Abstract
Laminin 332, composed of the α3, β3 and γ2 chains, is an epithelial-basement membrane specific laminin variant. Its main role in normal tissues is the maintenance of epithelial-mesenchymal cohesion in tissues exposed to external forces, including skin and stratified squamous mucosa. After being secreted and deposited in the extracellular matrix, laminin 332 undergoes physiological maturation processes consisting in the proteolytic processing of domains located within the α3 and the γ2 chains. These maturation events are essential for laminin 332 integration into the basement membrane where it plays an important function in the nucleation and maintenance of anchoring structures. Studies in normal and pathological situations have revealed that laminin 332 can trigger distinct cellular events depending on the level of its proteolytic cleavages. In this review, the biological and structural characteristics of laminin 332 domains are presented and we discuss whether they trigger specific functions.
Keywords: laminin 332, LG45 domain, LE domain, syndecan, basement membrane, keratinocyte
Laminins are large extracellular glycoproteins that are important components of all basement membranes. They are involved in several biological processes, including self polymerization, binding to the extracellular matrix (ECM) and cellular interactions.1,2 All laminins are composed of three different gene products, termed α, β and γ chains assembled into a cross-shaped heterotrimer αβγ. The three chains assemble within the endoplasmic reticulum through their C-terminal domains to form a triple stranded α-helical coiled coil rod.3,4 Sixteen laminin isoforms of different subunit composition selected from five individual α chains (α1 to α5), three β chains (β1 to β3) and three γ chains (γ1 to γ3), are known with variable cell and tissue specific expression, and they are differentially recognized by cellular receptors.5 All laminin α chains possess a large globule at the carboxyl-terminal end that consists of five similar domains LG1 to LG5 each containing about 200 residues.6,7
Laminin 332 and Its Maturation Events
Laminin 332, composed of the α3, β3 and γ2 chains, is an epithelial-basement membrane specific variant. The α3 chain is encoded by the LAMA3 gene, which has two transcript variants α3A and α3B. As the truncated LAMA3A variant is expressed and incorporated into laminin 332 heterotrimers,8 we will refer to the α3Aβ3γ2 trimer as representing laminin 332 in this review. Its main role in normal tissues is the maintenance of epithelial-mesenchymal cohesion in tissues exposed to external forces including skin and stratified squamous mucosa.9,10 To fulfill this function, laminin 332 undergoes several physiological post-translational processing events allowing its supramolecular integration into the basement membrane. In other situations such as wound healing or carcinoma, additional cleavages may take place delivering pro-migratory signals to cells therefore compromising the adhesive function of the protein.
Laminin 332 processing and integration into the skin basement membrane
In skin, laminin 332 is synthesized by keratinocytes as a high molecular weight precursor protein of 460 kDa. After secretion and deposition into the ECM, the α3 and γ2 chains undergo maturation events consisting in specific processing to smaller forms.4,11 The 190–200 kDa α3 chain (α3200) can be successively processed at both C- and N-terminal extremities producing 165 (α3165) and 145 kDa (α3145) maturation products. The 155 kDa γ2 chain (γ2155) is processed in the N-terminal region leading to a 105 kDa product (γ2105).
Processing of the α3 chain consists of cleavage of the C-terminal globular domains 4 and 5 (LG45) within the spacer between LG3 and LG4 (Fig. 1A and B).12,13 In vitro studies using human laminin 332 have revealed that enzymes involved in the processing include plasmin,12 MMP-2, MT1-MMP and the C-proteinase family of enzymes, especially mammalian tolloid (mTLD) and the bone morphogenic protein 1 (BMP-1).14,15 In addition, thrombin was shown to have the potency to cleave LG45 specifically.16 Alternatively, there may be other mechanisms that control the rate of laminin α3 LG45 processing, such as the tissue plasminogen proteolytic cascade.12 N-terminal amino acid sequencing of human LG45 purified from the conditioned medium of either primary or immortalized human keratinocytes and of the human gastric adenocarcinoma cells STKM-1 revealed that the α3 chain is cleaved between Q1337 and D1338 within the hinge region between LG3 and LG4 suggesting that a proteolytic cleavage site of the LG45 domain matches the minimal consensus sequence LLQD (Fig. 1B).17,18 It is not known what endopeptidase catalyzes the hydrolysis of this cleavage sequence, and the existence of additional proteolytic cleavage sites has been suspected both in the hinge and the adjacent regions of the LG3 and LG4 domains.7,13,14,19 This hypothesis is compatible with the spacer length of the laminin α3 chain, which is longer than those of the α1, α2 and α5 chains.7 Punctual mutations within the spacer region and/or deletion of the sequence LLQD did not protect the α3 LG3-LG4 linker from cleavage.14,19 As proteolytic processing of LG domains occurs in all α chains except α1, the substitution of the α3 LG3-LG4 hinge with that of the α1 chain was an elegant way to render the laminin α3 chain uncleavable.19 This was also the case when a 46 amino-acid portion within the α3 LG3-LG4 hinge was deleted.16 These LG45-uncleavable α3 chain constructs were expressed in human19 or mouse16 skin keratinocytes deficient for expression of the α3 chain, and in both cases the resulting LG45-uncleavable heterotrimeric laminin 332 was deposited in the ECM. Information gained from the structure of the mouse α2 chain LG45 domain pair revealed an unusual path of the LG3-LG4 linker as it forms an inter-domain disulfide bridge to LG5.20 Being integrated into the LG45 tandem the linker is therefore an integral part of the structure, a feature that is conserved in all laminin α chains. It is tempting to speculate that proteinases may have easy access to the LG3-LG4 cleavage sites located within a well-exposed linker.7
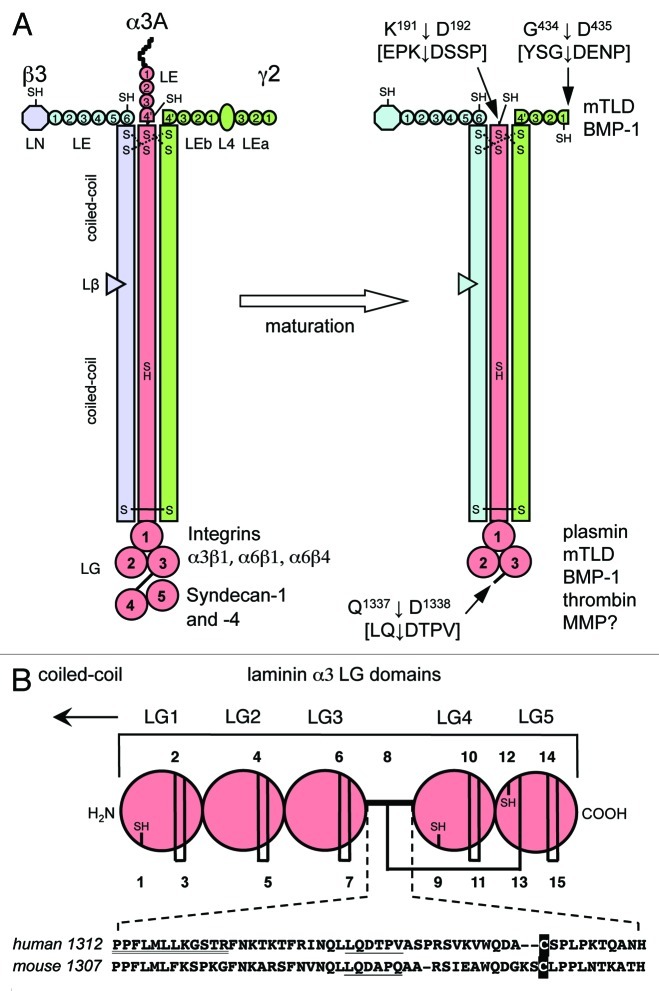
Figure 1. Structure of human laminin 332 and its physiological maturation process. (A) Laminin 332 is composed of three subunits α3A, β3 and γ2. Each chain is composed of different domains that are indicated. Domain L4 in the γ2 chain corresponds to an LE domain with a ~180 residues insert between the third and fourth cysteine of the canonical 8-Cys pattern. LE4' and LEb4' of the α3A and γ2 chain are truncated LE repeats containing only the first four and six cysteines, respectively, of the pattern. Based on the odd number of cysteines, β3LN, α3ALE4', β3LE6 and γ2LEb1 (after maturation), and the coiled coil of α3 have free SH groups as indicated. The coiled coil is stabilized by disulfide bonds at the N-terminus (dotted lines) though the connectivity is yet unknown. The large LG structure located at the C-terminal end of the α3 chain contains five repeating LG domains. The first three repeats (LG1–3) interact with α3β1, α6β1 and α6β4 integrins while the last two (LG45) contain binding sites for syndecan-1 and -4. Laminin 332 is synthesized as a precursor molecule that undergoes maturation by proteolytic processings at the α3Α chain N- and C-terminus as well as at the γ2 chain N-terminal extremity. The cleavage sites are indicated by arrows as well as enzymes involved identified so far. (B) Schematic structures of LG. The five LG domains contain numerous cysteines that are numbered. Disulfide bridged and free cysteines (SH) within the human sequence are indicated. At the bottom, an alignment of the human and mouse LG3-LG4 linker is shown. Each sequence displays the same cleavage site (underlined) and a cysteine (printed in inverse font) that forms an inter-domain disulfide bond to LG5.20 The sequence P1312PFLMLLKGSTR supposed to be crucial for integrin α3β1 binding58 is double underlined.
Further cleavage occurs in the N-terminal LE region of the α3 chain releasing the full short arm which might be important for laminin 332 function (Fig. 1A).14,21 A mutation causing an N-terminal deletion of 226 amino acids in the human α3A isoform was identified in the laryngo-oncho-cutaneous syndrome, a rare autosomal recessive disorder characterized by chronic production of vascularized granulation tissue.22 Cultured keratinocytes obtained from patients revealed that heterotrimeric laminin 332 carrying the mutant α3 chain is assembled and secreted. The mutated laminin was regularly expressed in the skin basement membrane and only very subtle ultrastructural changes were seen such as focal widening of the lamina lucida in places where hemidesmosome plaques were smaller. These studies suggest that the laminin α3 N-terminal domain may be a key regulator of the granulation tissue response during wound healing.
