SUMMARY
Chronic Hepatitis B virus (HBV) infection afflicts millions worldwide with cirrhosis and liver cancer. HBV e-antigen (HBeAg), a clinical marker for disease severity, is a non-particulate variant of the protein (core antigen, HBcAg) that forms the building-blocks of capsids. HBeAg is not required for virion production, but is implicated in establishing immune tolerance and chronic infection. Here, we report the crystal structure of HBeAg, which clarifies how the short N-terminal propeptide of HBeAg induces a radically altered mode of dimerization relative to HBcAg (~140° rotation), locked into place through f ormation of intramolecular disulfide bridges. This structural switch precludes capsid assembly and engenders a distinct antigenic repertoire, explaining why the two antigens are cross-reactive at the T-cell level (through sequence identity) but not at the B-cell level (through conformation). The structure offers insight into how HBeAg may establish immune tolerance for HBcAg while evading its robust immunogenicity.
INTRODUCTION
Hepatitis B virus (HBV) infection remains a major source of acute and chronic liver disease worldwide. More than 360 million people have chronic HBV infection, which results in one million deaths annually, primarily due to cirrhosis and liver cancer. Over the four decades since the discovery of HBV, striking advances have been made in our understanding of the molecular biology, immunology, and pathogenesis of infection. However, certain aspects of HBV biology remain elusive. One such concern is the structure of the viral e-antigen (HBeAg), as well as its functional role in HBV infection. While HBeAg has no demonstrated role in the viral replication cycle (Chang et al., 1987; Chen et al., 1992; Schlicht et al., 1987), the antigen has long been a key clinical marker for viral replication, infectivity, disease severity, and response to treatment (Elgouhari et al., 2008). Further, HBeAg (or an equivalent) exists in all members of the Hepadnaviridae family, suggesting an evolutionarily conserved and therefore important function (Revill et al., 2010).
The HBV capsid protein (HBcAg; core-antigen) comprises a 149-residue assembly domain and a 34-residue arginine-rich domain (Figure 1A). The assembly domain forms dimers with a central four-helix bundle and flanking α-helices that assemble into icosahedral capsids of two sizes, with the four-helix bundles projecting as spikes (Packianathan et al., 2010; Wynne et al., 1999). HBeAg consists of the ten N-terminal residues (the propeptide: SKLCLGWLWG) appended to the assembly domain with the C-terminus at residue 149. (Figure 1A) (Ou et al., 1986; Standring et al., 1988; Takahashi et al., 1983). Translation of the C gene from an alternative upstream start codon yields a protein with a 29-residue signal peptide which routes it to the ER, where it is processed to the 10-residue propeptide (Standring et al., 1988). However, despite possessing an intact assembly domain, HBeAg does not assemble into capsids and is secreted by infected liver cells in non-particulate form.
Figure 1. Structure of HBeAg.
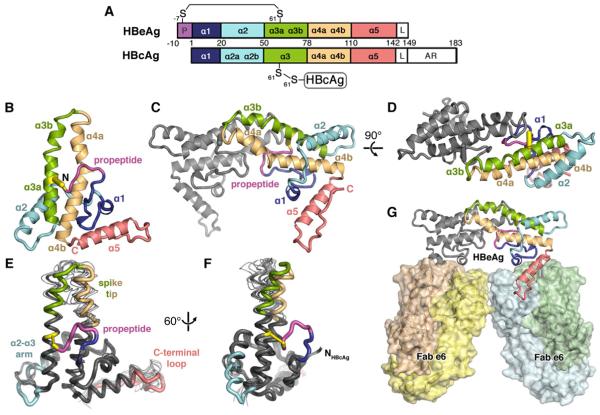
(A) Domain schematic of HBeAg and HBcAg, which share the core domain but differ in that HBeAg retains the N-terminal propeptide (P; magenta) and lacks the RNA-binding arginine-rich domain (AR). The linker region (L) is present, but disordered in crystal structures of both antigens.
(B) Ribbon diagram of HBeAg monomer, colored blue to red from N- to C-terminus, with the propeptide (magenta) shown forming an intramolecular disulfide (yellow) between C(-7) and C61. Secondary structure elements are entirely α-helical (α1-α5); propeptide is irregular coil.
(C, D) Ribbon diagram of the HBeAg dimer. viewed axially. Front subunit is colored according to the scheme in A; rear subunit is colored gray. Hairpins of the α3b and α4a helices from each subunit form the dimer interface, supported by the propeptides intercalated between them.
(E, F) Superposition of HBeAg and HBcAg monomers (thick and thin ribbon, respectively) (Packianathan et al., 2010; Wynne et al., 1999).
(G) Crystal asymmetric unit depicts HBeAg dimer complexed with two Fab e6 molecules (shown as molecular surface) binding at α5 and the C-terminal loop (red).
Much evidence suggests that HBeAg can modulate the host immune response to favor chronic infection following perinatal transmission (the most common form of HBV transmission worldwide) and prevent severe liver injury during adult infections (Chen et al., 2005; Chen et al., 2004; Milich and Liang, 2003; Ou, 1997; Visvanathan et al., 2007; Yang et al., 2006). The epidemiological evidence is persuasive: more than 90% of infants born to mothers who are HBeAg-positive HBV carriers also develop chronic infection, whereas those born to HBeAg-negative mothers rarely progress to chronicity (Terazawa et al., 1991). While the molecular mechanisms underlying these processes are unclear, it has been shown that HBeAg can downregulate the inflammatory response directed at HBcAg, while itself averting robust immunogenicity (Chen et al., 2005; Chen et al., 2004; Milich and Liang, 2003). Further, HBeAg (but not HBcAg) can cross the placenta from mother to child (Schodel et al., 1993), consistent with data suggesting that HBeAg may induce clonal tolerance against HBcAg and HBeAg in utero (Chen et al., 2004; Milich et al., 1990). While the connection between HBeAg and chronic infection is not fully understood, infections with HBV strains that do not express HBeAg (due to mutation in pre-C region) lead to a much higher frequency of fulminant hepatitis and acute liver failure (Fagan et al., 1986; Liang et al., 1991). In these circumstances, the lack of immune modulation by HBeAg is thought to lead to an unregulated and overwhelming immune response to HBcAg. To develop a fundamental understanding of the functional distinction between HBcAg and HBeAg, knowledge of their respective structures is essential.
