SUMMARY
The essential helicase-like protein Sen1 mediates termination of RNA Polymerase II (Pol II) transcription at snoRNAs and other non-coding RNAs in yeast. A mutation in the Pol II subunit Rpb1 that increases the elongation rate increases readthrough transcription at Sen1-mediated terminators. Termination and growth defects in sen1 mutant cells are partially suppressed by a slowly transcribing Pol II mutant and are exacerbated by a faster transcribing Pol II mutant. Deletion of the nuclear exosome subunit Rrp6 allows visualization of non-coding RNA intermediates that are terminated but not yet processed. Sen1 mutants or faster transcribing Pol II increase the average lengths of pre-processed snoRNA, CUT, and SUT transcripts, while slowed Pol II transcription produces shorter transcripts. These connections between transcription rate and Sen1 activity support a model whereby kinetic competition between elongating Pol II and Sen1 helicase establishes the temporal and spatial window for early Pol II termination.
INTRODUCTION
Eukaryotic gene expression requires efficient transitions between the stages of the transcription cycle: initiation, elongation, and termination. Initiation involves the recruitment and accurate positioning of RNA Polymerase II (Pol II) at gene promoters to begin polymerization of RNA transcripts. In elongation phase, RNA synthesis proceeds with the assistance of processivity and chromatin modifying factors. Termination releases the nascent RNA transcript from the polymerase and dissociates Pol II from the DNA template.
In S. cerevisiae, termination of Pol II transcription can be mediated by at least two mechanisms: the Sen1-dependent termination pathway and the poly(A)-dependent pathway (Kim et al., 2006). The Sen1 pathway principally operates at smaller (≤500 bp) non-coding genes such as small nuclear RNA (snRNAs), small nucleolar RNAs (snoRNAs), and cryptic unstable transcripts (CUTs). This pathway is dependent on the essential 5′ to 3′ ATP-dependent Superfamily 1 helicase Sen1 (Rasmussen and Culbertson, 1998; Steinmetz et al., 2001). In contrast, the poly(A) pathway acts at protein-coding mRNAs and some non-coding transcripts, which are generally longer than 0.5 kb. RNAs subject to this pathway undergo cleavage, polyadenylation, and Pol II termination by the Rat1/Xrn2 exonuclease (Connelly and Manley, 1988; Kim et al., 2004). However, gene length alone is not the sole determinant as to which pathway mediates Pol II termination, as some genes utilize both early and poly(A) termination regimes to varying degrees (Kawauchi et al., 2008; Rondon et al., 2009; Marquardt et al., 2011). For example, Sen1 regulates some protein-coding genes via transcription attenuation mediated by premature termination (Arigo et al., 2006; Kim and Levin, 2011). Senataxin, the human ortholog of Sen1, has been proposed to function in mRNA termination by assisting the action of Xrn2 (Skourti-Stathaki et al., 2011).
Sen1 operates in conjunction with the proteins Nrd1 and Nab3 to mediate Pol II termination (Steinmetz et al., 2001). Nrd1 and Nab3 contain RNA Recognition Motif (RRM) domains that bind specific RNA sequences that are often enriched in Sen1 pathway target genes (Steinmetz et al., 2006a). Nrd1 also contains a CTD Interacting Domain with specificity for phosphorylated serine 5 residues (Ser5P) in the C-terminal domain (CTD) of the Pol II subunit Rpb1, a mark enriched during early elongation (Conrad et al., 2000; Vasiljeva et al., 2008). The prevailing model for early termination is that a combination of Nrd1/Nab3 motifs in the nascent transcript and higher levels of Ser5P on the Pol II CTD recruit the Nrd1/Nab3 complex during early elongation, which in turn delivers Sen1 to the nascent RNA (Carroll et al., 2004; Gudipati et al., 2008). Sen1 also interacts with the Pol II CTD, with a preference for Ser2 phosphorylation (Chinchilla et al., 2012). This interaction, as well as observations that Sen1 immunoprecipitates with numerous non-coding transcripts (Ursic et al., 2004) and crosslinks to many transcripts with no apparent sequence specificity (Creamer et al., 2011), suggests that Sen1 contacts both Pol II and the nascent RNA transcript.
Several mechanisms have been proposed for how Sen1 might mediate termination. One model is that Sen1 translocates 5′ to 3′ along the RNA until it runs into Pol II and/or breaks contacts between the polymerase and transcript to catalyze termination (Kim et al., 1999; Steinmetz et al., 2006a). In this model, Sen1 functions analogously to Rho helicase in prokaryotes (Brow, 2011; Richardson, 2006). Alternatively, Sen1 might strip proteins from the transcript, allowing access to a true termination factor, perhaps Pcf11 (Zhang et al., 2005). Finally, it has been suggested that R-loop formation between the transcript and template DNA strand is an intermediate that must be disrupted by Sen1 as part of the termination mechanism (Mischo et al., 2011; Skourti-Stathaki et al, 2011).
One prediction of the Rho-like model is that termination should occur over a region downstream of the termination signals (Zhu and von Hippel, 1998), rather than at a discrete site as seen with eukaryotic Pol III or bacterial intrinsic terminators. Furthermore, this termination window would be affected by the rate of transcription, factors that influence Pol II kinetics and passage through chromatin, as well as the rate at which Sen1 translocates along the RNA. Such a kinetic competition model of termination operates in bacteria, where polymerase elongation rate influences the termination efficiency of both intrinsic and Rho-dependent terminators (Bar-Nahum et al., 2005; Jin et al., 1992). Here we show that a faster elongating Pol II mutant shifts the early termination window further downstream and increases readthrough at non-coding genes. Conversely, slowly transcribing Pol II shifts termination towards the 5′ end of non-coding genes. Sen1 termination functions are in kinetic competition with Pol II elongation, as a slow Pol II mutation partially suppresses termination defects of two Sen1 mutants, while fast Pol II transcription exacerbates these phenotypes. Our findings indicate that a balance between Pol II elongation and the antagonizing action of Sen1 defines the window for efficient termination. Perturbations of this balance can lead to readthrough transcription into downstream genes with potentially deleterious effects on their expression.
RESULTS
Faster Pol II Elongation Causes Termination Defects at snoRNA Genes
To test the impact of Pol II elongation rate on early termination efficiency in S. cerevisiae, we analyzed two RPB1 mutants originally isolated in a screen for mutations causing sensitivity to the drug 6-azauracil (6-AU) (Malagon et al., 2006). These mutants show differential elongation rates in vitro, with rpb1-N488D being slower and rpb1-E1103G faster than wild-type (WT) Pol II (Malagon et al., 2006). In agreement with its in vitro properties, the rpb1-N488D (“slow Pol II”) mutant displays a slower transcription rate in vivo (Jimeno-Gonzalez et al., 2010). Residue N488 is close to the Mg2+ and NADFDGD motif in the Pol II active site (Malagon et al., 2006). Residue E1103 is located in the Rpb1 trigger loop, which plays a major role in NTP selectivity and incorporation (Cramer et al., 2001). In addition to increased elongation rate, rpb1-E1103G (“fast Pol II’) mutants have slightly decreased fidelity in vitro and in vivo (Kaplan et al., 2008; Kireeva et al., 2008). Analogous mutations in the E. coli RNA Polymerase trigger loop also increase elongation rate and decrease fidelity, underscoring a conserved function for this feature (Bar-Nahum et al., 2005).
