Abstract
Atherosclerosis is a complex disease resulting from the interaction of multiple genes. We have used the Ldlr knockout mouse model in an interspecific genetic cross to map atherosclerosis susceptibility loci. A total of 174 (MOLF/Ei × B6.129S7-Ldlrtm1Her) × C57BL/6J-Ldlrtm1Her backcross mice, homozygous for the Ldlr null allele, were fed a Western-type diet for 3 months and then killed for quantification of aortic lesions. A genome scan was carried out by using DNA pools and microsatellite markers spaced at ≈18-centimorgan intervals. Quantitative trait locus analysis of individual backcross mice confirmed linkages to chromosomes 4 (Athsq1, logarithm of odds = 6.2) and 6 (Athsq2, logarithm of odds = 6.7). Athsq1 affected lesions in females only whereas Athsq2 affected both sexes. Among females, the loci accounted for ≈50% of the total variance of lesion area. The susceptible allele at Athsq1 was derived from the MOLF/Ei genome whereas the susceptible allele at Athsq2 was derived from C57BL/6J. Inheritance of susceptible alleles at both loci conferred a 2-fold difference in lesion area, suggesting an additive effect of Athsq1 and Athsq2. No associations were observed between the quantitative trait loci and levels of plasma total cholesterol, high density lipoprotein cholesterol, non-high density lipoprotein cholesterol, insulin, or body weight. We provide strong evidence for complex inheritance of atherosclerosis in mice with elevated plasma low density lipoprotein cholesterol and show a major influence of nonlipoprotein-related factors on disease susceptibility. Athsq1 and Athsq2 represent candidate susceptibility loci for human atherosclerosis, most likely residing on chromosomes 1p36–32 and 12p13–12, respectively.
The genetics of atherosclerosis has been the focus of intense investigation. A subset of cases is caused by uncommon Mendelian mutations that predispose individuals to atherosclerosis (1–3). The mutated genes include low density lipoprotein receptor (LDLR) (4), cystathionine β-synthase (5), and, in some cases, ATP-binding cassette-A1 (6–8) among others. Identification of these genes has shed light on biochemical pathways involved in atherogenesis and provided the basis for current therapeutic interventions. However, the common forms of atherosclerosis are multifactorial in origin. Attempts to map the common susceptibility loci have been hampered by genetic heterogeneity, polygenic inheritance, incomplete pedigrees, and environmental influences. The fact that few of the genomewide linkage studies have reported loci with large effects points to the existence of multiple loci, each having small to moderate effects (9–12). The modest nature of susceptibility gene effects will likely require extremely large sample sizes or very densely spaced genetic markers for successful linkage mapping (13).
Mouse models offer significant advantages for genetic dissection of complex diseases. The ability to perform selective breeding, produce many offspring, determine inheritance of alleles without ambiguity, and control the environment is a critical factor. Early studies of murine atherosclerosis indicated that there was a clear genetic component. Inbred strains of mice exhibited a spectrum of aortic fatty streak lesion areas after the feeding of atherogenic diets high in cholesterol, fat, and cholic acid (14–16). A number of susceptibility loci (Ath1–8) were reported based on phenotypic analyses of recombinant inbred strains derived from “resistant” and “susceptible” parents (17–20). Although these studies were instrumental in pointing out strain-specific variations, none of the loci have been confirmed by more rigorous analyses of large genetic crosses.
A shortcoming of the diet-fed, inbred mouse model (in terms of carrying out quantitative genetic studies) is that aortic lesion development is minimal even in susceptible strains. Recently, Dansky et al. (21) showed that the strain-related differences in susceptibility could be accentuated when a gene-targeted disease model was used. Thus, C57BL/6J mice homozygous for the apolipoprotein E knockout allele exhibited 7- to 9-fold greater aortic root lesion area relative to FVB/NJ mice homozygous for the allele without any overlap of the phenotypic values (21). To provide candidate susceptibility loci for human atherosclerosis, we have performed a genome scan of an interspecific cross by using the Ldlr knockout model (22). In this model, feeding of a Western-style diet results in elevated plasma LDL levels (similar to levels in humans) and development of human-like complicated fibrous plaques (23). Two significant susceptibility loci were localized to chromosome (Chr) 4 and Chr 6. The effects of these loci were independent of common risk factors for human disease including plasma lipoprotein levels, plasma insulin levels, and body weight.
Materials and Methods
Mice.
MOLF/Ei (MOLF) and B6.129S7-Ldlrtm1Her (formerly C57BL/6J-Ldlrtm1Her; hereafter referred to as B6-Ldlr0) were purchased from The Jackson Laboratory. MOLF females were mated with B6-Ldlr0 males to produce F1 mice. Female F1s were backcrossed to B6-Ldlr0 males to produce N2 mice homozygous for the Ldlr knockout allele. N2 mice were weaned onto standard laboratory chow (PicoLab Rodent 20, #5053) at 21 days of age and switched to a Western-style diet at 8–12 weeks of age. The Western diet contained 21% (wt/wt) butterfat and 0.15% (wt/wt) cholesterol (Harlan Teklad Adjusted Calories TD 88137, Madison, WI). Mice were bled after 2 weeks and 3 months of Western diet feeding and killed at the 3-month time point. The breeding colony was produced and maintained in a specific pathogen-free environment. All mice were given ad libitum access to food and water and maintained on a standard 12-h light-dark cycle throughout the study. All experimental protocols were approved by the Institutional Animal Care and Research Advisory Committee.
