Abstract
In plant development, cell-to-cell signaling is mediated by mobile signals, including transcription factors and small RNA molecules. This communication is essential for growth and patterning. Short-range movement of signals occurs in the extracellular space via the apoplastic pathway or directly from cell-to-cell via the symplastic pathway. Symplastic transport is mediated by plant specific structures called plasmodesmata, which are plasma membrane-lined pores that traverse the cell walls of adjacent cells thus connecting their cytoplasms. However, a thorough understanding of molecules moving via plasmodesmata and regulatory networks relying on symplastic signaling is lacking. Traffic via plasmodesmata is highly regulated, and callose turnover is known to be one mechanism. In Arabidopsis, plasmodesmata apertures can be regulated in a spatially and temporally specific manner with the icals3m, an inducible vector system expressing the mutated CalS3 gene encoding a plasmodesmata localized callose synthase that increases callose deposition at plasmodesmata. We discuss strategies to use the icals3m system for global analyses on symplastic signaling in plants.
Keywords: symplastic communication, mobile signal, Arabidopsis, root development, plasmodesmata, callose
Positional Information Determines Cell Fate in Plants
Continuous growth and development is an important characteristic of plants. As plants are sessile, continuous plastic growth is a mechanism of adaptation to environmental conditions and maintaining fitness. The root system of Arabidopsis consists of a primary root formed during embryogenesis and several orders of postembryonic lateral roots originating from the primary root. In the root apical meristem there is an area called the stem cell niche, where self-renewing stem cells called initials surround the quiescent center (QC), a small group of mitotically less active organizing cells necessary for the maintenance of the initials.1 The initials divide asymmetrically to produce a new initial that remains next to the QC and another daughter cell that eventually differentiates. The daughters then undergo divisions in the meristematic zone located above the stem cell niche to increase their number. After the meristem, the cells enter the elongation zone, where divisions cease and strong expansion begins. Finally, in the differentiation zone, cells acquire their specific fates. Since plant cells do not move, the daughters of the initials are organized in cell files that form lineages where mature cells are located further from the tip. Thus, the age of a cell can be determined by its position along the longitudinal axis. The final radial organization of the root consists of concentric layers of distinct tissues along the radial axis (Fig. 1). The vasculature with its conductive tissues xylem and phloem and intervening procambium are located in the center of the root. The xylem axis develops bilaterally differentiating into central metaxylem with pitted cell walls and peripheral protoxylem with spiral cell walls. Surrounding the vasculature is the pericycle, the site for lateral root initiation. The vasculature and pericycle, together forming the stele, are surrounded by two layers of ground-tissue, a single layer of endodermis and cortex. The outermost layer is the epidermis with alternating hair cells and non-hair cells.2
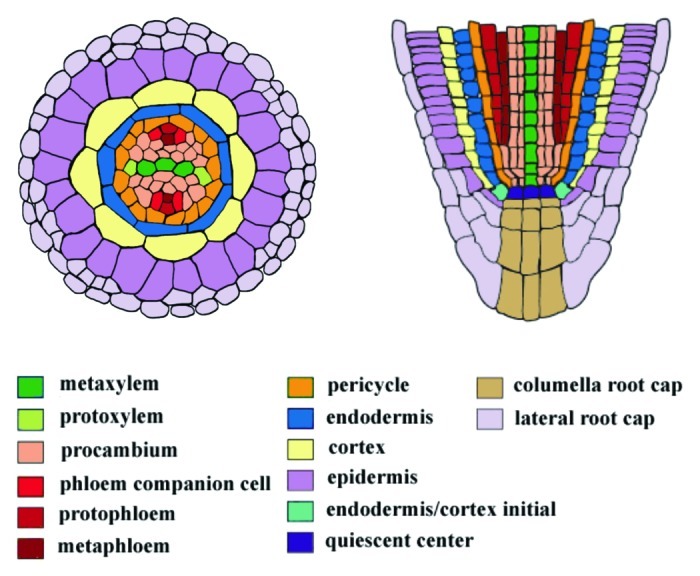
Figure 1. Cell types in the Arabidopsis root. Radial (left) and longitudinal (right) sections of the Arabidopsis root tip. Cell types are marked by different colors. Modified from Miyashima et al.9 and Carlsbecker et al.8
In plants, the fate of cells is largely determined by their spatial context rather than by their cell lineages. Genetic studies and the removal of QC cells by laser ablation first showed that the maintenance of the initials in an undifferentiated state depends on signals from the QC cells.3,4 Furthermore, ablation of cortex/endodermal initials has shown that adjacent cells can replace their function.5 Thus, positional information that is exchanged between cells predominantly coordinates pattern formation during development and underlies the flexibility of plant growth. Cell-to-cell communication is mediated by moving of regulatory molecules. Mobile signals include phytohormones, transcription factors and small RNAs, which vary in their mechanisms of action and the developmental outcomes they produce.
