Abstract
Laminins are large molecular weight glycoproteins constituted by the assembly of three disulfide-linked polypeptides, the α, β and γ chains. The human genome encodes 11 genetically distinct laminin chains. Structurally, laminin chains differ by the number, size and organization of a few constitutive domains, endowing the various members of the laminin family with common and unique important functions. In particular, laminins are indispensable building blocks for cellular networks physically bridging the intracellular and extracellular compartments and relaying signals critical for cellular behavior, and for extracellular polymers determining the architecture and the physiology of basement membranes.
Keywords: adhesion, coil-coiled domain, integrin, laminin, laminin N-terminal (LN) domain, laminin-type epidermal growth factor-like (LE) domain
Introduction
Basement membranes are specialized extracellular matrices holding cells and tissues together, a property largely due to their content in laminins. Laminins are glycoproteins with both common and specific functions. One common and most important function of laminins is to interact with receptors anchored in the plasma membrane of cells adjacent to basement membranes. In doing so laminins regulate multiple cellular activities and signaling pathways. Structurally, laminins are composed of a few independently folded, distinct domains, which number, location and size, as well as the interactions that they develop with other molecular components of the basement membranes, vary from one laminin member to another. Every basement membrane contains at least one, sometimes several, members of the laminin family, which structural diversity determines, to a large extent, the unique physiological functions of the various basement membranes of the body.
This review summarizes the structural and molecular basis for both common and unique functions of laminins.
The Oldest Member in the Laminin Family is in Its Thirties
In 1979, a large molecular weight (about 850 kDa) non-collagenous glycoprotein was isolated from both a basement membrane-rich tumor transplantable to mouse [the Engelbreth-Holm-Swarm (EHS) sarcoma] and basement membrane-producing cells.1,2 This new glycoprotein was purified in quantities sufficient for biochemical, structural and immunological characterization and it was given the name laminin.1,3 Biochemical analysis showed that the glycoprotein laminin is constituted by three disulfide-linked polypeptides, initially called A, B1 and B2, with electrophoretic migration mobilities corresponding to 220 and 440 kDa.1,2 Observation of purified laminin molecules by electron microscopy after rotary shadowing indicated that the three chains assemble to form an asymmetrical cross-shaped structure, with a long arm of about 77 nm carrying a large globule at its end, and three short arms, two of 34 nm and one of 48 nm, each being terminated by a globular domain.4,5 Between the center of the cross and the ends of the 34 and 48 nm-short arms, there are one and two additional globules, respectively.4,5
Following the discovery of related but not identical material from various cells and tissues,6,7 it was realized that the molecule isolated from the EHS sarcoma was not unique, and likely to be the first member of a new protein family. To distinguish between the diverse members, they were first given various names,6,7 and then numbered in the order of their discovery.8 The three constitutive subunits initially designated with A, B1 and B2 were renamed with the Greek letters α, β and γ. Sequencing of the subunits provided evidence for distinct polypeptides at the amino acid level. Following the chronological order of their identification, the polypeptides were designated by adding numbers to the Greek letters α, β and γ (α1, α2, … β1, β2, ... γ1, γ2, …). Finally to avoid confusion and facilitate transfer of information, there was a consensus to simplify the laminin nomenclature.9 Now each laminin isoform is designed by its chain composition, i.e., the heterotrimer composed of α1, β1 and γ1 chains is known as laminin 111. It is the prototype of the family and the best characterized laminin isoform.
Eleven Genetically Distinct Laminin Chains
The development of cloning and automated sequencing techniques permitted to identify rapidly genetically different laminin subunits in human9 and many other species (for review see ref. 10). In the human genome, 11 genes distributed on chromosomes 1, 3, 6, 7, 9, 18 and 20, code for five α, three β and three γ laminin subunits (Table 1). At the protein level, laminin subunits have different sizes, with predicted molecular masses ranging from 129,572 Da for the smallest laminin β3 chain to 399,737 Da for the largest laminin α5 chain (Table 1). The laminin α3 chain exists as two polypeptides of different sizes, α3A and α3B, with predicted masses of 189,335 and 366,649 Da, respectively (Table 1). The actual sizes of laminin subunits are, however, larger due to post-translational modifications of the polypeptides, essentially by glycosylation.11-13 For instance molecular masses deduced from the electrophoretic migration mobility are approximately 400 kDa for the laminin α1 chain and 140 kDa for the laminin β3 chain.
Table 1. Chromosome location of human laminin genes and sizes of the corresponding polypeptides.
| Gene name | Chromosome localization | Chain name | Amino acids number* | Molar mass (predicted) |
|---|---|---|---|---|
| LAMA1 |
18p11.31 |
α1 |
3,075 |
337,084 |
| LAMA2 |
6q22-q23 |
α2 |
3,122 |
343,905 |
| LAMA3** |
18q11.2 |
α3A |
1,713 |
189,335 |
| α3B |
3,333 |
366,649 |
||
| LAMA4 |
6q21 |
α4 |
1,823 |
202,524 |
| LAMA5 |
20q13.2-q13.3 |
α5 |
3,695 |
399,737 |
| LAMB1 |
7q22 |
β1 |
1,786 |
198,038 |
| LAMB2 |
3p21 |
β2 |
1,798 |
195,981 |
| LAMB3 |
1q32 |
β3 |
1,172 |
129,572 |
| LAMB4*** |
7q22-q31.2 |
β4 |
1,161 |
193,540 |
| LAMC1 |
1q31 |
γ1 |
1,609 |
177,603 |
| LAMC2 |
1q25.31 |
γ2 |
1,193 |
130,976 |
| LAMC3 | 9q31-q34 | γ3 | 1,575 | 171,227 |
See http://uniprot.org for more detailed information and relevant references providing sequence data and analysis. *The number of amino acids includes the signal peptide. **The LAMA3 gene encodes two transcripts, the short α3A and the long α3B, sharing identical C-terminus. ***Presumably a pseudogene. Transcripts have not been found.
There are more than 50 theoretically possible heterotrimeric associations between all the α, β and γ chains; however, 16 only have been suggested (Fig. 1) and even a smaller number have been definitively proven to exist and purified.9 The laminin chains show tissue and cell specific distribution, with variations between development and pathological states14-16. The laminin α1 chain is expressed already at the 2-cell embryonic stage, and it disappears progressively from most basement membranes during development to the adult organism. It commonly associates with the β1 and γ1 chain, so that the laminin 111 heterotrimer is rather ubiquitous in the embryo, while it is restricted to a small subset of basement membranes in the adult. In contrast, heterotrimers containing the α5 chain, especially laminins 511 and 521, are the most ubiquitous isoforms in the adult organism14 (and in this issue). Besides α5 chain-containing isoforms, laminins 211 and 221 are present in basement membranes of skeletal and cardiac muscles15,16 and Holmberg and Durbeej, this issue), while laminins 411 and 421 are abundant in endothelial basement membranes (Yousif et al., this issue). Laminin 332 is specific for the basal lamina underlying epithelial cells (Rousselle and Beck, this issue).
