 1 shows two fusion genes that link three of the M. genitalium genes, which are functionally related in that they encode sequential steps in glycolysis. Another seven of the links apparently relate genes encoding subunits of the same complex. Fusion links among three genes (MG016, MG017, and MG018) that are all annotated as putative RNA helicases suggest that these genes are also of the same cellular function. It is important to remember that the genes being linked are not similar in sequence.
For the five fusion links between pairs of M. genitalium genes in which the annotation for either gene or both genes is not available, the fusion method is able to suggest a putative function. A link between a gene of unknown function (MG328) and a gene associated with the structure of the cell membrane (MG386) suggests that the former is also related to the biogenesis of the cell membrane. Because the pathogenicity of M. genitalium is caused by host–parasite interactions, MG328 is potentially an important gene for further study.
Functional Cascade: Example of Fusion Links in Saccharomyces cerevisiae.
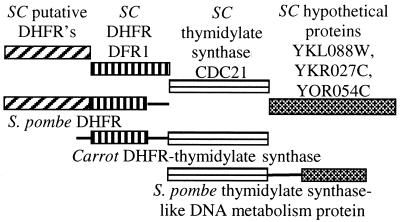
Fig. 2 shows an example of fusion links among S. cerevisiae genes that illustrates several features of fusion links including those among sequential steps in a biochemical pathway, a functional prediction, and the discovery of a misleading annotation.
Figure 2.
Functional cascade: a gene may inherit a misleading functional annotation because it shares a domain with a gene, the annotated function of which arises from a domain other than the shared one. In this case, the S. cerevisiae (SC) genes YHR049W, YMR222C, and YOR280C are annotated as putative dihydrofolate reductases (DHFRs), because they have sequence similarity to the N-terminal domain of a Schizosaccharomyces pombe DHFR, although the DHFR activity belongs to the C-terminal domain. DFR1 and CDC21 act in sequential steps in folate biosynthesis and are found fused in a carrot gene. The S. cerevisiae DHFRs with misleading putative annotations are likely to be involved in folate biosynthesis, but not as DHFRs.
Fusion of Sequential Steps in a Biochemical Pathway.
We find a number of fusion links among genes encoding sequential or nearly sequential steps in biochemical pathways. In this case, the S. cerevisiae dihydrofolate reductase (DFR1) and thymidylate synthase (CDC21) genes encode sequential steps in the biosynthesis of folate, and they are found fused in the bifunctional carrot protein dihydrofolate reductase–thymidylate synthase.
Functional Prediction.
CDC21 is fused in the S. pombe thymidylate synthase-like DNA metabolism protein with portions of hypothetical proteins YKL088W, YKR027C, and YOR054C, implicating the latter genes in folate or pyrimidine metabolism. Future biochemical experiments with these genes can be directed toward these putative functions. The involvement of YOR045C and YKL088W in transcription is supported by other methods (9).
Misleading Annotation.
Genes are sometimes annotated by transitivity, i.e., a gene of unknown function inherits the function of a gene with which it shares a domain, when the function ascribed does not pertain to the common domain. This can result in an incorrect functional annotation (17, 18) as in the assignment of DHFR activity to the three S. cerevisiae genes YHR049W, YMR222C, and YOR280C (Fig. 2). These genes match the first half of an S. pombe gene labeled as a DHFR, this activity pertains to the second half of the protein, and the two halves are not homologous. Through their association with DHFR, the fusion implicates the three proteins in folate biosynthesis but not as DHFRs.
Global Views of Fusion Links in Whole Genomes According to Functional Categories.