To confirm rpb1-E1103G Pol II transcribes faster in vivo, the relative elongation rates of WT and mutant strains were tested in a GAL1-YLR454 shutoff experiment. This assay uses chromatin immunoprecipitation (ChIP) to measure the time-dependent loss of Pol II after gene repression, allowing assessment of relative elongation rates (Mason and Struhl, 2005). As assayed by ChIP of Pol II subunit Rpb3, rpb1-E1103G shows a more rapid loss of Pol II density from promoter and coding regions relative to WT Rpb1. Therefore, in accordance with its in vitro properties, rpb1-E1103G Pol II exhibits faster transcription in vivo (Figure 1A).
Figure 1. The Fast Pol II rpb1-E1103G Mutant Increases Readthrough at snoRNA Genes.
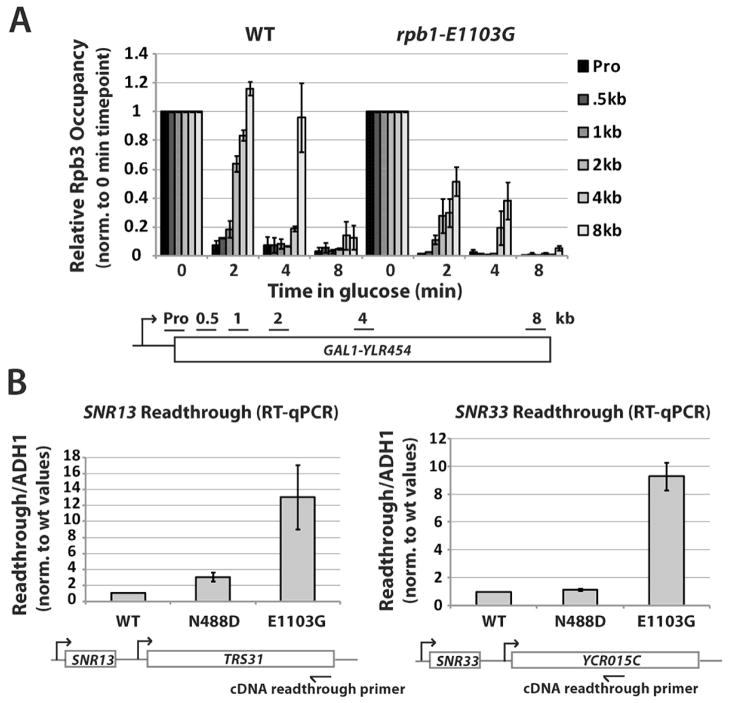
(A) ChIP assay to measure in vivo elongation rate. Graphs show occupancy of Pol II subunit Rpb3 in WT (YSB2716) and rpb1-E1103G (YSB2717) strains containing a GAL1-YLR454 construct, before (0 min) and after glucose addition (2, 4, 8 min) to shut off GAL1 promoter-driven transcription. Primer locations are indicated in gene schematic. Rpb3 occupancy values are expressed relative to the 0 min timepoint (pre-glucose) after normalization to a non-transcribed telomeric region. Error bars represent standard error from two independent experiments.
(B) Quantitative Reverse Transcription PCR analysis (RT-qPCR) of SNR13 and SNR33 readthrough transcript levels in WT (GRY3020), rpb1-N488D (GRY3027), and rpb1-E1103G (GRY3028) strains grown at 30°C relative to ADH1 control transcripts. Error bars represent standard error from three independent experiments.
To assess the efficiency of termination in slow and fast Pol II strains, levels of readthrough transcription at two snoRNA genes were measured. Reverse transcription followed by quantitative PCR (RT-qPCR) analysis of rpb1-N488D cells showed little to no increase in readthrough at SNR33 but a small increase at SNR13. Analysis of fast Pol II cells reveals a 13-fold increase in readthrough at SNR13 and a nearly 10-fold increase at SNR33 (Figure 1B). Sequencing of cDNAs made from snR13 readthrough transcripts revealed no mutations in Nrd1 or Nab3 sites (data not shown), indicating the 3- to 5-fold increase of misincorporation in rpb1-E1103G cells (Kaplan et al., 2008; Kireeva et al., 2008) does not cause the termination defect. Therefore, an Rpb1 mutation that increases the rate of elongation also causes significant readthrough defects, suggesting an important role for Pol II kinetics in Sen1-dependent termination.
Mapping the SNR33 Termination Window
Northern blot or RT-qPCR analyses of snoRNA species can show steady-state levels of mature RNAs that have undergone 3′ end processing, as well as readthrough transcripts that failed to terminate normally and instead utilized a downstream polyadenylation site. However, the intermediate RNA species that are properly terminated but not yet processed are short-lived and therefore not typically seen in standard assays. Because snoRNAs are synthesized as precursor transcripts that are 3′ trimmed by the exosome, the terminated but pre-processed snoRNA transcripts are stabilized by deletion of the nuclear exosome subunit Rrp6 (van Hoof et al., 2000). Similarly, transcriptome analyses of rrp6Δ cells revealed widespread cryptic unstable transcripts (CUTs) that are normally rapidly degraded by the exosome (Neil et al., 2009; Xu et al., 2009).
Northern blot analysis of SNR33 expression in rrp6Δ or other exosome mutant backgrounds shows two major species. The mature snoRNA is approximately 180 nucleotides long, whereas the terminated, pre-processed transcripts appear as a heterogeneous population of approximately 350–600 bases long (van Hoof et al., 2000) (Figure 2A, 2B; lane 1). Targeted digestion with RNase H and oligo(dT) (Figures 2C, S1A; lane 1 vs. 6) ruled out the possibility that the larger species are due solely to oligo(A) or poly(A) tail addition, although the observation that the smear resolved into more discrete bands does suggest oligoadenylation, as expected for an exosome substrate (van Hoof et al., 2000; Wlotzka et al., 2011). Three observations argue that the smear is not caused by a slower rate of degradation by exosome, which requires a free 3′ RNA end of a terminated transcript as a substrate. First, a slower core exosome would be expected to create a smear continuing all the way down to the mature species. Second, the window of putative termination products agrees with the downstream boundary of Pol II occupancy as assayed by both Pol II ChIP and NET-Seq experiments (Churchman and Weissman, 2011; Kim et al., 2006; Steinmetz et al., 2006b). Third, longer exposure of the blots shows that the smear of pre-processed transcripts is seen even when cells contain Rrp6 (Figures 2D, lane 1; S1B, lane 1; S2, lane 1). Therefore, we conclude that the 3′ heterogeneity arises from termination occurring stochastically in a region downstream of the mature 3′ end, with rrp6Δ allowing a steady-state view of the termination window.
Figure 2. Termination Window Mapping of Pre-Processed SNR33 Transcripts in WT, Slow, and Fast RNA Pol II strains.