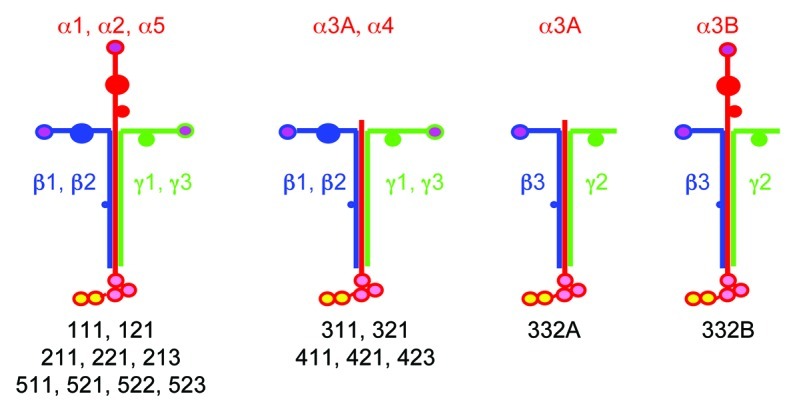
Figure 1. Known and/or predicted laminin heterotrimers. Eleven genes encode five α, three β and three γ chains in the human genome. There are two transcripts for the laminin α3 chain, one short α3A and one long α3B transcript. In theory, there are more than 50 possible heterotrimeric αβγ assemblies, but only those shown in the figure have been isolated or experimentally predicted.
The diverse patterns of laminin α chain expression endow basement membranes with molecular heterogeneity, likely contributing common and unique functions. At this point it should be noted that the notion evolving from initial immunological studies suggesting that the new component isolated from the EHS sarcoma was ubiquitous of basement membranes3 had to be revised. It is now definitely established that although all basement membranes contain laminin, most of the laminin molecules present in the adult human organism are not identical to laminin 111 isolated from the EHS sarcoma. Except for laminin 332, most other laminin isoforms are immunologically related to laminin 111 because they contain either the β1 or the γ1 chains, or both (Fig. 1). It explains why antisera raised against laminin 111 purified from the EHS sarcoma stain all basement membranes, even if laminin α1 chain is absent.
An Organized Patchwork of a Few Structurally Distinct Domains Forms Laminins
Although laminin chains differ at the level of the amino acid sequences, the polypeptide chains fold into several kind of unique or multiple copies of independent structural domains whose overall arrangement in a cross-shaped pattern is conserved among laminin isoforms. The laminin short arms of the cross are contributed by the separately folded N-terminal regions of every α, β and γ chains, while all three chains form the stem of the long arm.
The long arm of laminins: a coiled-coil stem and a globular domain
In each laminin heterotrimer, the long arm is formed by amino acid stretches of similar lengths (561 to 591 residues depending on the chain) located at the C-terminus of the β and γ chains and the adjacent portion of the α chain. These stretches are rich in heptad repeats of non-polar amino acids and fold in an α-helical coiled-coil structure.17,18 An extra 31–33 residues insert between the heptad repeats of all three β chains to form a knob, the β knob, popping out of the long arm. Sequences within the coiled-coil domains of laminin chains contain information for selective chain recognition, and ionic interactions between these domains determine the specificity of chain assembly.19-22 The coiled-coil stretches direct intracellular assembly of α, β and γ chains together, processing through different steps. First, dimerization between β and γ chains takes place, followed by incorporation of an α chain by folding a large C-terminal portion of the three subunits in a triple-stranded α-helical coiled-coil structure stabilized by disulfide bridges.
The C-terminus of the laminin α chains is 865–900 residues longer than that of the β and γ chains and it forms the large laminin globular or LG domain at the end of the long arm.23 This C-terminal extension of the α chain is folded into five sub-domains, LG1 to LG5, of about 160 to 200 residues each. The LG1 to LG3 trio is separated by a short stretch of amino acids from the LG4-LG5 pair. The crystal structure has been determined for recombinant LG1 to LG3 trio of the laminin α2 chain24 and LG4-LG5 pair of the laminin α1 and α2 chains.25,26 It indicates that LG domains adopt β-sandwich folds, with canonical calcium binding sites. Moreover, LG2 and LG3 interact through a substantial interface.24
The laminin short arms: divergent mosaics of globular domains and rod-like arrays
In contrast to the highly conserved structure of the long arm of laminins, the short arms considerably diverge (Fig. 2). They are contributed by the N-terminus of α, β and γ chains folding separately into three types of structural domains: the laminin N-terminal (LN), the laminin-type epidermal growth (EGF) factor-like (LE) and the laminin IV (L4/LF) domains.9
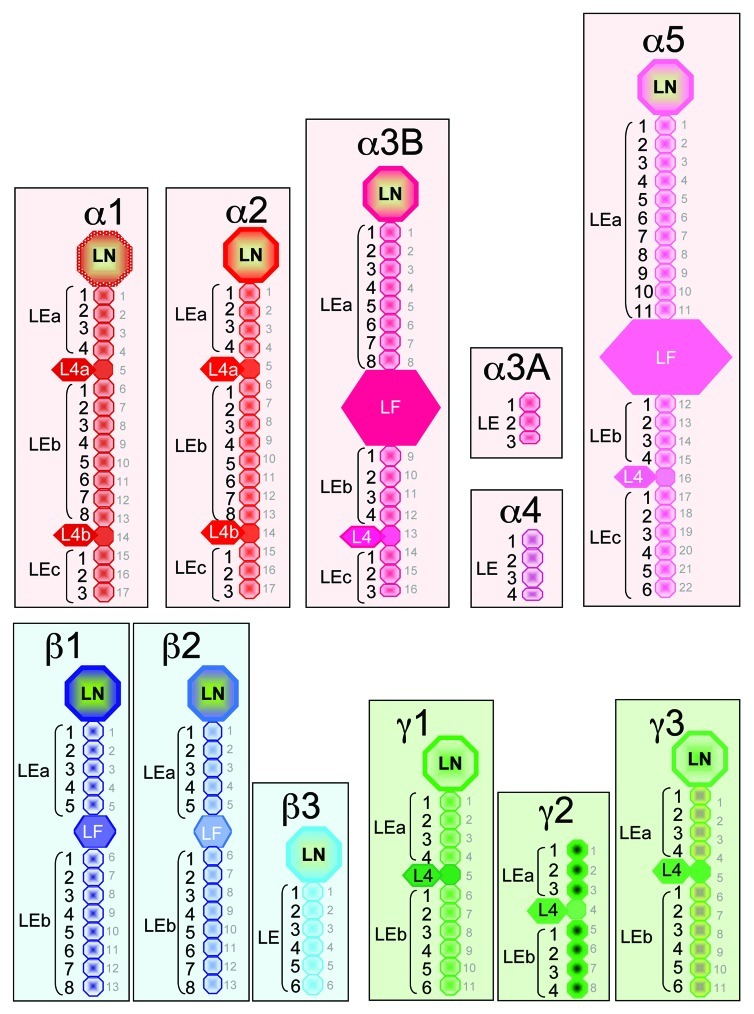
Figure 2. Schematic representation of the highly diverging N-terminus of laminin chains. Three basic structural domains found in the N-terminus of the laminin chains are the LN (laminin N-terminal), LE (laminin-type epidermal growth (EGF) factor-like) and L4/LF (laminin IV) domains. The names (in black), location and size of the various domains are indicated for the different laminin chains. Each L4 domain corresponds to a long stretch of residues inserted between cysteine residues 3 and 4 of one LE domain. In this case the L4 and LE domains should be considered as one single domain which is emphasized by using the same color. In contrast the LF domains are located between two LE repeats as in the α3B, α5, β1 and β2 chains. When L4 or LF globular domains interrupt the arrays of repeating LE domains, they determine stretches named LEa, LEb and LEc. The numbering of the LE domains starts with 1 for each stretch (LEa1, LEa2, …; LEb1, LEb2, etc…). For example, the N-terminus of the laminin α1 chain starts with one LN domain, followed by a first array of four LE domains (LEa1 to LEa4), then one globular domain L4a (consisting of a long stretch of residue inserted within one LE domain), followed by a second array of eight LE domains (LEb1 to LEb8), another globular domain L4b (consisting of a long stretch of residue inserted within one LE domain), and finally a stretch of three LE domains (LEc1 to LEc3). In contrast the N-terminus of the laminin α3A chain consists of three LE domains only. This nomenclature adopted in 20059 is indicated to the left of each diagram. It should be noted that in the UniProt (http://uniprot.org) data bank, numbering of the LE domains is different. It is based on the total number of LE domains as indicated with the small gray lettering right to the various diagrams.
