Abstract
The heterotrimeric laminins are a defining component of all basement membranes and self-assemble into a cell-associated network. The three short arms of the cross-shaped laminin molecule form the network nodes, with a strict requirement for one α, one β and one γ arm. The globular domain at the end of the long arm binds to cellular receptors, including integrins, α-dystroglycan, heparan sulfates and sulfated glycolipids. Collateral anchorage of the laminin network is provided by the proteoglycans perlecan and agrin. A second network is then formed by type IV collagen, which interacts with the laminin network through the heparan sulfate chains of perlecan and agrin and additional linkage by nidogen. This maturation of basement membranes becomes essential at later stages of embryo development.
Keywords: laminin, collagen IV, nidogen, agrin, perlecan, dystroglycan
Introduction
Basement membranes (BMs) are cell-associated extracellular matrices that cover the basal aspect of epithelial and endothelial cells and surround muscle, fat and Schwann cells. They are defined morphologically by their characteristic appearance in electron micrographs and biochemically by their typical components, which include laminins, type IV collagen, nidogen and the heparan sulfate proteoglycans (HSPGs) perlecan and agrin.1,2 BMs are evolutionarily ancient and appear to have been required for the emergence of tissues and differentiated cells. The typical BM proteins consequently are found in all metazoa.3 The myriad functions of BMs in embryo development, tissue homeostasis and human disease have been reviewed elsewhere.1,4-6 Here, we discuss the architecture of BMs and how they are assembled, concentrating on the critical role of laminins in this process.
In electron micrographs BMs appear as electron-dense material in close apposition with the cell surface. Conventional fixation produces a dense layer (lamina densa) separated from the plasma membrane by a translucent layer (lamina lucida), while the milder method of freeze substitution results in a homogeneous appearance of the BM.7 Most BMs are 50–100 nm thick, but some specialized BMs are thicker, e.g., the extra-embryonic Reichert’s membrane and the kidney glomerular BM. Studying the ultrastructure of thin BMs in situ is difficult. The most detailed images have been obtained from the extracellular matrix of the mouse Engelbreth-Holm-Swarm (EHS) sarcoma, which is rich in BM proteins and resembles a thick BM. Quick-freeze deep-etch electron microscopy revealed the EHS extracellular matrix as a three-dimensional meshwork with an average pore size of ~10 nm.8 Removal of type IV collagen by collagenase digestion revealed a more open network with a strut length of ~30 nm, which resembled closely the network obtained by polymerizing purified EHS laminin in vitro.9 The molecular interpretation of this laminin network was greatly helped by structure-function studies with proteolytic EHS laminin fragments (see below).
All laminins are heterotrimers composed of one of five α chains, one of three β chains and one of three γ chains (in mammals). Out of all possible combinations, a total of 16 laminin isoforms have been characterized biochemically. EHS laminin has the chain composition α1β1γ1 and is now referred to as laminin-111.10 Laminins appear as cross-shaped molecules in rotary shadowing electron micrographs.11,12 The long arm of the cross (~80 nm length) is an α-helical coiled coil formed from all three chains, whereas the three short arms (35–50 nm) are composed of one chain each (Fig. 1A). At the distal end of the long arm, the α chain adds five laminin G-like (LG) domains that contain the major cell-adhesive sites of laminin. The homologous short arms are composed of a distal laminin N-terminal (LN) domain that is followed by tandem repeats of laminin-type epidermal growth factor-like (LE) domains, interspersed with globular domains of unknown structure. As discussed below, the LN domains are essential for laminin polymerization and BM assembly. Eight of the 16 laminin isoforms do not have a full complement of LN domains10 and this is predicted to limit their ability to form homopolymers.

Figure 1. Domain structure and self-assembly of laminin-111. (A) Schematic drawing of the laminin-111 heterotrimer. The three short arms of the cross-shaped molecule have a common domain structure and consist of laminin N-terminal (LN) domains, laminin-type epidermal growth factor-like (LE) domains, and L4 domains, as indicated for the α1 chain. The α1 chain uniquely contains five laminin G-like (LG) domains. LG1-3 likely interact with the C-terminal residues of the γ1 chain. (B) The three-arm interaction model of laminin self-assembly. The ternary nodes in the network are formed by the N-terminal regions of one α, one β and one γ chain. The long arm of the laminin heterotrimer is not involved in network formation.
Laminin Polymerization in Solution
The first mechanistic insights into the process of laminin polymerization came from studies using purified EHS laminin-111, which forms a gel at physiological temperature and in the presence of calcium ions.13,14 Limited proteolysis of laminin-111 yielded several defined fragments.15 The only fragment that retained the ability to polymerize contained all three short arms (C1–4 fragment);16 fragments containing less than three short arms (E1, E1′ and E4) did not polymerize but inhibited polymerization of the C1–4 fragment; and a short-arm fragment lacking the LN domains (P1′) did not inhibit polymerization.17 In direct binding studies, most fragments interacted very weakly or not at all, with the single exception of E1′ (short arms of α1 and γ1) and E4 (short arm of β1). Finally, electron micrographs showed that the short arms interacted through their globular LN domains, with a strong preference for ternary interactions involving one α1, one β1 and one γ1 short arm. These observations were synthesized into the “three-arm interaction model” of laminin polymerization (Fig. 1B).18 Soon after, the dy2J muscular dystrophy mouse was shown to have a destabilizing deletion in the LN domain of the laminin α2 chain,19 which resulted in truncated laminin-211/221 heterotrimers that were unable to polymerize in vitro.20 These and other studies firmly established that the LN domains in laminins are essential for polymerization.
The recombinant production of full-length heterotrimeric laminins and monomeric short arm fragments enabled structural and mechanistic experiments that had not been possible with the limited set of proteolytic fragments. The crystal structure of the laminin α5 LN-LEa1-2 fragment revealed that the LN domain is a β-sandwich with several elaborate loops that is attached like the head of a sunflower to a stalk made up of the LE domains (Fig. 2A).21 The N-terminal segment of the LN domain interacts intimately with the first LE domain, explaining why LN domains cannot be produced in isolation.22,23 The seemingly rigid LE domain tandem does not have a conventional hydrophobic core, but is stabilized by a ladder of disulfide bonds linked in a [1–3, 2–4, 5–6, 7–8] pattern; this linkage pattern was first described for the laminin γ1 LEb2-4 fragment.24 The α5 LN-LEa1-2 crystal structure is in good agreement with a low-resolution structure of the entire short short arm of the γ1 chain.25 An analysis of surface conservation identified a patch on the α5 LN domain that was shown by mutagenesis to be involved in laminin network interactions,21 but how the α chain interacts with the other two chains in the laminin network is currently not known.