In human skin laminin 332, the processing of the γ2155 chain leads to the γ2105 subunit and was shown to be achieved by BMP-1 and mTLD metalloproteinases.11,14,15,23,24 The N-terminal processing is complex as it occurs within a disulfide-linked loop of the L4 domain followed by reshuffling of disulfide bonds for release of the cleaved fragments.24 The immunohistochemical analysis of laminin 332 in mTLD/BMP-1-deficient mouse skin revealed a strong expression of the precursor γ2 chain confirming that these enzymes are involved in the γ2 chain maturation in vivo.25 In contrast, the processing of the LG45 domain in the α3 chain seemed, at least partially, unaffected reinforcing the hypothesis that several enzymes are involved in this maturation process.
Laminin 332 was shown to be the major component of anchoring filaments in skin9 where it mediates cell adhesion via interaction of the α3 carboxyl-terminal LG1–3 triplet domain with both α3β1 and α6β4 integrins,26-29 while the N-terminal short arms connect to basement membrane components. Laminin 332 can be incorporated into the basement membrane through at least two mechanisms. The first involves cross-linking of laminin 332 with laminin 311 (α3β1γ1) in the skin basement membrane.21 The complex of laminin 311 with laminin 332 is most likely to derive from an interaction of domain LN in the β3 chain of laminin 332 with domain LE of the α3 chain short arm in laminin 311 (see below). As seen on rotary shadowing electron micrographs of the complex, the LN domain of the short arm interacts with a laminin 311 domain near the intersection of the laminin 311 short arms.21 These complexes are most likely stabilized by a disulfide bridge between an unpaired cysteine in domain LN of the β3-chain and domain LE of processed α3 in laminin 311. A similar complex between laminin 332 and laminin 321 (α3β2γ1) is found in the basement membrane of the amnions. According to the 3-arm interaction hypothesis of laminin polymerization,30 the dimers could self-associate. The second mechanism reports a direct interaction between anchoring filaments and anchoring fibrils. Anchoring fibrils are disulfide bond stabilized dimers of type VII collagen.31 Monomeric laminin 332 as well as the laminin 332/311 dimer directly bind the N-terminal globular domain NC1 of type VII collagen.10,32 The interaction is likely to occur within the short arm of the β3 and/or γ2 subunit.32
Other processing events in laminin 332
In addition to the laminin 332 maturation aimed at regulating the skin basement membrane structural integrity, other studies have reported two specific migration-inducing MMP-2 cleavages of the laminin γ2 N-termini producing γ2 chain fragments of 100 and 80 kDa.33 These γ2 processing could also be carried by MT1-MMP in colon and breast carcinoma cells.34 Both enzymes, that were first shown to cleave rat laminin 332, were proposed to also cut down human laminin 332 in both physiological and pathological situations such as tumorigenesis.35,36 MMP-3, -12, -13, -19 and -20 were also shown to process the γ2 chain inducing epithelial cell migration.37,38 Migration events might therefore result from the interaction of newly exposed laminin domains with cell surface signaling receptors. Rat recombinant γ2LEb1-LEb4' was shown to bind and activate the EGF receptor to stimulate cell migration.39 Studies with cancer cells have reported that MMP-7, MT1-MMP and hepsin cleave laminin β3 leading to increased migration.40-42 The molecular mechanisms of laminin 332 in squamous carcinoma have been reviewed in detail by Marinkovich.43
Laminin 332 Sequence-Structure Relationship
The structure of the heterotrimeric laminin 332 molecule consisting of disulfide-linked α3, β3 and γ2 chains was first revealed by rotary shadowing electron microscopy, which showed a rod 107 nm in length terminated by one large and two small globules at opposite ends9 which correspond to the C-terminal LG and N-terminal LN and LE/L4 domains, respectively (Fig. 1A). This length is consistent with the later determined sequences which indicate that ca. 570 residues per chain contribute to the three stranded α-helical coiled coil structure giving a length of ~80 nm, and ~7 tandem LE domains accounting for a further 20 nm. Circular dichroism spectroscopy revealed an α-helical content of ~30%44 similar to that determined for laminin 11145 suggesting that most of this secondary structure contribution relates to the coiled coil. Assembly of the laminin 332 trimer occurs in the endoplasmic reticulum where first a disulfide linked β3-γ2 dimer is formed to which the α3 chain aligns via the coiled coil. Trimer formation is a prerequisite for transport to the Golgi complex and secretion but does not require N-linked glycosylation.4
Laminin β3 LN domain
So far no high resolution structures of laminin 332 domains have been solved. However, based on sequence similarity, some insight into the molecular organization of various domains can be anticipated from homology modeling. Recently, the crystal structure of the N-terminal LN domain together with four adjacent LE modules of the mouse laminin β1 chain has been solved (pdb code: 4aqs).46 The corresponding regions of human laminin β3 can be fitted to this structure (Fig. 2) suggesting that LN (residues 18–248; accession number NP_000219) forms a β-sandwich consisting of eight β-strands folded into a jelly roll motif. The first LE domain (res. 249–314) is tightly integrated into LN, which starts with a disulfide-bonded reverse turn connected by C21–C26, and the N-terminal region (res. 18–28) associates to LE1. Pro28, which is conserved in all laminin LN sequences, fits into a pocket of LE1. Further disulfide bonds can be expected between C47-C69, C56-C66 and C153-C173 (Fig. 2A). This leaves C67, which is unique for β3 LN domains and surface accessible (Fig. 2B), in a reduced state making it available for covalently binding to other proteins like laminin 331. The N-linked glycosylation sites found within β1 LN at N120 and in γ1 LN at N58/N132 are absent in the β3 chain. The critical residues D106 and T114 of γ1 LN involved in calcium binding,46 which might be important for laminin polymerization, are replaced by S70 and R76 in the β3 chain indicating that this site will not be functional.
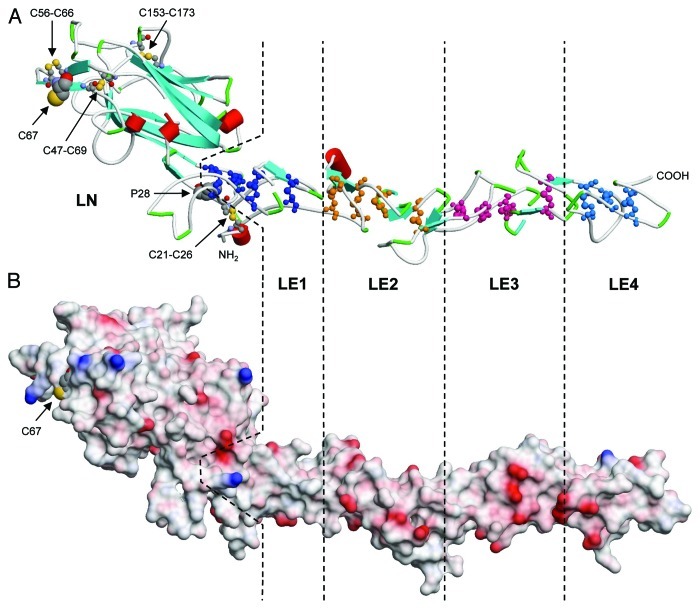
Figure 2. Structural model of the human laminin β3 LN-LE1–4 domains. (A) The structure shows residues Q18 to Q464 as a cartoon with red cylinders and blue arrows indicating α-helices and β-strands, respectively. Disulfide bonded cysteines within the LE domains are depicted in ball-and-stick presentation using different colors. Domain boundaries are indicated by dashed lines. P28 fits into a pocket shared by the LN and LE1 domain, and the N-terminal residues are interacting with LE1. Disulfides and the free cysteine residue of the LN domain are indicated. (B) The electrostatic surface potential is represented in the same orientation and on the same scale as in (A). Negative and positive potentials are shown in red and blue, respectively. Homology based modeling was performed using Swiss-Model106 in manual alignment mode with the mouse β1 structure (pdb code 4aqs46) as a template.
Laminin 332 LE and L4 domains
A common feature of LE modules is their connectivity of the eight cysteine residues as C1-C3, C2-C4, C5-C6 and C7-C8. The loop formed between C5-C6 usually consists of eight residues with a glycine in position six. In human laminin 332, the following LE domains deviate from the 8-Cys pattern (Fig. 1A): β3-LE6 preceding the coiled coil contains one extra cysteine (C572) between C7 and C8 whereas the corresponding region within α3A-LE4' is truncated after the fourth cysteine (C185; accession number AAA59483) followed by an irregular cysteine (C199) just before C202/C205, which are presumably forming intrachain disulfides at the N-terminus of the coiled coil. The α3A chain is processed just after LE4' between K191 and D192.14 LEb1 following domain L4 of the human γ2 chain is missing the fifth cysteine of the 8-Cys pattern and does not contain the glycine residue N-terminal to the sixth cysteine (C442; accession number NP_005553). This probably results in an increased mobility of the loop between C431 and C442 allowing for protease susceptibility. Indeed, major processing occurs by cleavage of the G434-D435 site (Fig. 1A).23
The L4 domain separating LEa3 and LEb1 can be regarded as a specialized LE domain with a large, ~180 residues long insert between the third (C196) and fourth (C382) cysteine of the regular pattern. In case of the γ2 chains, the L4 insert is highly conserved from human down to Xenopus and zebrafish, and contains one unique cysteine absent in all other L4 domains. For L4-LEb1 it was shown that this C309 is covalently linked with the last cysteine C459 of LEb1, which contains an unusual S-S bridge between C442-C445 corresponding to C6-C7. After cleavage by BMP-1 between G434 and D435, protein disulfide isomerase was required to release the fragments.24 It remains unclear whether after processing C459 stays in a reduced state, or whether disulfide reshuffling might form the usual C7-C8 connectivity and leaves C6, i.e., C442, with a free SH group.24 The mouse L4-LEb1 domain pair was modified probably via one or both of the two putative NxT acceptor sites for N-linked glycosylation, which are also found in the human sequence (N342 and N362). The N342xT site is conserved down to zebrafish. Mouse L4-LEb1 binds heparin, nidogen-1 (but not -2), and fibulin-1 and -2; it remains unclear whether these interactions occur between L4 or LEb1, or whether epitopes from both are required.24
In conclusion, besides the β3 LN domain, each of the three irregular LE modules, α3A-LE4', β3-LE6 and γ2-LEb1 after processing, have one free cysteine which could participate either in interchain disulfide bonding or covalent binding to other proteins like laminins 311 and/or 321. Based on the canonical cysteine pattern, the most likely residues are α3A-C199, β3-C572 and γ2-C442 or γ2-C459.