Except for the α3A, α4 and γ2 chains, laminin subunits have one globular LN domain at the N-terminal end. Depending on the chain, LN domains are 228 to 259 residues long and they adopt a globular fold. The crystal structure of recombinant LN domain of the α5, β1 and γ1 chains have been recently resolved.27,28 It shows that the respective polypeptides are similarly arranged in a β-sandwich with elaborate loop regions that differ between laminin chains, and that, together with adjacent LE domains, the shape resembles the head and stalk of a flower.27,28
The LE domains comprise 41 to 70 residues each, containing 8 cysteine residues, except in a few cases. Disulfide bonds between cysteine pairs 1 and 3, 2 and 4, and 5 and 6, determine the formation of loops a to c like in EGF.29 An additional loop d between cysteine pair 7 and 8, specific for laminin, provides distinct contact with loop b of the next LE domain.29 This characteristic is thought to be important for the limited flexibility of the arrays typically formed by repetitive LE domains. In addition, the α3, α4, α5, β1, β2 and γ2 chains contain one truncated LE domain each (15 to 35 residues) besides the classical LE domains. Finally, the arrays of LE domains determine spacers of various lengths between other type of domains, and it can be highly divergent from one chain to another, with 3 LE domains only in α3A, and up to 22 in α5 (Fig. 2).
One or two globular domains interrupt the rod-like arrays of repeating LE domains in some of the laminin chains (Fig. 2). These globular domains, known as domain IV in an older nomenclature,8 were recently renamed L4 and LF for laminin 4 and laminin four, respectively.9 The L4 and LF globular domains delimit two or three stretches in the LE arrays, referred to as LEa, LEb and LEc.9 The L4 domain consists of an extra-long stretch of residues (169 to 204) inserted between cysteine residues 3 and 4 in one LE domain, thereby considerably enlarging loop b, and the LE domain itself. This is the case for the L4a domain between LEa4 and LEb1, and the L4b domain between LEb8 and LEc1 in the α1 and α2 chain; for the L4 domains between LEb4 and LEc1 in the α3B chain and between LEb4 and LEc1 in the α5 chain, as well as for the L4 domains of the γ chains (Fig. 2). In early time, the second kind of domain IV was recognized in the β chains as an independent globular domain intercalated between two LE units. It is now called LF to differentiate it from the L4 domain inserted in loop b of one LE unit. A single copy of this domain is present in β1 and β2 chains (219 and 217 amino acid residues, respectively). Interestingly, the α3B and α5 chains contain also one independent LF globular domain intercalated between two LE units, which size (470 and 587 amino acid residues, respectively) is much larger than that of L4 or LF domains in other laminin chains.
To summarize, the number and distribution of the LN, LE and L4/LF domains forming the short arm of laminins substantially differ between the 11 different chains. A full set of domains (LN, LE and L4/LF) form the N-terminus of the α1, α2, α3B and α5 chains, while the N-terminus of the α3A and α4 chains are devoid of LN and L4/LF domains and they solely consist of a very short stretch of 3 or 4 LE domains. One LN domain is also present at the N-terminus of the β and γ chains, except in γ2. The β1 and β2 chains, but not the β3, contain one globular LF domain, while all three γ chains have one globular L4 domain inserted in loop b of an LE domain (Fig. 2).
Essential Functions of Laminins
Several of the structurally related laminin domains have nearly identical or similar functions in every laminin isoform. As a general rule, the C-terminal ends of laminins interact with proteins anchored in the plasma membranes of cells or microorganisms, thereby relaying biochemical and mechanical signals between intracellular and extracellular molecular networks (Fig. 3). The N-terminal ends of laminins are involved in interactions mainly with other extracellular matrix molecules present in basement membranes (Fig. 3). Thereby, they are an integral part of the complex extracellular molecular networks important for the architecture and physiology of basement membranes. Some not yet well-defined parts of laminins are thought to interact with small molecules such as growth factors and cytokines. This function is thought to be important for sequestration and storage of these small molecules and for regulating their distribution, activation, and presentation to cells.