Bidirectional Signaling Coordinates Radial Development
In Arabidopsis root, a bidirectional signaling pathway was recently identified operating between the stele and endodermis that directs their patterning (Fig. 2). A GRAS family transcription factor SHORTROOT (SHR) is transcribed in the stele and moves from the stele to the endodermis to activate the transcription of SCARECROW (SCR).6,7 Carlsbecker et al.8 demonstrated that SHR and SCR located in the endodermis are needed for correct specification of protoxylem and metaxylem. SHR, together with SCR, activates transcription of MIR165A and MIR166B in the endodermis, and the miRNAs move radially to the stele periphery to cleave the mRNAs of PHABULOSA (PHB), a Class III homeodomain leucine zipper (HD-ZIP III) TF, and other members of the family, resulting in protoxylem differentiation. shr and scr mutants have abnormal xylem patterning in which metaxylem is formed in the protoxylem position. phb-7d gain of function mutant with a point mutation in the miR165/166 target site forms similarly ectopic metaxylem. Conversely, inducing MIR165A expression in the stele reduces PHB expression and leads to ectopic protoxylem development.8 Miyashima et al.9 further identified miR166A as a third miRNA expressed in the endodermis in a SCR-dependent manner, and that MIR166A and MIR166B are expressed in the QC in addition to the endodermis. Importantly, the authors also demonstrated that miR165 restricts the PHB expression domain and xylem differentiation in a dose-dependent manner. The miRNAs are additionally required for proper ground-tissue and pericycle development. miRNA-dependent restriction of PHB expression is required for the proper expression of a C2H2 zinc finger protein JACKDAW (JKD) in the ground-tissue, which is necessary to restrict SHR movement. In phb-1d roots with ectopic PHB expression SHR movement is not inhibited by JKD, leading to the development of an extra cortex layer. In addition, PHB restriction mediated by the miRNAs is necessary for pericycle differentiation, as in both phb-1d and scr-3 the expression of two pericycle identity markers, AHP6 and SKOR, is reduced in the pericycle domain.9 Thus, stele and the ground-tissue exchange information via mobile SHR and miR165/6 species to coordinate each other’s development.
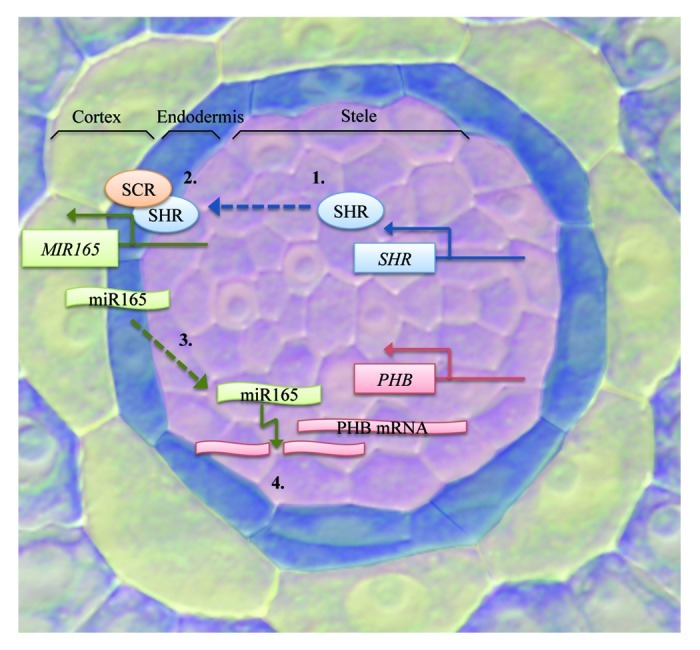
Figure 2. Bidirectional signaling pathway between the stele and the ground-tissue. SHR transcription occurs in the stele (1), after which the protein moves outwards from the stele to the endodermis to activate miRNA transcription together with SCR (2). The miRNAs move to the stele periphery (3) and downregulate PHB expression by targeting its mRNA transcripts for cleavage (4). Stele is highlighted in pink, endodermis in blue and cortex in yellow.