Laminin 332 coiled coil and Lβ domain
Carboxyl terminal to the LE modules, the three laminin chains have a C-x-x-C motif (α3A: C202- C205; β3: C581-C582; γ2: C609-C612) which cysteines are conserved in all laminins and most likely serve for interchain covalent linkage of the heterotrimer.The actual disulfide pattern has not yet been established. The following ~570 residues associate via formation of an α-helical coiled coil represented on the sequence level by a 3–4 heptad pattern of hydrophobic residues.3 As it is the case for all β chains, the β3 coiled coil is interrupted by domain Lβ ~200 residues away from the C-x-x-C motif containing six cysteines (human β3: C788-C818) probably forming intrachain disulfide bonds. The α3 chain coiled coil contains one cysteine (human α3: C551) about 50 nm from the C-x-x-C motif, which has no adjacent counterpart in the other two chains. The coiled coil region is terminated by cysteines close to the C-terminus (human β3: C1171; γ2: C1184), which based on similarity to the β1 and γ1 chains most probably form an interchain disulfide bond.48
Besides its structural importance for assembly, it has recently been found that in the absence of the LG domains the laminin 111 coiled coil has anti-adhesive properties. When cultured on such truncated laminin 111, cell adhesion and spreading were inhibited; genes compatible with a pro-migratory and pro-invasive function like MMPs and various matricellular proteins were upregulated.47 No comparable studies have yet been performed for laminin 332, but an antagonistic interplay between coiled-coil and LG domains (see below) could be of importance in cancer metastasis.
Laminin α3 LG domains
The large globule seen in electron micrographs at the tip of the long arm consists of five LG domains formed by the α chains. Domains LG4-LG5 are separated from LG1 to LG3 by a flexible linker (Fig. 1B). As these domains are involved in many interactions (see below), particular interest has been focused on their structural characterization (for a review, see ref. 7). X-ray structures have been solved for the mouse α5 LG1-LG3 triplet (pdb code: 2wjs),49 mouse α1 and α2 LG45 pair (2jd4,50 1dyk,20 1okq51), and mouse α2 LG5 (1qu052) which can be used to model the corresponding α3 LG1-LG5 domains (Fig. 3). LG domains fold to a β-sandwich consisting of 14 strands, where the two sheets form a concave and convex interface. Close to the C-terminus, each LG sequence contains two cysteines ~30 residues apart, which form intradomain disulfide bonds (Fig. 3A and C). In contrast to electron micrographs suggesting close proximity of LG1 to LG3,53,54 the α2 LG1–3 structure shows LG1 dissociated from the LG2-LG3 pair.49 Previous work suggested that trimerization of the α with β and γ chains is required for the LG domain to exert its integrin binding activity, and that specifically a glutamic acid two residues apart from the C-terminus of the γ chain is essential.55 This residue is indeed highly conserved in the γ1 and γ2, but not γ3 subunits. A lack of integrin α3β1 binding activity in the absence of at least short β3 and γ2 segments has been shown for α3 LG1-LG3.54,56 The absence of such crucial parts of the β and γ chains could result in the open LG1-LG2 conformation (Fig. 3A and B) and might not reflect the conformation of the native molecule.49
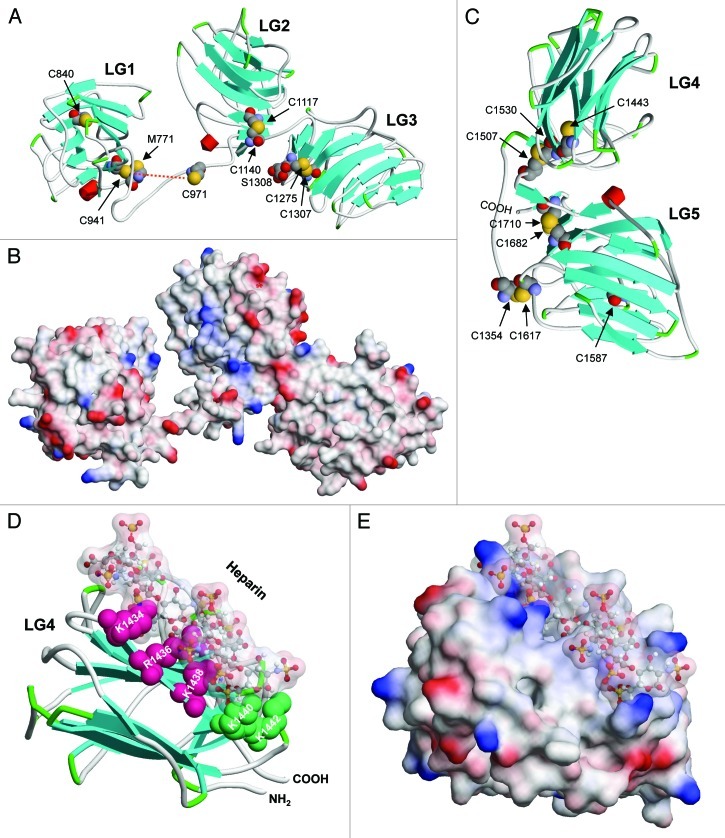
Figure 3 (See previous page). Structural models of human laminin α3 LG domains. (A) The structure of LG1 to LG3 encompassing residues M771 to S1308 is shown as a cartoon as described for Figure 2. All cysteine residues are depicted in space filling (CPK) style. The disulfide bridge between C941 and C971 (red dotted line) could not be correctly modeled as the template sequence has a shorter loop between the corresponding residues. All other cysteines form disulfide bridges except for C840 in LG1 which side chain points into the inner space of the β-sandwich. (B) The electrostatic surface potential is represented in the same orientation and on the same scale as in (A). Negative and positive potentials are shown in red and blue, respectively. (C) A model of LG45 corresponding to residues C1354 to Q1713 is shown with all cysteines in CPK style. C1354 from the loop region connecting LG3 and LG4 forms an interdomain disulfide bond with C1617. C1443 in LG4 and C1587 in LG5 are pointed to the space between the β sheets of the sandwich structures and are not surface accessible, whereas C1507-C1530 and C1682-C1710 form disulfide bonds. (D) A model of LG4 interacting with a heparin-like carbohydrate chain consisting of four GlcNS(6S)-IdoA(2S) disaccharides (ball-and-stick representation with a surface envelope) is shown. The side chains of K1434, R1436, and K1438, which were shown to be critically involved in syndecan-1 interaction are depicted in red, whereas those of K1440 and K1442 involved in syndecan-4 interaction are shown in green.59 The ε-amino and guanidinium groups of the lysine and arginine residues, respectively, point to the sulfate groups of the carbohydrate chains. (E) The surface view of the model shown in (D) indicates the tight packing of the heparin fragment into the groove formed by the concave surface of the LG4 β-sandwich with positive surface potential spaced to fit the distance of sulfate groups. The template structures used for modeling were mouse laminin α2 LG1-LG3 (pdb code: 2wjs)49 and mouse laminin α1 LG4-LG5 (2jd4, chain B).50 Docking of a heparin fragment (1hpn)60 to LG4 was performed with PatchDock.107
The LG3 and LG4 domains are connected by a flexible linker, which was realized very early by the proteolytic susceptibility of mouse laminin 111.45 Based on the C- and N-terminal residues, which can be resolved in the mouse α5 LG1–3 (2wjs49) and α1 LG4–5 (2jd450) structures, respectively, for human α3 the termini of the corresponding loop can be defined as N1310 and D1352 (Fig. 1B). Close to the middle, α3 LG is processed between Q1337 and D1338.17,57 This leaves a segment P1312PFLMLLKGSTR attached to LG3 which, based on a recombinantly overexpressed LG and synthetic peptide approach, was reported as crucial for integrin α3β1 binding.58 C-terminally truncated peptides, however, abolished this activity. The difference of this part in mouse α3 (PPFLMLFKSPKG, Fig. 1B), however, would suggest that this interaction is rather species specific.
Based on the structures of the mouse α1 and α2 LG4–5 pairs, modeling predicts that human α3 C1354 within the loop region forms a disulfide bond with C1617 in LG5 (Fig. 3C). Corresponding cysteines within the loop and LG5 are found in all laminin α chains down to Drosophila and C. elegans. Interestingly, human α3 LG4 (C1443) and α1 LG3 contain further cysteines in a similar position, which are not conserved in mouse and rat. Within the model (Fig. 3C), the C1443 and C1587 SH groups point into the hydrophobic core of the β sandwich.
The crystal structures of mouse α5 LG1, LG2, and α1, α2 LG4, LG5 contain tightly bound calcium or magnesium ions interacting with pairs of aspartate residues.20,49-51 Our human α3 LG models (Fig. 3) suggest that these cation binding sites are not conserved. Although within LG1 and LG2 D827 and D1090, respectively, are present, their partner residues are changed to N910 and R1023. Within LG4 and LG5, the corresponding residues are replaced by S1413/S1481 and K1584/S1654. This does not exclude that α3 LG domains might contain other divalent cation binding sites, but so far no conclusive experimental data exist.
Due to their physiological relevance, many studies have been performed to elucidate the interaction of laminins with proteoglycans. As a model for the complex glycosaminoglycan chains, most experiments are performed with heparin. We have recently reported on the specific interaction of human α3 LG45 with syndecans and found that K1434, R1436 and K1438 are crucial for syndecan-1 binding, whereas K1440 and K1442 show a preference for syndecan-4 (see below).59 Using the modeled α3 LG4 domain and a truncated heparin-like structure (pdb code 1hpn),60 docking simulations predict that the carbohydrate chain aligns to the concave surface of LG4 with the positively charged residues filling the grooves formed by the repeating sulfates (Fig. 3D and E).