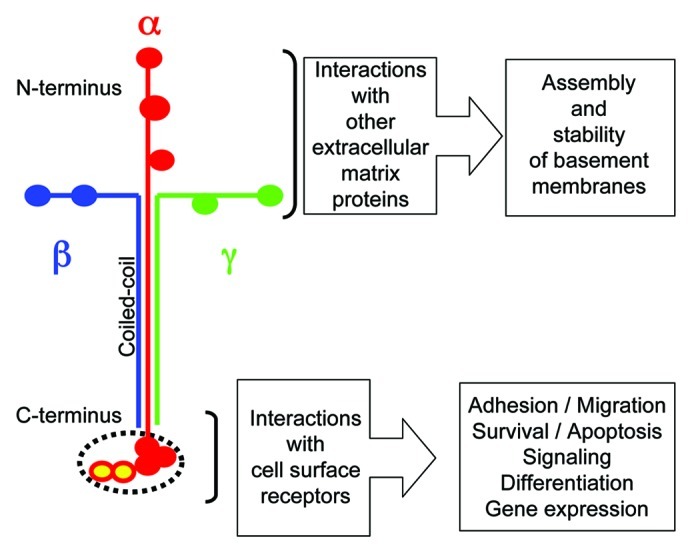
Figure 3. Mapping of the major functions of laminins. The laminin short arms (N-terminus) are involved in architectural function within the basement membrane, while the end of the long arm (C-terminus) is typically involved in cellular interactions.
Laminins and Cell Adhesion-Promoting Activity
Very early on multiple biological activities were described for laminin isolated from the EHS sarcoma, in particular it was shown to be a cell adhesion molecule.30 Enzymatic dissection of laminin 111 from the EHS sarcoma into fragments identified several cell binding sites (see ref. 9 for the nomenclature of laminin fragments). The first fragment identified having cell adhesion-promoting activity is fragment P1 obtained by pepsin treatment of EHS laminin, and it originates from the center of the cross.9,31-34 The activity is due to an RGD sequence in the LEb8 domain of the laminin α1 chain, which is not accessible to cells in the intact chain because it is masked by domain L4b.35 Additional non-cryptic cell binding sites exist on the intact laminin short arms of at least some isoforms.36,37 They are ligands for the α1β1 and α2β1 integrins, which are classical collagen receptors, and they have been mapped to the LN domain of the laminin α1 and α2 chains.38-41 The biological relevance of these interactions remains to be elucidated.
Two other fragments from the long arm have either heparin-dependent (fragment E3) or -independent (fragment E8) cell binding activity.42-44 Fragment E3 with high affinity for heparin corresponds to the LG4–5 pair and it contains the binding sites for α-dystroglycan, syndecans and galactosylsulfatides.25,26,43,44 The crystal structure of the laminin α2 LG4-LG5 domain localizes a basic surface region between calcium sites suitable for binding α-dystroglycan and heparin.25 Fragment E8 consists of the LG1-LG3 domains attached to the end of the long arm (Fig. 4), and it interacts with integrins expressed by a large variety of cells.42,45 All newly identified laminin isoforms, either purified from tissues or cells or expressed as recombinant proteins, display integrin-mediated cell adhesion to the LG domains.46-55 Depending on the laminin isoform, the interactions are mediated by one of the four different, so-called laminin-binding integrins, α3β1, α6β1, α7β1 and α6β4.42-55 The specificity of the integrin-laminin interaction is determined by the laminin α chain.56 For example α6β1 has variable affinity for all laminin isofoms, α3β1 and α6β4 bind nearly exclusively to laminins containing the α3 and α5 chains, and α7β1 has preference for isoforms with the α2 and α5 chains.56

Figure 4. The integrin binding site at the C-terminus of the laminin long arm. (A) The major integrin binding site on laminins is formed by the LG1 to LG3 domains and the extremity of coiled-coil fold formed by the α, β and γ chains. It is thought that the C-terminus of the γ chain is needed to stabilize a conformation of the LG1-LG3 trio compatible with integrin binding. (B) Representation of the amino acid sequences at the C-terminus of the three laminin γ chains. The γ1 and γ2 chains contains a glutamic acid residue (E, highlighted green) at the third position from the carboxyl termini. The residue is thought to be important for maintaining integrin binding activity of the structure shown in (A).59 The laminin γ3 chain is shorter and lacks the glutamic acid residue at the same position. Recombinant fragment engineered as shown in A and containing the γ3 chain have no integrin binding activity.60
Integrin binding to the C-terminus of laminins requires both an intact LG1-LG3 trio and the coiled-coil fold of the laminin α, β, and γ chains. Separation of the LG1-LG3 trio from the long arm stem57 or unfolding of the coiled-coil structure abolishes cell adhesion-promoting activity.57,58 The molecular basis of this feature involves a glutamic acid residue (Fig. 4) at the third position from the carboxyl termini of the laminin γ1 and γ2 chain,59 which is absent in the shorter laminin γ3 chain.60 Interestingly, a recombinant laminin heterotrimer containing the γ3 chain has no integrin binding activity.60 It suggests that the γ chain either binds directly to the integrin, or it is required for maintaining the spatial organization of the LG1-LG3 trio into a biologically active conformation, perhaps by stabilizing the interactions between LG1, LG2 and LG3.24 Recently, a global analysis of N-linked glycosylation sites shows that the β-sandwich faces of the LG1 domain are free of carbohydrate modifications in all five laminin α chains, indicating that these surfaces may harbor the integrin binding site.24 The C-terminal region of laminin β chains may modulate also the integrin binding affinities of laminins.61
Integrin-mediated interactions with the C-terminus of laminins are important for several cellular activities by activating specific signaling networks governing adhesion, migration, differentiation, survival and many other aspects of cell behavior reviewed in the other papers of this special focus of Cell Adhesion & Migration.
The Short Arms of Laminins Guide Basement Membrane Assembly and Organization
The major task of laminins’ short arms is to participate in basement membrane assembly and organization by interacting with other components of the basement membranes, including laminins themselves. As it should be anticipated from the differences and similarities in size, number and layout of structural domains in the laminin short arms, both highly divergent and common interactions have been recognized for the diverse isoforms of laminins. For instance isoforms such as laminins 111, 211 and 511 form the classical network of laminin polymers, following a process involving the LN domains at each end of three full length short arms (ref. 62 and Hohenester and Yurchenco, this issue). These laminin polymers are connected by the heparan sulfate proteoglycan perlecan63-65 and by nidogen (entactin)66,67 to collagen IV polymers, forming supramolecular networks important for basement membrane stability. Nidogen, which has been isolated from the EHS sarcoma as a stable complex with laminin 111,68 has affinity for a specific LE domain in the laminin γ169-72 and laminin γ3 chains.73 It has affinity also for the laminin γ2 chain L4-LEb1 domains, part of which is enzymatically removed in mature laminin 332.74 It may explain why in transgenic mice deficient in nidogen, basement membranes containing the laminin γ2 chain, such as that at the epidermal-dermal junction, are apparently normal, while those underlying, for instance, endothelial cells and containing the laminin γ1 chain are defective.75 It could be that at the dermal-epidermal junction, an interaction between nidogen and the laminin γ2 chain is needed, but dispensable for targeting newly synthesized proteins at the correct anatomical location, but it is not needed for the architectural and mechanical integrity of mature basement membranes.