Transport Pathways
Plants have specific mechanisms to exchange information between cells. Long-distance transport occurs via the two conductive tissues, xylem and phloem, whereas for short-range movement of molecules plants use apoplastic/transcellular and symplastic pathways. In apoplastic transport, a molecule is secreted to the extracellular space between cell walls, called the apoplast, where it moves without entering the target cell. In the transcellular transport, molecules moving in the apoplast are transported into the target cells through the plasma membrane by various mechanisms. Polar auxin transport is an example of a trans-cellular transport pathway, where auxin is polarly transported by efflux and influx carriers.10 The symplastic transport is a unique pathway found only in plants mediated by plasmodesmata (PD), plant-specific structures that enable direct communication between cells (Fig. 3). PD are plasma membrane-lined pores that provide cytoplasmic connection by traversing cell walls of adjacent cells. Inside the pore runs a desmotubule which is a tube of tightly packed endoplasmic reticulum (ER). The intervening space between the desmotubule and the plasma membrane, called the cytoplasmic sleeve, contains proteins that are attached to both membranes. These proteins divide the passage into microchannels that are thought to control the movement of molecules.11-13 PD build a cytoplasmic network throughout the plant body that connects most cell types. PD not only provide a direct route from cell-to-cell, which overcomes the limitations of the rigid cell walls surrounding all plant cells, but also integrate local movement of molecules with long distance transport by functioning in the loading and unloading of phloem. Water and solutes and larger molecules including proteins and RNA move via PD. In addition to endogenous molecules, viruses and some other pathogens have evolved to use PD as their pathway to spread inside plants.11-13
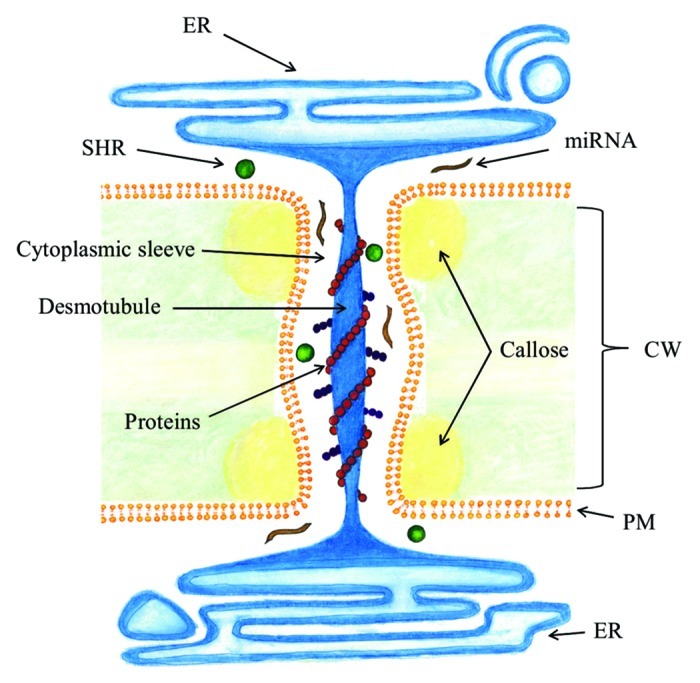
Figure 3. Plasmodesmata. Plasmodesmata connect cytoplasms of adjacent cells by traversing the cell wall. Appressed endoplasmic reticulum, called the desmotubule, runs through the plasma membrane-lined pore. Molecules move via the cytoplasmic sleeve between the desmotubule and plasma membrane. Callose is deposited at the neck region in the cell wall. CW, cell wall; PM, plasma membrane; ER, endoplasmic reticulum.