Atherosclerotic Lesion Measurements.
Anesthetized mice were killed by cervical dislocation. The hearts were perfused with 0.9% NaCl by cardiac intraventricular canalization. Then, the hearts and aortic root were dissected and fixed in 10% formalin. The aortic root was sectioned and stained with oil red O, and lesion areas were quantified as described (24).
Plasma Lipoprotein and Insulin Measurements.
Mice were bled in the middle of the light cycle after a 5- to 6-h fast. Retro-orbital bleeding was performed under Forane anesthesia (Baxter, Deerfield, IL). Blood was collected directly into heparinized capillary tubes (Becton Dickson). Plasma was separated from cells by centrifugation and stored at −70°C. Isolation of high density lipoprotein (HDL) cholesterol by chemical precipitation (HDL reagent, Sigma), as well as enzymatic measurements of cholesterol and triglycerides (Wako Pure Chemical, Osaka), were carried out according to the manufacturers' instructions. Non-HDL cholesterol was calculated by subtracting HDL cholesterol from total cholesterol. Insulin was measured by using a commercially available ELISA kit (Crystal Chem, Chicago).
DNA Extraction and Ldlr Genotyping.
DNA was extracted from tail tips by a quick alkaline lysis protocol (25). Briefly, the tail tips were incubated in 50 mM NaOH for 1 h at 95°C, vortexed, and neutralized in 1 M Tris (pH 8). Cellular debris was pelleted by centrifugation, and the supernatant was used for PCR amplification of Ldlr alleles. Primer sequences and a protocol for Ldlr genotyping were obtained from http://www.jax.org/resources/documents/imr/protocols/Ldlr_KO.html (8/5/1998). Ldlr typings were confirmed by measuring plasma cholesterol levels.
DNA Pooling and Genome Scan.
DNA was quantified, in quadruplicate, by spectrophotometry. Equal amounts of DNA were pooled from 10–15 mice in the top or bottom 20% of the phenotypic ranges. Separate pools were made for males and females. The final concentration of DNA in the pools was 100–150 ng/μl, such that each individual sample was represented at a concentration of 10 ng/μl in a pool. Microsatellite markers (26, 27) were typed by PCR amplification using primers purchased from Research Genetics (Huntsville, AL) following the manufacturer's protocol. PCR products were separated on 7% Long Ranger polyacrylamide (FMC) gels and scored by using a LI-COR (Lincoln, NE) model 4000S automated DNA sequencer and gene imagir version 3.55 software (Scanalytics, Billerica, MA). Parental and F1 DNA samples were run alongside the pools as controls.
Testing of Candidate Linkages by Formal Linkage Analysis of the Backcross Panel.
Markers exhibiting a biased representation of alleles in the DNA pools (significantly different from the expected Mendelian distribution of 75% B6, 25% MOLF alleles for an unlinked marker) subsequently were subject to linkage analysis using the panel of 174 individual backcross samples. In addition, flanking markers were typed to confirm positive (linkage) or negative (no linkage) results by using the complete panel of individuals. For positive results, chromosomal linkage maps with multiple markers were constructed to refine the localization of the quantitative trait locus (QTL), as described (28). Linkage analysis was performed by using MAP MANAGER QTB28PPC as described for backcrosses (29, 30). Due to the strong effect of sex on atherosclerosis and lipoprotein phenotypes, all analyses were performed separately for males and females. Similar results were obtained by using raw or square root-transformed lesion area data. A logarithm of odds (LOD) score of 3.3 was used as the threshold for “significant” linkage (31).
Statistical Analysis.
ANOVA was performed by using STATVIEW 5.0 (Abacus Concepts, Berkeley, CA) for Macintosh computers.
Results
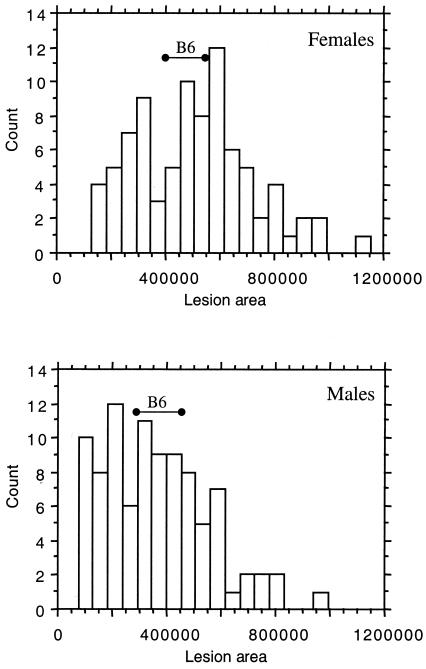
The distribution of aortic fatty streak lesion areas among 174 [(MOLF × B6.Ldlr0) × B6.Ldlr0] backcross mice homozygous for the Ldlr knockout allele (Mbc-Ldlr0), and the range of values in a set of B6-Ldlr0 controls, is shown in Fig. 1. Female Mbc-Ldlr0 mice exhibited 28% larger mean lesion areas than males (mean ± SD: 5.1 ± 2.2 × 105 vs. 3.7 ± 1.9 × 105 μm2/section, respectively, P < 0.0001). However, there was a broad distribution of lesion values among both female and male Mbc-Ldlr0 mice. The range of lesion areas observed for the B6-Ldlr0 controls was centered around the middle of the distribution curves for both female and male Mbc-Ldlr0 mice, suggesting the presence of both resistance and susceptibility alleles within the B6 genome.