Functions of LG45 in Precursor α3
A potential function for the tandem LG45 domains was initially suspected based on the ability of laminin 332 to trigger distinct cellular events depending on the level of processing of its α3 chain. A form of laminin 332 that lacked LG45 was found in mature basement membranes, where it was shown to play an important function in the nucleation and maintenance of anchoring structures through α3β1 and α6β4 integrin interactions.61-63 In contrast, laminin 332 with intact LG45 (α3200) was found in migratory/remodelling situations such as epidermal repair.64-66 Indeed, laminin 332 with an α3200 chain was found in the ECM of keratinocytes migrating on collagen I or after stimulation by TGF-β1.67,68 In vivo, epidermal injury activates the transcription and deposition of laminin 332 into the provisional matrix by the leading keratinocytes in the process of epidermal outgrowth and migration at the wound edge.8,61,69 Noteworthy, α3200 laminin 332 is found in this provisional matrix but is absent from mature basement membranes.16,61,62 Recently, laminin 332 comprising an α3200 chain was proposed to be involved in the invasion of squamous cell carcinomas in vivo.70 Polyclonal antibodies targeting the LG45 domains induced squamous cell carcinoma apoptosis in vivo and thus inhibited tumor proliferation.70
Function in laminin 332/311 deposition into the ECM
A function for LG45 in the deposition of laminin 332 in the ECM has been first hypothesized by Carter and coworkers,16 who showed that exogenous human α3200 laminin 332, but not α3165 laminin 332, was trapped by cultured LAMA3 deficient mouse keratinocytes and deposited within their ECM. Moreover, when these keratinocytes were transfected with constructs encoding either an LG45-uncleavable or a precleaved α3 chain, they deposited a larger amount of the LG45-uncleavable laminin 332 into their ECM as compared with the LG45 pre-cleaved laminin 332 that was preferentially secreted into the culture medium. The importance of LG45 in laminin 332 deposition was confirmed by a study reporting that a mutant laminin 332 lacking the LG45 domain, stably expressed in laminin 332 null keratinocytes derived from a patient with junctional epidermolysis bullosa (JEB) with underlying LAMA3 gene mutations, was less retained in the ECM but rather found in the culture medium as compared with the WT.70 Co-expression of LG45 in these α3165 laminin expressing keratinocytes enhanced laminin 332 deposition into the ECM, reinforced keratinocyte adhesion to the ECM and decreased migration. In these cells, LG45 co-localized with the α6 integrin subunit in stable adhesion contacts in a manner comparable to the LG45 domain of WT laminin 332 keratinocytes.70
Another study, however, found no difference in laminin 332 deposited by keratinocytes expressing either LG45 uncleavable or pre-cleaved laminin 332.19 Skin equivalents epithelialized with keratinocytes expressing the α3165 laminin 332 displayed a dermal-epidermal junction identical to that obtained with wild type keratinocytes suggesting that α3165 laminin 332 was properly deposited into the basement membrane or at least in an amount sufficient to allow dermal-epidermal cohesion.19 Besides, skin equivalents designed with keratinocytes expressing the LG45 uncleavable laminin 332 revealed absence of hemidesmosomes accompanied by an appreciable thickening of the lamina lucida likely secondary to the reduced cohesion of the tissue.19 These experiments show that LG45 cleavage appears to be a prerequisite for hemidesmosome formation in skin equivalent models.
These studies show that LG45 has the ability to integrate into the keratinocyte ECM either on its own or when present in the α3200 chain. It targets α6β4 integrin containing stable adhesion contacts suggesting that it may play a role in α6β4 integrin clustering in vitro. Although LG45 removal decreased laminin 332 deposition in the ECM, it did not fully prevent it, suggesting that other mechanisms may contribute in controlling laminin 332 deposition.
Defective α3 and γ2 processing was seen in cylindromatosis, a rare genetic human disorder characterized by the occurrence of multiple irregular benign epithelial tumors in the upper dermis.63 These nodules display a dramatically enlarged basement membrane (up to 4.3 µm thickness) as well as ultrastructural abnormalities as no clear lamina densa could be detected. The different integrin receptors are found in improper ratios as β1 integrins are upregulated while α6β4 integrin expression and hemidesmosome numbers are decreased.63 Despite a massive accumulation of laminin 332 throughout the entire basal lamina, a thin labeling of the LG45 domain detected at the interface between cylindroma cells and the basal lamina suggests that the LG45 processing may have been delayed. It is not clear whether laminin 332 accumulation in the basement membrane is related to this processing defect. Only one study reported a missense mutation in the LAMA3 gene affecting LG4 in a patient with a mild non-Herlitz JEB phenotype.71 This mutation resulting in G1506E triggers an imperfect local protein folding that, without impairing trimerization of the coiled coil, causes laminin 332 intracellular accumulation within the endoplasmic reticulum. Only a small amount of the laminin 332 harboring the mutated α3 chain is secreted and physiologically processed thus providing partial adhesion functions and explaining the mild phenotype. Therefore structural changes caused by mutations of this highly conserved residue throughout laminin LG4 domains highlights potential important functions of LG45 in laminin secretion.
When a laminin α3 cDNA was transfected into HT1080 cells, the exogenous α3 chain was assembled with the endogenous β3/γ2 and β1/γ1 to produce laminin 332 and laminin 311 heterotrimers.72 Out of the two laminin isoforms found in the culture medium, laminin 311 was found primarily with a precursor α3200 chain while the laminin 332 was found with an α3165 chain.72 Sigle et al.16 showed that exogenous human α3200 laminin 311 was not trapped by cultured LAMA3 deficient mouse keratinocytes nor deposited within their ECM like α3200 laminin 332 (see above). When these keratinocytes were transfected with constructs encoding an LG45-uncleavable α3 chain, they deposited large amount of LG45-uncleavable laminin 332 within their ECM, and the LG45-uncleavable laminin 311 was found exclusively in the culture medium.16 These results suggest that the two laminin isoforms integrate within the basement membrane through different mechanisms. That α3200 laminin 311 was not deposited within the ECM first shows that LG45 could not fulfil this event on its own. It also reinforces the hypothesis that a protein domain present in laminin 332 but absent in laminin 311, possibly in the γ2 chain,73 may contribute to the laminin deposition.
Since laminin 311 forms a complex with laminin 332 in vivo, it is tempting to speculate that laminin 311 deposition within the ECM might may depend on its covalent association with laminin 332. Most interestingly, co-polymers of perlecan with mature laminin 311 were identified in ECM formed by alveolar epithelial cells.74,75 Through nidogen linkage to the laminin 311 γ1 chain, perlecan was proposed to nucleate formation of laminin 311 fibrils.
Function in cell adhesion and migration
It has been proposed that precursor laminin 332, together with integrin α3β1, plays a central role in the polarization and migration of cells.12,66 Processing of the laminin 332 α chain was suggested to alter its binding ability to various integrins by differential exposure of binding sites/sequences to integrin receptors. Cell adhesion studies have shown that immobilized purified α3200 laminin 332 promotes adhesion of keratinocytes, but to a lesser extent than that obtained with mature laminin 332.76 Further analysis revealed that the α3β1 integrin was the major receptor to interact with α3200 laminin 332, while both α3β1 and α6β4 integrins were involved in cell adhesion to α3165 laminin 332.76 These findings correlate with previous results showing that cells plated on an α3200 laminin 332 rich matrix failed to form α6β4 integrin-dependent hemidesmosomes, whereas plasmin cleavage of the LG45 domain inhibited migration and promoted hemidesmosome formation.12 These data were corroborated by studies showing that α3200 laminin 332 is a preferential ligand for integrin α3β1 and/or α6β1, whereas processed α3 LG domains are the preferred ligand for integrin α6β4.65 Besides, the analysis of the binding capacity of soluble recombinant α3β1 integrin to a mini-laminin 332 comprising the five LG domains has revealed an increased affinity for integrin binding when LG45 was cleaved off.56 Information gained from the structural model of the LG predicts that LG1–3 have the shape of a cloverleaf from which LG45 extends.7 The specific length of the LG3-LG4 linker results in LG5 being closer to the LG1–3 cloverleaf than LG4 suggesting that the integrin-binding domain may be differently exposed in α3200 and α3165 laminin 332. Other studies have contradicted this hypothesis as both α6β4 and α3β1 integrins were found to interact with an LG45-uncleavable laminin 33216 and were colocalized in precursor laminin 332 trails left behind migrating cells.57 Moreover, α6β4 integrin containing stable anchoring complexes77 were detected in cultured keratinocytes expressing precursor- or LG45-uncleavable laminin 33216,19 in which LG45 and α6β4 integrins colocalized.70 Although the β4 integrin was correctly expressed and localized at the apical side of the epidermal basal layer, LG45-uncleavable laminin 332 expressing keratinocytes failed to form hemidesmosomes when seeded over a dermal equivalent model.19
Other studies have reported that soluble LG45 may have a function when released from laminin 332. Experiments have shown that adding either recombinant α3LG45 or LG4 to the culture medium could induce migration of keratinocytes in a MMP-9-dependent manner.78 In similar experiments, LG4 induced activation of pro-migratory MMP-1.79 Moreover, Ras/IkBα transformed keratinocytes that lacked the LG45 domain showed a deficiency in MMP-9 and MMP-1 expression. This deficiency was reversed by replacing LG45 through retroviral transduction.70
Binding sites within α3 LG45
The mechanism underlying the function of the LG45 domain in laminin 332 remains poorly understood. Several heparin-binding sites have been identified in the α3LG45 sequence (Table 1). Some of these conferred heparin-dependent cell adhesion properties, which suggested that this region in laminin 332 could interact with a heparan sulfate proteoglycan receptor.29,59 An early report described the characterization of a heparin-binding synthetic peptide corresponding to residues K1398PRLQFSLDIQT derived from the murine α1 LG4 sequence.80 This peptide induces adhesion of the melanoma cells B16F10 through an heparan sulfate proteoglycan type receptor that remains to be identified. A peptidic screening of the murine α3 LG45 domains allowed identification of three heparin binding sites with cell adhesion properties.81 Peptidic screening of the human sequence lead to the identification of a motif that included residues N1412SFMALYLSKGR, which was shown to induce syndecan-2 and -4 mediated cell adhesion, neurite outgrowth and MMP-1 and -9 secretion.78,79,82,83 Further work suggested that this motif also induced keratinocyte migration by triggering syndecan-4 clustering and subsequent β1 integrin activation.84 Later, three novel heparin-binding sites were identified based on cross-linking the native protein to heparin beads.85 Recently, through a site-directed mutagenesis approach to alter the most critical basic residues in a recombinant LG45 protein, we identified a unique heparin-binding site surrounded by a track of converging low affinity, positively charged residues (Fig. 3D and E).49 We further showed that this K1433KLRIKSKEK sequence region, which matches one of the sites identified by Vives et al.85 harbours distinctive syndecan-1 and -4 interaction sites (Table 1). Besides, our group has reported that syndecan-1 is the cellular receptor involved in cell adhesion to the α3 LG45 domain57,86 and this interaction may participate in keratinocyte migration by supporting the formation of actin-based cellular protrusions (Fig. 4A and B).76,87 The development of these membrane protrusions remarkably requires dephosphorylation of tyrosine residues in the cytoplasmic tail of syndecan-1, a condition essential for syntenin-1 recruitment.87,88 In the epidermis, syndecan-1 is located in the pericellular region of keratinocytes and displays a modest expression in the basal cell layer, which becomes increasingly intense in the suprabasal layers. Remarkably, syndecan-1 is strongly induced in wound edge keratinocytes during wound healing.89-91 This elevated expression in keratinocytes appears to be specific to syndecan-1 as no changes have been detected for other syndecans.92 Moreover, syndecan-1 deficient mice display a defect in keratinocyte proliferation and migration during wound healing.93 In light of these data, syndecan family members stimulate interest as potential laminin 332 co-receptors. Recent data has provided evidence that when human squamous carcinoma A431 cells are plated on laminin 332, syndecan-1 forms a complex with the α6β4 integrin. This triggers Fyn-mediated phosphorylation of the β4 integrin cytoplasmic tail activating PI3K- and Akt-mediated signaling, protecting the cells against apoptosis.94 This is of particular interest as LG45 in α3200 laminin 332 was proposed as an essential PI3K pathway activation promoter.70
Table 1. Heparin binding sites identified within human and mouse LG45 domains.