An analysis of fusion links in other genomes, similar to that in M. genitalium, is complicated by the fact that the number of links within a genome grows exponentially relative to the number of genes in that genome (Table 1). For example, there are 664 fusion links among the genes of Vibrio cholerae, which poses the daunting task of classifying them individually. To detect patterns of fusion links, we tabulated the number of fusion links between all possible pairs of functional categories as annotated by the COGs database. The function correlation tables (Fig. 3) show the distribution of fusion links within and among functional categories along with the statistical significance (henceforth referred to as “significance”; see Methods), i.e., the number of standard deviations above the expected mean, in the genomes of Xylella fastidiosa and Deinococcus radiodurans. These results exclude self-fusions (see Methods), avoiding a potential bias in favor of the significance of the elements in the diagonal, which represent intracategory fusion links.
Table 1.
Statistics of fusion links in 30 microbial genomes
| A
|
B
|
C
|
D
|
E
|
F
|
G ± H
|
I
|
J
|
|---|---|---|---|---|---|---|---|---|
| Genome | Genes in genome | Genes in COGs | Genes in links | Fusion links | Sens. (%) | Expect ± SD (%) | No. SD | Spec. (%) |
| M. genitalium | 468 | 320 | 33 | 16 | 90 | 10 ± 10 | 8 | 84 |
| U. urealyticum | 604 | 335 | 27 | 14 | 91 | 13 ± 10 | 8 | 84 |
| M. pneumoniae | 680 | 356 | 36 | 18 | 100 | 11 ± 8 | 10 | 85 |
| R. prowazekii | 836 | 568 | 64 | 35 | 86 | 15 ± 6 | 11 | 89 |
| C. trachomatis | 877 | 536 | 59 | 36 | 70 | 13 ± 6 | 9 | 89 |
| T. pallidum | 1036 | 559 | 54 | 29 | 79 | 16 ± 10 | 6 | 89 |
| C. pneumoniae | 1051 | 547 | 63 | 38 | 74 | 13 ± 6 | 10 | 90 |
| H. pylori J99 | 1492 | 866 | 125 | 76 | 76 | 16 ± 5 | 12 | 91 |
| A. aeolicus | 1560 | 1028 | 236 | 163 | 69 | 13 ± 3 | 19 | 92 |
| H. pylori 26695 | 1578 | 880 | 150 | 88 | 77 | 14 ± 4 | 17 | 91 |
| C. jejuni | 1634 | 1031 | 193 | 128 | 70 | 14 ± 4 | 14 | 91 |
| B. burgdorferi | 1637 | 554 | 49 | 26 | 88 | 22 ± 8 | 8 | 89 |
| H. influenzae | 1671 | 1196 | 222 | 128 | 76 | 15 ± 3 | 19 | 92 |
| M. jannaschii | 1745 | 881 | 235 | 143 | 75 | 15 ± 3 | 16 | 90 |
| P. abyssi | 1767 | 962 | 231 | 125 | 67 | 16 ± 4 | 12 | 90 |
| T. maritime | 1858 | 1140 | 300 | 184 | 74 | 26 ± 3 | 13 | 91 |
| M. thermoautotrophicum | 1873 | 964 | 281 | 137 | 72 | 19 ± 3 | 16 | 90 |
| P. horikoshii | 2080 | 891 | 213 | 103 | 62 | 17 ± 4 | 10 | 90 |
| N. meningitidis | 2081 | 1151 | 223 | 138 | 72 | 15 ± 3 | 16 | 91 |
| A. fulgidus | 2418 | 1257 | 365 | 193 | 56 | 17 ± 3 | 12 | 90 |
| A. pernix | 2722 | 837 | 138 | 72 | 63 | 12 ± 5 | 11 | 89 |
| X. fastidiosa | 2766 | 1187 | 219 | 131 | 70 | 12 ± 3 | 22 | 92 |
| Synechocystis | 3168 | 1586 | 543 | 297 | 63 | 18 ± 3 | 17 | 92 |
| D. radiodurans | 3192 | 1618 | 426 | 229 | 65 | 20 ± 2 | 18 | 92 |
| V. cholerae | 3829 | 2146 | 664 | 315 | 60 | 20 ± 2 | 20 | 92 |
| M. tuberculosis | 3927 | 1855 | 706 | 302 | 62 | 19 ± 2 | 21 | 92 |
| B. subtilis | 4123 | 2160 | 679 | 356 | 57 | 25 ± 2 | 16 | 92 |
| E. coli | 4224 | 2598 | 791 | 402 | 67 | 22 ± 2 | 17 | 92 |
| P. aeruginosa | 5567 | 3265 | 930 | 458 | 60 | 32 ± 2 | 13 | 92 |
| S. cerevisiae | 5942 | 1779 | 421 | 135 | 59 | 28 ± 4 | 6 | 89 |
| Total | 68406 | 35053 | 8676 | 4515 | ||||
| Average | 72 | 90 |
In each genome (A), a subset (C) of all genes (B) have a functional category in the COGs database. A subset of genes (D) is present in COGs and involved in fusion links (E). The sensitivity of the fusion method (F) is compared to the mean (G) and standard deviation (H) of the expected distribution. The significance (I) is the number of standard deviations by which the observed ratio exceeds the expected. The specificity of the fusion method is also calculated (J).