Figure 1.
Distribution of fatty streak lesion areas among 174 Mbc-Ldlr0 mice grouped by sex. Mice were fed a Western-type diet for 3 months. Values are expressed as μm2/section. Solid horizontal bars represent the range of values for sex- and age-matched B6-Ldlr0 controls (n = 6 for each sex).
To rule out an effect of Apoa2, previously reported to have major effects on HDL cholesterol levels and aortic lesion susceptibility in other genetic crosses (32, 33), we typed the closely linked microsatellite marker D1Mit206 in the panel of 174 Mbc-Ldlr0 mice. No linkage was detected for HDL cholesterol or atheroslerosis susceptibility (data not shown). The lack of association between lesion areas and genotype at the Apoa2-linked marker suggested the presence of novel susceptibility loci segregating among the Mbc-Ldlr0 mice.
To detect candidate linkages for lesion susceptibility, we performed a genome scan by using a DNA pooling strategy. The mean lesion areas in Mbc-Ldlr0 mice selected for the “low” pools were 2.3 × 105 and 1.4 × 105 μm2/section for females and males, respectively. The mean lesion areas for the “high” pools were 7.0 × 105 and 6.5 × 105 μm2/section for females and males, respectively. A total of 88 polymorphic markers were typed, resulting in an average marker spacing of ≈18 centimorgans (cM). DNA pooling usually can detect linkage within 30 cM of an allele that is preferentially represented in affected individuals (34, 35).
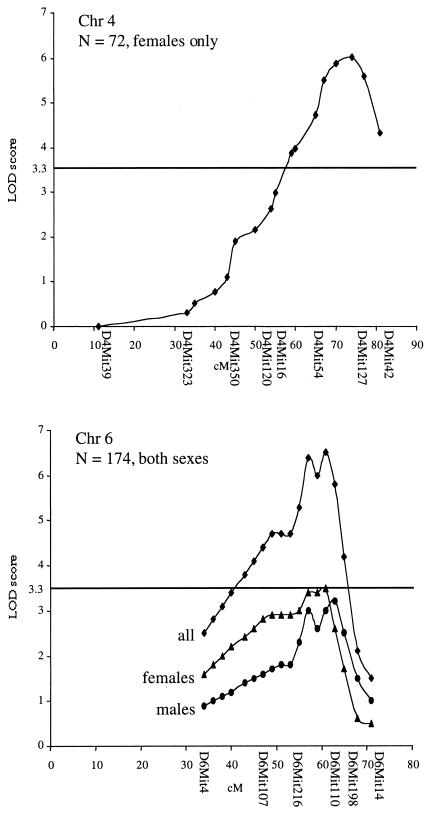
Two candidate loci were confirmed by linkage analysis by using the complete panel of 174 backcross mice (Table 1). The loci have been designated Athsq1 (Chr 4) and Athsq2 (Chr 6) for atherosclerosis susceptibility QTL 1 and 2. Athsq1 was supported by a peak LOD score of 6.2 near D4Mit127 (≈77 cM distal to the centromere, as listed in the Mouse Genome Database, MGD) (Fig. 2). Linkage was detected in females only, explaining 32% of the total variance of atherosclerotic lesion areas among females. Athsq2 was supported by a peak LOD score of 6.7 near D6Mit110 (62 cM distal to the centromere, as listed in MGD) (Fig. 2). The Chr 6 locus exhibited similar linkage in females (LOD = 3.5, explaining 16% of the variance) and males (LOD = 3.2, explaining 14% of the variance). Female and male LOD plots were coincident, indicating that a single QTL underlies the linkage in both sexes. Confidence intervals defined by a one-unit decrease in the peak LOD score were estimated to be ≈10 cM for both Athsq1 and Athsq2.
Table 1.
Linkage of lesion susceptibility QTLs to Chr 4 and Chr 6 in Mbc-Ldlr0 mice
| Chr | cM | LOD (%VAR)
|
LOD combined (n = 174) | QTL symbol | |
|---|---|---|---|---|---|
| Males (n = 92) | Females (n = 72–82) | ||||
| 4 | 77 | — | 6.2 (32%) | — | Athsq1 |
| 6 | 62 | 3.2 (14%) | 3.5 (18%) | 6.7 | Athsq2 |
cM, distance from the centromere in centimorgans. LOD, logarithm of the odds ratio for linkage; %VAR, an estimate of the percent of the total variance of lesion area explained by the locus.
Figure 2.
LOD score plots for Chr 4 and Chr 6 lesion susceptibility QTLs. The y axis indicates LOD scores; the x axis indicates position along the chromosome (distance from the centromere in cM). Microsatellite markers typed in Mbc-Ldlr0 mice are indicated below the x axis. LOD scores were calculated and plotted at 2-cM intervals by using map manager qt software. The significance threshold of P = 0.05 for a backcross is indicated by a solid line at LOD = 3.3.