Figure 2. Crystal structures of laminin fragments. Polypeptide chains are shown in cartoon representation, disulfide bonds as yellow sticks and calcium ions as magenta spheres. The N- and C-termini are labeled. (A) Crystal structure of the laminin α5 LN-LEa1-2 fragment.21 Three residues whose mutation abolishes the inhibitory activity of α5 LN-LEa1-2 on laminin-111 polymerization21 are shown as pink sticks. The dotted line indicates a loop region that is disordered in the α5 LN-LEa1-2 crystal structure; its general location can be inferred from structures of the related netrin G proteins.102,103 (B) Putative structure of the laminin LG1-5 region, assembled from crystal structures of the α2 LG1-3 and α1 LG4-5 fragments.33,34 The dotted lines indicate linker regions that were not resolved by the crystal structures. In the intact laminin heterotrimer, the LG1 domain is predicted to interact tightly with the LG2-3 pair (see text). Basic residues in LG4 whose mutation reduces α-DG, heparin and sulfatide binding34 are shown as pink sticks. The calcium ion in LG4 is essential for α-DG binding.54 (C) Crystal structure of the laminin γ1 LEb2-4 fragment (blue) bound to domain G3 of nidogen-1 (magenta).89 Two residues whose mutation abolishes nidogen-1 binding88 are shown as pink sticks.
A comprehensive analysis of laminin short arm interactions detected binary interactions with dissociation constants in the 0.01–1 μM range for the majority of α-α, α-β, α-γ and β-γ pairings, and the authors concluded that the laminin network might be less regular than in the three-arm model.23 However, a more recent study failed to detect the reported α5-α5, α5-β1 and α5-γ1 interactions and found only a weak interaction between the β1 and γ1 short arms. Consistent with the three-arm model, a stable complex was observed only when all three short arms (α5, β1 and γ1) were added together.21 Furthermore, in experiments with recombinant laminin-111 heterotrimers, deletion of any single LN domain or replacement of β1 LN or γ1 LN with α1 LN abolished polymerization.26 Finally, it was shown that a chimeric protein in which the laminin α1 LN-LEa1-4 region was fused to the laminin γ1 chain-binding region of nidogen (αLNNd) could restore polymerization to a laminin lacking α1 LN, but not to one lacking β1 LN or γ1 LN.27 Collectively, these data strongly support the three-arm model.
A remaining open question is whether, in a ternary network node, the LN domains interact with each other (as shown in Fig. 1B), with the LE stalks of another chain, or both. Because the LN domains cannot be produced without adjacent LE domains,22,23 this question will have to be addressed by mutagenesis or structure determination of a complete network node.
Laminin Binding to the Cell Surface
Laminin polymerization in vivo does not occur in solution but at the cell surface, to which laminins are anchored through direct or indirect interactions with cellular receptors (Fig. 3). Confinement of laminin to two dimensions through cell surface anchorage promotes polymerization by increasing the surface laminin concentration above that of the surrounding milieu. This effect was first demonstrated in vitro using planar lipid bilayers containing sulfated glycolipids28 and has since been studied extensively on cultured cells.26,27,29,30
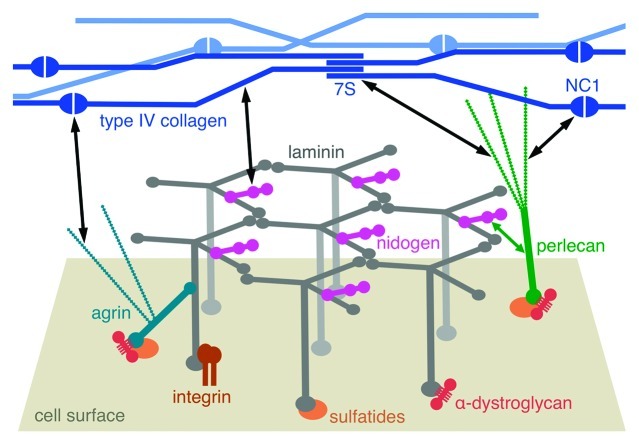
Figure 3. Schematic drawing of the molecular structure of a basement membrane. The laminin network is anchored to the cell surface by interactions of the long arms with cellular receptors (integrins, α-dystroglycan and sulfated glycolipids/sulfatides). Collateral interactions are made with the heparan sulfate proteoglycans agrin and perlecan. An independent network is formed by type IV collagen, through interactions of its N-terminal 7S and C-terminal NC1 domains, as well as through lateral associations of the triple helices. The laminin and collagen networks are linked by nidogen and heparan sulfates (black double-headed arrows).
Cellular receptors predominantly bind to the five LG domains at the C-terminal end of the long arm of laminins. In negatively stained electron micrographs of laminin-111, the LG1-3 region of laminins has the appearance of a cloverleaf to which is flexibly attached the LG4-5 pair.31 The LG domain has a lectin-like β-sandwich fold and many LG domains have a calcium ion bound to one edge of the sandwich.32 Crystal structures are available of the LG1-3 region of the laminin α2 chain33 and the LG4-5 regions of the α1 and α2 chains (Fig. 2B).34,35 The LG1-3 structure is more open than the cloverleaf seen in the electron micrographs, presumably due to the absence of the coiled coil (see below). The LG4-5 structures show a seemingly rigid V-shaped arrangement of the two LG domains. The long linker between LG3 and LG4 is disulfide-bonded to LG5, which results in LG4 being the distal domain in the LG1–5 structure (Fig. 2B).31
Integrin binding requires the LG1-3 region in the context of the heterotrimer,36-38 while α-dystroglycan (α-DG), heparan sulfates and sulfated glycolipids (sulfatides) mostly bind to the LG4-5 region.15,37,39-41 The major laminin-binding integrins are α3β1, α6β1, α7β1 and α6β4; their specificities for the different laminin isoforms have been determined.42 Experiments with recombinant laminin-511 heterotrimers have shown that the γ1 chain tail, and in particular a glutamic acid in the third position from the C-terminus, is required for integrin binding.38,43 It is not known whether the γ1 chain tail interacts directly with the integrin or is required to maintain a LG1-3 conformation that is competent for integrin binding. C-terminal truncation of the γ2 chain in laminin-332 results in a more open LG1-3 structure and abrogates integrin binding, suggesting that the γ chain tail might have a structural role in maintaining a competent LG1-3 structure.44 Indeed, in the α2 LG1-3 crystal structure (without a stabilizing γ chain tail) the LG1 domain was found to be dissociated from the LG2-3 pair.33 The structure determination of a heterotrimeric, integrin-binding, laminin fragment represents a formidable challenge, but will ultimately be needed to understand how the γ chain participates in integrin binding.