HBcAg and HBeAg have been viewed as serologically distinct (Conway et al., 1998; Imai et al., 1982; Salfeld et al., 1989). However, a recent analysis of a panel of six monoclonal antibodies found four to cross-react with both antigens, albeit with markedly differing affinities, and one each specific for HBcAg and HBeAg (Watts et al., 2010). The HBeAg-specific Fab e6, was found to form a stable complex with the recombinantly-expressed HBeAg dimer. We have now crystallized the complex and determined its structure. Here we develop this information to explore the long-standing question of how the 10-residue propeptide retained by HBeAg transforms the protein’s propensity to assemble, its antigenic character, and its apparent ability to modulate the immune response to favor viral persistence.
RESULTS
Here we report the crystal structure of recombinantly expressed HBeAg and Fab e6 to a resolution of 3.3 Å, solved by molecular replacement, exploiting a 2.5 Å structure for the Fab e6 alone, which we determined separately. The Fab fragment facilitated crystallization (Figure S1). Even at 3.3 Å resolution, the key features of HBeAg are firmly established (Table 1 and Figure S1).
TABLE I. Data Collection and Refinement Statistics.
| Crystal data | HBeAg - e6 Fab complex | Fab e6 |
| Space group | Triclinic P1 | Monoclinic C2 |
| Unit cell parameters (Å, °) | a = 66.7, b = 75.8, c = 88.7, | a = 124.1, b = 68.2, c = 236.6, |
| α = 96.8, β = 103.8, γ = 116.0 | α = 90.0, β = 96.3, γ = 90.0 | |
| x2003; HBeAg dimer molecules per ASU | 1 | 0 |
| e6 Fab molecules per ASU | 2 | 4 |
| Vm (Å3 Da−1) | 2.92 | 2.60 |
| Solvent content (%) | 57.9 | 52.7 |
| Data collection statistics | ||
| Resolution (Å) | 46.2 - 3.34 (3.43 – 3.34) | 31.1 – 2.52 (2.58 – 2.52) |
| Rwork (%) | 11.6 (73.7) | 9.6 (91.1) |
| I/δ(I) | 6.4 (1.0) | 7.7 (1.3) |
| Completeness (%) | 92.6 (88.8) | 99.5 (99.6) |
| Mean multiplicity | 1.8 (1.6) | 3.4 (3.5) |
| Refinement statistics | ||
| No. of total reflections | 34,840 | 223,579 |
| No. of unique reflections | 19,664 | 66,469 |
| No. of reflections used in refinement | 19,651 | 66,468 |
| Rwork / Rfree (%) | 23.3/23.4 | 18.1/21.9 |
| Reflections used for Rfree (|Fo| > 0) (%) | 5.0 | 5.0 |
| Ramachandran plot (%) | ||
| Most favored | 96.0 | 96.9 |
| Allowed | 3.7 | 3.0 |
| Outliers | 0.3 | 0.1 |
| No. of protein atoms | 9,076 | 13,480 |
| Average B-factor (Å2) | 111 | 62 |
| Wilson B-factor (Å2) | 76 | 58 |
| R.m.s. deviations | ||
| bond lengths (Å) | 0.008 | 0.010 |
| bond angles (°) | 1.19 | 1.27 |
| B-factor of bonded atoms (Å2) | 0.8 | 1.7 |
| NCS related HBeAg coordinates (Å) | 0.03 | -- |
| NCS related HBeAg B-factors (Å2) | 6.6 | -- |
| NCS related Fab e6 coordinates (Å) | 0.09 | 0.64 |
| NCS related Fab e6 B-factors (Å2) | 7.5 | 14.1 |
| Targeted Refine. Fab coords (Å) | 0.50 | -- |
| Targeted Refine. Fab B-factors (Å2) | 13.0 | -- |
HBeAg and HBcAg monomer folds are similar
The structure shows HBeAg as a dimer with each subunit bound to an e6 Fab (Figure 1G), consistent with the finding that HBeAg is dimeric (Steven et al., 2005) and the reported stoichiometry of the complex (Watts et al., 2010). Overall, the monomer fold is similar to that of HBcAg: helices α1 and α2 and the loop between them encircle an amphipathic hairpin of kinked helices (α3 and α4), followed by α5 and a proline-rich C-terminal loop (Figure 1B) (Packianathan et al., 2010; Wynne et al., 1999). The respective core domains superpose with an RMSD of 1.6 Å for the 141 corresponding Cαs, with variability most affecting the spike apex, the loop between α2 and α3a, and the C-terminal loop. However, the greatest difference is the propeptide in HBeAg (Figure 1E, 1F) which adopts a loop structure that forms an intramolecular disulfide bond between C(-7) and C61 (Figure 1B). These disulfides in HBeAg (two per dimer)—versus the single inter-molecular C61-C61 disulfide that stabilizes the HBcAg dimer—has been reported to be critical for the secretion of HBeAg (Nassal and Rieger, 1993; Schodel et al., 1993; Wasenauer et al., 1992).
HBeAg and HBcAg form radically different dimers
HBcAg dimerizes through the pairing, in parallel, of two helical hairpins to form a four-helix bundle (Figure 2A, right). The interface between the hairpins is largely hydrophobic and rather flat (Packianathan et al., 2010; Wynne et al., 1999). Flanking the four-helix bundle are salt bridges and hydrogen bonds between the charged, polar residues of the N-terminal strand and the α2-α3 loop. In contrast, in HBeAg, the propeptide loop makes stabilizing hydrophobic contacts with the central part of the α3-α4 surface of its own polypeptide chain, where it sterically blocks the formation of an HBcAg-like dimer (Figure 1C). Instead, HBeAg dimerization involves (part of) the same molecular surfaces but with a relative rotation of ~140° between the h airpins (Figure 2A, left). In HBeAg, the two α3b helices are nearly anti-parallel and make an array of hydrophobic contacts as they pair to form a ridge in the dimer interface. In contrast, the α4a helices do not interact with each other, but instead interact with the exterior surface of the partner’s propeptide, also via hydrophobic contacts. In the view along the two-fold axis of the HBeAg dimer (Figure 1D), it is apparent that the α4a helices are too far apart to interact and that the intercalated propeptide loop completes the dimer interface.