Fusion links tend to involve genes of the same functional category. Fig. 3 shows that fusions are nonrandom events, intracategory fusion links generally account for the most significant number of fusion links, and fusion links between some pairs of categories occur less frequently than expected by chance. Some illustrative examples are energy production (C), nucleotide transport and metabolism (F), DNA replication, recombination, and repair (L), and amino acid transport and metabolism (E).
In D. radiodurans, for example, we find five fusion links between genes involved in DNA replication, recombination, and repair (L), which has a significance of 10. These genes are found fused to genes in another functional category no more frequently than expected by chance alone in D. radiodurans, with the exception of one fusion event with a gene involved in translation (J), which has a significance of 2 because of the small size of the latter category.
Although both genomes demonstrate the overall dominance of intracategory links, specific intracategory links differ among the genomes in their frequency and significance. For example, X. fastidiosa genes involved in amino acid transport and metabolism (E) are only found fused with genes in the same category, whereas D. radiodurans genes in that category frequently fuse with genes involved in other functions.
Fusion Links in 30 Microbial Genomes.
To further characterize fusion links as well as to determine whether the observed results were species-dependent, we identified the fusion links among genes in 30 microbial genomes (Table 1). Altogether, we found 10,073 fusion links among 10,540 genes. After removing the 5,558 pairs of which either or both link partners did not belong to a COG, we were left with 4,515 links. For each genome, we determined the fraction of fusion links that are composed of two genes of the same functional category. We estimated the significance of this fraction by comparing it with a random distribution (see Methods). In every genome, we found that the number of intracategory fusion links was significantly greater than expected; on average, this significance was 13 standard deviations greater than expected by chance alone. The significance of intracategory links increases linearly with the number of genes in the organism (Table 1). This corroborates the notion that intracategory links are significant, because larger genomes have more fusion links and thus offer better statistical sampling.
To view the function correlation tables of all genomes simultaneously, we normalized the tables for individual genomes (as in Fig. 3) by dividing the number of intercategory and intracategory fusion links by the total number of fusion links in that genome. We then superimposed these normalized tables onto one table (Fig. 4). This global view confirms the dominance of intracategory links in terms of both frequency and significance. Illustrative examples include DNA replication (L), signal transduction (T), and energy production and conversion (C). Fusion links among amino acid transport and metabolism (E) proteins are the most frequent, averaging 15 per genome. In addition, fusion links between many pairs of categories occur less frequently than expected by chance such as amino acid transport and metabolism (E) with coenzyme metabolism (H). The significant combinations of different functional categories include signal transduction (T) with cell motility and secretion (N) and with posttranslational modification (O). Many of the intercategory links relate genes with strong functional associations (see Discussion).
Figure 4.