In the laminin α5 chain, the LG1-3 region also binds to the Lutheran blood group antigen/basal cell adhesion molecule (Lu/B-CAM), a cell surface protein consisting of five immunoglobulin-like domains.45,46 The binding determinants for Lu/B-CAM on laminin-511 are similar to those for integrins and the binding sites for the two receptors are overlapping.
The laminin receptor dystroglycan consists of a membrane-spanning β subunit and an extracellular α subunit, which are derived from a single gene product by post-translational cleavage. Dystroglycan is part of the dystrophin glycoprotein complex, which has important roles in the nervous system and in maintaining the integrity of the skeletal muscle membrane.47 Laminin binds to the highly glycosylated α subunit of dystroglycan in a calcium- and carbohydrate-dependent manner.39,48 Among the several O-linked carbohydrate modifications found on α-DG, the laminin-binding modification remained elusive for a long time and a breakthrough was achieved only recently. Building on the genetic analysis of dystroglycanopathies (i.e., diseases resulting from defective α-DG modification), a series of elegant biochemical experiments revealed that laminin binding requires a phosphorylated O-mannosyl core to which the glycosyltransferase LARGE adds a chain of alternating xylose (Xyl) and glucuronic acid (GlcA) moieties.49,50 Overexpression of LARGE also modifies other glycan cores, but whether these modifications bind laminin in a physiological setting is not clear.51-53 The α-DG binding sites in laminins generally are located in the LG4-5 region, but the α2 chain contains an additional binding site in the LG1-3 region.39-41 The calcium ion in LG4 and several adjacent basic residues are required for α-DG binding to the α1 chain;34,54 it is notable that these binding determinants are located at the very tip of the LG1-5 region (Fig. 2B). The calcium ion has an incomplete coordination sphere and a GlcA carboxylate group of α-DG most likely binds directly to the ion.32 How the (presumed) specificity for Xyl-GlcA chains is achieved is not known. The remaining two receptors for direct laminin anchorage, heparan sulfates and sulfated glycolipids, bind to basic surface patches in the LG4-5 region, which are overlapping but not identical with the binding sites for α-DG.34,54-57
Laminin Polymerization at the Cell Surface
Laminins are essential for BM assembly. The first two BMs to assemble are those of the mouse embryonic plate and Reichert’s membrane in the peri-implantation period. Assembly of the BM of the embryonic plate, initially residing between visceral endoderm and inner cell mass, is followed by the formation of a central cavity and differentiation of inner cell mass to a polarized pseudostratified epithelial layer, the epiblast. Genetic ablation of either the laminin γ1 or β1 chain was found to prevent laminin heterotrimer formation and BM assembly, causing early (E5.5 in mouse) embryonic lethality.58,59 Ablation of expression of the other key BM components, on the other hand, was found to cause developmental defects that presented only later (E10 to perinatal period) in development and did not prevent formation of most BMs in tissues.60-64 The unique role of laminins for BM assembly and the role of BM in the mediation of differentiation was recapitulated in vitro. Embryonic stem (ES) cells grown as small suspended aggregates form an outer endoderm layer that secretes laminin-111 and laminin-511, nidogen, perlecan and type IV collagen. These components are assembled into a thick sub-endodermal BM followed by formation of a polarized epiblast layer from the underlying previously undifferentiated ES cells.65 ES cells null for Lamc1, unlike the wild-type counterpart, failed to assemble a BM and did not convert ES cells into epiblast.66,67 BM assembly could be restored by the addition of laminin-111 to the culture medium. However, this assembly and accompanying cell polarization was prevented with laminin proteolytic fragments that inhibit either polymerization or LG domain-mediated cell adhesion.67 In later studies examining BM assembly on cultured Schwann cells, it was found that laminin-111 from which all LG domains were deleted was unable to assemble an ECM.26 Thus, genetic studies, supported by in vitro observations, established that laminins are uniquely required for ECM assembly.
The studies also implied that laminins must first bind to the cell surface, allowing recruitment of the other laminin-binding components into the nascent self-assembling BM. As described above, the principal cell-binding sites for laminins (i.e., integrins, α-DG, heparan sulfates and sulfated glycolipids) are found in the LG domains. Since BMs are found in tissues in which either integrin or dystroglycan gene expression has been ablated, it seemed unlikely that these receptor classes are essential for laminin assembly (for discussions, see refs. 66 and 68). Integrins α6β1, α7β1, and α3β1 and α-DG have a unique role in establishing firm anchorage of the LG domains to the actin cytoskeleton, however. A number of integrin-associated proteins (integrin-linked kinase, vinculin, talin and filamin) bind to F-actin, either directly or indirectly,69 and the cytoplasmic domain of β-dystroglycan is connected to F-actin through the cytoskeletal proteins dystrophin and utrophin.70-72
One way in which laminins may adhere to cell surfaces independently of integrins or α-DG is provided by sulfated glycolipids (sulfated glycoceramides and presumably other related lipids). These molecules are present in the outer leaflet of the plasma membrane of Schwann, renal and other cells and bind with substantial affinity to the LG4 domain of laminin-111 and the LG3 and LG4-5 domains of laminin-211.30
Collateral Linkage of Laminin to the Cell Surface
In addition to the direct laminin-receptor interactions described so far, there exist other, indirect, mechanisms whereby laminins can be tethered to the cell surface. An important connection is provided by agrin, a HSPG present in many BMs.73 Its N-terminal domain binds to the laminin γ1 chain within the coiled coil region of the long arm,74,75 and its C-terminal LG domains bind to α-DG.73 These interactions are of high affinity and the agrin bridge has been shown to augment laminin anchorage both in vitro and in vivo.27,76 The biological functions of agrin are regulated by alternative splicing of its LG3 domain.73 The “non-neural” splice variant is responsible for agrin’s contribution to BM formation and stability, whereas the “neural” splice variant plays an important role in the formation of neuromuscular junctions by mediating the binding of agrin to low-density lipoprotein receptor-related protein 4 and thereby activating the muscle-specific receptor tyrosin kinase, MuSK.77-79 The other major HSPG of BMs, perlecan, also has the potential to provide a bridge between laminin and the cell surface. Like agrin, perlecan contains α-DG-binding LG domains,40 and it may connect to laminin directly through its heparan sulfate chains or indirectly through nidogen.