Figure 2. Comparison of HBeAg and HBcAg structures.
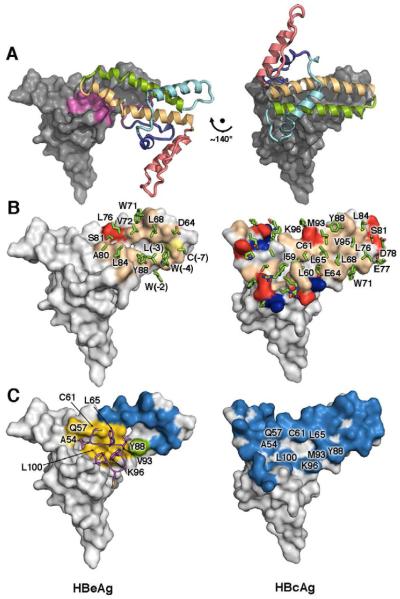
(A) The conformational switch. Propeptide density sterically interferes to block formation of the HBcAg dimer interface, allowing HBeAg to form a different dimer interface using the same surfaces, but ~140° rotated. The gray surface-rende red subunits of each dimer are shown in the same orientation, providing a frame of reference in which the relative rotation between the colored ribbon subunits is evident.
(B) Dimer interface comparison. HBeAg employs molecular mimicry to form a similar, hydrophobic dimer interface with α3/α4 hairpins, in an inverted orientation relative to HBcAg. Arrays of hydrophobic groups, and even the positioning of aromatics such as Y88 and W71 in HBcAg are replicated by different residues, such as W71 and W(-4) in HBeAg.
(C) The propeptide mediates hydrophobic contacts within elements that would otherwise already be buried in the context of the HBcAg dimer.
Molecular mimicry between the HBeAg and HBcAg dimer interfaces
In the HBeAg dimer is a constellation of hydrophobic residues that mimics many of the hydrophobic interactions at the HBcAg dimer interface. For example, propeptide residues W(-4), L(-3), W(-2) in HBeAg make dimer contacts analogous to those of L68, W71, and L76 in HBcAg (Figure 2B). These three propeptide residues form a hydrophobic cluster accounting for >50% of the surface area buried at the interface. Along the α3b ridge, another hydrophobic patch involves L68, V72, L76, and W71 of both dimer subunits, whereas in HBcAg these residues interact with L84, Y88, M93, and V95 (Figure 2B). However, the HBeAg dimer interface is much smaller than that of HBcAg, burying 1640 vs. 3970 Å2, has poorer shape complementarity (0.61 versus 0.70) (Lawrence and Colman, 1993), and makes fewer predicted hydrogen bonds, consistent with biophysical data showing that the HBeAg dimer is stable but less so than the HBcAg dimer (melting temperatures 51 and 65°C, respective ly) (Watts et al., 2011). Despite this difference in stability, the propeptide introduces steric hindrance that blocks HBeAg from adopting the HBcAg conformation. Hydrophobic regions that are buried in the HBcAg dimer— but would otherwise be exposed in the HBeAg conformation—are shielded by the propeptide (Figure 2C). This leaves many of the flanking polar residues that participate in dimer-dimer capsid interactions in HBcAg, exposed in HBeAg.
Unlike oxidized HBeAg, the reduced protein can assemble into capsids
Previous work has shown that, under certain in vitro conditions, HBeAg can be induced to form capsids (Watts et al., 2011). On the other hand, the crystal structure of HBeAg reveals a conformation that is incompatible with capsid assembly. Given the presence of the HBeAg-specific disulfide bridge, we hypothesized that the protein’s oxidation state may affect its state of assembly. To test this idea, HBeAg samples were analyzed by analytical ultracentrifugation sedimentation velocity (AUC) using material verified to be dimeric and completely oxidized, as well as the same material but pre-treated with reductant. Oxidized HBeAg remains dimeric (single homogenous sedimenting boundary ~2.5S), whereas reduced HBeAg contains about 30% of a fast-sedimenting component (>40S), consistent with high-molecular weight protein (Figure 3A, B). Negative-stain electron microscopy (EM) confirmed the presence of capsids in reductant-treated samples together with some other polymeric structures, of which there was no sign in the oxidized sample (Figure 3C, D). Consistent with these results, it has been shown that an HBeAg mutant in which C(-7) is substituted to A forms dimers with the same intermolecular C61-C61 disulfide bond seen in HBcAg dimers. This change increases the protein’s melting temperature to 62°C (similar to the value observed for HBcAg) (Watts et al., 2011), suggesting that in the absence of the C(-7)-C61 disulfide bridge, the propeptide is displaced, allowing HBeAg to adopt an HBcAg-like conformation. This inference is supported by previous studies that HBeAg could form capsid-like particles (Watts et al., 2011), and we have now demonstrated that this occurs only when the disulfide is disrupted by reduction (Figure 3).
Figure 3. Centrifugation and electron microscopy experiments on HBeAg in reduced and oxidized forms.
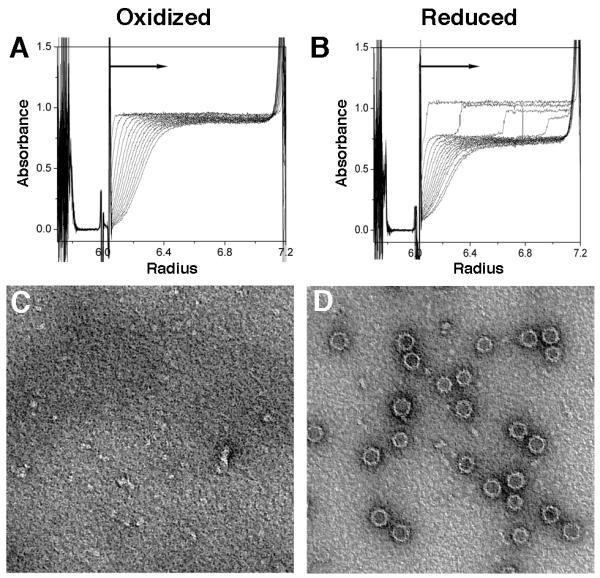
(A, B) Sedimentation velocity analyses performed on (A) oxidized and (B) reduced HBeAg using a Beckman Optima XL-I analytical ultracentrifuge, absorption optics, an An-60 Ti rotor, and standard double-sector centerpiece cells. Measurements at 20 °C were taken at 45,000 rpm for 3 hours with data collection at 10 minute intervals. The profiles show protein absorbance at 280 nm as a function of radial distance.