| Laminin a3 LG45 heparin binding sites | Method used for identification | Binding partner | References |
|---|---|---|---|
|
Human |
|
|
|
| N1412SFMALYLSKGR |
Peptide |
Syndecan-2 Syndecan-4 |
82 |
| L1428GTDGKKLRIKSKEKCNDG |
LG45/heparin cross-link |
Heparin |
85 |
| K1433KLRIK |
Directed mutagenesis |
Syndecan-1 |
59 |
| R1436IKSKEK |
Directed mutagenesis |
Syndecan-4 |
59 |
| L1488GSPPSGKPKSL |
LG45/heparin cross-link |
Heparin |
85 |
| V1610TPKQSLCD |
LG45/heparin cross-link |
Heparin |
85 |
|
Mouse |
|
|
|
| K1336ARSFNVNQLLQD |
Peptide |
Unknown receptor |
81 |
| K1398PRLQFSLDIQT |
Peptide |
Unknown receptor |
81 |
| D1629GQWHSVTVSIK | Peptide | Unknown receptor | 81 |
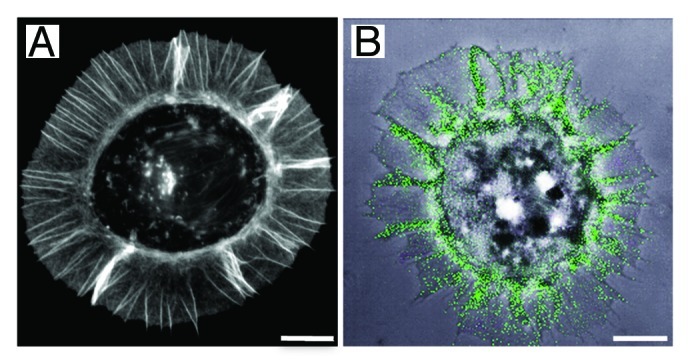
Figure 4. Syndecan-1 mediated cell adhesion to laminin 332 LG45 domain. (A) Fibrosarcoma HT1080 cells were plated on surfaces coated with purified LG45 domain for 1 h, fixed and stained with phalloidin-FITC. The analysis of the actin staining revealed an organization of the cytoskeleton in radial arrays of microspikes, protrusive adhesion structures known to be involved in transient cell-substratum interactions. (B) Confocal image of HT1080 cells expressing GFP-syndecan-1 plated on the LG45 domain. The phase contrast image shows the typical induced cell shape characterized by the formation of spike-like actin-based protrusive structures. The receptor syndecan-1 (green) promotes cell adhesion to the laminin fragment and aligns along the actin filaments, which are organized into parallel bundles. Bars, 10 µm.
Functions of Precursor γ2 N-Terminal Domain
It has been shown that in human skin basement membrane, the N-terminal domain of the γ2 chain is absent suggesting that the γ2 processing could have an important physiological role.11,63 In contrast to these findings, antibodies specific for the released L4 domain were shown to label the basal epidermal cells in mouse skin.24 A positive signal was also found in the basement membranes of early stages of skin development suggesting that the precursor γ2 chain or its released N-terminal domain expression might persist in vivo.24 The analysis of mTLD/BMP-1 deficient mice revealed that although defaults in skin cohesion were not clinically apparent, ultrastructural examination of the basement membrane showed places where anchoring structures were disconnected at the level of the lamina densa with presence of rudimentary hemidesmosomes.15,25 A strong expression of the precursor γ2 chain suggested that γ2 chain maturation takes part in the basement membrane cohesion and stability. Besides, defective γ2 processing was found in the rare inherited cylindromatosis disease (see above), in which abnormal basement membranes and excessive ectopic expression of collagen VII were found.63,96 The defective processing of the γ2 chain may have resulted in a deficient interaction with its basement membrane partners collagen VII causing defaults in anchoring fibrils linkage.32,95
Numerous studies conducted with normal breast epithelial and carcinoma cells have shown that cell migration-inducing functions of the γ2 chain rely on its proteolysis.35 Recently, it has been reported that a syndecan-1 interaction with the N-terminal domain in precursor γ2 could negatively regulate β4 integrin phosphorylation leading to enhanced carcinoma cell adhesion and decreased motility97 reinforcing the idea that the precursor γ2 chain does not support cell migration. In contrast, other reports have suggested that precursor γ2 may play a role in keratinocyte migration during the wound healing process. An interaction of its N-terminal portion with α2β1 integrin was shown to occur during TGF-β1 induced keratinocyte migration in vitro and was proposed to take place at the wound margin of the skin healing process in vivo.68 Keratinocytes at the edge of wounds made in early passage cultures shown to co-express the laminin 332 γ2 chain and the cell cycle inhibitor p16INK4a, displayed increased directional motility.98 Additional studies revealed that this keratinocyte hypermotility/growth arrest response relied on both the precursor state of the laminin 332 γ2 chain and the participation of a serum co-factor through a TGFβ receptor I dependent mechanism.99 This apparent contradiction in experimental findings is likely due to tissue specific differences. A study indicated that the L4 module mediates the integration of laminin 332 into the extracellular matrix through intermolecular interactions.73
The question arises whether the heparin-binding domains located within the α3 LG45 domains and the N-terminal portion of the γ2 chain are involved in laminin 332 deposition. Heparin was shown to inhibit laminin 332 deposition within the ECM suggesting that a heparan sulfate proteoglycan might be involved in binding to both precursor α3 and γ2.13 Null mutation in syndecan-1100 and -4101 has not reported defects in laminin deposition, which suggests that any of these two receptors is solely responsible for this mechanism. Generation of double syndecan-1 and -4 knockout mice would reveal whether both these receptors are implicated. Future studies should examine whether a heparan sulfate proteoglycan cell surface receptor or a basement membrane components are is implicated in precursor laminin 332 deposition. Interestingly, a recent study has reported that the perlecan colocalizes with laminin 332 at the wound margin of healing full thickness wounds generated in mice suggesting proximity of these two components during basement membrane remodelling.102 However, as perlecan was shown not to be essential for matrix assembly in perlecan-null mouse embryos,103 its presence in the ECM might not be an absolute requirement for laminin 332 deposition. Considering the important function of laminin 332 in skin homeostasis, several extracellular ligands might be involved in its deposition in the basement membrane.
Conclusions
Comprehension of the physiological significance of laminin 332 maturation events remains an open issue. Numerous studies have focused on the LG45 and γ2 N-terminal domains suggesting their involvement in diverse important functions such as laminin secretion, deposition and/or retention within the ECM; others have suggested a participation in epithelial cell adhesion and migration processes. However, their exact function remains unknown. While in vitro data sometimes disagree due to the cellular system’s diversity, in vivo models firmly demonstrate that defects in laminin 332 maturation impede correct basement assembly as well as hemidesmosome formation. Molecular interaction abnormalities occur at the level of cellular receptors and basement membrane components causing both functional and organisational anomalies. Molecular characterization and structural definition of laminin domains open interesting and every so often unexpected questions. From its identification in the 1990s9,26,104,105 to nowadays, laminin 332 has stimulated incessantly increasing interest in the scientific community due to its multifunctional properties and its involvement in human physiological processes and pathologies.
Acknowledgments
We thank Dr. François Letourneur (CGMC, Lyon) for his help in designing the syndecan-1 GFP constructs. Original work by the authors was financially supported by the Agence Nationale pour la Recherche grants Chemispike and adhesioMab, the Association pour la Recherche sur le Cancer and the Ligue Nationale contre le Cancer. We acknowledge the contribution of the PLATIM platform of SFR Biosciences Gerland-Lyon Sud (UMS344/US8).