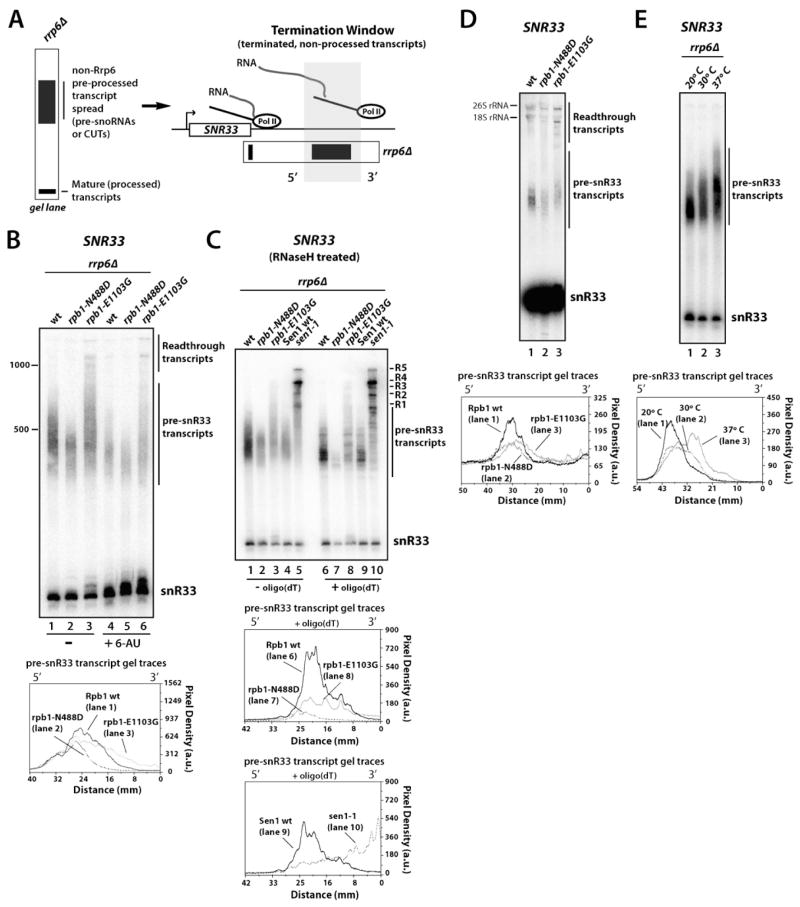
(A) Schematic illustrating Termination Window Mapping analysis. Pre-processed terminated snoRNA transcripts (pre-snoRNAs) and cryptic unstable transcripts (CUTs) accumulate in rrp6Δ cells and offer a visual readout of the termination window.
(B) Northern blot analysis of SNR33 from RPB1 (WT, YSB2697), slow rpb1-N488D (YSB2698), and fast rpb1-E1103G (YSB2699) strains also containing rrp6Δ. Cells were grown at 30°C in the presence or absence of 6-AU (75 μg/ml). Relative densitometry comparisons of pre-snR33 transcripts from indicated lanes are shown below.
(C) Northern blot analysis of SNR33 was performed as in part B, with the addition of isogenic Sen1 (YSB2859) and sen1-1 (YSB2860) strains containing rrp6Δ grown at 30°C. Before electrophoresis, RNA was treated with RNase H in the absence or presence of oligo(dT). R1-R5; readthrough/extended transcripts that increase in the sen1-1 background.
(D) Northern blot analysis of SNR33 from RRP6 strains carrying RPB1 (GRY3020), slow rpb1-N488D (GRY3027), and fast rpb1-E1103G (GRY3028) alleles.
(E) Northern blot analysis of SNR33 from an rrp6Δ (YSB2697) strain grown at 20°C, 30°C, and 37°C. See also Figure S1.
To examine whether Pol II elongation rate influences the region and efficiency of termination, rrp6Δ was introduced into RPB1, rpb1-N488D, and rpb1-E1103G strains and SNR33 transcripts were analyzed by Northern blot (Figure 2B–D). Strikingly, Pol II speed correlates with pre-snR33 transcript lengths. The rpb1-N488D mutant has a shorter distribution of pre-snR33 transcripts relative to cells containing WT Rpb1. The decreased spread length indicates that more termination events occur earlier upstream in the slow Pol II background. Similar results are obtained with another slow Pol II mutant, the rpb2-10 allele of the RPB2 subunit (Powell and Reines, 1996) (Figure S1B, C). Conversely, the fast rpb1-E1103G mutant leads to a longer set of pre-snR33 transcripts and greater readthrough levels. RNase H digestion with oligo(dT) suggests that the different Pol II enzymes generate the same set of 3′ ends, but their relative usage shifts with Pol II speed (Figure 2C). The readthrough species decrease in size upon oligo(dT)/RNase H treatment (data now shown), consistent with termination triggered by downstream polyadenylation sites. Interestingly, the upstream edge of the termination window is similar in all three strains, suggesting there could be a termination initiating event (for example, Nrd1/Nab3 binding or a certain CTD phosphorylation pattern) that is independent of elongation rate (Figure 2B). The same effects of Pol II speed on the termination window were seen on the SNR9 gene (Figure S1A, S2).
Further validation that our assay measures effects of elongation rate on termination was seen in three additional experiments. First, RPB1, rpb1-N488D, and rpb1-E1103G cells were treated with 6-AU, a drug that causes nucleotide depletion and thereby retards elongation by triggering pausing (Mason and Struhl, 2005). In all three strains, 6-AU treatment reduces overall transcription levels, but also shifts the peak of pre-snR33 termination products further upstream (Figure 2B). Second, depletion of uracil from the growth media, which decreases Pol II elongation rate in vivo (Mason and Struhl, 2005) leads to a reduction in the termination windows, similar to the effects of the slow Pol II (Figure S1E, F). The decreased abundance of pre-snR33 transcripts with 6-AU, uracil depletion, or rpb1-N488D mutation likely reflects the loss of Pol II processivity linked to decreased elongation rate, as previously suggested (Jimeno-Gonzalez et al., 2010; Mason and Struhl, 2005). Finally, higher temperatures increase the elongation rate of E. coli RNA Polymerase (Abbondanzieri et al., 2005). In accordance, the length of pre-processed snoRNA transcripts increases with temperature (Figure 2E, S1D), consistent with faster elongation leading to a downstream shift in termination.
Mutations in Sen1 Exhibit Opposing Interactions with Fast and Slow RPB1 alleles
If Sen1 functions by binding the RNA transcript and tracking behind Pol II, the relative translocation rates of Sen1 and Pol II should determine whether and how quickly Sen1 would trigger termination. This prediction was tested by combining functionally compromised mutants of Sen1 with slow and fast transcribing polymerases. The sen1-1 allele is a G1747D point mutation in motif IV of helicase domain 2, believed to function in single-stranded RNA/DNA binding (Figure 3A) (Bleichert and Baserga, 2007; Winey and Culbertson, 1988). The sen1-E1597K mutation resides in motif II of helicase domain 1, a region involved in NTP binding and hydrolysis to power translocation (Figure 3A) (Bleichert and Baserga, 2007; Steinmetz and Brow, 1996). The fact that both mutations reside in conserved motifs and display severe early termination defects strongly suggests that translocase activity is required in vivo.