The frequency of PD between cells and their permeability is controlled throughout development in a dynamic manner. PD apertures can alternate between closed, open and dilated states. In the closed state, PD are completely sealed from all traffic. In the open state, small molecules less than 1 kDa can diffuse through, whereas in the dilated state, diffusion of larger molecules is possible. The extent of the dilation determines the size exclusion limit (SEL), which is the upper size limit of molecules that can pass through PD. The SEL varies between 30–50 kDa in most growing tissues.14,15
Callose Regulates the SEL of PD
PD are structurally complex and their molecular composition as well as mechanisms regulating their permeability are not well known, since their purification is very difficult and mutants with altered PD structure/transport are often lethal.16,17 However, recent studies have increased this understanding. Regulation of molecular traffic through PD has been associated with modifications in PD structure and callose deposition. Callose is a b-1,3 glucan polymer synthesized from UDP-glucose, which is involved in various biological processes in plants. Callose is synthesized in a number of locations, including cell plates of dividing cells, phloem sieve plates and at PD, and is deposited during normal growth and development, as well as in response to various mechanical and physiological stresses, such as wounding, chemical exposure and pathogen invasion.18-20 Callose turnover at PD is a key component in controlling PD permeability. Its deposition at PD is thought to physically constrict the aperture, which reduces the SEL, and in some cases blocks it altogether from traffic. The deposition and turnover of callose are very dynamic, and controlled by the joint action of b-1,3-glucanase and b-1,3-glucan synthase, which degrade and synthesize callose, respectively.21,22
There are 12 callose synthase (CALS) genes encoded in the Arabidopsis genome.23 CALS genes code for large plasma membrane localized proteins, several of which are known to produce callose in specific physiological and developmental processes. The most recent finding is CALS3 (At5g13000) that participates in the biosynthesis of callose in the cell wall surrounding PD. CALS3 is strongly expressed in the stele and in and around the stem cell niche. cals3-1d and cals3-2d are allelic gain-of-function mutations in the CALS3 gene that were identified in a genetic screen for altered vascular patterning. Their phenotype is strikingly similar to that of phb-7d and other mutants with defects in the bidirectional signaling pathway mediated by SHR and miR165/166. cals3-d mutations display ectopic metaxylem formation in the root and are shorter than wild-type roots, similar to shr and phb-7d. Also, those mutants were found to have impaired phloem unloading as GFP driven by a companion cell specific promoter SUC2 diffused in more restricted manner compared with wild-type roots. These results suggested that the SEL of PD is altered in these mutants. Furthermore, the cals3-d mutations lead to reversible overproduction of callose at PD, which is thought to be caused by altered activity of the CALS3 protein. Root development in cals3-1d mutants was partially rescued by driving a β-1,3-glucanase from the CALS3 promoter.24
icals3m: A Tool to Regulate Symplastic Trafficking
Based on the discovery of cals3-d mutants, we have designed a tool to study symplastic signaling networks by regulating PD permeability in a spatially and temporally specific manner.24 icals3m is a tissue-specific inducible vector system that enables us to overexpress mutated cals3 (cals3m) gene that contains both cals3-1d and cals3-2d mutations. When the transgene is activated, callose production is enhanced in tissue specific manner, leading to reduced PD apertures. For inducible expression, icals3m is controlled by the XVE system activated only with estradiol, enabling the expression of the transgenes to be regulated temporally.25
The icals3m system when expressed in the vasculature or QC is capable of blocking SHR movement, confirming that the protein moves via PD, and that protein traffic from cell-to-cell can be regulated with icals3m.24 Moreover, microRNA movement was shown to be inhibited by icals3m. The mobility of miRNAs was investigated by combining two genetic tools, icals3m and nlsYFP_targetMIR165mu, a nuclear localized YFP marker targeted by mutated version of miR165 (miR165mu) (Fig. 4). First, UAS::icals3m construct was introduced into the ground-tissue specific enhancer trap line (J0571) established by J. Haseloff (www.plantsci.cam.ac.uk/Haseloff), and previously used by Carlsbecker et al.8 The UAS promoter is activated by GAL4-VP16 protein transcribed in the ground-tissue of this enhancer-trap line. In addition to activating icals3m, the line contains GFP marker (Fig. 4A). pSPR1::nlsYFP_targetMIR165mu/UAS::MIR165mu was then introduced into J0571; UAS::icals3m (Fig. 4B). MIR165Amu was placed under the UAS promoter (UAS::165Amu) to express it in the ground-tissue, whereas the coding sequence of miR165mu-targeted nuclear-localized YFP was driven by the broadly-expressed SPIRAL1 (SPR1) promoter,26 (pSPR1::nlsYFP_165mu_tgt) to express it throughout the root.