In contrast to progress made in defining the interactions of classical laminins with other extracellular matrix components, the knowledge is missing or fragmentary concerning the mechanism(s) how laminin isoforms lacking a full set of LN domains, such as those containing the α3A and α4 chains, are integrated in the basement membrane framework. Laminin 332 with rudimentary N-terminus and only one copy of the LN domain on the β3 chain provides the core of a unique and biologically important network anchoring the epidermis to the dermis.76 Although truncated, the N-terminal region of laminin 332 associates with other extracellular components, including at least laminin 311,77 collagen VII,78-80 collagen XVII81 and fibulin.74,82 It is not known whether these interactions are sufficient to maintain anchorage of the N-terminal regions of laminin 332 within the extracellular matrix of the epidermal-dermal basement membrane, or whether additional interactions are required.
Finally, interactions of fibulins with the L4 domain of γ chains have been reported74,82 and the knowledge about potential functions of the L4/LF domains is very limited.
Conclusion and Open Questions
Over the past 30 years the knowledge on laminins has expanded tremendously. The cell adhesion-promoting activity of laminin isoforms is now well characterized, also at the structural level. However, the specificity, if any, of the signaling pathways activated by the different laminin-binding integrins is not known. Integration of classical laminins in the basement membrane framework is documented into details, but information is missing for laminins having truncated short arms. Furthermore, it remains to be determined whether the diverse interactions described in vitro for LN domains—i.e., with other laminin LN domain for polymerization, with heparan sulfate-containing domains of perlecan, and with collagen-binding α2β1 and α1β1 integrins—also occur in vivo, whether they are simultaneous or mutually exclusive and what is their biological significance. Also, it would be important in future studies to determine how the diverse oligosaccharides present on laminin chains modify laminin interactions and functions, and whether glycosylation of laminin chains adds a further degree of specificity to the different basement membranes.
Acknowledgments
The author acknowledges financial support from the German Federal Ministry for Education and Research (Network Epidermolysis bullosa, 01GM0832, and Network Stem Cell Therapy for Severe Skin Fragility Syndromes, 01GN0972), the Marga und Boll Stiftung and the Köln Fortune program from the Medical Faculty of the University of Cologne. The author apologizes for having selected a few references only among more than 20,000 entries retrieved by searching PubMed with “laminin.”
Glossary
Abbreviations:
- LE
laminin-type epidermal growth factor-like
- LG
laminin globular
- LN
laminin N-terminal
- L4
LF, laminin domain IV
- EHS
Engelbreth-Holm-Swarm
- Da
dalton
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/22826
References
- 1.Timpl R, Rohde H, Robey PG, Rennard SI, Foidart JM, Martin GR. Laminin--a glycoprotein from basement membranes. J Biol Chem. 1979;254:9933–7. [PubMed] [Google Scholar]
- 2.Chung AE, Jaffe R, Freeman IL, Vergnes JP, Braginski JE, Carlin B. Properties of a basement membrane-related glycoprotein synthesized in culture by a mouse embryonal carcinoma-derived cell line. Cell. 1979;16:277–87. doi: 10.1016/0092-8674(79)90005-9. [DOI] [PubMed] [Google Scholar]
- 3.Rohde H, Wick G, Timpl R. Immunochemical characterization of the basement membrane glycoprotein laminin. Eur J Biochem. 1979;102:195–201. doi: 10.1111/j.1432-1033.1979.tb06280.x. [DOI] [PubMed] [Google Scholar]
- 4.Engel J, Odermatt E, Engel A, Madri JA, Furthmayr H, Rohde H, et al. Shapes, domain organizations and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix. J Mol Biol. 1981;150:97–120. doi: 10.1016/0022-2836(81)90326-0. [DOI] [PubMed] [Google Scholar]
- 5.Bruch M, Landwehr R, Engel J. Dissection of laminin by cathepsin G into its long-arm and short-arm structures and localization of regions involved in calcium dependent stabilization and self-association. Eur J Biochem. 1989;185:271–9. doi: 10.1111/j.1432-1033.1989.tb15112.x. [DOI] [PubMed] [Google Scholar]
- 6.Leivo I, Engvall E. Merosin, a protein specific for basement membranes of Schwann cells, striated muscle, and trophoblast, is expressed late in nerve and muscle development. Proc Natl Acad Sci U S A. 1988;85:1544–8. doi: 10.1073/pnas.85.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter DD, Shah V, Merlie JP, Sanes JR. A laminin-like adhesive protein concentrated in the synaptic cleft of the neuromuscular junction. Nature. 1989;338:229–34. doi: 10.1038/338229a0. [DOI] [PubMed] [Google Scholar]
- 8.Burgeson RE, Chiquet M, Deutzmann R, Ekblom P, Engel J, Kleinman H, et al. A new nomenclature for the laminins. Matrix Biol. 1994;14:209–11. doi: 10.1016/0945-053X(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 9.Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, et al. A simplified laminin nomenclature. Matrix Biol. 2005;24:326–32. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Fahey B, Degnan BM. Origin and evolution of laminin gene family diversity. Mol Biol Evol. 2012;29:1823–36. doi: 10.1093/molbev/mss060. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara S, Shinkai H, Deutzmann R, Paulsson M, Timpl R. Structure and distribution of N-linked oligosaccharide chains on various domains of mouse tumour laminin. Biochem J. 1988;252:453–61. doi: 10.1042/bj2520453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arumugham RG, Hsieh TC, Tanzer ML, Laine RA. Structures of the asparagine-linked sugar chains of laminin. Biochim Biophys Acta. 1986;883:112–26. doi: 10.1016/0304-4165(86)90142-X. [DOI] [PubMed] [Google Scholar]
- 13.Kumar AP, Nandini CD, Salimath PV. Structural characterization of N-linked oligosaccharides of laminin from rat kidney: changes during diabetes and modulation by dietary fiber and butyric acid. FEBS J. 2011;278:143–55. doi: 10.1111/j.1742-4658.2010.07940.x. [DOI] [PubMed] [Google Scholar]