Basement Membrane Maturation
Type IV collagen is common to all mammalian BMs throughout development and adulthood. It is the only other component apart from laminin that forms a polymer.8,80,81 In the invertebrate C. elegans, type IV collagen deposition into a BM follows that of laminins by several developmental stages.82,83 Mutations in the Gly-X-Y repeats of type IV collagen were temperature-sensitive and found to result in late embryonic lethality.84 Genetic ablation of type IV collagen in mice resulted in lethality between E10 and E11, substantially due to a disruption of Reichert’s membrane.62 Prior to that failure, tissues containing type IV collagen-deficient BMs appeared normal. Thus, type IV collagen incorporation into BMs can be considered a maturation step that provides structural stability, which becomes critical in later development.
Nidogen-1 (entactin) is a glycoprotein that binds with high affinity to the LEb3 domain of the laminin γ1 chain and additionally contains binding sites for perlecan and type IV collagen.85-87 The crystal structure of a minimal laminin-nidogen complex showed that a critical loop in LE3b88 is inserted into the central depression of the β-propeller of the nidogen G3 domain (Fig. 2C).89 In keeping with its ability to form multiple interactions, nidogen-1 was found to efficiently bridge type IV collagen to laminin assembled on a Schwann cell surface.26 However, the hypothesis that nidogen serves as the major bridge between the laminin and type IV collagen networks2 has not been supported by genetic and developmental evidence.60,90,91 Thus, there must be another general mechanism to accomplish this important function, acting in concert with nidogen or in the absence of a nidogen bridge.
A recent study on the epidermal BM has shown that it is heparan sulfates that serve this function.92 Fragmentation of the BM revealed separable laminin-enriched and type IV collagen-enriched fractions connected through heparan sulfates. The evidence implicated perlecan as playing a major role, binding to the laminin-enriched fraction through its core protein. However, since perlecan-deficient mice have BMs containing type IV collagen in most tissues,63 it seems unlikely that perlecan is the sole substitute for nidogen. The most likely compensating candidate is agrin. In this model (Fig. 3), the heparan sulfate chains of both perlecan and agrin would extend from the nidogen-containing laminin network and bind type IV collagen, most likely at its 7S and NC1 domains.93,94
Non-Polymerizing Laminins
Laminin-3A32, -3A11, -3A21, -411 and -421 are laminins that cannot polymerize because they do not possess a full complement of LN domains. Laminin-3A32 (“A” denotes a short splice-variant while “B” denotes the full-length variant), an epithelial laminin and the most extensively studied, binds to several integrins through its LG domains (notably to α6β4 and α3β1 integrin) and to type VII collagen, forming a connecting bridge between cellular hemidesmosomes and stromal anchoring fibrils.95 Laminin-3A11 can similarly bind to the cell surface but lacks binding to type VII collagen. Its accumulation in BMs, like that of laminin-411, may depend upon its interactions with perlecan, agrin and nidogen.
Concluding Remarks
Over three decades of laminin research have elucidated many functions of these fascinating molecules, but important questions remain. The difficulty of imaging BMs in situ, without harmful extraction from tissues, has prevented a clearer understanding of BM architecture and our current model is largely based on a multitude of indirect clues rather than on direct observation. Recent advances in cryo-electron tomography of vitrified tissue sections have produced detailed views of native desmosomes,96 for example, but whether this technique can be applied to the study of BMs remains to be seen. An intriguing aspect is that the thickness of a typical BM is of the same order as the dimensions of a single laminin molecule, which makes it unlikely that laminins are standing erect on the cell surface (as shown for clarity in Fig. 3). Conceivably, the short arms could even interact with the cell surface, which might help explain the poorly understood binding of α1β1 and α2β1 integrins to α chain short arms.22,97 Another question is whether we actually know all the molecular interactions that are important for BM assembly and maturation. Some of our current knowledge is based on early experiments with relatively crude protein preparations, and a search for additional interactions using modern recombinant and proteomic techniques might prove fruitful. In this context, one must also consider the possibility that pairwise interactions detected in vitro may not always be relevant in vivo.98 Finally, not all BMs are created equal and it will be important to study how the more specialized components, such as the netrins,99 Slits100 or Fras1/FREM proteins,101 are woven into the basic fabric of the BM.
Acknowledgments
E.H. acknowledges receipt of a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science (ref. 083942/Z/07/Z). P.D.Y. acknowledges support of a grant from the National Institutes of Health (R37-DK36425).