Figure 3. Sen1 Mutations Have Opposite Effects on Growth of rpb1-N488D and rpb1-E1103G Strains.
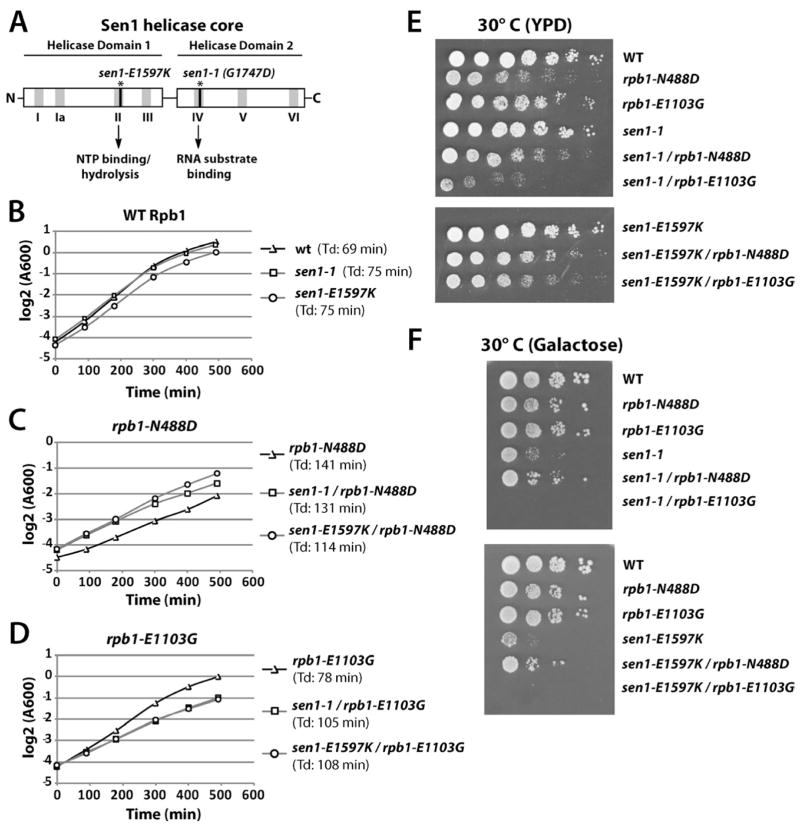
(A) Schematic of Sen1 C-terminal helicase domains (residues 1357-1822) with locations of sen1-E1597K and sen1-1 (sen1-G1747D) mutations.
(B) Cell growth curves of WT (YSB2700), sen1-1 (YSB2704), sen1-E1597K (YSB2707) strains; (C) rpb1-N488D (YSB2701), sen1-1/rpb1-N488D (YSB2705), sen1-E1597K/rpb1-N488D (YSB2708) strains; (D) rpb1-E1103G (YSB2702), sen1-1/rpb1-E1103G (YSB2706), sen1-E1597K/rpb1-E1103G (YSB2709) strains grown in YPD media at 30°C as measured by optical density at 600 nm (OD600) from a starting OD of ~0.05. Values were plotted on a log2 scale and the calculated doubling time (Td) of each strain is shown in parentheses.
(E) Spot growth assay of Rpb1 and Sen1 single and double mutant strains indicated in (B–D) grown on YPD media plates for 2 days at 30°C.
(F) Spot growth assay of strains indicated in (B–E) grown on 2% galactose/SC-uracil/x-gal media plates for 4 days at 30°C.
Epitope-tagged versions of sen1-1 and sen1-E1597K were introduced into RPB1, rpb1-N488D, and rpb1-E1103G strains and growth was assayed. The sen1-1 and sen1-E1597K mutations lead to minimal reduction of growth rate when grown in rich media at semi-permissive temperature (30°C) in strains expressing WT Rpb1 (Figure 3B, E). Interestingly, the slow growth phenotype of rpb1-N488D cells is partially suppressed by mutation of SEN1, particularly the sen1-E1597K allele, perhaps reflecting a resynchronization of Pol II and Sen1 activities (Figure 3C, E). In contrast, SEN1 mutation in the rpb1-E1103G background caused further decreases in growth rate (Figure 3D, E), perhaps due to the combined termination defects of each allele. As previously noted for the sen1-1 allele (Mischo et al., 2011), the two sen1 mutant strains grow slowly on media containing galactose, consistent with an important role for the Sen1 termination pathway in response to glucose deprivation (Darby et al., 2012). However, sen1 growth defects are markedly improved when combined with the slow rpb1-N488D polymerase and severely worsened with the rpb1-E1103G allele on galactose (Figure 3F). The synthetic genetic interactions when the fast Pol II and Sen1 mutants are combined could be due to increased termination defects.
To further examine the relationship between Sen1 function and Pol II elongation rate, levels of snoRNA readthrough transcription in the rpb1-N488D/sen1-1, rpb1-N488D/sen1-E1597K, rpb1-E1103G/sen1-1, and rpb1-E1103G/sen1-E1597K double mutant strains were compared to the single mutants. Because sen1-1 and sen1-E1597K mutations cause growth arrest at higher temperatures, RNA was isolated from cells grown at 30°C, a temperature at which termination defects are apparent but the cells are viable (Figure 3B–E). At SNR13, the rpb1-N488D mutant leads to a reproducible reduction in readthrough transcript levels in both sen1-1 and sen1-E1597K backgrounds (approximately 50% and 70% decreases, respectively; Figure 4A). In contrast, the fast rpb1-E1103G mutant causes elevated levels of SNR13 readthrough in the sen1 backgrounds, with the rpb1-E1103G/sen1-1 double mutant showing 2-fold more readthrough compared to sen1-1 with WT Rpb1 (Figure 4A). Similar results were seen with SNR3, SNR31, and SNR9 snoRNA genes (Figures 4B, 4C, S2).
Figure 4. Slow and Fast Pol II Mutants Differentially Affect snoRNA Termination in sen1 Mutant Strains.