- 14.Miner JH, Patton BL, Lentz SI, Gilbert DJ, Snider WD, Jenkins NA, et al. The laminin α chains: expression, developmental transitions, and chromosomal locations of α1-5, identification of heterotrimeric laminins 8-11, and cloning of a novel α3 isoform. J Cell Biol. 1997;137:685–701. doi: 10.1083/jcb.137.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falk M, Ferletta M, Forsberg E, Ekblom P. Restricted distribution of laminin α1 chain in normal adult mouse tissues. Matrix Biol. 1999;18:557–68. doi: 10.1016/S0945-053X(99)00047-5. [DOI] [PubMed] [Google Scholar]
- 16.Durbeej M. Laminins. Cell Tissue Res. 2010;339:259–68. doi: 10.1007/s00441-009-0838-2. [DOI] [PubMed] [Google Scholar]
- 17.Barlow DP, Green NM, Kurkinen M, Hogan BL. Sequencing of laminin B chain cDNAs reveals C-terminal regions of coiled-coil α-helix. EMBO J. 1984;3:2355–62. doi: 10.1002/j.1460-2075.1984.tb02140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsson M, Deutzmann R, Timpl R, Dalzoppo D, Odermatt E, Engel J. Evidence for coiled-coil α-helical regions in the long arm of laminin. EMBO J. 1985;4:309–16. doi: 10.1002/j.1460-2075.1985.tb03630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kammerer RA, Antonsson P, Schulthess T, Fauser C, Engel J. Selective chain recognition in the C-terminal α-helical coiled-coil region of laminin. J Mol Biol. 1995;250:64–73. doi: 10.1006/jmbi.1995.0358. [DOI] [PubMed] [Google Scholar]
- 20.Beck K, Dixon TW, Engel J, Parry DA. Ionic interactions in the coiled-coil domain of laminin determine the specificity of chain assembly. J Mol Biol. 1993;231:311–23. doi: 10.1006/jmbi.1993.1284. [DOI] [PubMed] [Google Scholar]
- 21.Nomizu M, Utani A, Beck K, Otaka A, Roller PP, Yamada Y. Mechanism of laminin chain assembly into a triple-stranded coiled-coil structure. Biochemistry. 1996;35:2885–93. doi: 10.1021/bi951555n. [DOI] [PubMed] [Google Scholar]
- 22.Macdonald PR, Lustig A, Steinmetz MO, Kammerer RA. Laminin chain assembly is regulated by specific coiled-coil interactions. J Struct Biol. 2010;170:398–405. doi: 10.1016/j.jsb.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E. Structure and function of laminin LG modules. Matrix Biol. 2000;19:309–17. doi: 10.1016/S0945-053X(00)00072-X. [DOI] [PubMed] [Google Scholar]
- 24.Carafoli F, Clout NJ, Hohenester E. Crystal structure of the LG1-3 region of the laminin α2 chain. J Biol Chem. 2009;284:22786–92. doi: 10.1074/jbc.M109.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tisi D, Talts JF, Timpl R, Hohenester E. Structure of the C-terminal laminin G-like domain pair of the laminin α2 chain harbouring binding sites for α-dystroglycan and heparin. EMBO J. 2000;19:1432–40. doi: 10.1093/emboj/19.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison D, Hussain SA, Combs AC, Ervasti JM, Yurchenco PD, Hohenester E. Crystal structure and cell surface anchorage sites of laminin α1LG4-5. J Biol Chem. 2007;282:11573–81. doi: 10.1074/jbc.M610657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain SA, Carafoli F, Hohenester E. Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region. EMBO Rep. 2011;12:276–82. doi: 10.1038/embor.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carafoli F, Hussain SA, Hohenester E. Crystal structures of the network-forming short-arm tips of the laminin β1 and γ1 chains. PLoS One. 2012;7:e42473. doi: 10.1371/journal.pone.0042473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stetefeld J, Mayer U, Timpl R, Huber R. Crystal structure of three consecutive laminin-type epidermal growth factor-like (LE) modules of laminin γ1 chain harboring the nidogen binding site. J Mol Biol. 1996;257:644–57. doi: 10.1006/jmbi.1996.0191. [DOI] [PubMed] [Google Scholar]
- 30.Kleinman HK, Cannon FB, Laurie GW, Hassell JR, Aumailley M, Terranova VP, et al. Biological activities of laminin. J Cell Biochem. 1985;27:317–25. doi: 10.1002/jcb.240270402. [DOI] [PubMed] [Google Scholar]
- 31.Rao CN, Margulies IMK, Tralka TS, Terranova VP, Madri JA, Liotta LA. Isolation of a subunit of laminin and its role in molecular structure and tumor cell attachment. J Biol Chem. 1982;257:9740–4. [PubMed] [Google Scholar]
- 32.Timpl R, Johansson S, van Delden V, Oberbäumer I, Höök M. Characterization of protease-resistant fragments of laminin mediating attachment and spreading of rat hepatocytes. J Biol Chem. 1983;258:8922–7. [PubMed] [Google Scholar]
- 33.Nurcombe V, Aumailley M, Timpl R, Edgar D. The high-affinity binding of laminin to cells. Assignation of a major cell-binding site to the long arm of laminin and of a latent cell-binding site to its short arms. Eur J Biochem. 1989;180:9–14. doi: 10.1111/j.1432-1033.1989.tb14608.x. [DOI] [PubMed] [Google Scholar]
- 34.Aumailley M, Nurcombe V, Edgar D, Paulsson M, Timpl R. The cellular interactions of laminin fragments. Cell adhesion correlates with two fragment-specific high affinity binding sites. J Biol Chem. 1987;262:11532–8. [PubMed] [Google Scholar]
- 35.Aumailley M, Gerl M, Sonnenberg A, Deutzmann R, Timpl R. Identification of the Arg-Gly-Asp sequence in laminin A chain as a latent cell-binding site being exposed in fragment P1. FEBS Lett. 1990;262:82–6. doi: 10.1016/0014-5793(90)80159-G. [DOI] [PubMed] [Google Scholar]
- 36.Goodman SL, Deutzmann R, von der Mark K. Two distinct cell-binding domains in laminin can independently promote nonneuronal cell adhesion and spreading. J Cell Biol. 1987;105:589–98. doi: 10.1083/jcb.105.1.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman SL, Aumailley M, von der Mark H. Multiple cell surface receptors for the short arms of laminin: α 1 β 1 integrin and RGD-dependent proteins mediate cell attachment only to domains III in murine tumor laminin. J Cell Biol. 1991;113:931–41. doi: 10.1083/jcb.113.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaff M, Göhring W, Brown JC, Timpl R. Binding of purified collagen receptors (α 1 β 1, α 2 β 1) and RGD-dependent integrins to laminins and laminin fragments. Eur J Biochem. 1994;225:975–84. doi: 10.1111/j.1432-1033.1994.0975b.x. [DOI] [PubMed] [Google Scholar]