(C, D) Negative-staining EM of assembly products of (C) oxidized and (D) reduced HBeAg. Both samples, in PBS +/− DTT, were buffer-exchanged into TBS to avoid precipitation. Both images are at the same magnification; capsids are ~32 nm in diameter.
DISCUSSION
It has been four decades since the first description of HBeAg (Magnius and Espmark, 1972), and three since its isolation from serum and the discovery that it is closely related to HBcAg (Ferns and Tedder, 1984; MacKay et al., 1981). Despite this close relation, the two antigens have proved to have profoundly different biophysical and functional properties (Chen et al., 2005; Chen et al., 2004; Milich and Liang, 2003; Nassal and Rieger, 1993; Steven et al., 2005; Wasenauer et al., 1992). The crystal structure of HBeAg reveals that its subunit has much the same fold as that of HBcAg (Wynne et al, 1998) but a radically different mode of dimerization, which explains the profound biophysical and antigenic differences between the two proteins. While there are many examples of alternative dimer interfaces in protein crystal packing, the phenomenon of alternative physiologically relevant modes of dimerization has few if any precedents (but see Silvan et al., 2012).
The propeptide sterically hinders formation of an HBcAg-like dimer
The HBeAg structure shows how the presence of the propeptide prevents formation of the HBcAg dimer interface by ordering against the inner α3-α4 hairpin surface and sterically blocking dimerization (Figure 2C). Remarkably, the HBeAg monomer circumvents this steric obstacle by using parts of the same molecular surfaces to form an alternative dimer, but with the subunits rotated ~140° relative to their orientation in HBcAg (Figure 2A). In this altered conformation, bulky, hydrophobic residues W(-4)L(-3)W(-2) in the propeptide shield hydrophobic groups on the opposing subunit’s surface that would otherwise be exposed (Figure 2B). Further, intercalation of the two propeptides between the α3-α4 hairpins of opposing subunits completes the HBeAg dimer interface, mimicking molecular interactions that take place in HBcAg. The importance of the propeptide is corroborated by the near complete conservation of its sequence among the mammalian Hepadnaviridae (Revill et al., 2010) (Figure S2).
The C(-7)-C61 disulfide bridge is essential for the HBeAg dimer structure
Our data (above) imply that the C(-7)-C61 intramolecular disulfide bridge present in HBeAg is crucial for maintaining the observed propeptide conformation. Evidence includes the propeptide’s lack of secondary structure and disorder of propeptide residues S(-10), K(-9), L(-8). We tested the influence of this disulfide on HBeAg structure by analyzing oxidized and reduced HBeAg samples by analytic ultracentrifugation and EM (Figure 3). When the disulfide bridge is disrupted, the HBeAg subunits revert to an HBcAg-like mode of association, and are able to form capsids. This reversion is consistent with calorimetry data indicating that the HBcAg dimer is more stable than the (oxidized) HBeAg dimer (melting temperature of 65° vs. 51° C) (Watts et al., 2011). This conclusion as to relative stability is further supported by the HBeAg dimer interface being markedly smaller than that of HBcAg (1640 vs. 3970 Å2). We conclude that once the disulfide is formed, it locks the propeptide into place. Alternatively, when it is disrupted, the subunits revert to the thermodynamically favored HBcAg conformation. The HBeAg dimer is assembly-incompetent because the subunits are arranged such that the inter-dimer contacts necessary for capsid assembly—between adjacent C-terminal loops and α2-α3 arms—cannot form (Ceres and Zlotnick, 2002).
Molecular switching between HBeAg and HBcAg explains the observed differences in antigenicity
The lack of a high-resolution structure for HBeAg has hampered understanding of its antigenicity. Some antibodies are cross-reactive between HBcAg and HBeAg, while others recognize only one, and the antigens are not considered cross-reactive at the B-cell level in natural infection (Baumeister et al., 2000; Imai et al., 1982; Milich and Liang, 2003; Salfeld et al., 1989; Steven et al., 2005; Watts et al., 2010). The human antibody response to HBcAg is primarily conformational and directed against the outer part of the capsid spike (residues 74-89) as well as the floor of the capsid around the threefold axis (Ferns and Tedder, 1986; Kandiah et al., 2012; Salfeld et al., 1989).
Cryo-EM analyses of Fab-labeled capsids have characterized the epitopes of six murine anti-HBcAg monoclonal antibodies—all conformational—on the capsid surface (Figure 4A) (reviewed in (Steven et al., 2005)). We have now mapped these epitopes onto the HBeAg crystal structure (Figures 4B and S3). HBcAg epitopes are typically juxtapositions of two or more loops from different subunits or discontinuous regions of the same subunit. In HBeAg, these loops are moved apart, leaving single loops with reduced affinity. For antibodies 3105 and F11A4, the reduction is by two and three orders of magnitude, respectively (Watts et al., 2010). Further, the epitope of anti-HBcAg antibody 3120 maps to the capsid floor around the three-fold axis, bridging between two adjacent dimers (Figure 4A). Such floor-binding antibodies do not bind to HBeAg, because formation of their composite epitope requires capsid assembly (Figure 4B). In addition to epitopes shared with HBcAg, the HBeAg structure reveals a large, accessible molecular surface that is likely to present antibody determinants unique to HBeAg. This surface exposes regions that are inaccessible in assembled capsids, including the epitope of the e6 antibody used in this study (binding to α5 and the adjacent C-terminal loop) (Figures 1G and S4), as well as new surfaces created from the rearrangement of the helical hairpins.
Figure 4. Antigenicity of HBeAg and HBcAg.