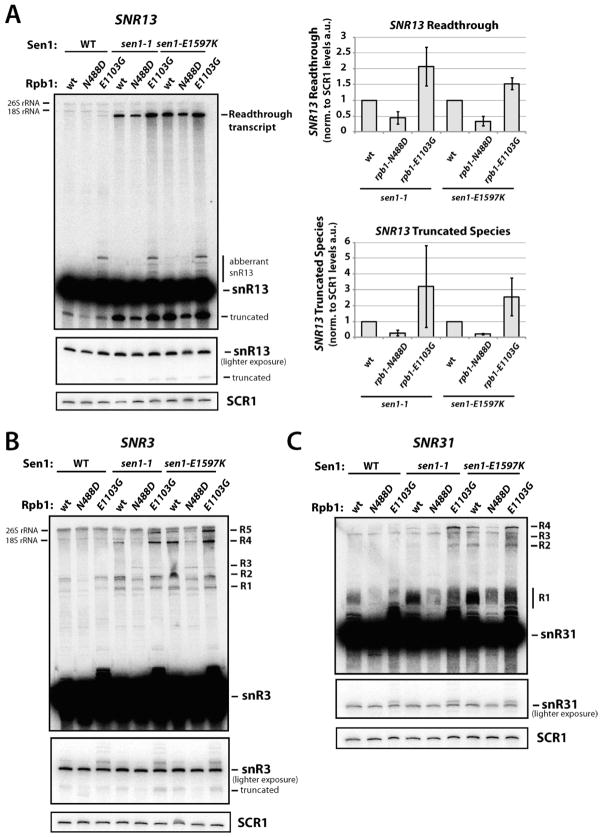
(A) Northern blot analysis of SNR13 with RNA from WT (YSB2700), rpb1-N488D (YSB2701), rpb1-E1103G (YSB2702), sen1-1 (YSB2704), sen1-1/rpb1-N488D (YSB2705), sen1-1/rpb1-E1103G (YSB2706), sen1-E1597K (YSB2707), sen1-E1597K/rpb1-N488D (YSB2708), and sen1-E1597K/rpb1-E1103G (YSB2709) strains grown at 30°C. The graphs show the relative levels of SNR13 readthrough transcripts (upper graph) and truncated snR13 transcripts (lower graph) relative to Pol III-transcribed SCR1 transcripts in arbitrary units (a.u.). Error bars represent standard error from two independent experiments.
(B) Northern blot analysis of SNR3 with RNA from indicated strains grown at 30°C. R1–R5; readthrough/extended transcripts that increase in sen1-1 and sen1-E1597K backgrounds, truncated; truncated snR3 transcript.
(C) Northern blot analysis of SNR31 from indicated strains grown at 30°C. R1–R4; readthrough/ extended transcripts that increase in sen1-1 and sen1-E1597K backgrounds. See also Figure S2.
The rpb1-E1103G mutation, in both WT and sen1 strains, also leads to aberrant transcripts of a slightly larger size than the mature snoRNA forms of snR13, snR3, snR31, and snR9 (Figures 4A–C, S2). How these transcripts form is unknown, but they appear at roughly equal levels in the presence of WT or mutant Sen1, indicating their formation is unrelated to Sen1 termination. Recently, rpb1-E1103G was shown to shift transcription start sites upstream at several mRNA promoters (Kaplan et al., 2012), suggesting these observed aberrant transcripts may be 5′ extended snoRNAs. In contrast, both Sen1 mutants increase levels of smaller species of snR13 and snR3 (Figures 4A, B). These are also seen in Nrd1 or Rrp6 mutants and previously characterized as a 5′-truncated form of the mature snR13 transcript (Rasmussen and Culbertson, 1998). The rpb1-N488D mutation reduces, while rpb1-E1103G increases, levels of this truncated species (Figure 4A). While the mechanism underlying the formation of the truncated transcript is unclear, it appears sensitive to Sen1 termination and Pol II speed.
Sen1 and Rpb1 Mutations Shift the Termination Window of a Broad Range of Non-Coding Transcripts
To visualize the termination window at SNR33 as in Figure 2, the RRP6 gene was deleted in Rpb1/Sen1 double mutant strains. Strains expressing sen1-1 and sen1-E1597K mutations not only produce readthrough species, but show a marked shift towards longer pre-snR33 transcripts (Figures 5A, 5B, 2C). The longer pre-snR33 transcripts in sen1-1 and sen1-E1597K cells correlate with higher levels of readthrough in sen1 cells, suggesting that more polymerases pass the downstream boundary of the Sen1 termination window and instead terminate via the poly(A) pathway. While pre-processed transcripts are strongly stabilized, readthrough transcripts are slightly increased in rrp6Δ cells (Figure S3B, C), perhaps reflecting the role ofRrp6 in quality control of polyadenylated transcripts (Rondon et al., 2009; Schmid et al., 2012).
Figure 5. Termination Window Mapping of Pre-Processed SNR33 and Other Non-coding Transcripts in WT, Slow, and Fast RNA Pol II strains.
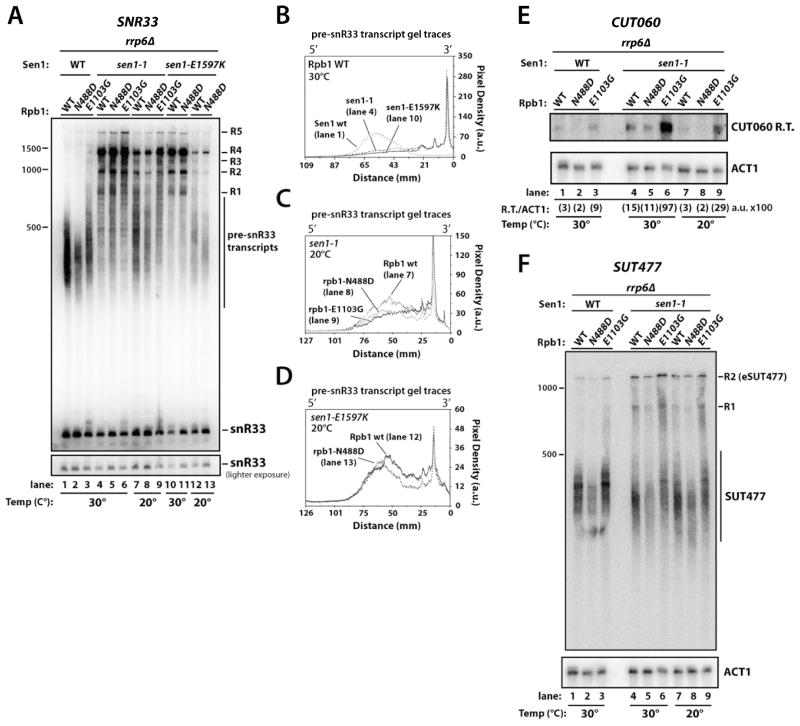
(A) Northern blot analysis of SNR33 with RNA from WT (YSB2697), rpb1-N488D (YSB2698), rpb1-E1103G (YSB2699), sen1-1 (YSB2711), sen1-1/rpb1-N488D (YSB2712), sen1-1/rpb1-E1103G (YSB2713), sen1-E1597K (YSB2714), and sen1-E1597K/rpb1-N488D (YSB2715), strains containing rrp6Δ grown at 30°C and 20°C. R1-R5; readthrough/extended transcripts that increase in sen1-1 and sen1-E1597K backgrounds.
(B, C, D) Relative densitometry comparisons of pre-snR33 transcripts from indicated lanes in part A.
(E) Northern blot analysis of CUT060 in indicated rrp6Δ strains grown at 30°C and 20°C. Shown here is the part of the gel corresponding to the CUT060 readthrough transcript (R.T.); the full blot including the terminated but un-degraded CUT transcripts is in Figure S3D. ACT1 mRNA serves as a loading control and the relative amount of readthrough is quantitated below each lane in arbitrary densitometry units (a.u.).
(F) Northern blot analysis of SUT447 in indicated rrp6Δ strains grown at 30°C and 20°C. R1, R2; readthrough/extended transcripts that increase in sen1-1 and sen1-E1597K backgrounds. eSUT447 is an extended SUT transcript (Marquardt et al., 2011).