The QTL effects on lesion areas and common risk factors for human atherosclerosis are shown in Tables 2 and 3. In females, inheritance of two copies of the B6-derived allele (BB) of Athsq1 resulted in 40% smaller mean lesion area relative to inheritance of one copy of the B6-derived and one copy of the MOLF-derived alleles (MB); no effect of genotype was observed in males (Table 2). Conversely, inheritance of the BB genotype at Athsq2 resulted in 28% (females) and 33% (males) larger mean lesion area relative to inheritance of the MB genotype (Table 3). Plasma total cholesterol, HDL cholesterol, and non-HDL-cholesterol levels after feeding of a Western-type diet for 2 weeks were tested for linkage to the atherosclerosis QTLs; no significant linkages were detected for any of the phenotypes. A small difference in mean HDL cholesterol levels was observed by ANOVA in mice grouped by genotype at Athsq1 (Table 2). However, the difference was not statistically significant after correcting for multiple testing. In addition, the atherosclerosis-resistant genotype was associated with lower HDL cholesterol levels. This finding is opposite to what would be expected if the mechanism for atherosclerosis susceptibility determination was through regulation of HDL cholesterol levels. No other effects of the QTLs on plasma cholesterol levels were observed. Similarly, no significant linkages were detected for triglycerides, body weight, or basal metabolic index (calculated as body weight divided by the squared nose to anus length; data not shown) at the atherosclerosis susceptibility QTLs.
Table 2.
Fatty streak lesion areas, plasma cholesterol levels, and fasting plasma insulin levels in Mbc-Ldlr0 mice grouped by genotype at D4Mit127
| Genotype | Lesion area, μm2/section | Total C, mg/dl | HDL-C, mg/dl | Non-HDL-C, mg/dl | Insulin, ng/ml |
|---|---|---|---|---|---|
| Females | |||||
| BB (n = 32) | 3.6 ± 1.8 × 105 | 344 ± 43 | 51 ± 13 | 295 ± 47 | 1.32 ± 1.0 (n = 15) |
| MB (n = 40) | 6.1 ± 2.0 × 105* | 341 ± 55 | 59 ± 16** | 284 ± 52 | 1.06 ± 0.67 (n = 7) |
| Males | |||||
| BB (n = 39) | 3.6 ± 2.2 × 105 | 384 ± 57 | 71 ± 17 | 311 ± 59 | 3.16 ± 1.66 (n = 27) |
| MB (n = 48) | 3.6 ± 2.0 × 105 | 366 ± 56 | 75 ± 15 | 291 ± 54 | 3.66 ± 2.61 (n = 10) |
BB, homozygous for C57BL/6J alleles; MB, heterozygous for C57BL/6J and MOLF alleles.
, P < 0.0001 vs. BB;
, P < 0.03 vs. BB. Values are mean ± SD.
Table 3.
Fatty streak lesion areas, plasma cholesterol levels, and fasting plasma insulin levels in Mbc-Ldlr0 mice grouped by genotype at D6Mit110
| Genotype | Lesion area, μm2/section | Total C, mg/dl | HDL-C, mg/dl | Non-HDL-C, mg/dl | Insulin, ng/ml |
|---|---|---|---|---|---|
| Females | |||||
| BB (n = 43) | 5.8 ± 2.0 × 105 | 342 ± 45 | 48 ± 16 | 292 ± 42 | 1.12 ± 0.79 (n = 16) |
| MB (n = 39) | 4.2 ± 2.1 × 105* | 341 ± 51 | 53 ± 14 | 286 ± 54 | 1.37 ± 1.12 (n = 7) |
| Males | |||||
| BB (n = 47) | 4.4 ± 1.8 × 105 | 379 ± 50 | 66 ± 17 | 312 ± 47 | 3.42 ± 2.21 (n = 17) |
| MB (n = 45) | 2.9 ± 1.8 × 105** | 364 ± 60 | 72 ± 12 | 287 ± 61 | 3.18 ± 1.77 (n = 20) |
BB, homozygous for C57BL/6J alleles; MB, heterozygous for C57BL/6J and MOLF alleles.
, P < 0.0009 vs. BB;
, P < 0.0002 vs. BB. Values are mean ± SD.
Epidemiological studies have shown an association between hyperinsulinemia and coronary atherosclerosis (36, 37), as well as clustering of cardiovascular disease risk factors (38–40). To test for an association between insulin levels and atherosclerosis susceptibility in our mouse model, we compared mean fasting insulin levels in a subset of Mbc-Ldlr0 mice grouped by genotype at the Chr 4 and Chr 6 QTLs. The mice had been fed the Western-type diet for 3 months. No significant associations were observed (Tables 2 and 3).
The combined effect of Athsq1 and Athsq2 was estimated by comparing mean lesion areas in mice grouped by genotype at both loci (Table 4). Mice carrying both susceptible genotypes, MB at Athsq1 and BB at Athsq2, exhibited 2-fold greater lesion area than mice carrying both resistant genotypes (mean ± SD: 6.6 ± 2.0 × 105 vs. 3.2 ± 1.8 × 105 μm2/section, respectively). Mice carrying one susceptible and one resistant genotype exhibited intermediate lesion areas. There was no evidence of interaction between the two loci by two-way ANOVA. These data are consistent with an additive effect of Athsq1 and Athsq2 on lesion susceptibility.
Table 4.