Glossary
Abbreviations:
- α-DG
α-dystroglycan
- BM
basement membrane
- ECM
extracellular matrix
- EHS tumor
Engelbreth-Holm-Swarm tumor
- ES cells
embryonic stem cells
- GlcA
glucuronic acid
- HSPG
heparan sulfate proteoglycan
- LE domain
laminin-type epidermal growth factor-like domain
- LG domain
laminin G-like domain
- LN domain
laminin N-terminal domain
- Lu/B-CAM
Lutheran blood group antigen/basal cell adhesion molecule
- Xyl
xylose
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/21831
References
- 1.Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3:a004911. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Timpl R, Brown JC. Supramolecular assembly of basement membranes. Bioessays. 1996;18:123–32. doi: 10.1002/bies.950180208. [DOI] [PubMed] [Google Scholar]
- 3.Hynes RO. The evolution of metazoan extracellular matrix. J Cell Biol. 2012;196:671–9. doi: 10.1083/jcb.201109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miner JH, Yurchenco PD. Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol. 2004;20:255–84. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]
- 5.Van Agtmael T, Bruckner-Tuderman L. Basement membranes and human disease. Cell Tissue Res. 2010;339:167–88. doi: 10.1007/s00441-009-0866-y. [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3:422–33. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 7.Miosge N. The ultrastructural composition of basement membranes in vivo. Histol Histopathol. 2001;16:1239–48. doi: 10.14670/HH-16.1239. [DOI] [PubMed] [Google Scholar]
- 8.Yurchenco PD, Ruben GC. Basement membrane structure in situ: evidence for lateral associations in the type IV collagen network. J Cell Biol. 1987;105:2559–68. doi: 10.1083/jcb.105.6.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yurchenco PD, Cheng YS, Colognato H. Laminin forms an independent network in basement membranes. J Cell Biol. 1992;117:1119–33. doi: 10.1083/jcb.117.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, et al. A simplified laminin nomenclature. Matrix Biol. 2005;24:326–32. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Beck K, Hunter I, Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J. 1990;4:148–60. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- 12.Engel J, Odermatt E, Engel A, Madri JA, Furthmayr H, Rohde H, et al. Shapes, domain organizations and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix. J Mol Biol. 1981;150:97–120. doi: 10.1016/0022-2836(81)90326-0. [DOI] [PubMed] [Google Scholar]
- 13.Paulsson M. The role of Ca2+ binding in the self-aggregation of laminin-nidogen complexes. J Biol Chem. 1988;263:5425–30. [PubMed] [Google Scholar]
- 14.Yurchenco PD, Tsilibary EC, Charonis AS, Furthmayr H. Laminin polymerization in vitro. Evidence for a two-step assembly with domain specificity. J Biol Chem. 1985;260:7636–44. [PubMed] [Google Scholar]
- 15.Ott U, Odermatt E, Engel J, Furthmayr H, Timpl R. Protease resistance and conformation of laminin. Eur J Biochem. 1982;123:63–72. doi: 10.1111/j.1432-1033.1982.tb06499.x. [DOI] [PubMed] [Google Scholar]
- 16.Bruch M, Landwehr R, Engel J. Dissection of laminin by cathepsin G into its long-arm and short-arm structures and localization of regions involved in calcium dependent stabilization and self-association. Eur J Biochem. 1989;185:271–9. doi: 10.1111/j.1432-1033.1989.tb15112.x. [DOI] [PubMed] [Google Scholar]
- 17.Schittny JC, Yurchenco PD. Terminal short arm domains of basement membrane laminin are critical for its self-assembly. J Cell Biol. 1990;110:825–32. doi: 10.1083/jcb.110.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yurchenco PD, Cheng YS. Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J Biol Chem. 1993;268:17286–99. [PubMed] [Google Scholar]
- 19.Xu H, Wu XR, Wewer UM, Engvall E. Murine muscular dystrophy caused by a mutation in the laminin α 2 (Lama2) gene. Nat Genet. 1994;8:297–302. doi: 10.1038/ng1194-297. [DOI] [PubMed] [Google Scholar]
- 20.Colognato H, Yurchenco PD. The laminin α2 expressed by dystrophic dy(2J) mice is defective in its ability to form polymers. Curr Biol. 1999;9:1327–30. doi: 10.1016/S0960-9822(00)80056-1. [DOI] [PubMed] [Google Scholar]
- 21.Hussain SA, Carafoli F, Hohenester E. Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region. EMBO Rep. 2011;12:276–82. doi: 10.1038/embor.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ettner N, Göhring W, Sasaki T, Mann K, Timpl R. The N-terminal globular domain of the laminin α1 chain binds to α1β1 and α2β1 integrins and to the heparan sulfate-containing domains of perlecan. FEBS Lett. 1998;430:217–21. doi: 10.1016/S0014-5793(98)00601-2. [DOI] [PubMed] [Google Scholar]
- 23.Odenthal U, Haehn S, Tunggal P, Merkl B, Schomburg D, Frie C, et al. Molecular analysis of laminin N-terminal domains mediating self-interactions. J Biol Chem. 2004;279:44504–12. doi: 10.1074/jbc.M402455200. [DOI] [PubMed] [Google Scholar]
- 24.Stetefeld J, Mayer U, Timpl R, Huber R. Crystal structure of three consecutive laminin-type epidermal growth factor-like (LE) modules of laminin γ1 chain harboring the nidogen binding site. J Mol Biol. 1996;257:644–57. doi: 10.1006/jmbi.1996.0191. [DOI] [PubMed] [Google Scholar]
- 25.Patel TR, Morris GA, Zwolanek D, Keene DR, Li J, Harding SE, et al. Nano-structure of the laminin γ-1 short arm reveals an extended and curved multidomain assembly. Matrix Biol. 2010;29:565–72. doi: 10.1016/j.matbio.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 26.McKee KK, Harrison D, Capizzi S, Yurchenco PD. Role of laminin terminal globular domains in basement membrane assembly. J Biol Chem. 2007;282:21437–47. doi: 10.1074/jbc.M702963200. [DOI] [PubMed] [Google Scholar]
- 27.McKee KK, Capizzi S, Yurchenco PD. Scaffold-forming and Adhesive Contributions of Synthetic Laminin-binding Proteins to Basement Membrane Assembly. J Biol Chem. 2009;284:8984–94. doi: 10.1074/jbc.M809719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalb E, Engel J. Binding and calcium-induced aggregation of laminin onto lipid bilayers. J Biol Chem. 1991;266:19047–52. [PubMed] [Google Scholar]