See also Figure S3.
Combining rpb1-N488D with sen1 mutations decreases the average length of pre-snR33 transcripts, particularly in cells grown at 20°C, signifying termination occurring earlier within the termination window (Figure 5A, C, D). The temperature effect likely reflects improved activity of the mutant Sen1 and/or decreased Pol II elongation rate at the lower temperature. Conversely, the rpb1-E1103G/sen1-1 double mutant has longer pre-snR33 transcripts compared to RPB1/sen1-1 cells (Figure 5A, C). The observation that the edge of the terminated transcript spread overlaps with some readthrough transcripts may suggest some competition between the Sen1 and poly(A) pathways. Despite multiple attempts, we were unable to introduce rrp6Δ into the rpb1-E1103G/sen1-E1597K background, suggesting a fully functional nuclear exosome may be essential in this double mutant. Analysis of SNR9 produced results very similar to those for SNR33 (Figure S3A).
The rpb1-N488D Pol II produces some uncapped transcripts that are degraded by Rat1 (Jimeno-Gonzalez et al., 2010). Since Rat1 participates in poly(A)-dependent termination (Kim et al., 2004), rat1-1 in RPB1 and rpb1-N488D cells was tested for effects on snoRNA termination. In contrast to Sen1 mutants, rat1-1 did not shift the SNR33 and SNR9 termination window, although some stabilization of pre-processed and readthrough SNR9 transcripts was seen in rat1-1/rrp6Δ relative to rrp6Δ cells at 37°C, perhaps suggesting some SNR9 transcripts are subject to degradation by Rat1 (Figure S3B, C). These termination window mapping results agree with our earlier ChIP and northern analyses that found no effect of RAT1 mutation on snoRNA termination (Kim et al., 2006).
With rrp6Δ in the Rpb1/Sen1 double mutant strains, we analyzed other small non-coding transcripts from the CUT or SUT (stable uncharacterized transcript) classes (Wyers et al., 2005; Xu et al., 2009). The CUT060 locus produces short Rrp6-sensitive transcripts whose termination is dependent on Nrd1 (Marquardt et al., 2011) and which migrate as a spread of roughly 200–400 nucleotides (Figure S3D). The rpb1-N488D mutant exhibits a shorter termination window, while the rpb1-E1103G strain shows a longer termination spread as well as more readthrough species (Figures 5E, S3D). The sen1-1 mutation also increased the spread size of CUT060 transcripts and readthrough levels, an effect partially reversed by combination with rpb1-N488D or exacerbated by the rpb1-E1103G mutation, with a 6- to 10-fold increase in CUT readthrough levels in the rpb1-E1103G/sen1-1 mutant over the sen1-1 strain (Figures 5E, S3D).
SUTs represent another class of small non-coding transcripts distinguishable from CUTs by a less stringent dependence on Nrd1 and at least partial resistance to nuclear exosome degradation (Marquardt et al., 2011; Xu et al., 2009). In rrp6Δ strains, the rpb1-N488D mutation decreases SUT477 transcript levels and average length while the rpb1-E1103G mutation increases the average SUT477 transcript size (Figure 5F).The sen1-1 mutation also increases the distal spread of SUT477 transcripts as well as levels of 3′ extended SUT477 transcripts (eSUTs) that accumulate in nrd1 mutants (Figure 5F) (Marquardt et al., 2011). The combined rpb1-E1103G and sen1-1 mutations produce more SUT477 transcripts with sizes between the SUT477 and eSUT477 transcripts, indicating that alterations in Pol II elongation rate and Sen1 function affect SUT expression and termination (Figure 5F).
DISCUSSION
Kinetic Competition Between Elongating Pol II and Sen1 helicase
Sen1 termination in S. cerevisiae operates primarily at small, non-coding RNA loci. Termination at these genes has been difficult to study because the transcript 3′ ends are rapidly processed or degraded by the nuclear exosome, which is physically linked to the Nrd1/Nab3/Sen1 complex (Vasiljeva and Buratowski, 2006). In strains lacking Rrp6, terminated but unprocessed transcripts are stabilized, allowing us to map termination windows in vivo. Utilizing this approach, we show that Sen1 termination is inherently sensitive to elongation rate, with faster elongating Pol II shifting termination downstream and slower Pol II leading to a shorter termination spread. Furthermore, a slow Pol II partially reverses the termination and growth defects of sen1 helicase domain mutants, while a faster Pol II intensifies them. These results strongly suggest that the termination window is determined by a kinetic competition between the polymerase and the Sen1 helicase (Figure 6).
Figure 6. Model of Kinetic Competition Between Pol II and Sen1.
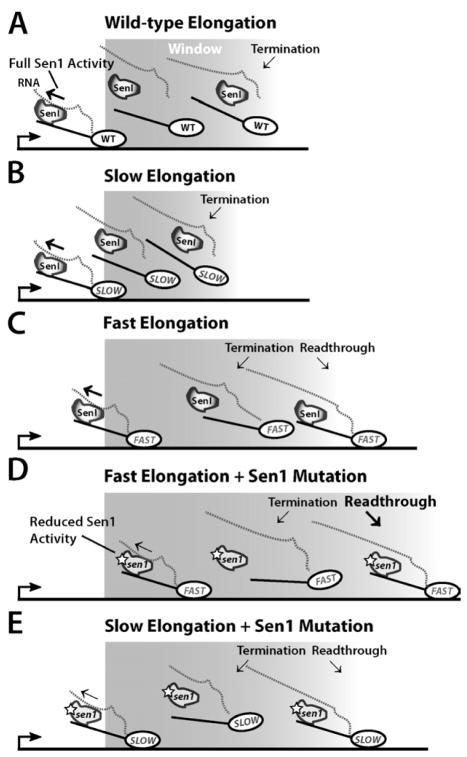
(A) A termination window, likely prescribed by Nrd1 and Nab3 RNA binding sites in the transcript and CTD phosphorylation patterns, in which normal Pol II elongation coupled with Sen1 bound to Pol II CTD and the RNA transcript leads to efficient termination (Chinchilla et al., 2012).
(B) Slow Pol II elongation shows window contraction with increased earlier termination events.
(C) Faster elongation in rpb1-E1103G leads to window expansion with more downstream termination events within the window and increased readthrough.
(D) Cells expressing both faster Pol II and mutations in Sen1 exhibit higher levels of window expansion and readthrough than single mutants and synergistic growth defects, indicative of a greater desynchronization between fast moving Pol II and less functional Sen1 molecules.
(E) Cells expressing both the slower Pol II (rpb1-N488D) and sen1 mutations have a less extended termination window and partial suppression of sen1 mutant phenotypes due to the slower elongation rate making it easier for the partially defective Sen1 to catch the polymerase.