Combined effects of Athsq1 and Athsq2 on lesion areas in 72 female Mbc-Ldlr0 mice
| QTL, genotype | Athsq2, BB | Athsq2, BM |
|---|---|---|
| Athsq1, MB | 6.6 ± 2.0 × 105 | 5.3 ± 2.0 × 105 |
| (n = 22) | (n = 16) | |
| Athsq1, BB | 4.1 ± 1.4 × 105 | 3.2 ± 1.8 × 105 |
| (n = 11) | (n = 19) |
BB, homozygous for C57BL/6J alleles; MB, heterozygous for C57BL/6J and MOLF alleles. Values are mean ± SD in μm2/section.
Discussion
We have used the Ldlr knockout model of atherosclerosis to map susceptibility loci to mouse Chr 4 (Athsq1) and Chr 6 (Athsq2). Athsq1 exhibited strong sex specificity, contributing to disease susceptibility in females but not males. Together, genotypes at Athsq1 and Athsq2 accounted for ≈50% of the total variance of lesion area among females. The DNA pooling strategy used in this study allows the detection of independent susceptibility loci that are common among individuals contributing to a pool. Thus, pooling by phenotype roughly corresponds to pooling by genotype. The inability to detect QTLs contributing to the remaining 50% of the genetic variation of lesion area in this cross is likely due to genetic heterogeneity, small gene effects, and gene–gene interactions. These results are consistent with complex inheritance of atherosclerosis susceptibility in the mouse model.
In previous studies, feeding an atherogenic diet to inbred strains of mice often resulted in marked decreases of HDL cholesterol levels in atherosclerosis-susceptible strains but not resistant strains (18, 20, 32, 33). This common finding led to the suggestion that genetic determinants of HDL cholesterol levels were responsible for the differences in atherosclerosis susceptibility. However, more recent studies of differential gene expression in macrophages and endothelial cells derived from resistant and susceptible strains point out that there are differences in a variety of pathways that could influence atherogenesis (41, 42).
In the current study, no significant associations were observed between Athsq1 or Athsq2 and plasma lipoprotein levels. These results suggest that in a hypercholesterolemic model of atherosclerosis, such as the Ldlr knockout model, variation in disease susceptibility is determined by factors independent of plasma lipoprotein levels. Similarly, genetic studies of atherosclerosis in the apolipoprotein E knockout model suggest a role for nonlipoprotein-related factors in determining the relative susceptibility of different mouse strains (21, 42, 43). The inability of cholesterol-lowering protocols to decrease risk of disease-related events in many susceptible humans has highlighted the need to develop novel therapeutic approaches. As such, the identification of nonlipoprotein-related factors—such as those involved in inflammation, LDL oxidation, and macrophage or endothelial cell function—is an area of intense investigation in the atherosclerosis field (44). Identification of the genes underlying Athsq1 and Athsq2 may shed light on novel pathways involved in atherogenesis.
The murine localizations of Athsq1 and Athsq2 can be used to predict the locations of human candidate susceptibility loci. Distal Chr 4 (Athsq1) and distal Chr 6 (Athsq2) exhibit extensive homologies with human Chr 1p36–32 and 12p13–12, respectively (Mouse Genome Database). The regions of homology flank the confidence interval for each QTL, contain mapping data for more than 50 orthologs per region, and do not overlap any other regions of homology. Thus, Chr 1p36–32 and Chr 12p13–12 are good candidates for focused linkage analyses with densely spaced markers. Single nucleotide polymorphisms covering the candidate regions have been identified (45, 46). These markers can be used in disease-association studies (47) to test the relevance of Athsq1 and Athsq2 in human atherosclerosis.
The current mouse maps (Mouse Genome Database) contain a number of candidate genes for both QTLs. A family of phospholipase A2 genes has been mapped to distal Chr 4. Overexpression of Pla2g2a in transgenic mice results in increased atherogenesis, possibly due to the accumulation of biologically active oxidized phospholipids (48, 49). Members of the tumor necrosis factor receptor superfamily also map to this region of Chr 4. Tnfr genes are involved in a diverse set of cell programs including proliferation, differentiation, and cell death (50). The results of linkage and association studies have implicated TNFRSF1B in insulin resistance, hypertension, and hypercholesterolemia (51), clear risk factors for atherosclerosis. In addition, increased levels of soluble TNFRSF1B have been associated with atherosclerosis in case-control studies (52, 53). Mouse genes mapped to the interval containing Athsq2, on Chr 6, include two additional Tnfr genes, Tnfrsf1a and Tnfrsf7 (formerly known as Cd27), and a Von Willebrand factor homolog. Cav, involved in the uptake of oxidized LDL via scavenger receptors, and its related homolog Cav-2 (54), have been mapped to Chr 6 by in situ hybridization; these genes should be not be considered strong candidates until the subchromosomal locations are known. Finally, numerous expressed sequence tags of unknown function have been mapped to distal Chrs 4 and 6; these sequences should be considered positional candidate genes.
A comparison of human and mouse gene maps has provided additional candidate genes for both QTLs. First, Schnyder's crystalline corneal dystrophy has been mapped to 1p34.1–36 by linkage analysis (55). The disease is characterized by accumulations of phospholipid, unesterified cholesterol, cholesterol ester, and apolipoproteins AI, AII, and E in the corneal stroma (56, 57). There is no clear association of plasma cholesterol levels with the disease; some affected individuals exhibit dyslipidemia but others have normal levels (58). The evidence suggests a local defect in HDL cholesterol metabolism or transport. It is interesting to speculate that a similar defect in cells in the arterial wall could give rise to increased foam cell formation and increased atherosclerosis. Although speculative, the homology between distal mouse Chr 4 and human 1p34–36 suggests that the same gene could perhaps underlie both Schnyder's corneal dystrophy and atherosclerosis susceptibility.