- 29.Colognato H, Winkelmann DA, Yurchenco PD. Laminin polymerization induces a receptor-cytoskeleton network. J Cell Biol. 1999;145:619–31. doi: 10.1083/jcb.145.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S, Liquari P, McKee KK, Harrison D, Patel R, Lee S, et al. Laminin-sulfatide binding initiates basement membrane assembly and enables receptor signaling in Schwann cells and fibroblasts. J Cell Biol. 2005;169:179–89. doi: 10.1083/jcb.200501098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E. Structure and function of laminin LG modules. Matrix Biol. 2000;19:309–17. doi: 10.1016/S0945-053X(00)00072-X. [DOI] [PubMed] [Google Scholar]
- 32.Hohenester E, Tisi D, Talts JF, Timpl R. The crystal structure of a laminin G-like module reveals the molecular basis of α-dystroglycan binding to laminins, perlecan, and agrin. Mol Cell. 1999;4:783–92. doi: 10.1016/S1097-2765(00)80388-3. [DOI] [PubMed] [Google Scholar]
- 33.Carafoli F, Clout NJ, Hohenester E. Crystal structure of the LG1-3 region of the laminin α2 chain. J Biol Chem. 2009;284:22786–92. doi: 10.1074/jbc.M109.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison D, Hussain SA, Combs AC, Ervasti JM, Yurchenco PD, Hohenester E. Crystal structure and cell surface anchorage sites of laminin α1LG4-5. J Biol Chem. 2007;282:11573–81. doi: 10.1074/jbc.M610657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tisi D, Talts JF, Timpl R, Hohenester E. Structure of the C-terminal laminin G-like domain pair of the laminin α2 chain harbouring binding sites for α-dystroglycan and heparin. EMBO J. 2000;19:1432–40. doi: 10.1093/emboj/19.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deutzmann R, Aumailley M, Wiedemann H, Pysny W, Timpl R, Edgar D. Cell adhesion, spreading and neurite stimulation by laminin fragment E8 depends on maintenance of secondary and tertiary structure in its rod and globular domain. Eur J Biochem. 1990;191:513–22. doi: 10.1111/j.1432-1033.1990.tb19151.x. [DOI] [PubMed] [Google Scholar]
- 37.Ido H, Harada K, Futaki S, Hayashi Y, Nishiuchi R, Natsuka Y, et al. Molecular dissection of the α-dystroglycan- and integrin-binding sites within the globular domain of human laminin-10. J Biol Chem. 2004;279:10946–54. doi: 10.1074/jbc.M313626200. [DOI] [PubMed] [Google Scholar]
- 38.Ido H, Nakamura A, Kobayashi R, Ito S, Li S, Futaki S, et al. The requirement of the glutamic acid residue at the third position from the carboxyl termini of the laminin γ chains in integrin binding by laminins. J Biol Chem. 2007;282:11144–54. doi: 10.1074/jbc.M609402200. [DOI] [PubMed] [Google Scholar]
- 39.Gee SH, Blacher RW, Douville PJ, Provost PR, Yurchenco PD, Carbonetto S. Laminin-binding protein 120 from brain is closely related to the dystrophin-associated glycoprotein, dystroglycan, and binds with high affinity to the major heparin binding domain of laminin. J Biol Chem. 1993;268:14972–80. [PubMed] [Google Scholar]
- 40.Talts JF, Andac Z, Göhring W, Brancaccio A, Timpl R. Binding of the G domains of laminin α1 and α2 chains and perlecan to heparin, sulfatides, α-dystroglycan and several extracellular matrix proteins. EMBO J. 1999;18:863–70. doi: 10.1093/emboj/18.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smirnov SP, McDearmon EL, Li S, Ervasti JM, Tryggvason K, Yurchenco PD. Contributions of the LG modules and furin processing to laminin-2 functions. J Biol Chem. 2002;277:18928–37. doi: 10.1074/jbc.M201880200. [DOI] [PubMed] [Google Scholar]
- 42.Nishiuchi R, Takagi J, Hayashi M, Ido H, Yagi Y, Sanzen N, et al. Ligand-binding specificities of laminin-binding integrins: a comprehensive survey of laminin-integrin interactions using recombinant α3β1, α6β1, α7β1 and α6β4 integrins. Matrix Biol. 2006;25:189–97. doi: 10.1016/j.matbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Ido H, Ito S, Taniguchi Y, Hayashi M, Sato-Nishiuchi R, Sanzen N, et al. Laminin isoforms containing the γ3 chain are unable to bind to integrins due to the absence of the glutamic acid residue conserved in the C-terminal regions of the γ1 and γ2 chains. J Biol Chem. 2008;283:28149–57. doi: 10.1074/jbc.M803553200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navdaev A, Heitmann V, Desantana Evangelista K, Mörgelin M, Wegener J, Eble JA. The C-terminus of the γ 2 chain but not of the β 3 chain of laminin-332 is indirectly but indispensably necessary for integrin-mediated cell reactions. Exp Cell Res. 2008;314:489–97. doi: 10.1016/j.yexcr.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 45.Udani M, Zen Q, Cottman M, Leonard N, Jefferson S, Daymont C, et al. Basal cell adhesion molecule/lutheran protein. The receptor critical for sickle cell adhesion to laminin. J Clin Invest. 1998;101:2550–8. doi: 10.1172/JCI1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kikkawa Y, Sasaki T, Nguyen MT, Nomizu M, Mitaka T, Miner JH. The LG1-3 tandem of laminin α5 harbors the binding sites of Lutheran/basal cell adhesion molecule and α3β1/α6β1 integrins. J Biol Chem. 2007;282:14853–60. doi: 10.1074/jbc.M611706200. [DOI] [PubMed] [Google Scholar]
- 47.Barresi R, Campbell KP. Dystroglycan: from biosynthesis to pathogenesis of human disease. J Cell Sci. 2006;119:199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- 48.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–23. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inamori K, Yoshida-Moriguchi T, Hara Y, Anderson ME, Yu L, Campbell KP. Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science. 2012;335:93–6. doi: 10.1126/science.1214115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida-Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, et al. O-mannosyl phosphorylation of α-dystroglycan is required for laminin binding. Science. 2010;327:88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patnaik SK, Stanley P. Mouse large can modify complex N- and mucin O-glycans on α-dystroglycan to induce laminin binding. J Biol Chem. 2005;280:20851–9. doi: 10.1074/jbc.M500069200. [DOI] [PubMed] [Google Scholar]