A glance at fusion links in 30 microbial genomes. This figure, in the same format as Fig. 3, shows the combined global views of all fusion links in the 30 microbial genomes. Each circle in the upper triangle indicates the number of fusion links between two categories in a genome, normalized to the total number of fusion links in that genome. The superimposed circles were “jittered” so that they can be seen. Each element in the lower triangle indicates the average number of fusion links between two categories, followed by the average significance. The categories are as described in the Fig. 3 legend. The figure was made by using spotfire (http://www.spotfire.com).
The intracategorical fusion links are over-represented in part because of fusion links among subunits of the same complex, which is consistent with the notion that fusion links identify protein–protein interactions (6, 7). We find examples of these interactions in translation (J), DNA replication (L), and energy (C) among others. One example is the fusion link between the alpha and beta E1 subunits of the thiamine pyrophosphate-dependent dehydrogenase complex.
Specificity and Sensitivity of the Fusion Method to Link Genes of the Same Functional Category.
To ascertain the predictive power of the fusion method to link genes of a common functional category, we sought to calculate its specificity and sensitivity. From the set of all possible gene pairs in a given genome, we determined the numbers of fusion-linked genes of common function (true positives), fusion-linked genes of different function (false negatives), non-fusion-linked genes of common function (false positives), and non-fusion-linked genes of different function (true negatives). We find that the average sensitivity, the ratio of true positives to the sum of true positives and false negatives, is 0.72, i.e., 72% of all fusion links involve genes of a common functional category.
For the 30 genomes considered here, we find that the probability that any two genes picked at random from the same genome will have an identical COG functional category is 0.088; this probability is close to the value expected if all 15 categories were of equal size (15/120 = 0.125). Combined with the fact that the number of fusion links is typically four orders of magnitude smaller than the total number of possible pairs of genes, this explains the high average specificity of 0.90, i.e., the ratio of true negatives to the sum of true negatives and false positives (pairs of genes of different COG functions).
Predictions from the Fusion Method in M. thermoautotrophicum.
Because the fusion method is able to link genes of common functional category accurately, it can be used to predict the functional category of genes of unknown function. Among the 30 genomes considered here, 671 genes ascribed to the COG categories general function prediction only (R) or function unknown (S) are linked to functionally categorized genes. Each of these links represents a hypothesis that the uncharacterized gene belongs to the same functional category as its fusion-link partner. In addition, we have detected 5,558 fusion links that could not be included in this analysis because one of the partners was not present in the COGs database. All fusion links, including the latter, can found at http://fusion.bu.edu.
To provide an idea of the type of predictions that can be made by using the fusion link method, we show the nine M. thermoautotrophicum genes of unknown function fusion-linked to M. thermoautotrophicum genes of known function (Table 2). We predict that each of these genes of unknown function belongs to the same functional category as its fusion-link partner. For example, MTH1425 (534 aa) is composed of a metal-dependent protease domain and a Ser/Tyr kinase domain, whereas MTH554 (188 aa) is unannotated. MTH554 is linked to the kinase domain of MTH1425 by a fusion gene in Caenorhabditis elegans (GenBank accession no. 3800951). The fusion gene is annotated only as a “conserved protein,” and therefore does not directly confer a putative function to MTH554, but it does imply a functional correlation between the two M. thermoautotrophicum genes; it is possible that MTH554 is involved in the same signaling cascade as MTH1425.
Table 2.