The second candidate gene identified by comparison of human and mouse gene maps is OLR1, the oxidized lipoprotein receptor 1 gene, mapped to human 12p13 (59). Oxidized LDL is believed to be an essential component of atherogenesis that induces endothelial dysfunction and accumulation of foam cells (60). OLR1 protein (also referred to as LOX-1) is a cell-surface receptor expressed in endothelial cells (61) and macrophages (62) among other cell types; the receptor specifically binds, internalizes, and degrades oxidized LDL but not native LDL (61). Recently, OLR1 was shown to be expressed in atheromatous intima (63, 64). The OLR1 gene is located within the human natural killer gene complex on Chr 12. Comparison of high-resolution physical maps of the human complex (65) and a homologous region of mouse Chr 4 (66) suggest that the mouse homolog of OLR1 (Olr1) is highly likely to map to the region of peak linkage for Athsq2. However, direct evidence implicating Olr1 in atherogenesis is not available, and further studies are needed to determine whether the expression or function of Olr1 differs between MOLF/Ei and C57BL/6J strains.
We have used QTL analysis to map two susceptibility loci for the complex disease atherosclerosis. Although a number of candidate genes have been suggested, actual identification of the genes underlying the QTLs remains an important hurdle. The recent successes of Vidal et al. (67) and Frary et al. (68) in identifying the genes responsible for natural resistance to infection and tomato fruit size QTLs, respectively, demonstrate that the hurdle is not insurmountable. Emerging information from the Human Genome Project and the mouse sequencing effort will increase the power of QTL analyses in the future.
Acknowledgments
We thank Anna Gorelik for excellent assistance with measurement of insulin levels. This work was supported by Public Health Service Individual National Research Service Award HL-09930 (to C.L.W.) and Program Project Grants HL-54591 and HL-22682.
Abbreviations
- Chr
chromosome
- cM
centimorgan
- HDL
high density lipoprotein
- LDL
low density lipoprotein
- Ldlr
LDL receptor gene
- LOD
logarithm of odds
- QTL
quantitative trait locus
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the Mouse Genome Database (accession no. J:69563).
References
- 1.Breslow J L. Annu Rev Genet. 2000;34:233–254. doi: 10.1146/annurev.genet.34.1.233. [DOI] [PubMed] [Google Scholar]
- 2.Keating M T, Sanguinetti M C. Science. 1996;272:681–685. doi: 10.1126/science.272.5262.681. [DOI] [PubMed] [Google Scholar]
- 3.Lifton R P. Science. 1996;272:676–680. doi: 10.1126/science.272.5262.676. [DOI] [PubMed] [Google Scholar]
- 4.Hobbs H H, Brown M S, Goldstein J L. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 5.Kraus J P, Janosik M, Kozich V, Mandell R, Shih V, Sperandeo M P, Sebastio G, de Franchis R, Andria G, Kluijtmans L A, et al. Hum Mutat. 1999;13:362–375. doi: 10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 6.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, et al. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 7.Brooks-Wilson A, Marcil M, Clee S M, Zhang L H, Roomp K, van Dam M, Yu L, Brewer C, Collins J A, Molhuizen H O, et al. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 8.Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette J C, Deleuze J F, Brewer H B, Duverger N, Denefle P, et al. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 9.Hixson J E, Blangero J. Ann N Y Acad Sci. 2000;902:1–7. doi: 10.1111/j.1749-6632.2000.tb06295.x. [DOI] [PubMed] [Google Scholar]
- 10.Rice T, Rankinen T, Province M A, Chagnon Y C, Perusse L, Borecki I B, Bouchard C, Rao D C. Circulation. 2000;102:1956–1963. doi: 10.1161/01.cir.102.16.1956. [DOI] [PubMed] [Google Scholar]
- 11.Shearman A M, Ordovas J M, Cupples L A, Schaefer E J, Harmon M D, Shao Y, Keen J D, DeStefano A L, Joost O, Wilson P W, et al. Hum Mol Genet. 2000;9:1315–1320. doi: 10.1093/hmg/9.9.1315. [DOI] [PubMed] [Google Scholar]
- 12.Aouizerat B E, Allayee H, Cantor R M, Davis R C, Lanning C D, Wen P Z, Dallinga-Thie G M, de Bruin T W, Rotter J I, Lusis A J. Am J Hum Genet. 1999;65:397–412. doi: 10.1086/302490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Risch N, Merikangas K. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 14.Qiao J-H, Xie P-Z, Fishbein M C, Kreuzer J, Drake T A, Demer L L, Lusis A J. Arterioscler Thromb. 1994;14:1480–1497. doi: 10.1161/01.atv.14.9.1480. [DOI] [PubMed] [Google Scholar]