- 52.Hu Y, Li ZF, Wu X, Lu Q. Large induces functional glycans in an O-mannosylation dependent manner and targets GlcNAc terminals on α-dystroglycan. PLoS One. 2011;6:e16866. doi: 10.1371/journal.pone.0016866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z, Zhang P, Hu H. LARGE expression augments the glycosylation of glycoproteins in addition to α-dystroglycan conferring laminin binding. PLoS One. 2011;6:e19080. doi: 10.1371/journal.pone.0019080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wizemann H, Garbe JH, Friedrich MV, Timpl R, Sasaki T, Hohenester E. Distinct requirements for heparin and α-dystroglycan binding revealed by structure-based mutagenesis of the laminin α2 LG4-LG5 domain pair. J Mol Biol. 2003;332:635–42. doi: 10.1016/S0022-2836(03)00848-9. [DOI] [PubMed] [Google Scholar]
- 55.Carulli S, Beck K, Dayan G, Boulesteix S, Lortat-Jacob H, Rousselle P. Cell surface proteoglycans syndecan-1 and -4 bind overlapping but distinct sites in laminin α3 LG45 protein domain. J Biol Chem. 2012;287:12204–16. doi: 10.1074/jbc.M111.300061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamashita H, Beck K, Kitagawa Y. Heparin binds to the laminin α4 chain LG4 domain at a site different from that found for other laminins. J Mol Biol. 2004;335:1145–9. doi: 10.1016/j.jmb.2003.11.047. [DOI] [PubMed] [Google Scholar]
- 57.Hozumi K, Suzuki N, Uchiyama Y, Katagiri F, Kikkawa Y, Nomizu M. Chain-specific heparin-binding sequences in the laminin α chain LG45 modules. Biochemistry. 2009;48:5375–81. doi: 10.1021/bi900542u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miner JH, Li C, Mudd JL, Go G, Sutherland AE. Compositional and structural requirements for laminin and basement membranes during mouse embryo implantation and gastrulation. Development. 2004;131:2247–56. doi: 10.1242/dev.01112. [DOI] [PubMed] [Google Scholar]
- 59.Smyth N, Vatansever HS, Murray P, Meyer M, Frie C, Paulsson M, et al. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J Cell Biol. 1999;144:151–60. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bader BL, Smyth N, Nedbal S, Miosge N, Baranowsky A, Mokkapati S, et al. Compound genetic ablation of nidogen 1 and 2 causes basement membrane defects and perinatal lethality in mice. Mol Cell Biol. 2005;25:6846–56. doi: 10.1128/MCB.25.15.6846-6856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gautam M, Noakes PG, Moscoso L, Rupp F, Scheller RH, Merlie JP, et al. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–35. doi: 10.1016/S0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- 62.Pöschl E, Schlötzer-Schrehardt U, Brachvogel B, Saito K, Ninomiya Y, Mayer U. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development. 2004;131:1619–28. doi: 10.1242/dev.01037. [DOI] [PubMed] [Google Scholar]
- 63.Costell M, Gustafsson E, Aszódi A, Mörgelin M, Bloch W, Hunziker E, et al. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol. 1999;147:1109–22. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arikawa-Hirasawa E, Watanabe H, Takami H, Hassell JR, Yamada Y. Perlecan is essential for cartilage and cephalic development. Nat Genet. 1999;23:354–8. doi: 10.1038/15537. [DOI] [PubMed] [Google Scholar]
- 65.Murray P, Edgar D. Regulation of programmed cell death by basement membranes in embryonic development. J Cell Biol. 2000;150:1215–21. doi: 10.1083/jcb.150.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yurchenco PD, Amenta PS, Patton BL. Basement membrane assembly, stability and activities observed through a developmental lens. Matrix Biol. 2004;22:521–38. doi: 10.1016/j.matbio.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 67.Li S, Harrison D, Carbonetto S, Fässler R, Smyth N, Edgar D, et al. Matrix assembly, regulation, and survival functions of laminin and its receptors in embryonic stem cell differentiation. J Cell Biol. 2002;157:1279–90. doi: 10.1083/jcb.200203073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Curr Pharm Des. 2009;15:1277–94. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brakebusch C, Fässler R. The integrin-actin connection, an eternal love affair. EMBO J. 2003;22:2324–33. doi: 10.1093/emboj/cdg245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winder SJ, Hemmings L, Maciver SK, Bolton SJ, Tinsley JM, Davies KE, et al. Utrophin actin binding domain: analysis of actin binding and cellular targeting. J Cell Sci. 1995;108:63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- 71.Rybakova IN, Patel JR, Ervasti JM. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol. 2000;150:1209–14. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sutherland-Smith AJ, Moores CA, Norwood FL, Hatch V, Craig R, Kendrick-Jones J, et al. An atomic model for actin binding by the CH domains and spectrin-repeat modules of utrophin and dystrophin. J Mol Biol. 2003;329:15–33. doi: 10.1016/S0022-2836(03)00422-4. [DOI] [PubMed] [Google Scholar]
- 73.Bezakova G, Rüegg MA. New insights into the roles of agrin. Nat Rev Mol Cell Biol. 2003;4:295–308. doi: 10.1038/nrm1074. [DOI] [PubMed] [Google Scholar]
- 74.Denzer AJ, Schulthess T, Fauser C, Schumacher B, Kammerer RA, Engel J, et al. Electron microscopic structure of agrin and mapping of its binding site in laminin-1. EMBO J. 1998;17:335–43. doi: 10.1093/emboj/17.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kammerer RA, Schulthess T, Landwehr R, Schumacher B, Lustig A, Yurchenco PD, et al. Interaction of agrin with laminin requires a coiled-coil conformation of the agrin-binding site within the laminin γ1 chain. EMBO J. 1999;18:6762–70. doi: 10.1093/emboj/18.23.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moll J, Barzaghi P, Lin S, Bezakova G, Lochmüller H, Engvall E, et al. An agrin minigene rescues dystrophic symptoms in a mouse model for congenital muscular dystrophy. Nature. 2001;413:302–7. doi: 10.1038/35095054. [DOI] [PubMed] [Google Scholar]