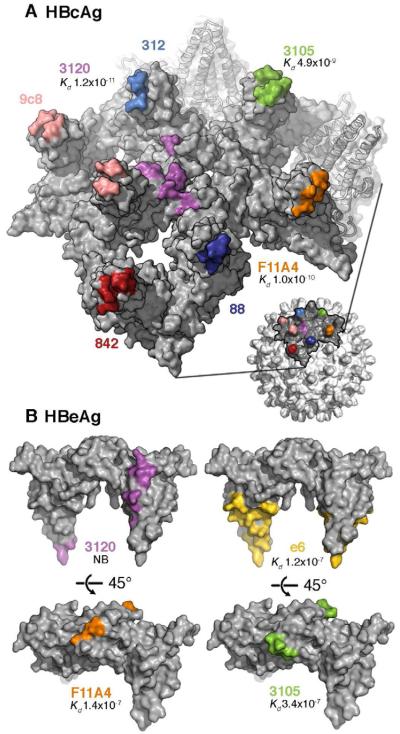
(A) Partial surface of an HBcAg capsid with the epitopes for several anti-HBc/eAg antibodies mapped in colors. Most of these epitopes reside around the spike tips, either on one subunit of the dimeric spike or bridging both subunits. Mab 3120 binds to the floor around the 3-fold and 5-fold symmetry axes.
(B) The same epitopes (matched colors) mapped on the HBeAg dimer. Affinity constants determined by surface plasmon resonance (Kd values (Watts et al., 2010)) are given, where available. A 2 - 3 order of magnitude drop is observed for Mabs F11A4 and 3105 on HBeAg relative to HBcAg, consistent with the constituent loops, which are close together on HBcAg, being well separated on HBeAg.
Alternative dimerization of HBcAg and HBeAg may underlie their apparent “split” immune tolerance
HBcAg and HBeAg appear to be regulated independently by the immune system, resulting in a significantly more immunogenic (HBcAg) or tolerogenic (HBeAg) T cell response (Chen et al., 2005; Milich et al., 1997a; Milich and McLachlan, 1986; Milich et al., 1997b; Vanlandschoot et al., 2003). One aspect of this regulation is the observation that HBcAg, but not HBeAg, can directly bind to and activate B cells without requirement for T cell support, leading to a robust humoral and cytotoxic T cell response (Lazdina et al., 2001; Milich et al., 1997a; Milich and McLachlan, 1986; Milich et al., 1997b). How this takes place is unclear, although it has been proposed that structural differences between the antigens are responsible: the array of spike-resident epitopes on the capsid surface (Figure 4A) may crosslink B cell receptors, activating the B cells and initiating the cascade (Milich et al., 1997a; Milich et al., 1997b; Vanlandschoot et al., 2003). As HBeAg has a conformation that precludes capsid formation and the clustering of B-cell receptors, this may explain its inability to activate B cells as well as its reduced immunogenicity compared to HBcAg. However, it is known that HBcAg and HBeAg remain cross-reactive at the T cell level, due to sequence identity (as T cell activation involves MHC-presentation of short antigenic peptides) (Milich and Liang, 2003). This duality may engender an immune response in which T cell cross-reactivity is necessary for inducing clonal tolerance to HBcAg (Chen et al., 2005; Chen et al., 2004; Vanlandschoot et al., 2003), while the antigenic switching allows HBeAg to avert the robust immune response that HBcAg elicits.
The molecular switch that relates the pairing of core domains in HBeAg and HBcAg explains many of the fundamental biophysical and antigenic differences between them (Figure 5) and has implications for their respective immunological properties in the context of chronic HBV infection. There are probably other levels of regulation in which HBeAg also takes part to establish chronicity, including, for example, modulation of the innate and adaptive immune responses via direct interaction with host immune proteins (Lang et al., 2011; Purvina et al., 2012; Visvanathan et al., 2007; Yang et al., 2006). A long-standing perception of HBeAg has been that it is a monomeric protein with an N-terminal propeptide that happens to be requisite for HBeAg functionality. Our data have shown that HBeAg is not a monomer, but a dimer, and that the crucial feature of its propeptide is its strategically positioned Cys residue that dictates the structure of the HBeAg dimer. This structure may, we conjecture, confer an ability to interact directly with host immune proteins in ways that HBcAg cannot. The crystal structure now provides a framework upon which further study can fully elucidate the role of HBeAg in HBV persistence.
Figure 5. Antigenic switching of HBV capsid protein.
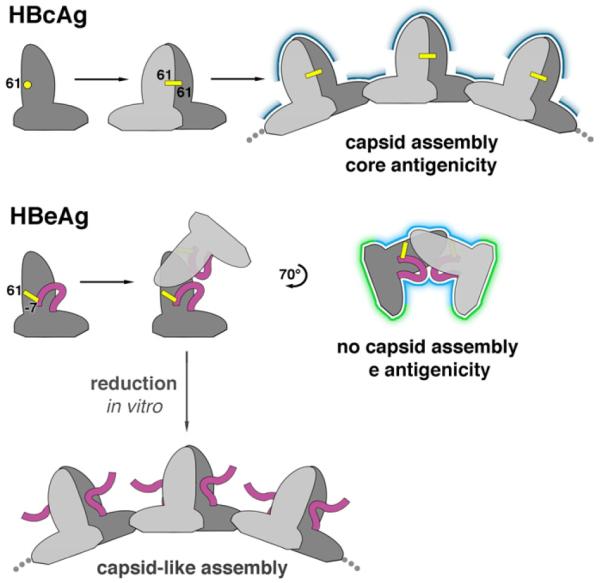
HBcAg and HBeAg polypeptides share the same monomer fold. When HBcAg dimerizes, an inter-molecular disulfide bridge forms between C61 of each subunit. In the HBeAg dimer, however, two intra-molecular disulfides form between C61 and propeptide residue C(-7). Once locked into place, the propeptides block HBcAg dimer formation. Instead, the HBeAg subunits dimerize in an entirely different quaternary arrangement. As a result, HBeAg dimers cannot form the dimer-dimer contacts employed in capsid assembly. The molecular switch also explains how HBeAg and HBcAg are antigenically distinct: surfaces presenting conformational epitopes on HBcAg (dark blue) are altered in HBeAg (light blue) and antibody-inaccessible surfaces on the interior of capsids are exposed in HBeAg (green). Antibodies that bind to only one subunit within the shared surfaces (light and dark blue) may be cross-reactive for both antigens. If HBeAg is subjected to reducing conditions in vitro, these disulfides are disrupted and capsid-like assemblies can form.