Glossary
Abbreviations:
- ECM
extracellular matrix
- LG
laminin globular domain
- pre-laminin
precursor-laminin
- MMP
matrix metalloproteinase
- mTLD
mammalian tolloid
- BMP-1
bone morphogenic protein 1
- JEB
junctional epidermolysis bullosa
- pdb
protein data bank
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/23132
References
- 1.Domogatskaya A, Rodin S, Tryggvason K. Functional diversity of laminins. Annu Rev Cell Dev Biol. 2012;28:523–53. doi: 10.1146/annurev-cellbio-101011-155750. [DOI] [PubMed] [Google Scholar]
- 2.Miner JH, Yurchenco PD. Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol. 2004;20:255–84. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]
- 3.Beck K, Dixon TW, Engel J, Parry DA. Ionic interactions in the coiled-coil domain of laminin determine the specificity of chain assembly. J Mol Biol. 1993;231:311–23. doi: 10.1006/jmbi.1993.1284. [DOI] [PubMed] [Google Scholar]
- 4.Matsui C, Wang CK, Nelson CF, Bauer EA, Hoeffler WK. The assembly of laminin-5 subunits. J Biol Chem. 1995;270:23496–503. doi: 10.1074/jbc.270.40.23496. [DOI] [PubMed] [Google Scholar]
- 5.Durbeej M. Laminins. Cell Tissue Res. 2010;339:259–68. doi: 10.1007/s00441-009-0838-2. [DOI] [PubMed] [Google Scholar]
- 6.Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, et al. A simplified laminin nomenclature. Matrix Biol. 2005;24:326–32. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E. Structure and function of laminin LG modules. Matrix Biol. 2000;19:309–17. doi: 10.1016/S0945-053X(00)00072-X. [DOI] [PubMed] [Google Scholar]
- 8.Ryan MC, Tizard R, VanDevanter DR, Carter WG. Cloning of the LamA3 gene encoding the α 3 chain of the adhesive ligand epiligrin. Expression in wound repair. J Biol Chem. 1994;269:22779–87. [PubMed] [Google Scholar]
- 9.Rousselle P, Lunstrum GP, Keene DR, Burgeson RE. Kalinin: an epithelium-specific basement membrane adhesion molecule that is a component of anchoring filaments. J Cell Biol. 1991;114:567–76. doi: 10.1083/jcb.114.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rousselle P, Keene DR, Ruggiero F, Champliaud MF, Rest M, Burgeson RE. Laminin 5 binds the NC-1 domain of type VII collagen. J Cell Biol. 1997;138:719–28. doi: 10.1083/jcb.138.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marinkovich MP, Lunstrum GP, Burgeson RE. The anchoring filament protein kalinin is synthesized and secreted as a high molecular weight precursor. J Biol Chem. 1992;267:17900–6. [PubMed] [Google Scholar]
- 12.Goldfinger LE, Stack MS, Jones JC. Processing of laminin-5 and its functional consequences: role of plasmin and tissue-type plasminogen activator. J Cell Biol. 1998;141:255–65. doi: 10.1083/jcb.141.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsubota Y, Yasuda C, Kariya Y, Ogawa T, Hirosaki T, Mizushima H, et al. Regulation of biological activity and matrix assembly of laminin-5 by COOH-terminal, LG4-5 domain of α3 chain. J Biol Chem. 2005;280:14370–7. doi: 10.1074/jbc.M413051200. [DOI] [PubMed] [Google Scholar]
- 14.Amano S, Scott IC, Takahara K, Koch M, Champliaud MF, Gerecke DR, et al. Bone morphogenetic protein 1 is an extracellular processing enzyme of the laminin 5 γ 2 chain. J Biol Chem. 2000;275:22728–35. doi: 10.1074/jbc.M002345200. [DOI] [PubMed] [Google Scholar]
- 15.Veitch DP, Nokelainen P, McGowan KA, Nguyen TT, Nguyen NE, Stephenson R, et al. Mammalian tolloid metalloproteinase, and not matrix metalloprotease 2 or membrane type 1 metalloprotease, processes laminin-5 in keratinocytes and skin. J Biol Chem. 2003;278:15661–8. doi: 10.1074/jbc.M210588200. [DOI] [PubMed] [Google Scholar]
- 16.Sigle RO, Gil SG, Bhattacharya M, Ryan MC, Yang TM, Brown TA, et al. Globular domains 4/5 of the laminin α3 chain mediate deposition of precursor laminin 5. J Cell Sci. 2004;117:4481–94. doi: 10.1242/jcs.01310. [DOI] [PubMed] [Google Scholar]
- 17.Tsubota Y, Mizushima H, Hirosaki T, Higashi S, Yasumitsu H, Miyazaki K. Isolation and activity of proteolytic fragment of laminin-5 α3 chain. Biochem Biophys Res Commun. 2000;278:614–20. doi: 10.1006/bbrc.2000.3851. [DOI] [PubMed] [Google Scholar]
- 18.Décline F, Okamoto O, Mallein-Gerin F, Helbert B, Bernaud J, Rigal D, et al. Keratinocyte motility induced by TGF-beta1 is accompanied by dramatic changes in cellular interactions with laminin 5. Cell Motil Cytoskeleton. 2003;54:64–80. doi: 10.1002/cm.10086. [DOI] [PubMed] [Google Scholar]
- 19.Baudoin C, Fantin L, Meneguzzi G. Proteolytic processing of the laminin α3 G domain mediates assembly of hemidesmosomes but has no role on keratinocyte migration. J Invest Dermatol. 2005;125:883–8. doi: 10.1111/j.0022-202X.2005.23881.x. [DOI] [PubMed] [Google Scholar]
- 20.Tisi D, Talts JF, Timpl R, Hohenester E. Structure of the C-terminal laminin G-like domain pair of the laminin α2 chain harbouring binding sites for α-dystroglycan and heparin. EMBO J. 2000;19:1432–40. doi: 10.1093/emboj/19.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Champliaud MF, Lunstrum GP, Rousselle P, Nishiyama T, Keene DR, Burgeson RE. Human amnion contains a novel laminin variant, laminin 7, which like laminin 6, covalently associates with laminin 5 to promote stable epithelial-stromal attachment. J Cell Biol. 1996;132:1189–98. doi: 10.1083/jcb.132.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean WH, Irvine AD, Hamill KJ, Whittock NV, Coleman-Campbell CM, Mellerio JE, et al. An unusual N-terminal deletion of the laminin α3a isoform leads to the chronic granulation tissue disorder laryngo-onycho-cutaneous syndrome. Hum Mol Genet. 2003;12:2395–409. doi: 10.1093/hmg/ddg234. [DOI] [PubMed] [Google Scholar]
- 23.Vailly J, Verrando P, Champliaud MF, Gerecke D, Wagman DW, Baudoin C, et al. The 100-kDa chain of nicein/kalinin is a laminin B2 chain variant. Eur J Biochem. 1994;219:209–18. doi: 10.1111/j.1432-1033.1994.tb19932.x. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki T, Göhring W, Mann K, Brakebusch C, Yamada Y, Fässler R, et al. Short arm region of laminin-5 γ2 chain: structure, mechanism of processing and binding to heparin and proteins. J Mol Biol. 2001;314:751–63. doi: 10.1006/jmbi.2001.5176. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki N, Labosky PA, Furuta Y, Hargett L, Dunn R, Fogo AB, et al. Failure of ventral body wall closure in mouse embryos lacking a procollagen C-proteinase encoded by Bmp1, a mammalian gene related to Drosophila tolloid. Development. 1996;122:3587–95. doi: 10.1242/dev.122.11.3587. [DOI] [PubMed] [Google Scholar]
- 26.Carter WG, Ryan MC, Gahr PJ. Epiligrin, a new cell adhesion ligand for integrin α 3 β 1 in epithelial basement membranes. Cell. 1991;65:599–610. doi: 10.1016/0092-8674(91)90092-D. [DOI] [PubMed] [Google Scholar]
- 27.Sonnenberg A, de Melker AA, Martinez de Velasco AM, Janssen H, Calafat J, Niessen CM. Formation of hemidesmosomes in cells of a transformed murine mammary tumor cell line and mechanisms involved in adherence of these cells to laminin and kalinin. J Cell Sci. 1993;106:1083–102. doi: 10.1242/jcs.106.4.1083. [DOI] [PubMed] [Google Scholar]
- 28.Rousselle P, Aumailley M. Kalinin is more efficient than laminin in promoting adhesion of primary keratinocytes and some other epithelial cells and has a different requirement for integrin receptors. J Cell Biol. 1994;125:205–14. doi: 10.1083/jcb.125.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima H, Takamura H, Miyagi Y, Kikkawa Y, Yamanaka N, Yasumitsu H, et al. Identification of integrin-dependent and -independent cell adhesion domains in COOH-terminal globular region of laminin-5 α 3 chain. Cell Growth Differ. 1997;8:979–87. [PubMed] [Google Scholar]
- 30.Cheng YS, Champliaud MF, Burgeson RE, Marinkovich MP, Yurchenco PD. Self-assembly of laminin isoforms. J Biol Chem. 1997;272:31525–32. doi: 10.1074/jbc.272.50.31525. [DOI] [PubMed] [Google Scholar]
- 31.Burgeson RE. Type VII collagen, anchoring fibrils, and epidermolysis bullosa. J Invest Dermatol. 1993;101:252–5. doi: 10.1111/1523-1747.ep12365129. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Marinkovich MP, Veis A, Cai X, Rao CN, O’Toole EA, et al. Interactions of the amino-terminal noncollagenous (NC1) domain of type VII collagen with extracellular matrix components. A potential role in epidermal-dermal adherence in human skin. J Biol Chem. 1997;272:14516–22. doi: 10.1074/jbc.272.23.14516. [DOI] [PubMed] [Google Scholar]
- 33.Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–8. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- 34.Koshikawa N, Giannelli G, Cirulli V, Miyazaki K, Quaranta V. Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J Cell Biol. 2000;148:615–24. doi: 10.1083/jcb.148.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guess CM, Quaranta V. Defining the role of laminin-332 in carcinoma. Matrix Biol. 2009;28:445–55. doi: 10.1016/j.matbio.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuruta D, Kobayashi H, Imanishi H, Sugawara K, Ishii M, Jones JC. Laminin-332-integrin interaction: a target for cancer therapy? Curr Med Chem. 2008;15:1968–75. doi: 10.2174/092986708785132834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirilä E, Sharabi A, Salo T, Quaranta V, Tu H, Heljasvaara R, et al. Matrix metalloproteinases process the laminin-5 γ 2-chain and regulate epithelial cell migration. Biochem Biophys Res Commun. 2003;303:1012–7. doi: 10.1016/S0006-291X(03)00452-2. [DOI] [PubMed] [Google Scholar]