In a kinetic competition model, the termination window may be opened by loading of Sen1 onto the emerging transcript. Like many RNA helicases (Jankowsky, 2011), Sen1 utilizes co-factors for targeting. Sen1 associates with the Nrd1/Nab3 complex, which recognizes specific RNA motifs found in target genes, as well as the Rpb1 CTD (Chinchilla et al., 2012). However, RNA binding by Sen1 is an essential first step in this model. Once loaded, Sen1 tracks along the nascent RNA in a 5′ to 3′ direction until it reaches the polymerase to either directly or allosterically promote transcript dissociation from the Pol II active site (Brow, 2011). The point of termination depends upon the relative rates of Sen1 and elongating Pol II, with the former needing to be slightly faster (Figure 6A). In agreement with a tracking model, Sen1 crosslinks to many sites along non-coding RNAs, including snR33 and snR9 (Jamonnak et al., 2011). We observe a window of termination spread over several hundred nucleotides, agreeing well with both Pol II ChIP and NET-Seq results (Churchman and Weissman, 2011; Kim et al., 2006; Steinmetz et al., 2006b). This extended zone of termination is likely due to stochastic variability in the rates of the two enzymes, and such heterogeneity rules out any models in which there is a discrete terminator sequence or RNA structure. However, RNase H/oligo(dT) experiments (Figures 2C, S1A) do suggest some preferred termination sites within the window. These may represent sites where Pol II is slowed in a locally restricted fashion, perhaps by specific sequences or proteins bound to the DNA, allowing Sen1 to catch up. Lending further support to a Rho-like model for Sen1, purified Sen1 can dissociate Pol II elongation complexes in vitro (O. Porrua and D. Libri, personal communication; H. Mischo, personal communication).
The model predicts that the termination window will be shifted by factors that change the rate of Pol II elongation. Indeed, 6-AU treatment, lower growth temperature, and uracil depletion all promote earlier termination, as do the slow rpb1-N488D and rpb2-10 polymerases (Figure 6B). In contrast, faster elongation in rpb1-E1103G cells leads to termination spreading further downstream (Figure 6C). Combining the rate-altered rpb1 alleles with Sen1 point mutations provides the strongest support for kinetic competition. Mutant Sen1 molecules may have difficulty catching up to Pol II, and the resulting readthrough is worsened by the faster elongating rpb1-E1103G Pol II (Figure 6D). Conversely, slowing elongation with rpb1-N488D Pol II partially compensates for the Sen1 mutations, presumably by allowing more time for the defective Sen1 to catch up and trigger termination (Figure 6E). This compensation is reflected in suppression of slow growth phenotypes (Figure 3C, E). Our observation that the termination window depends on a balance between Sen1 and Pol II argues that the rates of the two enzymes are uncoupled, even if the proteins are physically linked.
There is ample precedent for kinetic competition between elongating polymerase and a tracking factor. Termination at eukaryotic mRNAs requires cleavage at the polyadenylation site followed by co-transcriptional degradation of the downstream transcript by the 5′ to 3′ exonuclease Rat1/Xrn2. As seen for Sen1 terminators, Rat1-dependent termination occurs over several hundred nucleotides, consistent with the proposal that polymerase dissociation is somehow triggered when Rat1 catches up to the elongation complex (Kim et al., 2004). Furthermore, the slow rpb1-N488D allele partially suppresses the growth defects and mRNA termination defects of a rat1 mutant (Jimeno-Gonzalez et al., 2010). Even more so than Rat1, Sen1 activity may resemble termination by bacterial Rho, where kinetic competition between RNA polymerase and Rho helicase is well documented (Jin et al., 1992). Interestingly, heterologous expression of Rho in yeast causes growth defects that are worsened by slow Pol II variants, including rpb1-N488D, and alleviated by the rpb1-E1103G mutation, suggesting that slow Pol II is more susceptible to premature termination caused by Rho, while faster polymerases may outpace the tracking Rho helicase (Mosrin-Huaman et al., 2009).
Human Senataxin has been implicated in transcription termination and splicing (Skourti-Stathaki et al., 2011; Suraweera et al., 2009). Senataxin mutations lead to the progressive neurodegenerative diseases ataxia oculomotor apraxia 2 (AOA2) and amyotrophic lateral sclerosis type 4 (ALS4) (Lemmens et al., 2010), so it will be interesting to determine if Pol II transcription defects in neural cells contribute to disease causality.
Gene Regulation via Control of the Early Termination Window
Although the Sen1 pathway is responsible for 3′ end formation of small non-coding RNAs, Pol II crosslinking studies showed that a small number of mRNA genes are also affected by inactivating Sen1 (Steinmetz et al., 2006b). Furthermore, genome-wide ChIP of Nrd1 finds it at 5′ ends, and often other locations, within protein coding genes (Kim et al., 2010). Further analysis of specific Sen1-regulated mRNA genes has generally revealed at least two transcripts: one that is terminated soon after initiation by Sen1, and a second full-length mRNA that is cleaved and polyadenylated. One example of such regulation is the NRD1 gene itself, which is subject to negative feedback autoregulation (Arigo et al., 2006; Steinmetz et al., 2006a). High levels of Nrd1 protein bind to recognition motifs on the emerging transcript to trigger termination. At lower Nrd1 concentrations, Pol II bypasses the early termination window to produce full length mRNA. Another example is the FKS2 stress-response gene, which is regulated by Mpk1 (Kim and Levin, 2011). In non-inducing conditions, transcription from the FKS2 promoter terminates via Sen1 soon after initiation. Upon induction, Mpk1 binds to the Paf1 elongation complex to block Sen1 recruitment, leading to production of full-length FKS2 mRNA. In both examples, the choice between termination pathways, and thereby production of the encoded protein, can be controlled by regulating the access of Nrd1 or Sen1.
Histone modifications just downstream of the promoter, which are likely to affect Pol II rates in early elongation, may also influence the Sen1 termination pathway (Soares and Buratowski, 2012; Terzi et al., 2011). Additionally, elongation rates may vary from gene to gene, as recent studies show that Pol II speed in vivo can be highly dynamic (Maiuri et al., 2011). While control of polymerase passage through the early Sen1 termination window is used to regulate the expression of several yeast mRNAs as well as snoRNAs, CUTs, and SUTs, this system has obvious parallels to the more widespread Pol II “pausing” seen in higher eukaryotes (Buratowski, 2009; Nechaev and Adelman, 2011). In both cases, release of Pol II into full elongation is limiting, and these mechanisms may allow faster response times by bypassing the chromatin remodeling and initiation complex assembly steps that are rate-limiting at other promoters.
EXPERIMENTAL PROCEDURES
Yeast strains
Strain genotypes, constructions, and cell growth assays are listed in Table S1 and Supplemental Experimental Procedures.
Chromatin Immunoprecipitation (ChIP) Analysis
Chromatin preparations and qPCR analysis were performed as described previously (Keogh and Buratowski, 2004; Marquardt et al., 2011) and in Supplemental Experimental Procedures.