EXPERIMENTAL PROCEDURES
Protein preparation and crystallization
We used the construct Cp(-10)149, C48A, C107A, and refer to it here as HBeAg although it differs from wildtype HBeAg in having two Ala substitutions at C48 and C107, which do not form disulfide bridges (Wingfield et al., 1995). The HBeAg used for crystallization included the aforementioned mutations plus G123A (Watts et al., 2011). HBeAg and Fab e6 were produced as previously described (Watts et al., 2010), and to form the complex, the components were mixed at an expected stoichiometry of 1:2 (excess Fab was removed by size-exclusion chromatography). Crystallization trials were performed using the sitting drop vapor diffusion method at a protein concentration of 5.3 mg ml−1 in 20 mM HEPES pH 8.0 at 21 °C. Sitting drops were formed by mixing 100 nl of protein solution and 100 nl of reservoir solution (Walter et al., 2005). Plate-shaped crystals were grown using reservoir solution containing 20% PEG 6000 and 100 mM bicine pH 9.0. Rod-like crystals of Fab e6 crystals serendipitously grew from a preparation of the complex from which excess Fab had not been removed in reservoir solution containing 20% PEG 3350, 100 mM bis-Tris propane pH 6.5, and 200 mM potassium thiocyanate.
Structure solution and analysis
Diffraction data for HBeAg-e6 complex and Fab e6 were collected using synchrotron radiation from single crystals to a resolution of 3.3 Å and 2.5 Å, respectively. The data were integrated and scaled using xia2 (Winter, 2010). Initial phase information for the Fab e6 data were obtained via an automated Phaser (1994) molecular replacement (MR) search using a Fab structure library (Stanfield et al., 2006). Positional, TLS, and individual isotropic B-factor refinement on the Fab e6 structure were carried out in BUSTER (Bricogne et al., 2011) using fourfold NCS restraints, followed by iterative rebuilding of the hyper-variable loops and correction of the sequence. This Fab e6 structure, refined against 2.5 Å resolution data, was used as an MR model to obtain initial phase estimates for the HBeAg-Fab e6 data. Positional, group B-factor (one group per residue), TLS refinement were used for structural refinement, with twofold NCS-restraints and used local structure similarity restraints (LSSR) to the Fab structure applied to mitigate the limited resolution of the data (Smart et al., 2008). The Molprobity server (Chen et al., 2010) and validation tools in Coot (Emsley et al., 2010) informed the quality of the structure refinement process. Refinement statistics are given in Table 1, and final refined coordinates and structure factors have been deposited for Fab e6 and HBeAg-Fab e6 with the PDB with accession codes 3V6F and 3V6Z, respectively. PISA interface web server was utilized for buried surface area and interacting residue analysis of the HBeAg dimer interface and the HBeAg-Fab e6 epitope-paratope interface (Krissinel and Henrick, 2007). The Rapido server was used to determine structurally similar sub-domains within HBeAg and HBcAg monomers (Mosca et al., 2008). Secondary structure assignment of HBeAg was done using DSSP (Kabsch and Sander, 1983) and Stride (Heinig and Frishman, 2004). Molecular graphics were produced using Pymol (DeLano Scientific LLC).
Sedimentation velocity analysis and negative-stain electron microscopy
HBeAg was dialyzed against phosphate buffered saline (PBS) pH 7.2, plus 300 mM NaCl (total NaCl 450 mM). A sample treated with 10 mM DTT was also dialyzed against the same uffer, plus 2 mM DTT. Under either of these conditions, dimeric HBcAg (Cp149) readily and efficiently forms capsids. Following dialysis for ~24h, samples were analyzed by sedimentation velocity. Measurement of the height (UV absorbance) of the sedimenting boundaries allows the concentrations of the various species to be determined. The oxidized and reduced HBeAg samples, were also applied to glow-discharged, poly-lysine coated carbon grids at a concentration of ~0.25 mg/ml, stained with 1% uranyl acetate and observed at x35,000 magnification in a Philips CM-120 electron microscope.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ira Palmer and Joshua Kaufman for producing and purifying the proteins used in this study; Richard Tedder for supplying the original e6 hybridoma, Christoph Rader for advice on sequence determination of the e6 Fab, and Christian Siebold for support with data collection. We also thank Vincenzo Cerundolo, Matti Sällberg and E. Yvonne Jones for comments on the manuscript. This work was supported in part by the Intramural Research Programs of the National Institute for Arthritis and Musculoskeletal and Skin Diseases and the NIH-Oxford Scholars Program. D.I.S. is supported by the UK Medical Research Council and J.M.G. in part by SPINE2COMPLEXES LSHG-CT-2006-031220. The Wellcome Trust is acknowledged for administrative support (Grant 075491/Z/04). The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Atomic coordinates and structure factors for HBeAg-e6 Fab and e6 Fab have been deposited in the Protein Data Bank (http://www.pdb.org) with accession codes 3V6F and 3V6Z, respectively.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and Supplemental Experimental Procedures and can be found with this article online at .