- 15.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes P. Atherosclerosis. 1985;57:65–73. doi: 10.1016/0021-9150(85)90138-8. [DOI] [PubMed] [Google Scholar]
- 16.Roberts A, Thompson J S. Prog Biochem Pharmacol. 1977;14:298–305. [PubMed] [Google Scholar]
- 17.Paigen B. Am J Clin Nutr. 1995;62:458S–462S. doi: 10.1093/ajcn/62.2.458S. [DOI] [PubMed] [Google Scholar]
- 18.Paigen B, Nesbitt M N, Mitchell D, Albee D, LeBoeuf R C. Genetics. 1989;122:163–168. doi: 10.1093/genetics/122.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart-Phillips J L, Lough J, Skamene E. Clin Invest Med. 1989;12:121–126. [PubMed] [Google Scholar]
- 20.Paigen B, Mitchell D, Reue K, Morrow A, Lusis A J, LeBoeuf R C. Proc Natl Acad Sci USA. 1987;84:3763–3767. doi: 10.1073/pnas.84.11.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dansky H M, Charlton S A, Sikes J L, Heath S C, Simantov R, Levin L F, Shu P, Moore K J, Breslow J L, Smith J D. Arterioscler Thromb Vasc Biol. 1999;19:1960–1968. doi: 10.1161/01.atv.19.8.1960. [DOI] [PubMed] [Google Scholar]
- 22.Ishibashi S, Brown M S, Goldstein J L, Gerard R D, Hammer R E, Herz J. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masucci-Magoulas L, Goldberg I J, Bisgaier C L, Serajuddin H, Francone O L, Breslow J L, Tall A R. Science. 1997;275:391–394. doi: 10.1126/science.275.5298.391. [DOI] [PubMed] [Google Scholar]
- 24.Plump A S, Scott C J, Breslow J L. Proc Natl Acad Sci USA. 1994;91:9607–9611. doi: 10.1073/pnas.91.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truett G E, Walker J A, Truett A A, Mynatt R L, Heeger P, Warman M. BioTechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 26.Dietrich W, Katz H, Lincoln S E, Shin H S, Friedman J, Dracopoli N C, Lander E S. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love J M, Knight A M, McAleer M A, Todd J A. Nucleic Acids Res. 1990;18:4123–4130. doi: 10.1093/nar/18.14.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welch C L, Xia Y-R, Schechter I, Farese R, Mehrabian M, Mehdizadeh S, Warden C H, Lusis A J. J Lipid Res. 1996;37:1406–1421. [PubMed] [Google Scholar]
- 29.Manly K, Olson J M. Mamm Genome. 1999;10:327–334. doi: 10.1007/s003359900997. [DOI] [PubMed] [Google Scholar]
- 30.Paterson A H, Damon S, Hewitt J D, Zamir D, Rabinowitch H D, Lincoln S E, Lander E S, Tanksley S D. Genetics. 1991;127:181–197. doi: 10.1093/genetics/127.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lander E S, Kruglyak L. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 32.Machleder D, Ivandic B, Welch C, Castellani L, Reue K, Lusis A J. J Clin Invest. 1997;99:1406–1419. doi: 10.1172/JCI119300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehrabian M, Qiao J-H, Hyman R, Ruddle D, Laughton C, Lusis A J. Arterioscler Thromb. 1993;13:1–10. doi: 10.1161/01.atv.13.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Collin G B, Asada Y, Varnum D S, Nadeau J H. Mamm Genome. 1996;7:68–70. doi: 10.1007/s003359900017. [DOI] [PubMed] [Google Scholar]
- 35.Taylor B A, Navin A, Phillips S J. Genomics. 1994;21:626–632. doi: 10.1006/geno.1994.1323. [DOI] [PubMed] [Google Scholar]
- 36.Gaudet D, Vohl M C, Perron P, Tremblay G, Gagne C, Lesiege D, Bergeron J, Moorjani S, Despres J P. Circulation. 1998;97:871–877. doi: 10.1161/01.cir.97.9.871. [DOI] [PubMed] [Google Scholar]
- 37.Bavenholm P, Proudler A, Tornvall P, Godsland I, Landou C, de Faire U, Hamsten A. Circulation. 1995;92:1422–1429. doi: 10.1161/01.cir.92.6.1422. [DOI] [PubMed] [Google Scholar]
- 38.Meigs J B, Mittleman M A, Nathan D M, Tofler G H, Singer D E, Murphy-Sheehy P M, Lipinska I, D'Agostino R B, Wilson P W. J Am Med Assoc. 2000;283:221–228. doi: 10.1001/jama.283.2.221. [DOI] [PubMed] [Google Scholar]
- 39.Bonora E, Targher G, Zenere M B, Saggiani F, Cacciatori V, Tosi F, Travia D, Zenti M G, Branzi P, Santi L, Muggeo M. Eur J Clin Invest. 1997;27:248–254. doi: 10.1046/j.1365-2362.1997.1060658.x. [DOI] [PubMed] [Google Scholar]
- 40.Mykkanen L, Haffner S M, Ronnemaa T, Bergman R N, Laakso M. Am J Epidemiol. 1997;146:315–321. doi: 10.1093/oxfordjournals.aje.a009272. [DOI] [PubMed] [Google Scholar]