- 38.Sadowski T, Dietrich S, Koschinsky F, Ludwig A, Proksch E, Titz B, et al. Matrix metalloproteinase 19 processes the laminin 5 γ 2 chain and induces epithelial cell migration. Cell Mol Life Sci. 2005;62:870–80. doi: 10.1007/s00018-005-4478-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schenk S, Hintermann E, Bilban M, Koshikawa N, Hojilla C, Khokha R, et al. Binding to EGF receptor of a laminin-5 EGF-like fragment liberated during MMP-dependent mammary gland involution. J Cell Biol. 2003;161:197–209. doi: 10.1083/jcb.200208145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Udayakumar TS, Chen ML, Bair EL, Von Bredow DC, Cress AE, Nagle RB, et al. Membrane type-1-matrix metalloproteinase expressed by prostate carcinoma cells cleaves human laminin-5 β3 chain and induces cell migration. Cancer Res. 2003;63:2292–9. [PubMed] [Google Scholar]
- 41.Remy L, Trespeuch C, Bachy S, Scoazec JY, Rousselle P. Matrilysin 1 influences colon carcinoma cell migration by cleavage of the laminin-5 β3 chain. Cancer Res. 2006;66:11228–37. doi: 10.1158/0008-5472.CAN-06-1187. [DOI] [PubMed] [Google Scholar]
- 42.Tripathi M, Potdar AA, Yamashita H, Weidow B, Cummings PT, Kirchhofer D, et al. Laminin-332 cleavage by matriptase alters motility parameters of prostate cancer cells. Prostate. 2011;71:184–96. doi: 10.1002/pros.21233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marinkovich MP. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nat Rev Cancer. 2007;7:370–80. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- 44.Rousselle P, Golbik R, van der Rest M, Aumailley M. Structural requirement for cell adhesion to kalinin (laminin-5) J Biol Chem. 1995;270:13766–70. doi: 10.1074/jbc.270.23.13766. [DOI] [PubMed] [Google Scholar]
- 45.Ott U, Odermatt E, Engel J, Furthmayr H, Timpl R. Protease resistance and conformation of laminin. Eur J Biochem. 1982;123:63–72. doi: 10.1111/j.1432-1033.1982.tb06499.x. [DOI] [PubMed] [Google Scholar]
- 46.Carafoli F, Hussain SA, Hohenester E. Crystal structures of the network-forming short-arm tips of the laminin β1 and γ1 chains. PLoS One. 2012;7:e42473. doi: 10.1371/journal.pone.0042473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos-Valle P, Guijarro-Muñoz I, Cuesta AM, Alonso-Camino V, Villate M, Alvarez-Cienfuegos A, et al. The heterotrimeric laminin coiled-coil domain exerts anti-adhesive effects and induces a pro-invasive phenotype. PLoS One. 2012;7:e39097. doi: 10.1371/journal.pone.0039097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulsson M, Deutzmann R, Timpl R, Dalzoppo D, Odermatt E, Engel J. Evidence for coiled-coil α-helical regions in the long arm of laminin. EMBO J. 1985;4:309–16. doi: 10.1002/j.1460-2075.1985.tb03630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carafoli F, Clout NJ, Hohenester E. Crystal structure of the LG1-3 region of the laminin α2 chain. J Biol Chem. 2009;284:22786–92. doi: 10.1074/jbc.M109.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison D, Hussain SA, Combs AC, Ervasti JM, Yurchenco PD, Hohenester E. Crystal structure and cell surface anchorage sites of laminin α1LG4-5. J Biol Chem. 2007;282:11573–81. doi: 10.1074/jbc.M610657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wizemann H, Garbe JH, Friedrich MV, Timpl R, Sasaki T, Hohenester E. Distinct requirements for heparin and α-dystroglycan binding revealed by structure-based mutagenesis of the laminin α2 LG4-LG5 domain pair. J Mol Biol. 2003;332:635–42. doi: 10.1016/S0022-2836(03)00848-9. [DOI] [PubMed] [Google Scholar]
- 52.Hohenester E, Tisi D, Talts JF, Timpl R. The crystal structure of a laminin G-like module reveals the molecular basis of α-dystroglycan binding to laminins, perlecan, and agrin. Mol Cell. 1999;4:783–92. doi: 10.1016/S1097-2765(00)80388-3. [DOI] [PubMed] [Google Scholar]
- 53.Beck K, Hunter I, Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J. 1990;4:148–60. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- 54.Navdaev A, Heitmann V, Desantana Evangelista K, Mörgelin M, Wegener J, Eble JA. The C-terminus of the γ 2 chain but not of the β 3 chain of laminin-332 is indirectly but indispensably necessary for integrin-mediated cell reactions. Exp Cell Res. 2008;314:489–97. doi: 10.1016/j.yexcr.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 55.Ido H, Nakamura A, Kobayashi R, Ito S, Li S, Futaki S, et al. The requirement of the glutamic acid residue at the third position from the carboxyl termini of the laminin γ chains in integrin binding by laminins. J Biol Chem. 2007;282:11144–54. doi: 10.1074/jbc.M609402200. [DOI] [PubMed] [Google Scholar]
- 56.Künneken K, Pohlentz G, Schmidt-Hederich A, Odenthal U, Smyth N, Peter-Katalinic J, et al. Recombinant human laminin-5 domains. Effects of heterotrimerization, proteolytic processing, and N-glycosylation on α3β1 integrin binding. J Biol Chem. 2004;279:5184–93. doi: 10.1074/jbc.M310424200. [DOI] [PubMed] [Google Scholar]
- 57.Décline F, Okamoto O, Mallein-Gerin F, Helbert B, Bernaud J, Rigal D, et al. Keratinocyte motility induced by TGF-β1 is accompanied by dramatic changes in cellular interactions with laminin 5. Cell Motil Cytoskeleton. 2003;54:64–80. doi: 10.1002/cm.10086. [DOI] [PubMed] [Google Scholar]
- 58.Kim JM, Park WH, Min BM. The PPFLMLLKGSTR motif in globular domain 3 of the human laminin-5 α3 chain is crucial for integrin α3β1 binding and cell adhesion. Exp Cell Res. 2005;304:317–27. doi: 10.1016/j.yexcr.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 59.Carulli S, Beck K, Dayan G, Boulesteix S, Lortat-Jacob H, Rousselle P. Cell surface proteoglycans syndecan-1 and -4 bind overlapping but distinct sites in laminin α3 LG45 protein domain. J Biol Chem. 2012;287:12204–16. doi: 10.1074/jbc.M111.300061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulloy B, Forster MJ, Jones C, Davies DB. N.m.r. and molecular-modelling studies of the solution conformation of heparin. Biochem J. 1993;293:849–58. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lampe PD, Nguyen BP, Gil S, Usui M, Olerud J, Takada Y, et al. Cellular interaction of integrin α3β1 with laminin 5 promotes gap junctional communication. J Cell Biol. 1998;143:1735–47. doi: 10.1083/jcb.143.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldfinger LE, Hopkinson SB, deHart GW, Collawn S, Couchman JR, Jones JC. The α3 laminin subunit, α6β4 and α3β1 integrin coordinately regulate wound healing in cultured epithelial cells and in the skin. J Cell Sci. 1999;112:2615–29. doi: 10.1242/jcs.112.16.2615. [DOI] [PubMed] [Google Scholar]
- 63.Tunggal L, Ravaux J, Pesch M, Smola H, Krieg T, Gaill F, et al. Defective laminin 5 processing in cylindroma cells. Am J Pathol. 2002;160:459–68. doi: 10.1016/S0002-9440(10)64865-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryan MC, Lee K, Miyashita Y, Carter WG. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999;145:1309–23. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hintermann E, Quaranta V. Epithelial cell motility on laminin-5: regulation by matrix assembly, proteolysis, integrins and erbB receptors. Matrix Biol. 2004;23:75–85. doi: 10.1016/j.matbio.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 66.Frank DE, Carter WG. Laminin 5 deposition regulates keratinocyte polarization and persistent migration. J Cell Sci. 2004;117:1351–63. doi: 10.1242/jcs.01003. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen BP, Ryan MC, Gil SG, Carter WG. Deposition of laminin 5 in epidermal wounds regulates integrin signaling and adhesion. Curr Opin Cell Biol. 2000;12:554–62. doi: 10.1016/S0955-0674(00)00131-9. [DOI] [PubMed] [Google Scholar]
- 68.Decline F, Rousselle P. Keratinocyte migration requires α2β1 integrin-mediated interaction with the laminin 5 γ2 chain. J Cell Sci. 2001;114:811–23. doi: 10.1242/jcs.114.4.811. [DOI] [PubMed] [Google Scholar]
- 69.Kainulainen T, Hakkinen L, Hamidi S, Larjava K, Kallioinen M, Peltonen J, et al. Laminin-5 expression is independent of the injury and the microenvironment during reepithelialization of wounds. J Histochem Cytochem. 1998;46:353–60. doi: 10.1177/002215549804600309. [DOI] [PubMed] [Google Scholar]
- 70.Tran M, Rousselle P, Nokelainen P, Tallapragada S, Nguyen NT, Fincher EF, et al. Targeting a tumor-specific laminin domain critical for human carcinogenesis. Cancer Res. 2008;68:2885–94. doi: 10.1158/0008-5472.CAN-07-6160. [DOI] [PubMed] [Google Scholar]
- 71.Scaturro M, Posteraro P, Mastrogiacomo A, Zaccaria ML, De Luca N, Mazzanti C, et al. A missense mutation (G1506E) in the adhesion G domain of laminin-5 causes mild junctional epidermolysis bullosa. Biochem Biophys Res Commun. 2003;309:96–103. doi: 10.1016/S0006-291X(03)01533-X. [DOI] [PubMed] [Google Scholar]
- 72.Hirosaki T, Tsubota Y, Kariya Y, Moriyama K, Mizushima H, Miyazaki K. Laminin-6 is activated by proteolytic processing and regulates cellular adhesion and migration differently from laminin-5. J Biol Chem. 2002;277:49287–95. doi: 10.1074/jbc.M111096200. [DOI] [PubMed] [Google Scholar]