RNA isolation and Northern Blotting
25 ml cultures were grown at the indicated temperatures in synthetic complete media (SC) or SC –uracil media (SC-URA) to OD600 ~ 0.2–0.4. RNA was isolated by the hot phenol extraction method and quantified with a Nanodrop spectrophotometer. 0.5–5 μg of total RNA was separated on 6 or 8% polyacrylamide-7.5 M urea gels or 1.5% agarose-formaldehyde-MOPS gels. Polyacrylamide gels were transferred to nylon transfer membrane (Nytran SPC, Whatman) using a Bio-Rad TransBlot apparatus. Agarose gels were transferred to membranes via capillary blotting in 10X SSC for 18–24 hrs. RNA was crosslinked to membranes with a UV Stratalinker 1800. 10–20 μg of denatured DNA size standards (1 kb and 100 bp ladder, NEB) were loaded on gels for estimation of transcript size after staining membranes with methylene blue. Membrane hybridization was with single-stranded DNA 32P-radiolabeled probes prepared as previously described (Marquardt et al., 2011). Quantitation was performed with a Fujifilm BAS-2500 Phosphorimager and ImageGauge software (Fuji). RNase H/oligo(dT) analysis procedure is outlined in supplemental procedures. Primer sequences for probe generation are listed in Table S2.
Quantitative Reverse Transcription PCR Analysis (RT-qPCR)
10 μg RNA was isolated from triplicate yeast cultures and treated with DNase I (RQ1 DNase, Promega). Reverse transcription cDNA synthesis reactions were performed with 1.5 μg total RNA with gene specific primers according to manufacturer’s specifications (Superscript II, Invitrogen). Triplicate qPCR reactions from three biological replicates were performed on a Roche Lightcycler 480 and quantitation of snoRNA readthrough transcript levels relative to a control gene (ADH1) were calculated with the ΔΔcT method as previously described (Marquardt et al., 2011). Primer sequences for cDNA synthesis and PCR reactions are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Luis Soares, TaeSoo Kim, and other members of the Buratowski Lab for helpful discussions and suggestions; Hannah Mischo (CRUK), Odil Porrua and Domenico Libri (CGM/CNRS) for communicating unpublished results; and Jeffrey Strathern (NCI, Frederick), Kevin Struhl (HMS), David Brow (UW Madison), Doris Ursic and Michael Culbertson (UW Madison), and Torben Heick Jensen and Manfred Schmid (Aarhus University, Denmark) for strains and plasmids. D.Z.H. is an American Cancer Society Postdoctoral Fellow. S.M. is a Feodor Lynen Fellow of the Alexander von Humboldt Foundation. This work was supported by NIH grant GM56663 to S.B.
Footnotes
Supplemental information includes three figures and two tables and Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbondanzieri EA, Shaevitz JW, Block SM. Picocalorimetry of transcription by RNA polymerase. Biophys J. 2005;89:L61–63. doi: 10.1529/biophysj.105.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo JT, Carroll KL, Ames JM, Corden JL. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell. 2006;21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Brow DA. Sen-sing RNA Terminators. Mol Cell. 2011;42:717–718. doi: 10.1016/j.molcel.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla K, Rodriguez-Molina JB, Ursic D, Finkel JS, Ansari AZ, Culbertson MR. Interactions of Sen1, Nrd1, and Nab3 with Multiple Phosphorylated Forms of the Rpb1 C-Terminal Domain in Saccharomyces cerevisiae. Eukaryot Cell. 2012;11:417–429. doi: 10.1128/EC.05320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154:557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Creamer TJ, Darby MM, Jamonnak N, Schaughency P, Hao H, Wheelan SJ, Corden JL. Transcriptome-Wide Binding Sites for Components of the Saccharomyces cerevisiae Non-Poly(A) Termination Pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 2011;7:e1002329. doi: 10.1371/journal.pgen.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby MM, Serebreni L, Pan X, Boeke JD, Corden JL. The Saccharomyces cerevisiae Nrd1-Nab3 Transcription Termination Pathway Acts in Opposition to Ras Signaling and Mediates Response to Nutrient Depletion. Mol Cell Biol. 2012;32:1762–1775. doi: 10.1128/MCB.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- Jamonnak N, Creamer TJ, Darby MM, Schaughency P, Wheelan SJ, Corden JL. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in posttranscriptional RNA processing. RNA. 2011;17:2011–2025. doi: 10.1261/rna.2840711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-Gonzalez S, Haaning LL, Malagon F, Jensen TH. The yeast 5′-3′ exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol Cell. 2010;37:580–587. doi: 10.1016/j.molcel.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Jin DJ, Burgess RR, Richardson JP, Gross CA. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 1992;89:1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Jin H, Zhang IL, Belyanin A. Dissection of Pol II Trigger Loop Function and Pol II Activity-Dependent Control of Start Site Selection In Vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Buratowski S. Using chromatin immunoprecipitation to map cotranscriptional mRNA processing in Saccharomyces cerevisiae. Methods Mol Biol. 2004;257:1–16. doi: 10.1385/1-59259-750-5:001. [DOI] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Choe J, Seo YS. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry. 1999;38:14697–14710. doi: 10.1021/bi991470c. [DOI] [PubMed] [Google Scholar]
- Kim KY, Levin DE. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens R, Moore MJ, Al-Chalabi A, Brown RH, Jr, Robberecht W. RNA metabolism and the pathogenesis of motor neuron diseases. Trends Neurosci. 2010;33:249–258. doi: 10.1016/j.tins.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Maiuri P, Knezevich A, De Marco A, Mazza D, Kula A, McNally JG, Marcello A. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 2011;12:1280–1285. doi: 10.1038/embor.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, Strathern JN. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Hazelbaker DZ, Buratowski S. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription. 2011;2:145–154. doi: 10.4161/trns.2.3.16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosrin-Huaman C, Honorine R, Rahmouni AR. Expression of bacterial Rho factor in yeast identifies new factors involved in the functional interplay between transcription and mRNP biogenesis. Mol Cell Biol. 2009;29:4033–4044. doi: 10.1128/MCB.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Powell W, Reines D. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TP, Culbertson MR. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP. How Rho exerts its muscle on RNA. Mol Cell. 2006;22:711–712. doi: 10.1016/j.molcel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Poulsen MB, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz LM, Moore C, Jensen TH. Rrp6p controls mRNA poly(a) tail length and its decoration with poly(a) binding proteins. Mol Cell. 2012;47:267–280. doi: 10.1016/j.molcel.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, Buratowski S. Yeast Swd2 is essential due to antagonism between the Set1 histone methyltransferase complex and the Associated with Pta1 (APT) termination factor. J Biol Chem. 2012 doi: 10.1074/jbc.M112.341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Ng SB, Cloute JP, Brow DA. cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol Cell Biol. 2006a;26:2688–2696. doi: 10.1128/MCB.26.7.2688-2696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006b;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Suraweera A, Lim Y, Woods R, Birrell GW, Nasim T, Becherel OJ, Lavin MF. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum Mol Genet. 2009;18:3384–3396. doi: 10.1093/hmg/ddp278. [DOI] [PubMed] [Google Scholar]
- Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol Cell Biol. 2011;31:3569–3583. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, Chinchilla K, Finkel JS, Culbertson MR. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 2004;32:2441–2452. doi: 10.1093/nar/gkh561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Culbertson MR. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988;118:609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AQ, von Hippel PH. Rho-dependent termination within the trp t’ terminator. II. Effects of kinetic competition and rho processivity. Biochemistry. 1998;37:11215–11222. doi: 10.1021/bi972912s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