- 41.Friedman G, Ben-Yehuda A, Dabach Y, Hollander G, Babaey S, Ben-Maim M, Stein O, Stein Y. Arterioscler Thromb Vasc Biol. 2000;20:2459–2464. doi: 10.1161/01.atv.20.11.2459. [DOI] [PubMed] [Google Scholar]
- 42.Shi W, Wang N J, Shih D M, Sun V Z, Wang X, Lusis A J. Circ Res. 2000;86:1078–1084. doi: 10.1161/01.res.86.10.1078. [DOI] [PubMed] [Google Scholar]
- 43.Grimsditch D C, Penfold S, Latcham J, Vidgeon-Hart M, Groot P H, Benson G M. Atherosclerosis. 2000;151:389–397. doi: 10.1016/s0021-9150(99)00400-1. [DOI] [PubMed] [Google Scholar]
- 44.Glass C K, Wiztum J L. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 45.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane C R, Lim E P, Kalyanaraman N, et al. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 46.Wang D G, Fan J B, Siao C J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 47.Rubin E M, Tall A. Nature (London) 2000;407:265–269. doi: 10.1038/35025236. [DOI] [PubMed] [Google Scholar]
- 48.Ivandic B, Castellani L W, Wang X P, Qiao J H, Mehrabian M, Navab M, Fogelman A M, Grass D S, Swanson M E, de Beer M C, et al. Arterioscler Thromb Vasc Biol. 1999;19:1284–1290. doi: 10.1161/01.atv.19.5.1284. [DOI] [PubMed] [Google Scholar]
- 49.Leitinger N, Watson A D, Hama S Y, Ivandic B, Qiao J H, Huber J, Faull K F, Grass D S, Navab M, Fogelman A M, et al. Arterioscler Thromb Vasc Biol. 1999;19:1291–1298. doi: 10.1161/01.atv.19.5.1291. [DOI] [PubMed] [Google Scholar]
- 50.Lotz M. J Leukocyte Biol. 1996;60:1–7. doi: 10.1002/jlb.60.1.1. [DOI] [PubMed] [Google Scholar]
- 51.Glenn C L, Wang W Y, Benjafield A V, Morris B J. Hum Mol Genet. 2000;9:1943–1949. doi: 10.1093/hmg/9.13.1943. [DOI] [PubMed] [Google Scholar]
- 52.Fiotti N, Giansante C, Ponte E, Delbello C, Calabrese S, Zacchi T, Dobrina A, Guarnieri G. Atherosclerosis. 1999;145:51–60. doi: 10.1016/s0021-9150(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 53.Blann A D, McCollum C N. Inflammation. 1998;22:483–491. doi: 10.1023/a:1022346010304. [DOI] [PubMed] [Google Scholar]
- 54.Engelman J A, Zhang X, Galbiati F, Volonte D, Sotgia F, Pestell R G, Minetti C, Scherer P E, Okamoto T, Lisanti M P. Am J Hum Genet. 1998;63:1578–1587. doi: 10.1086/302172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shearman A M, Hudson T J, Andresen J M, Wu X, Sohn R L, Haluska F, Housman D E, Weiss J S. Hum Mol Genet. 1995;5:1667–1672. doi: 10.1093/hmg/5.10.1667. [DOI] [PubMed] [Google Scholar]
- 56.McCarthy M, Innis S, Dubord P, White V. Ophthalmology. 1994;101:895–901. [PubMed] [Google Scholar]
- 57.Weiss J S, Rodrigues M M, Kruth H S, Rajagopalan S, Rader D J. Ophthalmology. 1992;99:1072–1081. doi: 10.1016/s0161-6420(92)31848-2. [DOI] [PubMed] [Google Scholar]
- 58.Barchiesi B J, Eckel E H, Ellis P P. Surv Ophthalmol. 1991;36:1–22. doi: 10.1016/0039-6257(91)90205-t. [DOI] [PubMed] [Google Scholar]
- 59.Li X, Bouzyk M M, Wang X. Cytogenet Cell Genet. 1998;82:34–36. doi: 10.1159/000015059. [DOI] [PubMed] [Google Scholar]
- 60.Ross R. Nature (London) 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 61.Sawamura T, Kume N, Aoyama T, Moriwaki H, Hoshikawa H, Aiba Y, Tanaka T, Miwa S, Katsura Y, Kita T, et al. Nature (London) 1997;386:73–77. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- 62.Nagase M, Abe J, Takahashi K, Ando J, Hirose S, Fujita T. J Biol Chem. 1998;273:33702–33707. doi: 10.1074/jbc.273.50.33702. [DOI] [PubMed] [Google Scholar]
- 63.Kataoka H, Kume N, Miyamoto S, Minami M, Moriwaki H, Murase T, Sawamura T, Masaki T, Hashimoto N, Kita T. Circulation. 1999;99:3110–3117. doi: 10.1161/01.cir.99.24.3110. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida H, Kondratenko N, Green S, Steinberg D, Quehenberger O. Biochem J. 1998;334:9–13. doi: 10.1042/bj3340009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Renedo M, Arce I, Montgomery K, Roda-Navarro P, Lee E, Kucherlapati R, Fernandez-Ruiz E. Genomics. 2000;65:129–136. doi: 10.1006/geno.2000.6163. [DOI] [PubMed] [Google Scholar]
- 66.Depatie C, Lee S-H, Stafford A, Avner P, Belouchi A, Gros P, Vidal S M. Genomics. 2000;66:161–174. doi: 10.1006/geno.2000.6186. [DOI] [PubMed] [Google Scholar]
- 67.Vidal S, Tremblay M L, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frary A, Nesbitt T C, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert K B, Tanksley S D. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]