- 39.Colognato H, MacCarrick M, O’Rear JJ, Yurchenco PD. The laminin α2-chain short arm mediates cell adhesion through both the α1β1 and α2β1 integrins. J Biol Chem. 1997;272:29330–6. doi: 10.1074/jbc.272.46.29330. [DOI] [PubMed] [Google Scholar]
- 40.Ettner N, Göhring W, Sasaki T, Mann K, Timpl R. The N-terminal globular domain of the laminin α1 chain binds to α1β1 and α2β1 integrins and to the heparan sulfate-containing domains of perlecan. FEBS Lett. 1998;430:217–21. doi: 10.1016/S0014-5793(98)00601-2. [DOI] [PubMed] [Google Scholar]
- 41.Hozumi K, Ishikawa M, Hayashi T, Yamada Y, Katagiri F, Kikkawa Y, et al. Identification of cell adhesive sequences in the N-terminal region of the laminin α2 chain. J Biol Chem. 2012;287:25111–22. doi: 10.1074/jbc.M112.348151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonnenberg A, Linders CJ, Modderman PW, Damsky CH, Aumailley M, Timpl R. Integrin recognition of different cell-binding fragments of laminin (P1, E3, E8) and evidence that α 6 β 1 but not α 6 β 4 functions as a major receptor for fragment E8. J Cell Biol. 1990;110:2145–55. doi: 10.1083/jcb.110.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorokin L, Conzelmann S, Ekblom P, Aumailley M, Battaglia C, Timpl R. Monoclonal antibodies against laminin A chain fragment E3 and their effects binding to cells and proteoglycan and on kidney development. Exp Cell Res. 1992;201:137–44. doi: 10.1016/0014-4827(92)90357-E. [DOI] [PubMed] [Google Scholar]
- 44.Talts JF, Andac Z, Göhring W, Brancaccio A, Timpl R. Binding of the G domains of laminin α1 and α2 chains and perlecan to heparin, sulfatides, α-dystroglycan and several extracellular matrix proteins. EMBO J. 1999;18:863–70. doi: 10.1093/emboj/18.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aumailley M, Timpl R, Sonnenberg A. Antibody to integrin α 6 subunit specifically inhibits cell-binding to laminin fragment 8. Exp Cell Res. 1990;188:55–60. doi: 10.1016/0014-4827(90)90277-H. [DOI] [PubMed] [Google Scholar]
- 46.Carter WG, Ryan MC, Gahr PJ. Epiligrin, a new cell adhesion ligand for integrin α 3 β 1 in epithelial basement membranes. Cell. 1991;65:599–610. doi: 10.1016/0092-8674(91)90092-D. [DOI] [PubMed] [Google Scholar]
- 47.Brown JC, Wiedemann H, Timpl R. Protein binding and cell adhesion properties of two laminin isoforms (AmB1eB2e, AmB1sB2e) from human placenta. J Cell Sci. 1994;107:329–38. doi: 10.1242/jcs.107.1.329. [DOI] [PubMed] [Google Scholar]
- 48.Lindblom A, Marsh T, Fauser C, Engel J, Paulsson M. Characterization of native laminin from bovine kidney and comparison with other laminin variants. Eur J Biochem. 1994;219:383–92. doi: 10.1111/j.1432-1033.1994.tb19950.x. [DOI] [PubMed] [Google Scholar]
- 49.Rousselle P, Aumailley M. Kalinin is more efficient than laminin in promoting adhesion of primary keratinocytes and some other epithelial cells and has a different requirement for integrin receptors. J Cell Biol. 1994;125:205–14. doi: 10.1083/jcb.125.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verrando P, Lissitzky JC, Sarret Y, Winberg JO, Gedde-Dahl T, Jr., Schmitt D, et al. Keratinocytes from junctional epidermolysis bullosa do adhere and migrate on the basement membrane protein nicein through α 3 β 1 integrin. Lab Invest. 1994;71:567–74. [PubMed] [Google Scholar]
- 51.Kikkawa Y, Umeda M, Miyazaki K. Marked stimulation of cell adhesion and motility by ladsin, a laminin-like scatter factor. J Biochem. 1994;116:862–9. doi: 10.1093/oxfordjournals.jbchem.a124608. [DOI] [PubMed] [Google Scholar]
- 52.Kortesmaa J, Yurchenco P, Tryggvason K. Recombinant laminin-8 (α(4)β(1)γ(1)). Production, purification,and interactions with integrins. J Biol Chem. 2000;275:14853–9. doi: 10.1074/jbc.275.20.14853. [DOI] [PubMed] [Google Scholar]
- 53.Fujiwara H, Kikkawa Y, Sanzen N, Sekiguchi K. Purification and characterization of human laminin-8. Laminin-8 stimulates cell adhesion and migration through α3β1 and α6β1 integrins. J Biol Chem. 2001;276:17550–8. doi: 10.1074/jbc.M010155200. [DOI] [PubMed] [Google Scholar]
- 54.Doi M, Thyboll J, Kortesmaa J, Jansson K, Iivanainen A, Parvardeh M, et al. Recombinant human laminin-10 (α5β1γ1). Production, purification, and migration-promoting activity on vascular endothelial cells. J Biol Chem. 2002;277:12741–8. doi: 10.1074/jbc.M111228200. [DOI] [PubMed] [Google Scholar]
- 55.Sasaki T, Takagi J, Giudici C, Yamada Y, Arikawa-Hirasawa E, Deutzmann R, et al. Laminin-121--recombinant expression and interactions with integrins. Matrix Biol. 2010;29:484–93. doi: 10.1016/j.matbio.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishiuchi R, Takagi J, Hayashi M, Ido H, Yagi Y, Sanzen N, et al. Ligand-binding specificities of laminin-binding integrins: a comprehensive survey of laminin-integrin interactions using recombinant α3β1, α6β1, α7β1 and α6β4 integrins. Matrix Biol. 2006;25:189–97. doi: 10.1016/j.matbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Deutzmann R, Aumailley M, Wiedemann H, Pysny W, Timpl R, Edgar D. Cell adhesion, spreading and neurite stimulation by laminin fragment E8 depends on maintenance of secondary and tertiary structure in its rod and globular domain. Eur J Biochem. 1990;191:513–22. doi: 10.1111/j.1432-1033.1990.tb19151.x. [DOI] [PubMed] [Google Scholar]
- 58.Rousselle P, Golbik R, van der Rest M, Aumailley M. Structural requirement for cell adhesion to kalinin (laminin-5) J Biol Chem. 1995;270:13766–70. doi: 10.1074/jbc.270.23.13766. [DOI] [PubMed] [Google Scholar]
- 59.Ido H, Nakamura A, Kobayashi R, Ito S, Li S, Futaki S, et al. The requirement of the glutamic acid residue at the third position from the carboxyl termini of the laminin γ chains in integrin binding by laminins. J Biol Chem. 2007;282:11144–54. doi: 10.1074/jbc.M609402200. [DOI] [PubMed] [Google Scholar]