REFERENCES
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Baumeister MA, Medina-Selby A, Coit D, Nguyen S, George-Nascimento C, Gyenes A, Valenzuela P, Kuo G, Chien DY. Hepatitis B virus e antigen specific epitopes and limitations of commercial anti-HBe immunoassays. J Med Virol. 2000;60:256–263. [PubMed] [Google Scholar]
- Bricogne G, B E, M B, C F, P K, W P, P R, A S, O.S S, C V, et al. BUSTER. United Kingdom, Global Phasing Ltd.; Cambridge: 2011. [Google Scholar]
- Ceres P, Zlotnick A. Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry. 2002;41:11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- Chang C, Enders G, Sprengel R, Peters N, Varmus HE, Ganem D. Expression of the precore region of an avian hepatitis B virus is not required for viral replication. J Virol. 1987;61:3322–3325. doi: 10.1128/jvi.61.10.3322-3325.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Kew MC, Hornbuckle WE, Tennant BC, Cote PJ, Gerin JL, Purcell RH, Miller RH. The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J Virol. 1992;66:5682–5684. doi: 10.1128/jvi.66.9.5682-5684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Sallberg M, Hughes J, Jones J, Guidotti LG, Chisari FV, Billaud JN, Milich DR. Immune tolerance split between hepatitis B virus precore and core proteins. J Virol. 2005;79:3016–3027. doi: 10.1128/JVI.79.5.3016-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MT, Billaud JN, Sallberg M, Guidotti LG, Chisari FV, Jones J, Hughes J, Milich DR. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc Natl Acad Sci U S A. 2004;101:14913–14918. doi: 10.1073/pnas.0406282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Stahl SJ, Wingfield PT, Belnap DM, Kanngiesser U, Noah M, Steven AC. Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c and e-antigens. J Mol Biol. 1998;279:1111–1121. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- Elgouhari HM, Abu-Rajab Tamimi TI, Carey WD. Hepatitis B virus infection: understanding its epidemiology, course, and diagnosis. Cleve Clin J Med. 2008;75:881–889. doi: 10.3949/ccjm.75a.07019. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan EA, Smith PM, Davison F, Williams R. Fulminant hepatitis B in successive female sexual partners of two anti-HBe-positive males. Lancet. 1986;2:538–540. doi: 10.1016/s0140-6736(86)90112-1. [DOI] [PubMed] [Google Scholar]
- Ferns RB, Tedder RS. Monoclonal antibodies to hepatitis Be antigen (HBeAg) derived from hepatitis B core antigen (HBcAg): their use in characterization and detection of HBeAg. J Gen Virol. 1984;65(Pt 5):899–908. doi: 10.1099/0022-1317-65-5-899. [DOI] [PubMed] [Google Scholar]
- Ferns RB, Tedder RS. Human and monoclonal antibodies to hepatitis B core antigen recognise a single immunodominant epitope. J Med Virol. 1986;19:193–203. doi: 10.1002/jmv.1890190213. [DOI] [PubMed] [Google Scholar]
- Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32:W500–502. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Nomura M, Gotanda T, Sano T, Tachibana K, Miyamoto H, Takahashi K, Toyama S, Miyakawa Y, Mayumi M. Demonstration of two distinct antigenic determinants on hepatitis B e antigen by monoclonal antibodies. J Immunol. 1982;128:69–72. [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kandiah E, Watts NR, Cheng N, Cardone G, Stahl SJ, Heller T, Liang TJ, Wingfield PT, Steven AC. Cryo-EM study of Hepatitis B virus core antigen capsids decorated with antibodies from a human patient. J Struct Biol. 2012;177:145–151. doi: 10.1016/j.jsb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lang T, Lo C, Skinner N, Locarnini S, Visvanathan K, Mansell A. The hepatitis B e antigen (HBeAg) targets and suppresses activation of the toll-like receptor signaling pathway. J Hepatol. 2011;55:762–769. doi: 10.1016/j.jhep.2010.12.042. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lazdina U, Cao T, Steinbergs J, Alheim M, Pumpens P, Peterson DL, Milich DR, Leroux-Roels G, Sallberg M. Molecular basis for the interaction of the hepatitis B virus core antigen with the surface immunoglobulin receptor on naive B cells. J Virol. 2001;75:6367–6374. doi: 10.1128/JVI.75.14.6367-6374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang TJ, Hasegawa K, Rimon N, Wands JR, Ben-Porath E. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. N Engl J Med. 1991;324:1705–1709. doi: 10.1056/NEJM199106133242405. [DOI] [PubMed] [Google Scholar]
- MacKay P, Lees J, Murray K. The conversion of hepatitis B core antigen synthesized in E coli into e antigen. J Med Virol. 1981;8:237–243. doi: 10.1002/jmv.1890080404. [DOI] [PubMed] [Google Scholar]
- Magnius LO, Espmark JA. New specificities in Australia antigen positive sera distinct from the Le Bouvier determinants. J Immunol. 1972;109:1017–1021. [PubMed] [Google Scholar]
- Milich D, Liang TJ. Exploring the biological basis of hepatitis B e antigen in hepatitis B virus infection. Hepatology. 2003;38:1075–1086. doi: 10.1053/jhep.2003.50453. [DOI] [PubMed] [Google Scholar]
- Milich DR, Chen M, Schodel F, Peterson DL, Jones JE, Hughes JL. Role of B cells in antigen presentation of the hepatitis B core. Proc Natl Acad Sci U S A. 1997a;94:14648–14653. doi: 10.1073/pnas.94.26.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milich DR, Jones JE, Hughes JL, Price J, Raney AK, McLachlan A. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc Natl Acad Sci U S A. 1990;87:6599–6603. doi: 10.1073/pnas.87.17.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milich DR, McLachlan A. The nucleocapsid of hepatitis B virus is both a T-cell-independent and a T-cell-dependent antigen. Science. 1986;234:1398–1401. doi: 10.1126/science.3491425. [DOI] [PubMed] [Google Scholar]
- Milich DR, Schodel F, Hughes JL, Jones JE, Peterson DL. The hepatitis B virus core and e antigens elicit different Th cell subsets: antigen structure can affect Th cell phenotype. J Virol. 1997b;71:2192–2201. doi: 10.1128/jvi.71.3.2192-2201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca R, Brannetti B, Schneider TR. Alignment of protein structures in the presence of domain motions. BMC Bioinformatics. 2008;9:352. doi: 10.1186/1471-2105-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M, Rieger A. An intramolecular disulfide bridge between Cys-7 and Cys61 determines the structure of the secretory core gene product (e antigen) of hepatitis B virus. J Virol. 1993;67:4307–4315. doi: 10.1128/jvi.67.7.4307-4315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou JH. Molecular biology of hepatitis B virus e antigen. J Gastroenterol Hepatol. 1997;12:S178–187. doi: 10.1111/j.1440-1746.1997.tb00499.x. [DOI] [PubMed] [Google Scholar]