Fusion links in M. thermoautotrophicum involving one gene of unknown function
| Gene A | Gene B | COG (B) | Function (B) |
|---|---|---|---|
| MTH357_1 | MTH412_2, MTH845 | E | Transglutaminase-like enzymes, putative cysteine proteases |
| MTH1356 | MTH601_2 | C | Ferredoxin 2 |
| MTH861 | MTH614 | H | Hydroxymethylpyrimidine/phosphomethylpyrimidine kinase |
| MTH1258_2 MTH1835_2 | MTH1835_1, MTH1883 | G | 2-Phosphoglycerate kinase |
| MTH554 | MTH1425_2 | T | Serine/threonine protein kinases |
| MTH1172_2 | MTH1893 | P | Predicted Co/Zn/Cd cation transporters |
| MTH714_1 | MTH1363, MTH237, MTH317, MTH318, MTH351, MTH456, MTH514, MTH673, MTH928 | H | Cobalamin biosynthesis protein CobN and related Mg-chelatases |
| MTH1797 | MTH1345 | C | Formylmethanofuran dehydrogenase subunit E |
| MTH641 | MTH764 | L | Predicted EndoIII-related endonuclease |
Discussion
The hypothesis underlying this analysis is that a fusion gene in one organism can indicate an association between the independent genes in another organism, assuming that orthologous genes (19) have parallel functions in both organisms (2). Linking genes by way of fusion events, initially proposed by Marcotte et al. (6) and Enright et al. (7), can hint at direct physical interactions between proteins or a more general functional association such as between sequential members in a metabolic pathway. Because this method does not require sequence similarity between the genes, it was proposed by Marcotte et al. and Enright et al. that the fusion analysis could be a method that complements sequence similarity searches.
There may be many instances in which such inferences are not warranted. For example, the orthologous sequences as defined by the COGs database may have diverged to assume different functions despite significant sequence similarity (20). The orthologous genes may also be incomparable, because they operate in different cellular environments. In such instances, the fusion may not be a reliable predictor of a functional relation. In addition, the fusion protein may be composed of functionally unrelated proteins. In these cases, knowledge about the component domains may provide information about the fusion protein.
In this study, we determined the prevalence of common function in genes linked by fusion events in 30 microbial genomes. Two fusion-linked genes were said to be of common function if both belonged to the same functional category in the COGs database. Whereas the background probability that two genes are members of the same functional category is ≈9%, we find that on average 72% of pairs of fusion-linked genes consist of members of the same category (see Table 1, Average sens.). Views of the entire fusion-link repertoire of a genome in terms of the combinations of functional categories (Figs. 3 and 4) strikingly illustrate that intracategory fusion links are significant in comparison with a random model and frequently compared with intercategory fusion links. However, intercategory links do hint at a functional interaction. For example, a fusion between a Ser/Thr protein kinase and protein-disulfide isomerase suggests that the kinase domain regulates the posttranslational modification activity of the isomerase.
Many fusion genes fuse domains rather than entire genes. A particular advantage of the COGs database is that each of the domains of a protein is categorized separately when distinct conserved regions are present. Thus, when one domain of a multidomain gene is linked by a fusion to another gene or domain, the information about the fusion refers only to the domains. However, by extension of the notion that genes linked by fusion links are functionally related, information for one domain of a multidomain gene may suggest a function for the other domains or for the entire gene. We intend to study the functional correlations among the domains of multidomain microbial genes.
A universal property of microbial genomes is the considerable number of paralogous genes (21, 22), with implications for counting fusion links. For example, consider the paralogous genes A and A′ that are both fusion-linked to gene B. To avoid double-counting what is essentially a single link, we collapsed the links by allowing only a single link between any two COGs. We believe that the collapse of paralogs to one representative is an improvement to the fusion method as originally proposed by Marcotte et al. (6), and it obviates the consideration of “promiscuous” genes or domains.
In principle, fusion analysis can be used to predict the function of an unknown protein if it is fusion-linked to a partner of known function. The present study has generated 10,073 fusion links, 45% of which involve genes with functional annotations in the COGs database and are summarized in Table 1. The general conclusion that fusion links relate genes from the same functional category can be extended to the remaining links, which were excluded from the analysis. In addition, intercategory fusion links suggest cross-links between the proteins that may shed insight into their functions. We excluded from consideration the fusion links of homologous genes with the motivation of finding links that could not have been established through direct sequence comparisons. Thus, the frequency of intracategory fusion links in fact may be understated because of the exclusion of links between homologs.