- 60.Ido H, Ito S, Taniguchi Y, Hayashi M, Sato-Nishiuchi R, Sanzen N, et al. Laminin isoforms containing the γ3 chain are unable to bind to integrins due to the absence of the glutamic acid residue conserved in the C-terminal regions of the γ1 and γ2 chains. J Biol Chem. 2008;283:28149–57. doi: 10.1074/jbc.M803553200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taniguchi Y, Ido H, Sanzen N, Hayashi M, Sato-Nishiuchi R, Futaki S, et al. The C-terminal region of laminin β chains modulates the integrin binding affinities of laminins. J Biol Chem. 2009;284:7820–31. doi: 10.1074/jbc.M809332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schittny JC, Yurchenco PD. Terminal short arm domains of basement membrane laminin are critical for its self-assembly. J Cell Biol. 1990;110:825–32. doi: 10.1083/jcb.110.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woodley DT, Rao CN, Hassell JR, Liotta LA, Martin GR, Kleinman HK. Interactions of basement membrane components. Biochim Biophys Acta. 1983;761:278–83. doi: 10.1016/0304-4165(83)90077-6. [DOI] [PubMed] [Google Scholar]
- 64.Kleinman HK, McGarvey ML, Hassell JR, Martin GR. Formation of a supramolecular complex is involved in the reconstitution of basement membrane components. Biochemistry. 1983;22:4969–74. doi: 10.1021/bi00290a014. [DOI] [PubMed] [Google Scholar]
- 65.Garbe JH, Göhring W, Mann K, Timpl R, Sasaki T. Complete sequence, recombinant analysis and binding to laminins and sulphated ligands of the N-terminal domains of laminin α3B and α5 chains. Biochem J. 2002;362:213–21. doi: 10.1042/0264-6021:3620213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paulsson M, Aumailley M, Deutzmann R, Timpl R, Beck K, Engel J. Laminin-nidogen complex. Extraction with chelating agents and structural characterization. Eur J Biochem. 1987;166:11–9. doi: 10.1111/j.1432-1033.1987.tb13476.x. [DOI] [PubMed] [Google Scholar]
- 67.Aumailley M, Wiedemann H, Mann K, Timpl R. Binding of nidogen and the laminin-nidogen complex to basement membrane collagen type IV. Eur J Biochem. 1989;184:241–8. doi: 10.1111/j.1432-1033.1989.tb15013.x. [DOI] [PubMed] [Google Scholar]
- 68.Mann K, Deutzmann R, Aumailley M, Timpl R, Raimondi L, Yamada Y, et al. Amino acid sequence of mouse nidogen, a multidomain basement membrane protein with binding activity for laminin, collagen IV and cells. EMBO J. 1989;8:65–72. doi: 10.1002/j.1460-2075.1989.tb03349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerl M, Mann K, Aumailley M, Timpl R. Localization of a major nidogen-binding site to domain III of laminin B2 chain. Eur J Biochem. 1991;202:167–74. doi: 10.1111/j.1432-1033.1991.tb16358.x. [DOI] [PubMed] [Google Scholar]
- 70.Mayer U, Nischt R, Pöschl E, Mann K, Fukuda K, Gerl M, et al. A single EGF-like motif of laminin is responsible for high affinity nidogen binding. EMBO J. 1993;12:1879–85. doi: 10.1002/j.1460-2075.1993.tb05836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pöschl E, Fox JW, Block D, Mayer U, Timpl R. Two non-contiguous regions contribute to nidogen binding to a single EGF-like motif of the laminin γ 1 chain. EMBO J. 1994;13:3741–7. doi: 10.1002/j.1460-2075.1994.tb06683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pöschl E, Mayer U, Stetefeld J, Baumgartner R, Holak TA, Huber R, et al. Site-directed mutagenesis and structural interpretation of the nidogen binding site of the laminin γ1 chain. EMBO J. 1996;15:5154–9. [PMC free article] [PubMed] [Google Scholar]
- 73.Gersdorff N, Kohfeldt E, Sasaki T, Timpl R, Miosge N. Laminin γ3 chain binds to nidogen and is located in murine basement membranes. J Biol Chem. 2005;280:22146–53. doi: 10.1074/jbc.M501875200. [DOI] [PubMed] [Google Scholar]
- 74.Sasaki T, Göhring W, Mann K, Brakebusch C, Yamada Y, Fässler R, et al. Short arm region of laminin-5 γ2 chain: structure, mechanism of processing and binding to heparin and proteins. J Mol Biol. 2001;314:751–63. doi: 10.1006/jmbi.2001.5176. [DOI] [PubMed] [Google Scholar]
- 75.Mokkapati S, Baranowsky A, Mirancea N, Smyth N, Breitkreutz D, Nischt R. Basement membranes in skin are differently affected by lack of nidogen 1 and 2. J Invest Dermatol. 2008;128:2259–67. doi: 10.1038/jid.2008.65. [DOI] [PubMed] [Google Scholar]
- 76.Krieg T, Aumailley M. The extracellular matrix of the dermis: flexible structures with dynamic functions. Exp Dermatol. 2011;20:689–95. doi: 10.1111/j.1600-0625.2011.01313.x. [DOI] [PubMed] [Google Scholar]
- 77.Champliaud MF, Lunstrum GP, Rousselle P, Nishiyama T, Keene DR, Burgeson RE. Human amnion contains a novel laminin variant, laminin 7, which like laminin 6, covalently associates with laminin 5 to promote stable epithelial-stromal attachment. J Cell Biol. 1996;132:1189–98. doi: 10.1083/jcb.132.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen M, Marinkovich MP, Veis A, Cai X, Rao CN, O’Toole EA, et al. Interactions of the amino-terminal noncollagenous (NC1) domain of type VII collagen with extracellular matrix components. A potential role in epidermal-dermal adherence in human skin. J Biol Chem. 1997;272:14516–22. doi: 10.1074/jbc.272.23.14516. [DOI] [PubMed] [Google Scholar]
- 79.Rousselle P, Keene DR, Ruggiero F, Champliaud MF, Rest M, Burgeson RE. Laminin 5 binds the NC-1 domain of type VII collagen. J Cell Biol. 1997;138:719–28. doi: 10.1083/jcb.138.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen M, Marinkovich MP, Jones JC, O’Toole EA, Li YY, Woodley DT. NC1 domain of type VII collagen binds to the β3 chain of laminin 5 via a unique subdomain within the fibronectin-like repeats. J Invest Dermatol. 1999;112:177–83. doi: 10.1046/j.1523-1747.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 81.Tasanen K, Tunggal L, Chometon G, Bruckner-Tuderman L, Aumailley M. Keratinocytes from patients lacking collagen XVII display a migratory phenotype. Am J Pathol. 2004;164:2027–38. doi: 10.1016/S0002-9440(10)63762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Utani A, Nomizu M, Yamada Y. Fibulin-2 binds to the short arms of laminin-5 and laminin-1 via conserved amino acid sequences. J Biol Chem. 1997;272:2814–20. doi: 10.1074/jbc.272.5.2814. [DOI] [PubMed] [Google Scholar]