- Ou JH, Laub O, Rutter WJ. Hepatitis B virus gene function: the precore region targets the core antigen to cellular membranes and causes the secretion of the e antigen. Proc Natl Acad Sci U S A. 1986;83:1578–1582. doi: 10.1073/pnas.83.6.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packianathan C, Katen SP, Dann CE, 3rd, Zlotnick A. Conformational changes in the hepatitis B virus core protein are consistent with a role for allostery in virus assembly. J Virol. 2010;84:1607–1615. doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvina M, Hoste A, Rossignol JM, Lagaudriere-Gesbert C. Human hepatitis B viral e antigen and its precursor P20 inhibit T lymphocyte proliferation. Biochem Biophys Res Commun. 2012;417:1310–1315. doi: 10.1016/j.bbrc.2011.12.138. [DOI] [PubMed] [Google Scholar]
- Revill P, Yuen L, Walsh R, Perrault M, Locarnini S, Kramvis A. Bioinformatic analysis of the hepadnavirus e-antigen and its precursor identifies remarkable sequence conservation in all orthohepadnaviruses. J Med Virol. 2010;82:104–115. doi: 10.1002/jmv.21645. [DOI] [PubMed] [Google Scholar]
- Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J Virol. 1989;63:798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht HJ, Salfeld J, Schaller H. The duck hepatitis B virus pre-C region encodes a signal sequence which is essential for synthesis and secretion of processed core proteins but not for virus formation. J Virol. 1987;61:3701–3709. doi: 10.1128/jvi.61.12.3701-3709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel F, Peterson D, Zheng J, Jones JE, Hughes JL, Milich DR. Structure of hepatitis B virus core and e-antigen. A single precore amino acid prevents nucleocapsid assembly. J Biol Chem. 1993;268:1332–1337. [PubMed] [Google Scholar]
- Silvan U, Boiteux C, Sutterlin R, Schroeder U, Mannherz HG, Jockusch BM, Berneche S, Aebi U, Schoenenberger CA. An antiparallel actin dimer is associated with the endocytic pathway in mammalian cells. J Struct Biol. 2012;177:70–80. doi: 10.1016/j.jsb.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Smart OS, Brandl M, Flensburg C, Keller P, Paciorek W, Vonrhein C, Womack TO, Bricogne G. Refinement with Local Structure Similarity Restraints (LSSR) Enables Exploitation of Information from Related Structures and Facilitates use of NCS; Paper presented at: Annual Meeting of the American Crystallographic Association (Abstract TP13); 2008. [Google Scholar]
- Standring DN, Ou JH, Masiarz FR, Rutter WJ. A signal peptide encoded within the precore region of hepatitis B virus directs the secretion of a heterogeneous population of e antigens in Xenopus oocytes. Proc Natl Acad Sci U S A. 1988;85:8405–8409. doi: 10.1073/pnas.85.22.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Steven AC, Conway JF, Cheng N, Watts NR, Belnap DM, Harris A, Stahl SJ, Wingfield PT. Structure, assembly, and antigenicity of hepatitis B virus capsid proteins. Adv Virus Res. 2005;64:125–164. doi: 10.1016/S0065-3527(05)64005-5. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Machida A, Funatsu G, Nomura M, Usuda S, Aoyagi S, Tachibana K, Miyamoto H, Imai M, Nakamura T, et al. Immunochemical structure of hepatitis B e antigen in the serum. J Immunol. 1983;130:2903–2907. [PubMed] [Google Scholar]
- Terazawa S, Kojima M, Yamanaka T, Yotsumoto S, Okamoto H, Tsuda F, Miyakawa Y, Mayumi M. Hepatitis B virus mutants with precore-region defects in two babies with fulminant hepatitis and their mothers positive for antibody to hepatitis B e antigen. Pediatr Res. 1991;29:5–9. doi: 10.1203/00006450-199101000-00002. [DOI] [PubMed] [Google Scholar]
- Vanlandschoot P, Cao T, Leroux-Roels G. The nucleocapsid of the hepatitis B virus: a remarkable immunogenic structure. Antiviral Res. 2003;60:67–74. doi: 10.1016/j.antiviral.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Visvanathan K, Skinner NA, Thompson AJ, Riordan SM, Sozzi V, Edwards R, Rodgers S, Kurtovic J, Chang J, Lewin S, et al. Regulation of Toll-like receptor-2 expression in chronic hepatitis B by the precore protein. Hepatology. 2007;45:102–110. doi: 10.1002/hep.21482. [DOI] [PubMed] [Google Scholar]
- Walter TS, Diprose JM, Mayo CJ, Siebold C, Pickford MG, Carter L, Sutton GC, Berrow NS, Brown J, Berry IM, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasenauer G, Kock J, Schlicht HJ. A cysteine and a hydrophobic sequence in the noncleaved portion of the pre-C leader peptide determine the biophysical properties of the secretory core protein (HBe protein) of human hepatitis B virus. J Virol. 1992;66:5338–5346. doi: 10.1128/jvi.66.9.5338-5346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts NR, Conway JF, Cheng N, Stahl SJ, Steven AC, Wingfield PT. Role of the propeptide in controlling conformation and assembly state of hepatitis B virus e-antigen. J Mol Biol. 2011;409:202–213. doi: 10.1016/j.jmb.2011.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts NR, Vethanayagam JG, Ferns RB, Tedder RS, Harris A, Stahl SJ, Steven AC, Wingfield PT. Molecular basis for the high degree of antigenic cross-reactivity between hepatitis B virus capsids (HBcAg) and dimeric capsid-related protein (HBeAg): insights into the enigmatic nature of the e-antigen. J Mol Biol. 2010;398:530–541. doi: 10.1016/j.jmb.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Williams RW, Steven AC. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- Winter G. xia2: an expert system for macromolecular crystallography data reduction. Journal of Applied Crystallography. 2010;43:186–190. [Google Scholar]
- Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- Yang CY, Kuo TH, Ting LP. Human hepatitis B viral e antigen interacts with cellular interleukin-1 receptor accessory protein and triggers interleukin-1 response. J Biol Chem. 2006;281:34525–34536. doi: 10.1074/jbc.M510981200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