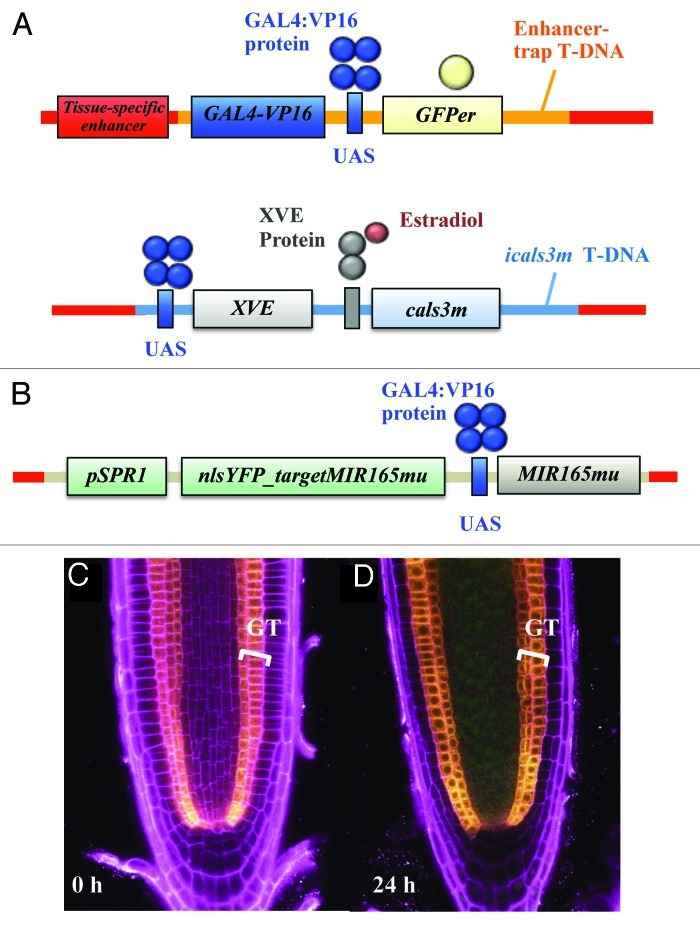
Figure 4. Analysis of miRNA movement with a miRNA sensor system. J0571; UAS::icals3m line expressing cals3m in an estradiol inducible manner and GFP constitutively in the ground-tissue (A) was introduced with a sensor system pSPR1::nlsYFP_targetMIR165mu/UAS::MIR165mu. The coding sequence of miR165mu-targeted nlsYFP was driven by the broadly-expressed SPR1 promoter, whereas the MIR165mu gene, which targets the nlsYFP_165mu-tgt, was placed under the UAS (B). In the resulting line (called J0571; UAS::icals3m/pSPR1::nlsYFP_targetMIR165mu/UAS::MIR165mu), YFP signal was not detected under non-induced condition (C). In contrast, after 24 h induction period, nlsYFP signal was detected throughout the stele (D), indicating that the miRNAs were not present to cleave the nlsYFP sequences and consequently YFP signal was not suppressed. GFP signal was detected in the ground-tissue layer of all of the roots, indicating that miR165mu transcription occurred in both conditions. Yellow, GFP signal; green, YFP signal; magenta, PI signal; GT, ground-tissue.