- 73.Gagnoux-Palacios L, Allegra M, Spirito F, Pommeret O, Romero C, Ortonne JP, et al. The short arm of the laminin γ2 chain plays a pivotal role in the incorporation of laminin 5 into the extracellular matrix and in cell adhesion. J Cell Biol. 2001;153:835–50. doi: 10.1083/jcb.153.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones JC, Lane K, Hopkinson SB, Lecuona E, Geiger RC, Dean DA, et al. Laminin-6 assembles into multimolecular fibrillar complexes with perlecan and participates in mechanical-signal transduction via a dystroglycan-dependent, integrin-independent mechanism. J Cell Sci. 2005;118:2557–66. doi: 10.1242/jcs.02395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamill KJ, Kligys K, Hopkinson SB, Jones JC. Laminin deposition in the extracellular matrix: a complex picture emerges. J Cell Sci. 2009;122:4409–17. doi: 10.1242/jcs.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bachy S, Letourneur F, Rousselle P. Syndecan-1 interaction with the LG4/5 domain in laminin-332 is essential for keratinocyte migration. J Cell Physiol. 2008;214:238–49. doi: 10.1002/jcp.21184. [DOI] [PubMed] [Google Scholar]
- 77.Carter WG, Kaur P, Gil SG, Gahr PJ, Wayner EA. Distinct functions for integrins α 3 β 1 in focal adhesions and α 6 β 4/bullous pemphigoid antigen in a new stable anchoring contact (SAC) of keratinocytes: relation to hemidesmosomes. J Cell Biol. 1990;111:3141–54. doi: 10.1083/jcb.111.6.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Momota Y, Suzuki N, Kasuya Y, Kobayashi T, Mizoguchi M, Yokoyama F, et al. Laminin α3 LG4 module induces keratinocyte migration: involvement of matrix metalloproteinase-9. J Recept Signal Transduct Res. 2005;25:1–17. doi: 10.1081/RRS-200047870. [DOI] [PubMed] [Google Scholar]
- 79.Utani A, Momota Y, Endo H, Kasuya Y, Beck K, Suzuki N, et al. Laminin α 3 LG4 module induces matrix metalloproteinase-1 through mitogen-activated protein kinase signaling. J Biol Chem. 2003;278:34483–90. doi: 10.1074/jbc.M304827200. [DOI] [PubMed] [Google Scholar]
- 80.Hoffman MP, Engbring JA, Nielsen PK, Vargas J, Steinberg Z, Karmand AJ, et al. Cell type-specific differences in glycosaminoglycans modulate the biological activity of a heparin-binding peptide (RKRLQVQLSIRT) from the G domain of the laminin α1 chain. J Biol Chem. 2001;276:22077–85. doi: 10.1074/jbc.M100774200. [DOI] [PubMed] [Google Scholar]
- 81.Urushibata S, Katagiri F, Takaki S, Yamada Y, Fujimori C, Hozumi K, et al. Biologically active sequences in the mouse laminin α3 chain G domain. Biochemistry. 2009;48:10522–32. doi: 10.1021/bi901421t. [DOI] [PubMed] [Google Scholar]
- 82.Utani A, Nomizu M, Matsuura H, Kato K, Kobayashi T, Takeda U, et al. A unique sequence of the laminin α 3 G domain binds to heparin and promotes cell adhesion through syndecan-2 and -4. J Biol Chem. 2001;276:28779–88. doi: 10.1074/jbc.M101420200. [DOI] [PubMed] [Google Scholar]
- 83.Kato K, Utani A, Suzuki N, Mochizuki M, Yamada M, Nishi N, et al. Identification of neurite outgrowth promoting sites on the laminin α 3 chain G domain. Biochemistry. 2002;41:10747–53. doi: 10.1021/bi020180k. [DOI] [PubMed] [Google Scholar]
- 84.Araki E, Momota Y, Togo T, Tanioka M, Hozumi K, Nomizu M, et al. Clustering of syndecan-4 and integrin β1 by laminin α 3 chain-derived peptide promotes keratinocyte migration. Mol Biol Cell. 2009;20:3012–24. doi: 10.1091/mbc.E08-09-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vivès RR, Crublet E, Andrieu JP, Gagnon J, Rousselle P, Lortat-Jacob H. A novel strategy for defining critical amino acid residues involved in protein/glycosaminoglycan interactions. J Biol Chem. 2004;279:54327–33. doi: 10.1074/jbc.M409760200. [DOI] [PubMed] [Google Scholar]
- 86.Okamoto O, Bachy S, Odenthal U, Bernaud J, Rigal D, Lortat-Jacob H, et al. Normal human keratinocytes bind to the α3LG4/5 domain of unprocessed laminin-5 through the receptor syndecan-1. J Biol Chem. 2003;278:44168–77. doi: 10.1074/jbc.M300726200. [DOI] [PubMed] [Google Scholar]
- 87.Sulka B, Lortat-Jacob H, Terreux R, Letourneur F, Rousselle P. Tyrosine dephosphorylation of the syndecan-1 PDZ binding domain regulates syntenin-1 recruitment. J Biol Chem. 2009;284:10659–71. doi: 10.1074/jbc.M807643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rousselle P, Letourneur F. Mysterious tasks of tyrosines in syndecan-1 cytoplasmic tail. ScientificWorldJournal. 2009;9:629–32. doi: 10.1100/tsw.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elenius K, Vainio S, Laato M, Salmivirta M, Thesleff I, Jalkanen M. Induced expression of syndecan in healing wounds. J Cell Biol. 1991;114:585–95. doi: 10.1083/jcb.114.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oksala O, Salo T, Tammi R, Häkkinen L, Jalkanen M, Inki P, et al. Expression of proteoglycans and hyaluronan during wound healing. J Histochem Cytochem. 1995;43:125–35. doi: 10.1177/43.2.7529785. [DOI] [PubMed] [Google Scholar]
- 91.Jaakkola P, Kontusaari S, Kauppi T, Määtä A, Jalkanen M. Wound reepithelialization activates a growth factor-responsive enhancer in migrating keratinocytes. FASEB J. 1998;12:959–69. doi: 10.1096/fasebj.12.11.959. [DOI] [PubMed] [Google Scholar]
- 92.Gallo R, Kim C, Kokenyesi R, Adzick NS, Bernfield M. Syndecans-1 and -4 are induced during wound repair of neonatal but not fetal skin. J Invest Dermatol. 1996;107:676–83. doi: 10.1111/1523-1747.ep12365571. [DOI] [PubMed] [Google Scholar]
- 93.Stepp MA, Gibson HE, Gala PH, Iglesia DD, Pajoohesh-Ganji A, Pal-Ghosh S, et al. Defects in keratinocyte activation during wound healing in the syndecan-1-deficient mouse. J Cell Sci. 2002;115:4517–31. doi: 10.1242/jcs.00128. [DOI] [PubMed] [Google Scholar]
- 94.Wang H, Leavitt L, Ramaswamy R, Rapraeger AC. Interaction of syndecan and α6β4 integrin cytoplasmic domains: regulation of ErbB2-mediated integrin activation. J Biol Chem. 2010;285:13569–79. doi: 10.1074/jbc.M110.102137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rousselle P, Keene DR, Ruggiero F, Champliaud MF, Rest M, Burgeson RE. Laminin 5 binds the NC-1 domain of type VII collagen. J Cell Biol. 1997;138:719–28. doi: 10.1083/jcb.138.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bruckner-Tuderman L, Pfaltz M, Schnyder UW. Cylindroma overexpresses collagen VII, the major anchoring fibril protein. J Invest Dermatol. 1991;96:729–34. doi: 10.1111/1523-1747.ep12470965. [DOI] [PubMed] [Google Scholar]
- 97.Ogawa T, Tsubota Y, Hashimoto J, Kariya Y, Miyazaki K. The short arm of laminin gamma2 chain of laminin-5 (laminin-332) binds syndecan-1 and regulates cellular adhesion and migration by suppressing phosphorylation of integrin β4 chain. Mol Biol Cell. 2007;18:1621–33. doi: 10.1091/mbc.E06-09-0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Natarajan E, Saeb M, Crum CP, Woo SB, McKee PH, Rheinwald JG. Co-expression of p16(INK4A) and laminin 5 gamma2 by microinvasive and superficial squamous cell carcinomas in vivo and by migrating wound and senescent keratinocytes in culture. Am J Pathol. 2003;163:477–91. doi: 10.1016/S0002-9440(10)63677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Natarajan E, Omobono JD, 2nd, Guo Z, Hopkinson S, Lazar AJ, Brenn T, et al. A keratinocyte hypermotility/growth-arrest response involving laminin 5 and p16INK4A activated in wound healing and senescence. Am J Pathol. 2006;168:1821–37. doi: 10.2353/ajpath.2006.051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stepp MA, Liu Y, Pal-Ghosh S, Jurjus RA, Tadvalkar G, Sekaran A, et al. Reduced migration, altered matrix and enhanced TGFbeta1 signaling are signatures of mouse keratinocytes lacking Sdc1. J Cell Sci. 2007;120:2851–63. doi: 10.1242/jcs.03480. [DOI] [PubMed] [Google Scholar]
- 101.Echtermeyer F, Streit M, Wilcox-Adelman S, Saoncella S, Denhez F, Detmar M, et al. Delayed wound repair and impaired angiogenesis in mice lacking syndecan-4. J Clin Invest. 2001;107:R9–14. doi: 10.1172/JCI10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Botta A, Delteil F, Mettouchi A, Vieira A, Estrach S, Négroni L, et al. Confluence switch signaling regulates ECM composition and the plasmin proteolytic cascade in keratinocytes. J Cell Sci. 2012;125:4241–52. doi: 10.1242/jcs.096289. [DOI] [PubMed] [Google Scholar]
- 103.Costell M, Gustafsson E, Aszódi A, Mörgelin M, Bloch W, Hunziker E, et al. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol. 1999;147:1109–22. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Verrando P, Blanchet-Bardon C, Pisani A, Thomas L, Cambazard F, Eady RA, et al. Monoclonal antibody GB3 defines a widespread defect of several basement membranes and a keratinocyte dysfunction in patients with lethal junctional epidermolysis bullosa. Lab Invest. 1991;64:85–92. [PubMed] [Google Scholar]
- 105.Miyazaki K, Kikkawa Y, Nakamura A, Yasumitsu H, Umeda M. A large cell-adhesive scatter factor secreted by human gastric carcinoma cells. Proc Natl Acad Sci U S A. 1993;90:11767–71. doi: 10.1073/pnas.90.24.11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 107.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Web Server issue):W363-7. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]