- 77.Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, et al. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135:334–42. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–97. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zong Y, Zhang B, Gu S, Lee K, Zhou J, Yao G, et al. Structural basis of agrin-LRP4-MuSK signaling. Genes Dev. 2012;26:247–58. doi: 10.1101/gad.180885.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Timpl R, Wiedemann H, van Delden V, Furthmayr H, Kühn K. A network model for the organization of type IV collagen molecules in basement membranes. Eur J Biochem. 1981;120:203–11. doi: 10.1111/j.1432-1033.1981.tb05690.x. [DOI] [PubMed] [Google Scholar]
- 81.Yurchenco PD, Furthmayr H. Self-assembly of basement membrane collagen. Biochemistry. 1984;23:1839–50. doi: 10.1021/bi00303a040. [DOI] [PubMed] [Google Scholar]
- 82.Graham PL, Johnson JJ, Wang S, Sibley MH, Gupta MC, Kramer JM. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J Cell Biol. 1997;137:1171–83. doi: 10.1083/jcb.137.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang CC, Hall DH, Hedgecock EM, Kao G, Karantza V, Vogel BE, et al. Laminin α subunits and their role in C. elegans development. Development. 2003;130:3343–58. doi: 10.1242/dev.00481. [DOI] [PubMed] [Google Scholar]
- 84.Guo XD, Johnson JJ, Kramer JM. Embryonic lethality caused by mutations in basement membrane collagen of C. elegans. Nature. 1991;349:707–9. doi: 10.1038/349707a0. [DOI] [PubMed] [Google Scholar]
- 85.Fox JW, Mayer U, Nischt R, Aumailley M, Reinhardt D, Wiedemann H, et al. Recombinant nidogen consists of three globular domains and mediates binding of laminin to collagen type IV. EMBO J. 1991;10:3137–46. doi: 10.1002/j.1460-2075.1991.tb04875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hopf M, Göhring W, Ries A, Timpl R, Hohenester E. Crystal structure and mutational analysis of a perlecan-binding fragment of nidogen-1. Nat Struct Biol. 2001;8:634–40. doi: 10.1038/89683. [DOI] [PubMed] [Google Scholar]
- 87.Mayer U, Nischt R, Pöschl E, Mann K, Fukuda K, Gerl M, et al. A single EGF-like motif of laminin is responsible for high affinity nidogen binding. EMBO J. 1993;12:1879–85. doi: 10.1002/j.1460-2075.1993.tb05836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pöschl E, Mayer U, Stetefeld J, Baumgartner R, Holak TA, Huber R, et al. Site-directed mutagenesis and structural interpretation of the nidogen binding site of the laminin γ1 chain. EMBO J. 1996;15:5154–9. [PMC free article] [PubMed] [Google Scholar]
- 89.Takagi J, Yang Y, Liu JH, Wang JH, Springer TA. Complex between nidogen and laminin fragments reveals a paradigmatic β-propeller interface. Nature. 2003;424:969–74. doi: 10.1038/nature01873. [DOI] [PubMed] [Google Scholar]
- 90.Willem M, Miosge N, Halfter W, Smyth N, Jannetti I, Burghart E, et al. Specific ablation of the nidogen-binding site in the laminin γ1 chain interferes with kidney and lung development. Development. 2002;129:2711–22. doi: 10.1242/dev.129.11.2711. [DOI] [PubMed] [Google Scholar]
- 91.Kim S, Wadsworth WG. Positioning of longitudinal nerves in C. elegans by nidogen. Science. 2000;288:150–4. doi: 10.1126/science.288.5463.150. [DOI] [PubMed] [Google Scholar]
- 92.Behrens DT, Villone D, Koch M, Brunner G, Sorokin L, Robenek H, et al. The epidermal basement membrane is a composite of separate laminin- or collagen IV-containing networks connected by aggregated perlecan, but not by nidogens. J Biol Chem. 2012;287:18700–9. doi: 10.1074/jbc.M111.336073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oberbäumer I, Wiedemann H, Timpl R, Kühn K. Shape and assembly of type IV procollagen obtained from cell culture. EMBO J. 1982;1:805–10. doi: 10.1002/j.1460-2075.1982.tb01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsilibary EC, Koliakos GG, Charonis AS, Vogel AM, Reger LA, Furcht LT. Heparin type IV collagen interactions: equilibrium binding and inhibition of type IV collagen self-assembly. J Biol Chem. 1988;263:19112–8. [PubMed] [Google Scholar]
- 95.Sugawara K, Tsuruta D, Ishii M, Jones JC, Kobayashi H. Laminin-332 and -511 in skin. Exp Dermatol. 2008;17:473–80. doi: 10.1111/j.1600-0625.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- 96.Al-Amoudi A, Díez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–7. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- 97.Colognato H, MacCarrick M, O’Rear JJ, Yurchenco PD. The laminin α2-chain short arm mediates cell adhesion through both the α1β1 and α2β1 integrins. J Biol Chem. 1997;272:29330–6. doi: 10.1074/jbc.272.46.29330. [DOI] [PubMed] [Google Scholar]
- 98.Bruckner P. Suprastructures of extracellular matrices: paradigms of functions controlled by aggregates rather than molecules. Cell Tissue Res. 2010;339:7–18. doi: 10.1007/s00441-009-0864-0. [DOI] [PubMed] [Google Scholar]
- 99.Schneiders FI, Maertens B, Böse K, Li Y, Brunken WJ, Paulsson M, et al. Binding of netrin-4 to laminin short arms regulates basement membrane assembly. J Biol Chem. 2007;282:23750–8. doi: 10.1074/jbc.M703137200. [DOI] [PubMed] [Google Scholar]
- 100.Xiao T, Staub W, Robles E, Gosse NJ, Cole GJ, Baier H. Assembly of lamina-specific neuronal connections by slit bound to type IV collagen. Cell. 2011;146:164–76. doi: 10.1016/j.cell.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pavlakis E, Chiotaki R, Chalepakis G. The role of Fras1/Frem proteins in the structure and function of basement membrane. Int J Biochem Cell Biol. 2011;43:487–95. doi: 10.1016/j.biocel.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 102.Brasch J, Harrison OJ, Ahlsen G, Liu Q, Shapiro L. Crystal structure of the ligand binding domain of netrin G2. J Mol Biol. 2011;414:723–34. doi: 10.1016/j.jmb.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seiradake E, Coles CH, Perestenko PV, Harlos K, McIlhinney RA, Aricescu AR, et al. Structural basis for cell surface patterning through NetrinG-NGL interactions. EMBO J. 2011;30:4479–88. doi: 10.1038/emboj.2011.346. [DOI] [PMC free article] [PubMed] [Google Scholar]