The analysis of miR165mu movement in J0571; UAS::icals3m/pSPR1::nlsYFP_targetMIR165mu/UAS::MIR165mu showed that icals3m expression in the ground-tissue is able to inhibit the movement of the miRNAs from the ground-tissue to the stele (Fig. 4C and D). This was visualized as YFP signal in the stele, which indicated that the miR165mu were not present to cleave their targeted YFP sequences.24 These results thus demonstrate for the first time that the miRNAs move via PD, and therefore provides new insights into the mechanisms of miRNA movement. This result is in accordance with previous suggestions that small RNAs use PD as their moving route. In addition, the data presented here indicate that the icals3m is very effective inhibitor of symplastic trafficking since it is now known the system not only blocks protein movement, but also miRNA movement. This observation provides additional value to the icals3m as a tool to study symplastic trafficking and proves its usefulness in visualization assays investigating differential movement of a molecule after PD closure.
The efficiency of the system to block diffusion of small molecules is, however, unclear. Thus, an important future experiment would be to quantify this aspect more precisely. The importance of PD in solute transport is also not well known. Even though water and some solutes can cross the membrane via specific transporter proteins, small molecules such as nutrients and hormones are assumed to move through PD. A recent study showed that symplastic solute transport is more extensive than previously thought.27 In the study the authors quantified PD flux in the root meristem, and concluded that the flux is 10-fold compared with previous measurements. Presumably with the measured flux rate, symplastic diffusion of sucrose is required for maintaining root growth. These results suggest that symplastic diffusion of solutes is very important, and can be a major route for sucrose transport in the meristem. The authors also assayed solute flux in an Arabidopsis line overexpressing PDCB1, a protein that promotes callose deposition at PD, and found out that flux is reduced by approximately one-half. This indicates that enhanced callose deposition at PD also suppresses solute movement. The effect of icals3m on solute transport is not known; however, these results raise the possibility that impaired solute diffusion might be occurring.
Toward Revealing Novel Symplastic Networks Using the icals3m
Although the data on signaling networks has increased, many questions still remain how developmental processes are regulated non-cell-autonomously. Thus, we envision identifying novel symplastic communication events that regulate development in Arabidopsis root. As described in the previous section, whether a known mobile molecule moves via the symplastic route can be investigated with icals3m. In addition, inhibiting symplastic communication using this system could be used to identify novel regulatory networks relying on symplastic signaling. Cell type- and tissue-specific icals3m plants enable us to block symplastic communication from the corresponding spatial domain in an inducible manner. icals3m expression in a tissue-specific manner combined with genome-wide expression analyses by microarray or RNA sequencing can reveal regulatory targets of symplastic signals. Especially tissue-enriched genes that are differentially expressed under callose induction are promising candidates involved in the cell specification and differentiation of that tissue. For example, after icals3m expression in the ground-tissue, differential expression of stele enriched genes is likely caused by blockage of a regulatory signal originating from the ground-tissue that is unable to transmit into the stele.
After collecting gene expression information following spatially and temporally specific inhibition of symplastic communication, expression profiles of different spatial domains can be compared with the root gene expression map28 to establish a global symplastic communication map of Arabidopsis root. This map would reveal valuable information on which genes are regulated by symplastic signals and how different tissues of the root, including the stele, ground-tissue and epidermis, regulate gene expression non-cell-autonomously through PD during the development of other root domains. On the other hand, using cell type specific lines within tissues reveals how the various cell types within tissues communicate with each other. Moreover, by looking at the number of affected genes, it is possible to determine which cell types and tissues are major sources and sinks of information signals. In addition, by performing functional analyses of the differentially expressed genes after icals3m expression, a deeper understanding of the processes regulated by symplastic signals can be obtained. It is likely that the differentially expressed genes include not only primary targets but also those more downstream. In order to reduce these secondary effects, a time-course analysis to optimize the induction time as short as possible is necessary.