Our results suggest that fusions are nonrandom events and therefore presumably confer a selective advantage; conversely, fusions occur much less frequently than expected randomly between certain functional groups, implying that those events are deleterious or neutral (assuming that the components precede the fusion in evolutionary history). West (23) has argued that diffusion is increasingly problematic for large cells, not because of distance but because of obstacles such as large protein complexes and the cytoskeleton. The physical proximity of multiple enzymes in the same metabolic pathway alleviates the problem of diffusion in a complex cell and inhibits side reactions (23). This notion is supported by the observation that some metabolic intermediates are found in the cytoplasm at concentrations lower than expected. The fusion of genes thus may reflect a strategy for coping with the increasing complexity of higher organisms by effectively compartmentalizing functionally related proteins. In addition, fusions simplify the regulation of transcription (23).
Although there are other methods for effectively compartmentalizing proteins such as direct interactions, scaffold proteins, and localization, this study indicates that fusions are nonrandom events that confer a selective advantage by linking functionally related proteins and therefore could serve as predictors of functional associations.
Acknowledgments
We thank Zhiping Weng and Carlos J. Camacho for stimulating discussions and John Aach and Nikos Reppas at Harvard Medical School for critical reading. We also thank our two anonymous reviewers. This work was supported in part by a National Science Foundation Integrative Graduate Education and Research Traineeship Program grant (to A.D.). I.Y. is supported by the Whitaker fellowship, and A.D. is supported by the Paul and Daisy Soros Fellowship for New Americans.
Abbreviations
- COG
cluster of orthologous groups
- DHFR
dihydrofolate reductase
References
- 1.Pellegrini M, Marcotte E M, Thompson M J, Eisenberg D, Yeates T O. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 3.Overbeek R, Fonstein M, D'Souza M, Pusch G D, Maltsev N. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dandekar T, Snel B, Huynen M, Bork P. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 5.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcotte E M, Pellegrini M, Ng H L, Rice D W, Yeates T O, Eisenberg D. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 7.Enright A J, Iliopoulos I, Kyrpides N C, Ouzounis C A. Nature (London) 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 8.Schwikowski B, Uerz P, Fields S. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 9.Marcotte E M, Pellegrini M, Thompson M J, Yeates T O, Eisenberg D. Nature (London) 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 10.Huynen M, Snel B, Lathe W, III, Bork P. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin M Y, Koonin E V. Nat Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- 12.Tsoka S, Ouzounis C A. Nat Genet. 2000;26:141–142. doi: 10.1038/79847. [DOI] [PubMed] [Google Scholar]
- 13.Tatusov R L, Galperin M Y, Natale D A, Koonin E V. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm L, Sander C. Bioinformatics. 1998;14:423–429. doi: 10.1093/bioinformatics/14.5.423. [DOI] [PubMed] [Google Scholar]
- 16.Burley S K, Almo S C, Bonanno J B, Capel M, Chance M R, Gaasterland T, Lin D, Sali A, Studier F W, Swaminathan S. Nat Genet. 1999;23:151–157. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- 17.Smith T F, Zhang X. Nat Biotechnol. 1997;15:1222–1223. doi: 10.1038/nbt1197-1222. [DOI] [PubMed] [Google Scholar]
- 18.Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. J Mol Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- 19.Fitch W M. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 20.Huynen M A, Bork P. Proc Natl Acad Sci USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynen M A, van Nimwegen E. Mol Biol Evol. 1998;15:583–589. doi: 10.1093/oxfordjournals.molbev.a025959. [DOI] [PubMed] [Google Scholar]
- 22.Yanai I, Camacho C J, DeLisi C. Phys Rev Lett. 2000;85:2641–2644. doi: 10.1103/PhysRevLett.85.2641. [DOI] [PubMed] [Google Scholar]
- 23.West I C. In: Channelling in Intermediary Metabolism. Agius L S, editor. Vol. 9. London: H. S. A; 1997. pp. 13–69. [Google Scholar]