To summarize, plant development is mainly controlled by positional signals, and the symplastic pathway is a major communication route in plants. Callose turnover is a central mechanism to regulate PD traffic, and the spatially and temporally controlled enhancement of callose deposition with icals3m is a very effective tool to block symplastic trafficking. At least protein and smRNA traffic are inhibited, but also small molecule transport may be affected. It is important to quantify exactly the reduction in PD diameter in order to understand if the movement of small molecules is inhibited. Finally, investigating genome-wide expression changes occurring after manipulating the symplastic connection by using icals3m system can be used to find novel symplastic networks that operate between different tissues and cell types of the root.
Acknowledgments
Financial support was provided by the Academy of Finland, University of Helsinki and Tekes. S.M. was supported by the Japan Society for the Promotion of Science.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/22126
References
- 1.Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Benfey PN, Scheres B. Root development. Curr Biol. 2000;10:R813–5. doi: 10.1016/S0960-9822(00)00814-9. [DOI] [PubMed] [Google Scholar]
- 3.van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–9. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 4.Scheres B. Plant cell identity. The role of position and lineage. Plant Physiol. 2001;125:112–4. doi: 10.1104/pp.125.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature. 1995;378:62–5. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- 6.Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–67. doi: 10.1016/S0092-8674(00)80865-X. [DOI] [PubMed] [Google Scholar]
- 7.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–11. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 8.Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–21. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138:2303–13. doi: 10.1242/dev.060491. [DOI] [PubMed] [Google Scholar]
- 10.Robert HS, Friml J. Auxin and other signals on the move in plants. Nat Chem Biol. 2009;5:325–32. doi: 10.1038/nchembio.170. [DOI] [PubMed] [Google Scholar]
- 11.Cilia ML, Jackson D. Plasmodesmata form and function. Curr Opin Cell Biol. 2004;16:500–6. doi: 10.1016/j.ceb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Lucas WJ, Lee JY. Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol. 2004;5:712–26. doi: 10.1038/nrm1470. [DOI] [PubMed] [Google Scholar]
- 13.Lucas WJ, Ham BK, Kim JY. Plasmodesmata - bridging the gap between neighboring plant cells. Trends Cell Biol. 2009;19:495–503. doi: 10.1016/j.tcb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Crawford KM, Zambryski PC. Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr Biol. 2000;10:1032–40. doi: 10.1016/S0960-9822(00)00657-6. [DOI] [PubMed] [Google Scholar]
- 15.Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC. Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:2227–31. doi: 10.1073/pnas.0409193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi K, Otegui MS, Krishnakumar S, Mindrinos M, Zambryski P. INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell. 2007;19:1885–97. doi: 10.1105/tpc.106.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burch-Smith TM, Zambryski PC. Loss of INCREASED SIZE EXCLUSION LIMIT (ISE)1 or ISE2 increases the formation of secondary plasmodesmata. Curr Biol. 2010;20:989–93. doi: 10.1016/j.cub.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuels AL, Giddings TH, Jr., Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–57. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parre E, Geitmann A. More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiol. 2005;137:274–86. doi: 10.1104/pp.104.050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen XY, Kim JY. Callose synthesis in higher plants. Plant Signal Behav. 2009;4:489–92. doi: 10.4161/psb.4.6.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bucher GL, Tarina C, Heinlein M, Di Serio F, Meins F, Jr., Iglesias VA. Local expression of enzymatically active class I beta-1, 3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 2001;28:361–9. doi: 10.1046/j.1365-313X.2001.01181.x. [DOI] [PubMed] [Google Scholar]
- 22.Zavaliev R, Ueki S, Epel BL, Citovsky V. Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma. 2011;248:117–30. doi: 10.1007/s00709-010-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verma DP, Hong Z. Plant callose synthase complexes. Plant Mol Biol. 2001;47:693–701. doi: 10.1023/A:1013679111111. [DOI] [PubMed] [Google Scholar]
- 24.Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell. 2011;21:1144–55. doi: 10.1016/j.devcel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Zuo J, Niu QW, Chua NH. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–73. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima K, Furutani I, Tachimoto H, Matsubara H, Hashimoto T. SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell. 2004;16:1178–90. doi: 10.1105/tpc.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutschow HL, Baskin TI, Kramer EM. Regulation of solute flux through plasmodesmata in the root meristem. Plant Physiol. 2011;155:1817–26. doi: 10.1104/pp.110.168187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–6. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]


